Abstract
Delta functions as a cell nonautonomous membrane-bound ligand that binds to Notch, a cell-autonomous receptor, during cell fate specification. Interaction between Delta and Notch leads to signal transduction and elicitation of cellular responses. During our investigations to further understand the biochemical mechanism by which Delta signaling is regulated, we have identified four Delta isoforms in Drosophila embryonic and larval extracts. We have demonstrated that at least one of the smaller isoforms, Delta S, results from proteolysis. Using antibodies to the Delta extracellular and intracellular domains in colocalization experiments, we have found that at least three Delta isoforms exist in vivo, providing the first evidence that multiple forms of Delta exist during development. Finally, we demonstrate that Delta is a transmembrane ligand that can be taken up by Notch-expressing Drosophila cultured cells. Cell culture experiments imply that full-length Delta is taken up by Notch-expressing cells. We present evidence that suggests this uptake occurs by a nonphagocytic mechanism.
INTRODUCTION
Delta(Dl) is a Drosophila neurogenic gene that encodes a cell surface protein believed to function as a membrane-bound ligand in intercellular signaling during development. Evidence suggests that Delta interacts with the receptor Notch in a signal transduction pathway that precludes or promotes cell fate specification in different developmental contexts (reviewed by Artavanis-Tsakonas et al., 1995; de la Pompa et al., 1997). Delta function is required for a number of developmental processes in Drosophila, beginning during oogenesis and continuing through metamorphosis. During embryogenesis, Delta is required during neuroblast determination (Lehmann et al., 1983), for correct specification of muscle cell fates within the mesoderm (Corbin et al., 1991; Bate et al., 1993), and for correct development of cell types within a variety of other tissues derived from each of the three embryonic germ layers (Hartenstein et al., 1992). Delta function is also required for many instances of cell fate specification during postembryonic development (Parody and Muskavitch, 1993). Delta function is necessary for the specification of cell types in the developing retina (Parks et al., 1995), the differentiation of cell types in the adult wing (Vässin and Campos-Ortega, 1987; Huppert et al., 1997), specification of the correct number of bristles in the head, abdomen, and thorax, and the differentiation of cell types that make up the adult bristle organs (Vässin and Campos-Ortega, 1987; Parks and Muskavitch, 1993).
The Delta extracellular domain consists of an amino-terminal domain similar to that of the Drosophila Serrate protein (Fleming et al., 1990; Thomas et al., 1991), followed by a tandem array of nine EGF-like repeats (Vässin et al., 1987; Kopczynski et al., 1988). The carboxy-terminal Delta intracellular domain is not significantly similar to any protein currently listed in protein databases. Initial structure–function analyses, using a Drosophila cultured cell aggregation assay (Fehon et al., 1990), have defined regions within Delta and Notch that are required for their interaction. This assay has been employed to show that Notch EGF-like repeats 11 and 12 are necessary and sufficient for aggregation with cells that express full-length Delta protein (Rebay et al., 1991). Analysis of truncated variants and chimeras in analogous experiments has revealed that the Delta amino terminus is sufficient for aggregation with cells that express Notch (Shepard and Muskavitch, unpublished data). In addition, it has been suggested that EGF-like repeats within the Delta extracellular domain play a role in the molecular interactions between Delta and Notch (Lieber et al., 1992).
The Delta protein exhibits dynamic subcellular trafficking in vivo. Delta is often found first on the cell surface and subsequently in subcellular vesicles that appear when expression is down-regulated (Kooh et al., 1993). For example, vesicular localization is discernible in neuroblasts shortly after their delamination within the embryonic ventral neurogenic ectoderm. These vesicles are found in the peripheral regions of cells, suggesting that they have entered the endocytic pathway. Vesicular localization of Delta protein has also been detected in embryonic mesoderm, wing imaginal discs, larval CNS, and retinal cells in the developing eye imaginal disc (Kooh et al., 1993).
The aggregation of Delta-expressing (Delta+) Drosophila cultured cells with Notch-expressing (Notch+) cells, as well as genetic and somatic mosaic analyses (reviewed by Muskavitch, 1994; Artavanis-Tsakonas et al., 1995), imply that the Delta–Notch signaling pathway is cell contact-dependent. Cell contact-dependent signaling is widely recognized as a mechanism by which localized cell communication establishes finely resolved tissue patterning during development, often through the action of membrane-anchored ligands (see Bosenberg and Massagué, 1993; and Fagotto and Gumbiner, 1996, for reviews). Membrane-anchored ligands elicit responses via two qualitatively distinct mechanisms. First, some ligands are active only as full-length, membrane-anchored molecules. For example, the Drosophila transmembrane ligand bride of sevenless (boss), which activates the tyrosine kinase receptor sevenless (sev), is only active as a full-length molecule (Hart et al., 1993). Neither a boss variant in which four of the seven transmembrane domains are deleted nor a secreted boss extracellular domain are capable of interacting with sevenless (Hart et al., 1993). Second, some membrane-anchored ligands also elicit responses after the proteolytic cleavage of the membrane-anchored ligand to generate a diffusible extracellular signal. This mechanism provides for short-range and long-range signaling by a cell. The members of the tumor necrosis factor (TNF) family of growth factors fall into this category. In the TNF family, the soluble and membrane-anchored ligand isoforms activate different receptors, and it has been suggested that the activation of these distinct receptors leads to different cellular responses (Grell et al., 1995). In addition, the cleavage of membrane-bound ligands to produce soluble, diffusible ligand activities can, in some cases, diminish or prevent adhesive interactions among cells (reviewed by Bosenberg and Massagué, 1993).
There is increasing evidence that proteases are key components in many signaling pathways in invertebrates. Proteolysis has recently been shown to be necessary for maturation and function of the Notch receptor in different species, implicating proteolytic processing as a prerequisite for Delta–Notch signal transduction. In mice, it has been reported that mNotch1 is proteolytically processed (Kopan et al., 1996) and that the intracellular domain subsequently translocates to the nucleus. The LNG repeats (three cysteine-rich Notch/Lin 12/Glp-1 repeats) within the Notch extracellular domain have been implicated in the regulation of this processing, and it has been suggested that ligand binding to the extracellular domain of mNotch1 may also regulate the processing of Notch (Kopan et al., 1996). More recently, in Drosophila and human tissue culture cells, it has been found that Notch is proteolytically cleaved during a maturation process necessary for the genesis of a functional Notch receptor (Blaumueller et al., 1997; Pan and Rubin, 1997).
Activation of a signaling pathway through ligand–receptor interaction is often followed by down-regulation of ligand–receptor complexes. Ligand-dependent internalization and entry into the endocytic pathway are features shared by several families of receptors, including the EGF and insulin receptor families (see Chang et al., 1993 and references within). Proteolysis has also been found to play a role in the down-regulation of certain classes of ligand-receptor complexes (reviewed by Authier et al., 1996), and complete or partial processing of ligands can occur in endosomes after the internalization of receptor-ligand complexes (Authier et al., 1996). Clathrin-coated vesicles mediate the internalization of many extracellular ligands (reviewed by Robinson et al., 1996). However, there are examples in which ligands are taken up by non–clathrin-coated vesicles, macropinocytosis, or phagocytosis (Robinson et al., 1996). Once ligand–receptor complexes enter the endocytic pathway, there are several possible destinations for the internalized molecules. Many classes of receptors are returned to the surface, some are transported to lysosomes for degradation, and in some cases, endocytosed proteins are sequestered in specialized compartments for later reuse (Robinson et al., 1996).
Several aspects of the cell biology of Delta and the biochemical mechanisms by which it becomes activated and down-regulated remain unclear. In the course of our investigations to understand these mechanisms further, we have identified four isoforms of Delta by immunoprecipitation from embryonic and larval extracts. We present evidence that these isoforms result from post-translational modification. To further characterize the domains present in these isoforms, we have generated an antibody to the Delta intracellular domain. Our immunolocalization data imply that Delta isoforms can exhibit distinguishable localization in vivo, an observation that correlates with the presence of multiple Delta isoforms in embryos and larvae. Finally, we present data demonstrating that Delta is a transmembrane ligand that can be taken up by neighboring Notch+ Drosophila cultured cells, by a nonphagocytic mechanism.
MATERIALS AND METHODS
Antibody Production and Immunohistochemistry
Polyclonal antibodies to the Notch extracellular domain were prepared using a 0.8 kilobase (kb) BstYI fragment (which encodes amino acids 237–501; Wharton et al., 1985) fused in frame into pGEX as described by Fehon et al. (1990). Inclusion bodies were prepared and used to immunize rats (Pocono Rabbit Farm and Laboratory, Canadensis, PA). mAbs to the Notch intracellular domain, C17.9C6 (MAb9C6), and the Delta extracellular domain, C594.9B (MAb9B, also known as MAb202) and C594.8A (MAb8A), were generated in the laboratory of Spyros Artavanis-Tsakonas (Yale University, New Haven, CT). The Delta mAbs, MAb9B and MAb8A, recognize an epitope in Delta EGF-like repeats 4 and/or 5 (our unpublished results). Guinea pig polyclonal antiserum to the Delta extracellular domain (GP581) is described by Huppert et al. (1997). For production of polyclonal antibodies to the Delta intracellular domain (C2), the Delta C2 fusion construct was generated by inserting sequences encoding amino acids 645–832 from the Delta intracellular domain (nucleotides [nt] 2072-nt 2636 of pDl1; Kopczynski et al., 1988) into pGEX-4T-3 (Pharmacia Biotech; Piscataway, NJ; construct generated by ATG Laboratories, Eden Prairie, MN). Expression of these fusion constructs and subsequent fusion protein purification were carried out by ATG Laboratories. The immunogen was prepared by standard methods and injected into mice and guinea pigs (mice, Cayman Chemical Company, Ann Arbor, MI; guinea pigs, Pocono Rabbit Farm and Laboratory). The mAb to the MYC epitope, 1–9E10.2 (MAb9E; American Type Culture Collection, Rockville, MD), was described by Evan et al. (1985). The mAb to the intracellular domain of the long form of Drosophila neuroglian (BP104, a gift from Allan Bieber, Purdue University, West Lafayette, IN) was described by Hortsch et al. (1990). The monoclonal antibody mAb-αboss1 (a gift from Helmut Krämer, The University of Texas Southwestern Medical Center, Dallas, TX) was described by Krämer et al. (1991).
For immunohistochemistry in embryos, antibodies in the C2 polyclonal antiserum that bind to Drosophila proteins other than Delta were removed by preadsorbing the antiserum against Drosophila S2 cells (Schneider, 1972) at a 1:500 dilution in PBS containing 1% normal goat serum (NGS) and 0.1% saponin. After preadsorbing for 1 h at room temperature, the diluted antiserum was removed and added to an equal volume of PBS containing 1% Triton-X 100 (TPBS). The Delta MAb9B ascites was diluted 1:1000 in PBS containing 0.5% Triton-X 100 before use. Diluted antiserum containing 10% NGS was incubated with blocked embryos as described by Kooh et al. (1993). Subsequent washes and incubation with secondary antibodies were performed as described by Kooh et al. (1993).
For immunohistochemistry in Drosophila S2 cells, antibodies were diluted in PBS containing 1% NGS and 0.1% saponin as follows: Notch rat-8 at 1:2000; MAb9C6 hybridoma supernatant, undiluted; GP581 at 1:5000; C2 at 1:1000; MAb9E hybridoma supernatant at 1:5 or undiluted; BP104 hybridoma supernatant at 1:10; mAb-αboss1 ascites at 1:3000.
Immunoprecipitations
Native protein extracts were prepared by Dounce homogenization of staged embryos, and first and second instar larvae, as described by Fehon et al. (1990), with the following modifications. Sequential triturations were omitted. Unless otherwise noted, at least 20 volumes of buffer, relative to tissue volume, were used during the extraction procedures. COMPLETE protease inhibitor tablets were used in the extraction procedures, as specified by the manufacturer (Boehringer Mannheim, Indianapolis, IN). Samples were first cleared at 10,000 × g and were either used immediately or frozen quickly with liquid nitrogen or with CO2/EtOH, and stored at −80°C. Just before immunoprecipitations, insoluble proteins were removed by spinning at 100,000 × g in an ultracentrifuge (large samples) or at high speed in a microcentrifuge (small samples). For preparation of native protein extracts from third instar larvae, larvae were frozen and ground with a mortar and pestle in liquid nitrogen, and then immediately extracted by Dounce homogenization as described for embryos. Delta was immunoprecipitated from extracts as described by Fehon et al. (1990). A total of 3.5–4.5 μg of MAb9B or MAb8A ascites were used to immunoprecipitate Delta molecules containing the extracellular domain.
SDS-PAGE and Western Blots
Immunoprecipitated samples were resuspended in an equal volume of 2× Laemmli sample buffer and separated by SDS-PAGE on 7.5% or 10% gels. Gels were electroblotted as described by Bisgrove et al. (1991). Western blots were blocked at room temperature for 1 h in PBS containing 0.05% Tween-20 (PBT)/5% nonfat dry milk/0.3% BSA. Antibodies were diluted in PBT containing 1% NGS and incubated with blots overnight at 4°C. The MAb9B ascites was used at 1:10,000 to 1:15,000, and whole mouse C2 or guinea pig C2 antiserum was used at 1:30,000 to 1:60,000. The MAb9E ascites was diluted 1:3000 and preadsorbed against S2 cells for 1 h before use. After three washes in PBT, blots were incubated with a peroxidase-conjugated goat-anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) at 1:18,000, in PBT/1% NGS, for 60 min at room temperature, and then washed three times in TPBS and three times in PBS. The ECL system (Amersham Life Science, Arlington Heights, IL) was used for the detection. Prestained and unstained molecular weight standards (Sigma, St. Louis, MO; Life Technologies, Inc., Gaithersburg, MD) were used to determine apparent molecular weights.
Insect Cell Culture
Drosophila S2 cells and derived stable cell lines were maintained as described by Fehon et al. (1990). The stable clonal Delta (Delta+) cell line used was described by Rebay et al. (1991). The stable clonal Notch (Notch+) cell line used was produced using the methods described by Rebay et al. (1991) and was a gift from Lucy Cherbas (Indiana University, Bloomington, IN). Stable transfected S2 cell mass populations programmed to express Delta variants under the control of the metallothionein promoter were produced as described by Bunch et al. (1988). Stable mass populations programmed to express full-length Delta protein with a C-terminal MYC epitope tag (Evan et al., 1985) fused at nt 2635 of pDl1 (referred to as DeltaWTNdeMYC), or the secreted Delta extracellular domain with a C-terminal FLAG epitope tag referred to as DeltaSEC1 (Hopp et al., 1988) fused at nt 1855 of pDl1 were created by the same technique. Constructs used in transient transfections include: DeltaNGIC (contains Delta nt 1–nt 2072 fused to neuroglian nt 3506–nt 3960; Bieber et al., 1989); DeltaDde (described by Shepard, 1991; Delta truncated at nt 2022 to include Delta amino acids 1–626 followed by a terminal His residue); pRMHa3-BossH6 (wild-type boss with a C-terminal His6 tag, a gift from Helmut Krämer, The University of Texas Southwestern Medical Center, Dallas, TX); and a wild-type neuroglian construct, pRMHa3-nrg180 (Hortsch et al., 1995). The schematic diagram in Figure 1 illustrates Delta variants and domains recognized by the various antibodies used in these experiments. Transfection, induction, fixation, and immunofluorescent labeling were carried out as previously described by Fehon et al. (1990), except that BBS (140 mM NaCl, 0.75 mM Na2HPO4, 25 mM N,N-bis[2-hydroxyethyl]-2-aminoethanesulfonic acid, pH 6.95) was used in place of HEPES-buffered saline. Briefly, protein expression in transfected cells was induced for 2 h with 1 mM CuSO4, after which 500 μl of Delta+ cells were mixed with 500 μl of Notch+ cells and aggregated with gentle rotation in microtiter plates for 4 h. The cells were placed onto poly-l-lysine (Sigma)-coated slides and allowed to adhere for 15 min. Cells were fixed for 15 min with 2% paraformaldehyde in PBS, rinsed three times with PBS, and then double-labeled with antibodies against Delta and Notch as described above. For the quantification of trans-endocytosis, the frequency of trans-endocytosis was recorded as the percentage of Notch+ cells in contact with Delta+ cells that contain Delta-positive internalized vesicles. Data were compiled from three separate sets of experiments.
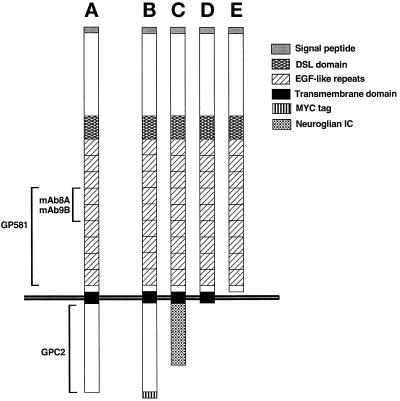
Figure 1.
Schematic representation of Delta variants expressed in Drosophila cultured cells and the domains recognized by various Delta antibodies. (A) DeltaWT, full-length Delta. (B) DeltaWTNdeMYC, full-length Delta. (C) DeltaNGIC, Delta extracellular and transmembrane domains fused to a neuroglian intracellular domain. (D) DeltaDde, Delta protein truncated near the inner face of the plasma membrane. (E) DeltaSEC1, secretable Delta extracellular domain.
Microscopy
Embryos were mounted in methyl salicylate (for HRP-conjugated secondary antibodies) or in 100% glycerol containing 1% n-propyl-gallate (for fluorochrome-conjugated secondary antibodies). Cultured cells were mounted in Gelutol containing 1% n-propyl-gallate. Images were collected using a Bio-Rad MRC 600 system (Bio-Rad, Richmond, CA) and an MRC-850 laser attached to a Nikon (Melville, NY) compound microscope. Images were subsequently transferred and processed in Adobe Photoshop and then transferred into Canvas 3.5 to assemble figures.
RESULTS
Delta Is Proteolytically Processed during Embryonic and Postembryonic Development
In light of recent studies implicating proteolytic processing as a prerequisite for Notch function (Kopan et al., 1996; Blaumueller et al., 1997; Pan and Rubin, 1997), as well as evidence that in many cases post-translational modification is necessary for ligand function (reviewed by Bosenberg and Massagué, 1993), we asked whether Delta is proteolytically processed. We have identified at least four embryonic protein species of the Delta protein in immunoprecipitations (Figure 2A) using antibodies against the Delta extracellular domain: a long form (L, ∼98 kDa), intermediate forms (I1, two or more bands, ∼92–96 kDa; and I2, ∼83 kDa) and a short form (S, ∼68 kDa). We refer to these Delta protein species as “isoforms” from this point on. The proportions of these four isoforms vary during embryonic development. The L isoform predominates in embryonic extracts during the first half of embryogenesis, whereas three of the four isoforms are present in approximately equal amounts in extracts prepared from animals completing embryogenesis. Immunoprecipitation of Delta from larval extracts reveals that these four isoforms are present in relatively equal amounts (Figure 2B). We also find that when embryos are extracted in 100 volumes or more of extraction buffer or when additional protease inhibitors (see MATERIALS AND METHODS) or 1% SDS is added to the extraction buffer, three of the isoforms (i.e., L, I2, and S) are still present in extracts (our unpublished results). Under these conditions, proteases should be either inhibited or sufficiently dilute that proteolysis should not occur during extraction, implying that the L, I2, and S species constitute native Delta isoforms that exist in vivo.
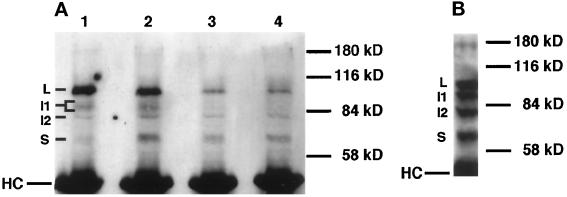
Figure 2.
Immunoprecipitation of Delta protein from detergent-soluble native extracts of staged embryos and larvae using an antibody against the Delta extracellular domain (MAb8A), detected with Delta MAb9B. (A) Staged embryonic extracts (6 mg wet weight/ml) immunoprecipitated using a mAb against the Delta extracellular domain (MAb8A). The same volume of extract was used for immunoprecipitation for each time point. Ages of pooled animals, in hours postoviposition (PO) at 25°C, for each lane are: lane 1, 0–6 h; lane 2, 7–12 h; lane 3, 13–18 h; lane 4, 19–24 h. The bracket next to “I1” indicates that more than one band is detected in the 92- to 96-kDa range. “HC” indicates the IgG heavy chain from the mouse ascites used in the immunoprecipitation. (B) Immunoprecipitation of Delta protein from third instar larval extracts (180 mg wet weight/ml) using MAb8A.
In Drosophila cultured cells programmed to express full-length Delta protein, the Delta L I1, I2, and S isoforms are found in cell extracts (Figure 3A), and Delta S is also found in the surrounding medium (our unpublished results). The apparent molecular weights of the embryonic and larval Delta S isoforms are substantially less than that of the full-length L isoform; yet these Delta S isoforms react with Delta-specific mAbs 9B and 8A, which are specific for the Delta extracellular domain (Figures 2 and 3A). We also find that embryonic Delta S has an apparent molecular weight very similar to that of DeltaSEC1, a secreted form of the Delta extracellular domain, truncated at amino acid 573, which we have expressed in Drosophila cultured cells (Figure 3B). These findings suggest that Delta S could be a proteolytically processed derivative of Delta L, cleaved within the Delta extracellular domain.
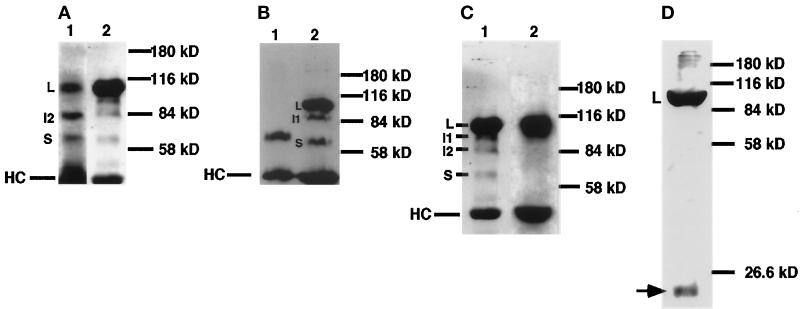
Figure 3.
Identification of Delta isoforms in cultured cells. (A and B) Immunoprecipitation of Delta protein from detergent-soluble native extracts of staged embryos and cultured Drosophila S2 cells using an antibody against the Delta extracellular domain (MAb8A), detected with MAb9B. (A) Comparison of Delta isoforms from embryos 7–12 h PO (lane 1) with Delta isoforms from cultured cells programmed to express full-length Delta protein (∼98 kDa, lane 2). (B) Comparison of Delta isoforms from cultured cells programmed to express a secreted form of the Delta extracellular domain (∼65 kDa, lane 1) with Delta isoforms from an extract of embryos 0–22 h PO (lane 2). (C) Comparison of Delta isoforms immunoprecipitated using MAb8A from cultured cells that express DeltaWTNdeMYC (lane 1) with Delta isoforms immunoprecipitated from DeltaWTNdeMYC+ cells with MAb9E, which binds to the MYC epitope at the C terminus of the intracellular domain (lane 2). (D) Total protein sample from DeltaWTNdeMYC+ cells, probed with MAb9E. Arrow indicates a protein species (∼22 kDa) that reacts with an antibody to the MYC tag at the C terminus of the Delta intracellular domain.
To test this hypothesis, we used the MYC epitope-specific MAb9E to immunoprecipitate Delta from a cell line that expresses full-length Delta with a C-terminal MYC epitope tag (i.e., DeltaWTNdeMYC+ cells). The Delta S isoform is not immunoprecipitated (Figure 3C), suggesting that the intracellular domain is missing from this isoform. In addition, mouse and guinea pig polyclonal antibodies against the Delta intracellular domain (see MATERIALS AND METHODS) do not react with the Delta S isoform but do react with Delta L, I1, and I2 on Western blots of samples containing the Delta isoforms immunoprecipitated using MAb8A (our unpublished results). Antibodies to the N-terminal domain of Delta (Klueg and Muskavitch, unpublished data) do react with Delta L, I1, I2, and S on Western blots of samples containing the Delta isoforms immunoprecipitated using MAb8A (our unpublished results). This implies that I1 and I2 arise by cleavage of Delta L within the intracellular domain, whereas Delta S appears to result from a cleavage that removes the majority, if not all, of the intracellular domain.
In Figure 3D, we show that MAb9E identifies a small protein species (∼22 kDa), which approximately corresponds to the predicted size of the Delta intracellular domain (i.e., Delta IC), in total protein samples from cells that express DeltaWTNdeMYC. Guinea pig polyclonal antibodies against the Delta intracellular domain also react with this 22-kDa protein species (our unpublished results), suggesting that the 22-kDa protein contains a substantial portion of the Delta intracellular domain. A similar band cannot be detected by immunoprecipitation of embryonic, larval, or cultured cell extracts because the predicted apparent molecular weight of the Delta intracellular domain is similar to that of the IgG light chains. These findings, and the similarity in the apparent molecular weight of the Delta S isoform with that of an engineered, secreted form of the Delta extracellular domain (Figure 3B), imply that the Delta S isoform and the Delta IC isoform arise via proteolytic cleavage of Delta within the extracellular domain, near the outer face of the plasma membrane.
To examine further the proposed proteolytic origins of these Delta isoforms, we altered the extraction conditions to determine whether proteolytic processing of Delta could occur in vitro. When extracts are prepared in small volumes of buffer (10 volumes or less), we find that the Delta S isoform predominates in extracts from later stages of embryonic development (Figure 4A). A comparison of Figure 4A with Figure 2A suggests that the Delta S isoform results from proteolytic processing. In addition, when we mix equal numbers of “late” stage embryos and “early” stage embryos before extraction, a loss of the Delta L isoform is apparent (Figure 4B, compare lane 2 with lanes 1 and 3). These “late” stage embryos [22–24 h postoviposition (PO)] contain a significant number of first instar larvae. In a similar experiment in which 19–21 h PO embryos are used, loss of the Delta L isoform is also observed (our unpublished results). When second instar larvae are mixed with early stage embryos (3–5 h PO) before extraction, some Delta L isoform appears to be converted into one or more of the shorter isoforms (Figure 4C, compare lane 2 with lane 1). Prolonged incubation on ice of these “mixed” extracts does not appear to lead to increased loss of the L isoform (our unpublished results).
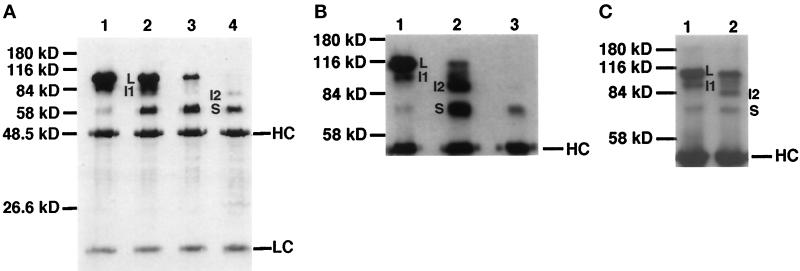
Figure 4.
Immunoprecipitation of Delta protein from detergent-soluble native extracts of staged embryos and larvae using MAb8A against the Delta extracellular domain, detected with MAb9B. (A) Immunoprecipitation of Delta from staged embryonic extracts (300 mg wet weight/ml). The same volume of extract was used for immunoprecipitation for each time point. Ages of pooled animals, in hours PO at 25°C, for each lane were: lane 1, 0–6 h; lane 2, 7–12 h; lane 3, 13–18 h; lane 4, 19–24 h. (B) Immunoprecipitation of Delta from mixed-stage embryonic extracts. Extracts from embryos 22–24 h PO mixed with embryos 2.5–4.5 h PO before extraction and immunoprecipitation are compared with extracts from the single samples (taken from the same egg collections used to prepare the mixed samples): lane 1, 2.5–4.5 h (110 mg/ml); lane 2, 2.5–4.5 h plus 22–24 h (220 mg/ml); lane 3, 22–24 h (110 mg/ml). (C) Immunoprecipitation of Delta from extracts of a mixture of embryos 3–5 h PO and second instar larvae compared with an extract made from embryos 3–5 h PO: lane 1, 3–5 h (45 mg/ml); lane 2, 3–5 h mixed with second instar larvae (a total of 120 mg/ml, of which 45 mg/ml was embryonic tissue 3–5 h PO and 72 mg/ml was second instar larval tissue). The contribution of Delta isoforms from second instar larvae is negligible, based on previous experiments in which it was determined that a minimum of 180 mg/ml of larvae are needed to detect larval Delta isoforms by immunoprecipitation (our unpublished results).
These data imply that when more concentrated extractions are performed, one or more proteolytic activities, present during later stages of embryogenesis and in young larvae, is capable of processing full-length Delta. This activity is released and is functional for a short time during the extraction procedure in concentrated extracts. The data presented in Figure 2 imply that this activity is present during earlier stages, but is either present at lower levels or functions with decreased efficiency because larger relative amounts of Delta S are not generated in 0–6 h PO concentrated extracts.
Multiple Delta Isoforms Exist and Exhibit Distinguishable Subcellular Localization in Vivo
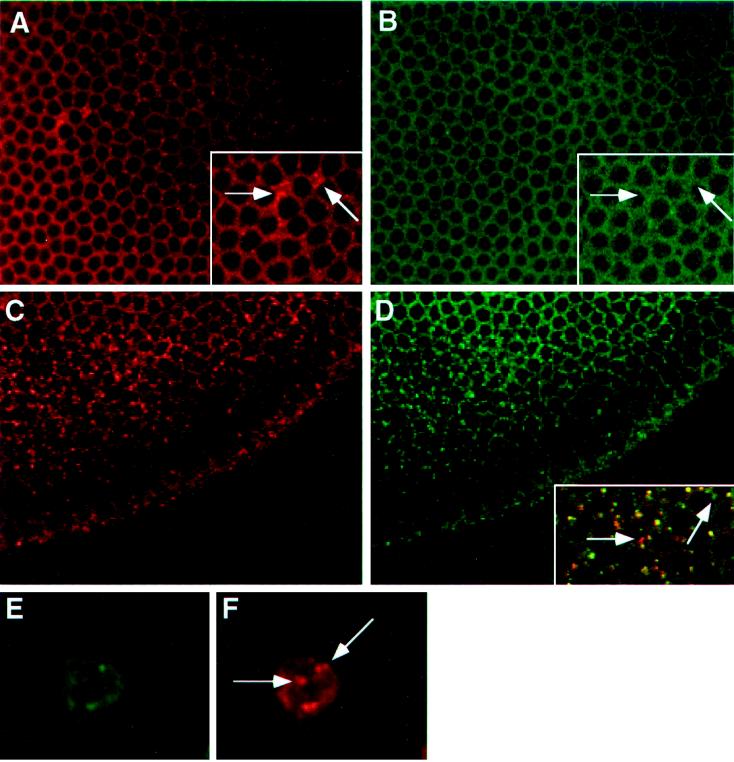
To provide further evidence that multiple Delta isoforms exist in vivo, we have examined the colocalization of the Delta extracellular and intracellular domains during the early stages of embryogenesis, using confocal microscopy. Analyses using antibodies specific for the Delta extracellular domain have shown that Delta localizes to plasma membranes during the precellular and cellular blastoderm stages (Kooh et al., 1993). Immediately before gastrulation, Delta becomes localized in vesicles within the ventral mesodermal anlage, presumably as a result of the down-regulation of Delta within this region (Kooh et al., 1993). Our colocalization studies in cellular blastoderm embryos reveal that antibodies to the Delta intracellular domain localize to plasma membranes (Figure 5B), as do antibodies to the Delta extracellular domain (Figure 5A). However, we detect some subtle differences at this stage in the distributions of Delta extracellular and intracellular domain epitopes. At high magnification, we observe occasional foci of intense staining at or near the plasma membrane with antibodies to the Delta extracellular domain. These foci are not detected with antibodies to the intracellular domain (compare the inset of Figure 5A with that of Figure 5B). Before gastrulation, within the mesodermal anlage, antibodies to the intracellular domain localize to vesicular structures (Figure 5D) as do antibodies to the extracellular domain (Figure 5C). Merging these images reveals that these two classes of Delta antibodies colocalize within a majority of these vesicular structures (Figure 5D, inset). However, a few vesicles appear to react with only antibodies specific for the extracellular or intracellular domain (Figure 5D, inset).
Figure 5.
Immunofluorescent localization of Delta extracellular and intracellular domains in cellular blastoderm embryos, and in cultured cells programmed to express DeltaWTNdeMYC. (A) Cellular blastoderm stained with MAb9B to the Delta extracellular domain. (B) Same embryo as in panel A, showing localization of the Delta intracellular domain (C2 guinea pig polyclonal antisera). Insets in panels A and B, a section of the larger panel presented at higher magnification, illustrate the difference between the localization of Delta extracellular and intracellular domains (arrows). (C) Cellular blastoderm embryo, immediately before gastrulation, showing localization of the Delta extracellular domain detected with MAb9B. At this stage, Delta is plasma membrane-associated, except in the mesodermal anlage where Delta accumulates in vesicles. (D) Same embryo as in panel C, showing localization of the Delta intracellular domain with C2 guinea pig polyclonal antisera. Inset in panel D shows a high magnification, merged image of the same area of panels C and D, to illustrate that Delta extracellular and intracellular domains colocalize in vesicular structures. However, some vesicular structures react with only one of the two domain-specific antibodies (arrows). (E and F) A cultured cell programmed to express DeltaWTNdeMYC. The Delta extracellular domain is detected with GP581 guinea pig polyclonal antiserum in panel E, and localization of the intracellular domain is assessed with MAb9E in panel F. Arrows in panel F indicate vesicles that reacts only with MAb9E.
We have also assessed subtle domain localization differences in DeltaWTNdeMYC+ cells. Colocalization experiments using MAb9E and antibodies to the Delta extracellular domain reveal that, within a single cell, there are occasionally vesicular structures that react with only one of these two classes of antibodies. For example, in Figure 5F, antibodies to the tagged intracellular domain localize to several vesicular structures. A subset of these vesicular structures do not react with antibodies to the Delta extracellular domain (Figure 5E). Collectively, these localization studies in embryos and cultured cells provide evidence for the existence and distinguishable localization of at least three Delta isoforms in vivo: Delta L, Delta S, and Delta IC.
Delta is a Transmembrane Ligand That Exhibits Intercellular Transfer
It has been reported previously that the Delta extracellular domain is taken up by adjacent Notch+ Drosophila cultured cells (Fehon et al., 1990). There has been one other report of the internalization of a membrane-bound ligand by adjacent cells in Drosophila. The transmembrane ligand boss, which interacts with the receptor sevenless, is completely internalized by neighboring sev+ cells in culture and in the course of cell fate specification during retinal development (Krämer et al., 1991; Cagan et al., 1992). Using antibodies to the Delta intracellular domain and the C-terminal MYC tag on DeltaWTNdeMYC+ cells, we have asked whether the entire Delta molecule is transferred into neighboring Notch+ cells and whether or not the trans-endocytosis of Delta by Notch+ cells is dependent on the Delta intracellular domain.
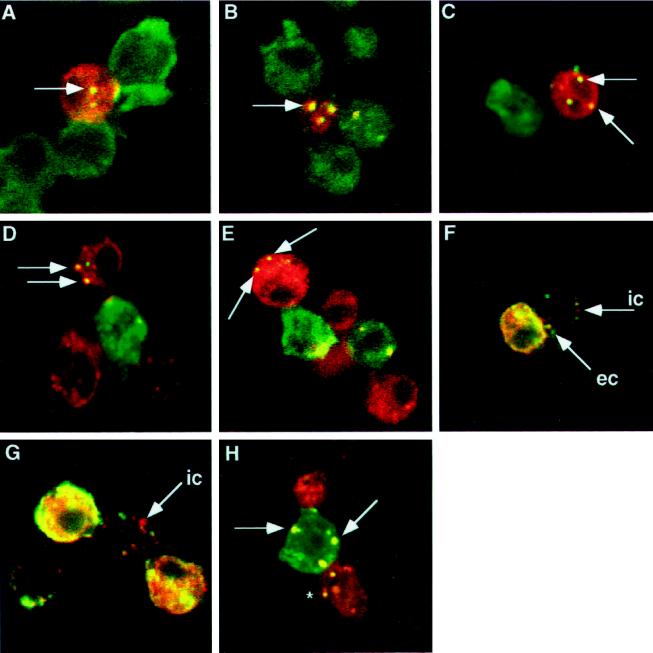
Using the Drosophila S2 cell aggregation assay, we have found that the extracellular domain and intracellular domain of Delta are taken up by neighboring Notch+ cells (Figure 6, A and B, and Table 1). The number of Notch+ cells, adjacent to one or more Delta+ cells, that contain vesicles positive for the Delta intracellular domain is 22%. This is similar to the number of Notch+ cells that contain vesicles positive for the Delta extracellular domain (27%, Table 1). In aggregates of DeltaWTNdeMYC+ cells and Notch+ cells, the Delta C-terminal MYC epitope is also taken up by Notch+ cells at similar frequencies (our unpublished results).
Figure 6.
Trans-uptake of Delta and Notch in Drosophila S2 cell aggregation assays. In panels A-C, antibodies to Delta are detected with fluorescein-conjugated secondary antibodies (green); antibodies to Notch are detected with Texas Red-conjugated secondary antibodies (red). These panels present merged confocal images. Colocalized Delta and Notch appear yellow. (A) Antibodies to the Delta extracellular domain colocalize with antibodies to the Notch intracellular domain in vesicular structures within a Notch+ cell (arrow). (B) Antibodies to the Delta intracellular domain colocalize with antibodies to the Notch intracellular domain in vesicular structures within a Notch+ cell (arrow). (C) Notch+ cells aggregated with cells that express DeltaNGIC, a Delta-neuroglian chimera, in which the Delta intracellular domain is replaced with the neuroglian intracellular domain. Antibodies to the Delta extracellular domain colocalize with antibodies to the Notch intracellular domain within vesicles in Notch+ cells (arrows). (D) In Notch+ cells aggregated with cells that express DeltaNGIC, antibodies to the neuroglian intracellular domain (green) colocalize with antibodies to the Notch extracellular domain (red) within vesicles in Notch+ cells (arrows). (E) In Notch+ cells aggregated with cells that express DeltaDde, which lacks the Delta intracellular domain, antibodies to the Delta extracellular domain (green) colocalize with antibodies to the Notch intracellular domain (red), within vesicles in Notch+ cells (arrows). (F and G) Delta–Notch aggregates stained with antibodies to the Delta extracellular domain (green) and antibodies to the Delta intracellular domain (red). This merged image shows Notch+ cells that contain vesicles (which appear yellow) that costain with the Delta extracellular and intracellular domain-specific antibodies. Some vesicles that are recognized by only the extracellular domain antibody (MAb9B, green vesicles, arrow labeled “ec”) or by only the intracellular domain antibody (C2 guinea pig polyclonal antiserum, red vesicles, arrows labeled “ic”) are also found. (H) In cells that express full-length Delta aggregated with cells that express full-length Notch, antibodies to the Notch intracellular domain (red) colocalize with antibodies to the Delta extracellular domain (green) in Delta+ cells (arrows). The asterisk indicates a Delta+ vesicle inside a Notch+ cell.
Table 1.
trans-Endocytosis of Delta into Notch+ cells
| Delta variant expressed | Delta domain detecteda | % trans-Endocytosisb (SE)c |
|---|---|---|
| DeltaWT | EC domaind | 27 (5)% |
| DeltaWT | IC domaine | 22 (2)% |
| DeltaNGICf | EC domain | 25 (2)% |
| DeltaNGIC | IC domain | 15 (4)% |
| DeltaDdeg | EC domain | 22 (2)% |
The domain in the Delta variant detected in Notch+ cells.
The frequency of trans-endocytosis was recorded as the percentage of Notch+ cells, in contact with Delta+ cells, that contain Delta-positive internalized vesicles.
SE for data are average for three independent experiments.
EC, extracellular.
IC, intracellular.
DeltaNGIC is a Delta variant in which the Delta intracellular domain is replaced with a segment of the neuroglian intracellular domain.
The DeltaDde variant lacks the Delta intracellular domain.
To determine whether the Delta intracellular domain is necessary for the trans-endocytosis of Delta by Notch+ cells, the Delta intracellular domain was replaced with a portion of the neuroglian intracellular domain (Bieber et al., 1989), another Drosophila Type I membrane protein. We find that the extracellular and intracellular domains of this chimera are taken up by neighboring Notch+ cells (Figure 6, C and D, and Table 1). When cells that express a Delta variant that lacks the Delta intracellular domain (i.e., DeltaDde) aggregate with Notch+ cells, the Delta extracellular domain is found in neighboring Notch+ cells (Figure 6E) at a frequency of 22% (Table 1). Therefore, we find that the entire Delta molecule is transferred to Notch+ cells and that trans-endocytosis of the Delta extracellular domain by Notch+ cultured cells is not dependent on the Delta intracellular domain.
When Notch+ cultured cells are mixed with cells programmed to express DeltaSEC1, a secreted form of the Delta extracellular domain, we do not observe Delta–Notch cell aggregates and we do not detect Delta-positive staining in Notch+ cells (our unpublished results). Therefore, either DeltaSEC1 does not bind to Notch in these assays, or it does bind to Notch and we are unable to detect it using our standard immunological methods. If DeltaSEC1 does bind to Notch+ cells, the concentrations of DeltaSEC1 accumulating within these cells are not sufficiently high for us to detect, or complexes between DeltaSEC1 and Notch are endocytosed and degraded more rapidly than complexes between membrane-anchored full-length Delta and Notch. In either case, DeltaSEC1-Notch complexes would escape our detection.
Based on our previous observation that Delta extracellular and intracellular domains can exhibit distinguishable localization in cultured cells programmed to express full-length Delta (Figure 5, E and F), we asked whether the Delta extracellular and intracellular domains can exhibit distinguishable localization after trans-endocytosis of Delta by Notch+ cells. In most cases, we find that antibodies to the Delta extracellular and intracellular domains colocalize to the same vesicular structures in Notch+ cells, after trans-endocytosis of Delta. However, we observe occasional differences. In Figure 6, F and G, we present examples of vesicles within Notch+ cells that stain with antibodies specific for the Delta intracellular domain, but not with extracellular domain-specific antibodies. Several vesicles that react with antibodies to both Delta domains are also evident in these cells.
During our analysis of the trans-endocytosis of Delta by Notch+ cultured cells, we noted that Notch staining is occasionally observed on neighboring Delta+ cells and within Delta+ cells in vesicular structures (Figure 6H). Antibodies against the Notch intracellular domain (Figure 6H) and extracellular domain (our unpublished results) stain vesicles in Delta+ cells within Delta+-Notch+ cell aggregates, implying that the entire Notch molecule can be transferred into neighboring Delta+ cells.
In the course of the trans-endocytosis experiments described above, we found that Delta+ cells and Notch+ cells frequently contain vesicles that are solely Delta- or Notch-positive, respectively. This indicates that Delta and Notch can be cleared from the surfaces of cultured cells in which they are expressed by a mechanism that does not depend on Delta–Notch interactions (e.g., constitutive endocytosis of cell surface proteins; Figure 6, D and H).
Intercellular Transfer of Delta and Notch in Cultured Cells Appears to Occur via a Nonphagocytic Mechanism
The observation that full-length Delta is taken up by Notch+ cells and that full-length Notch is taken up by Delta+ cells led us to investigate the mechanism by which these proteins are transferred. It has been previously noted, in mammalian cultured cells, that patches of membrane can be torn away and left behind at sites of adhesion between filopodia and substrates (Lazarides and Revel, 1979). The transfer of full-length proteins to adjacent cultured cells in Drosophila could involve phagocytosis of substantial portions of membranes at sites of adhesion. In Drosophila, phagocytosis has been proposed as one possible mechanism for internalization of boss–sev complexes in sev+ cells (Cagan et al., 1992; Krämer, 1993). Phagocytosis appears to be the mechanism that underlies membrane turnover in the rod outer segment in Xenopus laevis (Matsumoto et al., 1987). Because full-length Delta is taken up by Notch+ cells, we asked whether detectable amounts of Delta+ plasma membrane are transferred to Notch+ cells during the process of Delta trans-endocytosis.
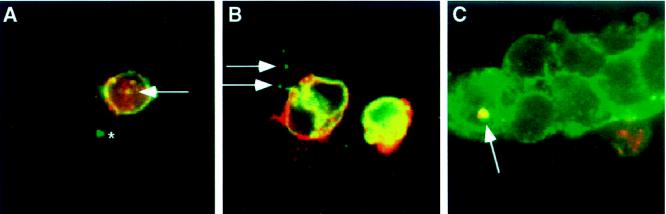
If phagocytosis or a similar mechanism mediates clearance of Delta–Notch complexes from the surfaces of Notch+ cells during Delta–Notch interactions in S2 cells, we would predict that large amounts of membrane from Delta+ cells would be taken up by Notch+ cells during this process. We used vectors supporting expression of either of two full-length Drosophila transmembrane proteins, boss (Krämer et al., 1991) or neuroglian (Hortsch et al., 1990), in cotransfections with an expression vector encoding full-length Delta to mark Delta+ cell membranes. This allowed us to follow the dynamics of bulk plasma membrane transfer during aggregation and trans-endocytosis. In experiments in which Drosophila S2 cells are programmed to express full-length Delta and boss, and then aggregated with Notch+ cells, boss is not detectable in Delta+ vesicles within Notch+ cells (Figure 7A). However, we do find that, on occasion, boss colocalizes with Delta in Delta+ vesicles within Delta+ cells (Figure 7A). In similar experiments, using neuroglian instead of boss to label plasma membranes, we do not observe colocalization of neuroglian in Delta+ vesicles in Notch+ cells (our unpublished results).
Figure 7.
Localization of plasma membrane proteins during intercellular transfer. (A) Delta–Notch aggregates in which boss is coexpressed on the cell surface with Delta. Aggregates are stained with antibodies to the Delta extracellular domain (GP581, green) and with antibodies to boss (mAb-αboss1, red). The asterisk indicates a Delta+ vesicle in an adjacent Notch+ cell; this vesicle is not boss+. Vesicular structures within Delta+ cells stain with Delta and boss antibodies (arrow). (B) Delta-Notch aggregates in which neuroglian (red) is expressed on the cell surface of Notch+ cells (green). Arrows indicate Notch+ vesicles in Delta+ cells; these vesicles are not neuroglian+. (C) Delta–Notch aggregates in which neuroglian (green) is expressed on the surfaces of Delta+ cells. Cells were also labeled for Notch (red). Arrow indicates a neuroglian+/Notch+ vesicle in a Delta+ cell.
Experiments in which Drosophila S2 cells programmed to express neuroglian and Notch are aggregated with Delta+ cells reveal that neuroglian does not colocalize with Notch in Notch+ vesicles within Delta+ cells (Figure 7B). Similar results are obtained when boss is used as the membrane marker (our unpublished results). In experiments with cells programmed to express Delta and neuroglian, neuroglian colocalizes with Notch in vesicles within Delta+ cells, suggesting that a substantial amount of Delta+ cell membrane is internalized when Notch is taken up by these cells (Figure 7C). In addition, in cells that express Notch and neuroglian, neuroglian colocalizes with Delta in Notch+ cells, suggesting that Notch+ plasma membrane is internalized when Delta is taken up by Notch+ cells. These results imply that substantial amounts of plasma membrane proteins proximal to Delta–Notch complexes are not cotransferred with ligand into receptor-bearing cells during trans-endocytosis of Delta or Notch. However, substantial amounts of plasma membrane-associated proteins are internalized, along with Delta–Notch complexes, during the cis-endocytic clearance of these ligand-receptor complexes from the surfaces of receptor-bearing cells.
DISCUSSION
We provide the first evidence that Delta is proteolytically processed and that multiple Delta isoforms exist in vivo. The Delta S isoform, which is present in embryos, larvae, and cultured cells, is comprised of a portion of the Delta extracellular domain. Delta S is secreted by cultured cells into the surrounding medium and has an apparent molecular weight similar to that of an engineered, secreted form of the Delta extracellular domain. It therefore appears that the Delta S isoform is generated after proteolytic cleavage within the Delta extracellular domain. The Delta IC isoform, which accumulates in cultured cells, is comprised of a portion of the Delta intracellular domain. Previous studies have demonstrated that a Delta IC domain fragment that includes the basic sequence KRKRKR, a putative stop transfer signal near the inner membrane face (Kopczynski et al., 1988), is metabolically stable and exhibits nuclear localization when overexpressed in vivo (Sun and Artavanis-Tsakonas, 1996). However, antibodies specific for the Delta intracellular domain do not stain nuclei in Delta+ cultured cells or embryos (Klueg and Muskavitch, unpublished data). This implies that the Delta IC isoform we observe in cultured cells either contains the Delta transmembrane domain or is generated by an additional proteolytic cleavage C-proximal to the KRKRKR sequence within the Delta intracellular domain. Mixing experiments involving extracts from different developmental stages suggest that one or more additional Delta isoforms (e.g., Delta I1 and I2) also arise via proteolysis. The probable origin of the Delta I1 and I2 isoforms as the result of additional proteolytic cleavages suggests that Delta may be subject to a number of processing events during development.
Cleavage of the full-length Delta protein to generate the Delta S isoform could be necessary for the activation of the Delta ligand, and a prerequisite for ligand-receptor binding. However, a number of findings argue against this hypothesis. Previous studies have shown that full-length Delta binds to Notch (Fehon et al., 1990), and work described in this paper reveals that full-length Delta is taken up by Notch+ cultured cells. Cleavage of the Delta ligand is therefore not a prerequisite for binding of Delta to the Notch receptor. We also find that Delta S accumulates in the medium from cultured cells programmed to express full-length Delta, even in the absence of Notch+ cells. This indicates that a processing mechanism that generates the Delta S isoform can operate before the interaction of Delta with Notch or to the clearance of Delta from the cell surface during the process of trans-endocytosis into Notch+ cells. Finally, expression of a Delta variant comprised of a secreted Delta extracellular domain has been shown to interfere with the signaling of endogenous Delta in vivo during retinal development (Sun and Artavanis-Tsakonas, 1997). Collectively, these observations imply that generation of the Delta S isoform is not a prerequisite for Notch binding or for the creation of an activated form of the Delta ligand.
The preponderance of evidence therefore supports the hypothesis that proteolytic cleavage occurs during Delta inactivation and contributes to the down-regulation of Delta–Notch signaling in vivo. Processing of internalized proteins in endocytic compartments is known to play a significant role in the down-regulation of several ligand classes, including EGF and insulin (reviewed by Authier et al., 1996). Once the full-length Delta protein enters the endocytic pathway, it could be proteolytically processed and the extracellular and intracellular domains subsequently transported to different compartments wherein they could meet different fates (e.g., degradation vs. recycling). The existence of Delta extracellular and intracellular domain-specific vesicles in cells within the embryonic mesodermal anlage, concomitant with clearance of Delta from the surfaces of these cells, supports the hypothesis that proteolytic processing is involved in Delta down-regulation in vivo. The fact that we detect differential localization of intracellular and extracellular domains in Notch+ cells after trans-endocytosis of Delta further supports the premise that proteolytic processing is involved in Delta down-regulation after ligand–receptor binding. We also detect apparent accumulations of one or more Delta species that react with antibodies against the extracellular domain, yet fail to react with antibodies against the intracellular domain, at or near cell surfaces in cellular blastoderm embryos. These observations imply that cleavage of the Delta intracellular domain from the full-length protein can occur at or near the cell surface. The mechanisms that underlie the differential localization of Delta isoforms may be complex, and further examination of processing events will require improved methods and markers for identifying different endocytic and exocytic compartments in Drosophila. Future identification of one or more proteolytic cleavage sites within Delta will enable us to create Delta variants that cannot be cleaved in vivo, and to assess the role(s) of Delta proteolysis in Delta–Notch signaling during development.
We provide the first evidence that the entire Delta molecule is transferred intercellularly and that this trans-endocytosis in cultured cells is not dependent on the Delta intracellular domain. These results imply that the domains involved in the molecular interaction between Delta and Notch are sufficient to initiate the trans-endocytosis of this transmembrane ligand, i.e., that the Delta intracellular domain is not required for this process in cultured cells. In C. elegans, a lag-2 variant in which a portion of the intracellular domain was replaced with β-galactosidase has been shown to be transferred intercellularly in vivo (Henderson et al., 1994). The Lag-2 extracellular domain contains a domain and two EGF-like repeats, reflecting similarity to Delta, and is thought to act as a ligand for the Notch-like receptor, Glp-1 (Yochem and Greenwald, 1989). Collectively, these data invite the speculation that intercellular transfer of Notch ligands may be intrinsic to Notch-mediated signaling processes or may be necessary for the down-regulation of the ligand–receptor complexes. The parallels between processing and subcellular localization of Delta isoforms in embryos and cultured cells imply that the Drosophila cultured cell system recapitulates at least some of the features of the Delta-Notch ligand–receptor interaction that operate during embryonic and metamorphic development. We are currently attempting to assess whether trans-endocytosis of Delta occurs during Drosophila development, in a manner analogous to that which we have discovered in cultured cells.
The uptake of Delta ligand by Notch receptor-bearing cells may be relevant solely to the inactivation of signal-receptor complexes and the down-regulation of signaling. However, it is tempting to speculate that such an intercellular transfer of ligand may be the basis for an activity of Delta that operates in a signal-receiving cell after endocytosis of the Delta-Notch complex. This speculation is encouraged, in part, by recent observations suggesting that endocytosis is required in cells that receive Notch-mediated signals, based on somatic mosaic analysis of shibire function in Drosophila development (Seugnet et al., 1997).
Furthermore, we note that the coendocytosis of Delta and Notch in cultured Drosophila cells is reminiscent of previous observations of Delta and Notch colocalization in vesicles within the larval retina (Kooh et al., 1993). This colocalization may reflect the down-regulation of Delta-Notch signal–receptor complexes during cell fate specification in the metamorphic retina. More recently, Krämer and Phistry (1996) have found that Delta and boss proteins appear to colocalize in vesicles in the developing retina. This colocalization could reflect some functional association between Delta and boss during retinal development. However, we favor the hypothesis that this latter observation reflects the coalescence of import vesicles in the endocytotic pathway within these cells, in the absence of genetic data suggesting the existence of a functional relationship between Delta and boss.
There has been past speculation that the intercellular transfer of membrane-bound ligands such as boss or Delta in Drosophila involves a phagocytic mechanism. We do not detect the trans-endocytosis from Delta+ cultured cells into Notch+ cells of plasma membrane proteins in addition to Delta. This suggests that a phagocytic mechanism, by which substantial portions of Delta+ total cell membrane at sites of intercellular apposition are taken up by Notch+ cells, does not mediate Delta trans-endocytosis in Drosophila S2 cells. However, our studies do not rule out the possibility that small amounts of lipid from Delta+ cells are taken up by Notch+ cells during Delta trans-endocytosis. There have been few studies assessing the membrane dynamics between adjacent cells during receptor-mediated endocytosis, as most studies of ligand-stimulated endocytosis in cultured cell systems have involved soluble ligands. Interestingly, there have been biophysical studies on the induced detachment of red blood cells that reveal that instead of breaking receptor–ligand bonds (e.g., antibody–antigen complexes), membrane-spanning antigen molecules are preferentially extracted from cell membranes (Evans et al., 1991; Xia et al., 1994). Such analyses suggest that the extraction of some membrane-spanning proteins (e.g., those not cross-linked to other proteins and polysaccharides through the cytoskeletal matrix) is energetically more favorable than the disruption of antigen–antibody interactions. Such membrane-spanning proteins are held in the membrane by intermolecular forces that are weaker than the bond strength calculated for antigen–antibody and other ligand–receptor complexes (e.g., Xia et al., 1994, and references within). The operation of such an “extraction” mechanism in vivo would intrinsically limit the number of proximate proteins within the lipid bilayer that would be taken up (nonspecifically) during the trans-endocytosis of ligand-receptor complexes. Alternatively, an “exclusion” mechanism could operate by which membrane-associated proteins not directly involved in a particular ligand-receptor interaction are actively or passively excluded from a region of the plasma membrane with a high local concentration of ligand-receptor complexes. Either mechanism could account for our observations that marker proteins are not transferred to adjacent cells during trans-endocytosis of Delta into Notch+ cells and Notch into Delta+ cells.
Finally, we find, to our surprise, that Notch is transferred by trans-endocytosis into Delta+ cultured cells, just as Delta is transferred by trans-endocytosis into Notch+ cultured cells. Furthermore, Notch extracellular and intracellular domain epitopes are transferred in this manner, implying that the full-length Notch protein may be transferred as a transmembrane ligand into Delta+ cells. It has been reported that Notch functions cell nonautonomously in some developmental contexts (Technau and Campos-Ortega, 1987; Baker and Schubiger, 1996), and it has been suggested that the Notch extracellular domain may function as a ligand during epidermal development (Baker and Schubiger, 1996). In lymphocytes, data suggest that the membrane proteins CD21 and CD23 may act as ligands and receptors for each other reciprocally, depending on context (reviewed in Bosenberg and Massagué, 1993). The ability of Delta and Notch to each function as ligand and receptor could constitute one basis for reciprocal neurogenic signaling during development among cells within equivalence groups and at developmental boundaries.
ACKNOWLEDGMENTS
The authors would like to thank Allan Bieber and Helmut Krämer for providing antibodies and constructs; Annette Parks and Scott Shepard for the use of unpublished constructs; and to Annette Parks and Bill Saxton for their editorial critiques. This work was supported by NIH grant GM-33291 (to M.A.T.M.) and American Cancer Society postdoctoral fellowship PF-4328 (to K.M.K.)
REFERENCES
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Authier F, Posner BI, Bergeron JJM. Endosomal proteolysis of internalized proteins. FEBS Lett. 1996;389:55–60. doi: 10.1016/0014-5793(96)00368-7. [DOI] [PubMed] [Google Scholar]
- Baker R, Schubiger G. Autonomous and nonautonomous Notch functions for embryonic muscle and epidermis development in Drosophila. Development. 1996;122:617–626. doi: 10.1242/dev.122.2.617. [DOI] [PubMed] [Google Scholar]
- Bate, M., Rushton, E., and Frasch, M. (1993). A dual requirement for neurogenic genes in Drosophila myogenesis. Dev. Suppl. 149–161. [PubMed]
- Bieber AJ, Snow PM, Hortsch M, Patel NH, Jacobs JR, Traquina ZR, Schilling J, Goodman CS. Drosophila neuroglian: a member of the immunogobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 1989;59:447–460. doi: 10.1016/0092-8674(89)90029-9. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Andrews ME, Raff RA. Fibropellins, products of an EGF repeat-containing gene, form a unique extracellular matrix structure that surrounds the sea urchin embryo. Dev Biol. 1991;146:89–99. doi: 10.1016/0012-1606(91)90449-d. [DOI] [PubMed] [Google Scholar]
- Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- Bosenberg MW, Massagué J. Juxtacrine cell signaling molecule. Curr Opin Cell Biol. 1993;5:832–838. doi: 10.1016/0955-0674(93)90032-l. [DOI] [PubMed] [Google Scholar]
- Bunch TA, Grinblat Y, Goldstein LSB. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan RL, Krämer H, Hart AC, Zipursky SL. The bride of sevenless and sevenless interaction: internalization of a transmembrane ligand. Cell. 1992;69:393–399. doi: 10.1016/0092-8674(92)90442-f. [DOI] [PubMed] [Google Scholar]
- Chang C-P, et al. Ligand-induced internalization of the epidermal growth factor receptor is mediated by the multiple endocytic codes analogous to the tyrosine motif found in constitutively internalized receptors. J Cell Biol. 1993;268:19312–19320. [PubMed] [Google Scholar]
- Corbin V, Michelson AM, Abmayr SM, Neel V, Alcamo E, Maniatis T, Young MW. A role for the Drosophila neurogenic genes in mesoderm differentiation. Cell. 1991;6:311–323. doi: 10.1016/0092-8674(91)90183-y. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, et al. Conservation of the Notch signaling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Berk D, Leung A. Detachment of agglutinin-bonded red blood cells. I. Forces to rupture molecular-point attachments. Biophys J. 1991;59:838–848. doi: 10.1016/S0006-3495(91)82296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Gumbiner BM. Cell contact-dependent signaling. Dev Biol. 1996;180:445–454. doi: 10.1006/dbio.1996.0318. [DOI] [PubMed] [Google Scholar]
- Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MA, T, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta. two EGF homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- Fleming RJ, Scottgale TN, Diederich RJ, Artavanis-Tsakonas S. The gene Serrate encodes a putative EGF-like transmembrane essential for proper ectodermal development in Drosophila melanogaster. Genes Dev. 1990;4:2188–2201. doi: 10.1101/gad.4.12a.2188. [DOI] [PubMed] [Google Scholar]
- Grell M, Douni E, Wajant H, Lohden M, Clauss M, Maxeiner W, Kollias G, Pfizenmaier K. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80-kDa tumor necrosis factor receptor. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- Hart AC, Krämer H, Zipursky SL. Extracellular domain of the boss transmembrane ligand acts as an antagonist of the sev receptor. Nature. 1993;361:732–736. doi: 10.1038/361732a0. [DOI] [PubMed] [Google Scholar]
- Hartenstein AY, Rugendorff A, Tepass U, Hartenstein V. The function of the neurogenic genes during epithelial development in the Drosophila embryo. Development. 1992;116:1203–1219. doi: 10.1242/dev.116.4.1203. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Hopp TP, Prickett KS, Price VL, Libby RT, March CJ, Cerretti DP, Urdal DL, Conlon PJ. A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio/Technology. 1988;6:1204–1210. [Google Scholar]
- Hortsch M, Bieber AJ, Patel NH, Goodman CS. Differential splicing generates a nervous system-specific form of Drosophila neuroglian. Neuron. 1990;4:697–709. doi: 10.1016/0896-6273(90)90196-m. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Wang YE, Marikar Y, Bieber AJ. The cytoplasmic domain of the Drosophila cell adhesion molecule neuroglian is not essential for its homophilic adhesive properties in S2 cells. J Cell Biol. 1995;270:18809–18817. doi: 10.1074/jbc.270.32.18809. [DOI] [PubMed] [Google Scholar]
- Huppert SS, Jacobsen TL, Muskavitch MAT. Feedback regulation is central to Delta-Notch signaling required for Drosophila wing vein morphogenesis. Development. 1997;124:3283–3291. doi: 10.1242/dev.124.17.3283. [DOI] [PubMed] [Google Scholar]
- Kooh PJ, Fehon RG, Muskavitch MAT. Implications of dynamic patterns of Delta and Notch expression for cellular interactions during Drosophila development. Development. 1993;117:493–507. doi: 10.1242/dev.117.2.493. [DOI] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski CC, Alton AK, Fechtel K, Kooh PJ, Muskavitch MA. Delta, a Drosophila neurogenic gene, is transcriptionally complex and encodes a protein related to blood coagulation factors and epidermal growth factors of vertebrates. Genes Dev. 1988;2:1723–1735. doi: 10.1101/gad.2.12b.1723. [DOI] [PubMed] [Google Scholar]
- Krämer H, Cagan RL, Zipursky SL. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature. 1991;352:207–212. doi: 10.1038/352207a0. [DOI] [PubMed] [Google Scholar]
- Krämer H. Patrilocal cell-cell interactions: sevenless captures its bride. Trends Cell Biol. 1993;3:103–105. doi: 10.1016/0962-8924(93)90165-w. [DOI] [PubMed] [Google Scholar]
- Krämer H, Phistry M. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J Cell Biol. 1996;133:1205–1215. doi: 10.1083/jcb.133.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarides E, Revel JP. The molecular basis of cell movement. Sci Am. 1979;240:100–113. doi: 10.1038/scientificamerican0579-100. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Jiménez F, Dietrich U, Campos-Ortega JA. On the phenotype and development of mutants of early neurogenesis in Drosophila melanogaster. Roux’s Arch Dev Biol. 1983;192:62–74. doi: 10.1007/BF00848482. [DOI] [PubMed] [Google Scholar]
- Lieber T, Wesley C, Alcamo E, Hassel B, Krane J, Campos-Ortega JA, Young MW. Single amino acid substitutions in EGF-like elements of Notch and Delta modify Drosophila development and affect cell adhesion in vitro. Neuron. 1992;9:847–859. doi: 10.1016/0896-6273(92)90238-9. [DOI] [PubMed] [Google Scholar]
- Matsumoto B, Defoe DM, Besharse JC. Membrane turnover in rod photoreceptors: ensheathment and phagocytosis of outer segment distal tips by pseudopodia of the retinal pigment epithelium. Proc R Soc Lond B Biol Sci. 1987;230:339–354. doi: 10.1098/rspb.1987.0023. [DOI] [PubMed] [Google Scholar]
- Muskavitch MAT. Delta-Notch signaling and Drosophila cell fate choice. Dev Biol. 1994;166:415–430. doi: 10.1006/dbio.1994.1326. [DOI] [PubMed] [Google Scholar]
- Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- Parody TR, Muskavitch MAT. The pleiotropic function of Delta during postembryonic development of Drosophila melanogaster. Genetics. 1993;135:527–539. doi: 10.1093/genetics/135.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Muskavitch MAT. Delta function is required for bristle organ determination and morphogenesis in Drosophila. Dev Biol. 1993;157:484–496. doi: 10.1006/dbio.1993.1151. [DOI] [PubMed] [Google Scholar]
- Parks AL, Turner FR, Muskavitch MAT. Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech Dev. 1995;50:201–216. doi: 10.1016/0925-4773(94)00336-l. [DOI] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas LF, Cherbas PT, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Robinson MS, Watts C, Zerial M. Membrane dynamics in endocytosis. Cell. 1996;84:13–21. doi: 10.1016/s0092-8674(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Seugnet L, Simpson P, Haenlin M. Requirement for dynamin during Notch signalling in Drosophila neurogenesis. Dev Biol. 1997;199:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- Shepard SB. A Genetic and Molecular Analysis of Interactions between the Drosophila melanogaster neurogenic genes Delta and Notch. Ph.D. Thesis. Bloomington, IN: Indiana University; 1991. [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- Sun X, Artavanis-Tsakonas S. The intracellular deletions of DELTA and SERRATE define dominant negative forms of the Drosophila Notch ligands. Development. 1996;122:2465–2474. doi: 10.1242/dev.122.8.2465. [DOI] [PubMed] [Google Scholar]
- Sun X, Artavanis-Tsakonas S. Secreted forms of DELTA and SERRATE define antagonists of Notch signaling in Drosophila. Development. 1997;124:3439–3448. doi: 10.1242/dev.124.17.3439. [DOI] [PubMed] [Google Scholar]
- Technau GM, Campos-Ortega JA. Cell autonomy of expression of neurogenic genes of Drosophila melanogaster. Proc Natl Acad Sci USA. 1987;84:4500–4504. doi: 10.1073/pnas.84.13.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas U, Speicher SA, Knust E. The Drosophila gene Serrate encodes and EGF-like transmembrane protein with a complex expression pattern in embryos and wing discs. Development. 1991;111:749–761. doi: 10.1242/dev.111.3.749. [DOI] [PubMed] [Google Scholar]
- Vässin H, Bremer KA, Knust E, Campos-Ortega JA. The neurogenic Delta of Drosophila melanogaster is expressed in neurogenic territories and encodes a putative transmembrane protein with EGF-like repeats. EMBO J. 1987;6:3431–3440. doi: 10.1002/j.1460-2075.1987.tb02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vässin H, Campos-Ortega JA. Genetic analysis of Delta, a neurogenic gene of Drosophila melanogaster. Genetics. 1987;116:433–445. doi: 10.1093/genetics/116.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus Notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Yochem J, Greenwald I. glp-1 and lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell. 1989;58:553–563. doi: 10.1016/0092-8674(89)90436-4. [DOI] [PubMed] [Google Scholar]
- Xia Z, Goldsmith HL, van de Ven TG M. Flow-induced detachment of red blood cells adhering to surfaces by specific antigen-antibody bonds. Biophys J. 1994;66:1222–1230. doi: 10.1016/S0006-3495(94)80906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]