Abstract
Whether the cell nucleus is organized by an underlying architecture analagous to the cytoskeleton has been a highly contentious issue since the original isolation of a nuclease and salt-resistant nuclear matrix. Despite electron microscopy studies that show that a nuclear architecture can be visualized after fractionation, the necessity to elute chromatin to visualize this structure has hindered general acceptance of a karyoskeleton. Using an analytical electron microscopy method capable of quantitative elemental analysis, electron spectroscopic imaging, we show that the majority of the fine structure within interchromatin regions of the cell nucleus in fixed whole cells is not nucleoprotein. Rather, this fine structure is compositionally similar to known protein-based cellular structures of the cytoplasm. This study is the first demonstration of a protein network in unfractionated and uninfected cells and provides a method for the ultrastructural characterization of the interaction of this protein architecture with chromatin and ribonucleoprotein elements of the cell nucleus.
INTRODUCTION
The presence of an organizing principal within the cell nucleus that is analagous to the cytoskeleton has been hotly contested over the years. Recently, it has become practical to study directly the dynamics and motion of chromatin and nonchromatin elements of the cell nucleus. Consequently, in the absence of a convincing demonstration of a nuclear skeleton within unfractionated nuclei, it is possible to determine whether biomolecules behave as if they are embedded in an organizing supramolecular structure or whether they are subject to substantial Brownian motion. The results of such studies are compelling. Chromatin (Abney et al., 1997; Marshall et al., 1997; Kanda et al., 1998; Zink et al., 1998; Sullivan et al., 1999) and nonchromatin structures such as nuclear speckles (Misteli et al., 1997) and foci enriched in transcription factors (Hendzel, Bisgrove, and Godbout, unpublished observations) are constrained from substantial Brownian motion, strongly supporting the view that there is a component or components of the cell nucleus that function to restrict the mobility of macromolecular complexes within the cell nucleus.
The cell nucleus has a number of compositionally distinct domains (for review, see Schul et al., 1998). This implies a mechanism for organizing biochemical components into discrete supramolecular structures. An example is the nonchromatin extranucleolar structure of the cell nucleus called the interchromatin granule cluster (IGC). This structure is more widely known by the nuclear speckles that are observed by indirect immunofluorescence when cells are stained with antibodies recognizing small nuclear ribonuclear proteins (RNPs) or SC-35 (for review, see Spector, 1993). The well-defined boundaries of nuclear speckles, when imaged by indirect immunofluorescence, indicates that they are discrete nuclear structures. Their large dimensions indicate that a physical continuity is maintained over relatively long distances. However, when these structures are imaged by standard electron microscopy, they appear as clusters of discrete 20- to 25-nm ribonucleoprotein particles. The question arises of what the basis is for the cluster integrity. In another example, transcriptional regulators exist in many hundred smaller nuclear foci that occur both in association and away from chromatin (van Steensel et al., 1995; Grande et al., 1997; Hendzel et al., 1998; Noordmans et al., 1998). It is compelling to invoke the presence of a protein architecture to organize these factors into the domains observed in both fixed (van Steensel et al., 1995; Grande et al., 1997) and unfixed cells (Htun et al., 1996; Misteli et al., 1997; Fejes-Toth et al., 1998; Sleeman et al., 1998; Hendzel, Bisgrove, and Godbout, unpublished observations).
A nucleus that spatially organizes biomolecules through specialization on an underlying architecture may be fundamentally different in the mechanisms that serve to transcribe, replicate, and repair DNA, process and export RNA, and transduce signals from that of a nucleus that is not ordered beyond the folding of chromatin. Consequently, it is essential to define and characterize such an architecture if one exists. The prospect that a definable protein architecture exists in the cell nucleus was first brought to light by biochemical fractionation experiments pioneered by Berezney and Coffey (1974, 1977). The original preparation used 2 M NaCl and nuclease digestion to extract a DNA-based structure. Berezney and Coffey introduced the term “nuclear matrix” to refer to this high-salt, DNase-resistant fraction of the cell nucleus. Despite the absence of direct evidence that these procedures generate nuclear structure through a precipitation of soluble nuclear components, experiments involving the isolation of nuclear structures have often been dismissed on this basis. In the face of mounting skepticism, methods involving radically different isolation procedures were developed (for review, see Martelli et al., 1996) and, perhaps surprisingly, were found to generate nuclear remnants with similar morphological properties to the original preparation of Berezney and Coffey (1974, 1977). The most elegant procedure involves encapsulating cells in agarose and digesting the chromatin with restriction endonucleases, followed by electroeluting the chromatin in “physiological buffers” (Jackson and Cook, 1985). This isolation procedure enables the visualization within the interchromatin space of an intermediate filament-like protein network, which closely resembles preparations by more harsh salt extraction procedures (Jackson and Cook, 1988; He et al., 1990).
If a network of protein and/or RNA is present in the cell nucleus, as supported by the extensive nuclear matrix literature, it is essential to develop an approach that is capable of ultrastructural analysis without disrupting individual components of the cell nucleus. This is particularly true for chromatin, which is the principal basis for the existence of the cell nucleus as a separate cellular compartment. To this end, we have been applying and optimizing an analytical microscopic technique for the study of nuclear components. Electron spectroscopic imaging (ESI) couples a conventional transmission electron microscope with an analytical imaging spectrometer. The technique is analogous to the resolution of different energies of light to extract compositional information (e.g., fluorescence microscopy) in light microscopy. The separation of electrons that vary in energy after interacting with a specimen can be exploited to extract compositional information from the electron microscope. Detailed descriptions of the applications of this method to the study of the structure and composition of the cell nucleus have been presented previously (Hendzel and Bazett-Jones, 1996; Bazett-Jones and Hendzel, 1999; Bazett-Jones et al., 1999). Using ESI, we have established that it is possible to resolve nucleic acids in nucleoprotein complexes and to quantify their mass and nucleic acid contents. This is achieved by imaging with electrons that have lost characteristic amounts of energy through interactions with phosphorus and nitrogen atoms of the specimen.
In this study, we apply phosphorus and nitrogen mapping to characterize the imaging properties of known protein-based components of the cell. Mitochondria, nuclear pores, the nuclear lamina, and cytoskeletal remnants are all shown to be characteristically rich in nitrogen but deficient in phosphorus. These components can be distinguished from the phosphorus-rich nucleic acid-containing structures of the cell nucleus. Having established the imaging properties of cellular components of known protein composition, we then addressed whether paraformaldehyde-fixed nuclei show evidence of a nuclear protein component. Our fixation methods are identical to the preparations most commonly used for indirect immunofluorescent analysis of nuclear organization. We provide evidence that the interchromatin space, including IGCs, is rich in complexes that are composed predominantly of protein. A structural role for this protein component is most strongly indicated within IGCs, where the granules are embedded and linked together by a protein-based architecture. This study provides the first evidence of a structural component of the cell nucleus in standard cytological preparations and in the absence of viral infection or nuclear fractionation.
MATERIALS AND METHODS
Tissue Culture and Fixation
Indian muntjac fibroblast, SK-N-SH neuroblastoma, and 10T½ cells were plated onto the polypropylene caps of sterile 50-ml conical centrifuge tubes and cultured under conditons recommended by the American Type Culture Collection (Vienna, VA). After 2 d in culture, when cells were at a cell density of 40–80% coverage of the growth surface, the cell cultures were fixed with 1% paraformaldehyde in PBS, pH 7.2, for 5 min at 22°C. After fixation, cells were rinsed with PBS and dehydrated through a graded ethanol series beginning at 35%. Incubations were 30–60 min per step; the final dehydration step used Quetol 651 and was performed overnight. The cells were then infiltrated with complete embedding medium through two changes of embedding medium over the course of 24 h. Finally, blocks were polymerized at 65°C for 2 d.
Electron Microscopy
Detailed descriptions of the electron microscopy procedure are presented elsewhere (Bazett-Jones and Hendzel, 1999; Bazett-Jones et al., 1999). Briefly, polymerized blocks were sectioned at ∼30 nm thickness using a diamond knife (Drukker, Cuijk, The Netherlands). The sections were placed directly on 1000-mesh uncoated copper grids. Electron micrographs were obtained using a Gatan (Pleasanton, CA) 14-bit slow-scan cooled charge-coupled device coupled to a Zeiss (Thornwood, NY) EM 902 transmission electron microscope containing a prism–mirror–prism spectrometer.
Quantification and Image Analysis
To quantify phosphorus and nitrogen concentrations, regions devoid of mass density were used to normalize 14-bit digital images such that the reference image and the element-enhanced image had equivalent backgrounds. After normalization, masks were drawn around individual structures or regions, and the net phosphorus and net nitrogen values were determined by subtracting the mass-dependent pre-edge image from the element-enhanced image. This yields values, in arbitrary units, for element densities within the regions sampled.
RESULTS
ESI of Thin Sections
ESI has been used to image and quantitatively analyze thin sections of cells prepared by aldehyde fixation. Previously, we have examined the organization of proteins and nucleic acids in nuclei of Triton X-100–extracted cell nuclei (Hendzel and Bazett-Jones, 1996). In these and subsequent studies (Hendzel et al., 1998, Bazett-Jones and Hendzel, 1999, Bazett-Jones et al., 1999), we have optimized methods for distinguishing protein- from nucleic acid-containing structures. The collection of an energy loss series enables a quantitative evaluation of the distribution of the elements phosphorus and nitrogen within structures of the specimen. The phosphorus-rich DNA and RNA structures are imaged well at energy losses above the phosphorus L2,3 ionization edge, which occurs at 132 electron volts (eV). In this region of the energy loss spectrum, it is more difficult to adequately image structures that are deficient in phosphorus, such as protein-based structures. They, however, can be imaged clearly by taking advantage of their nitrogen content. Because both protein and nucleic acid are nitrogen rich, the nitrogen signal quantitatively reflects the distribution of both of these cellular components. Because the nitrogen signal is present at a position in the spectra where a strong background is generated by the embedding medium because of a background carbon-specific signal, these structures are typically not well contrasted in the original image sets collected near the nitrogen energy loss peak. Nevertheless, this background can be minimized by selecting the appropriate embedding medium and by collecting images on sections less than 30 nm in thickness. The background that is present in the original images is virtually eliminated once net nitrogen images are generated. The net nitrogen images provide a relatively high contrast and quantitative morphological representation of the protein-rich structures that are difficult to image elsewhere in the energy loss spectrum.
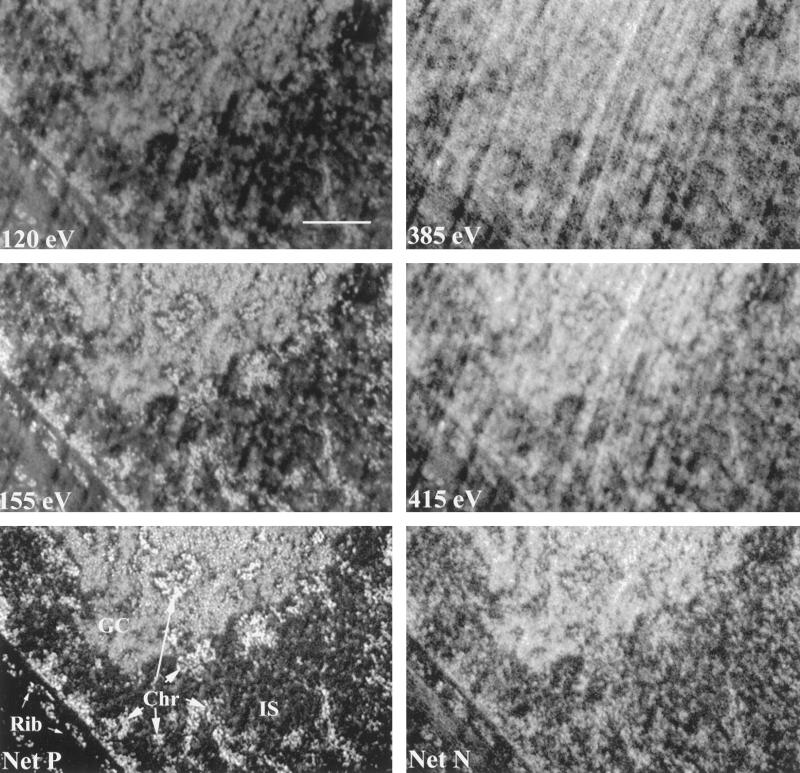
Figure 1 shows an energy loss series collected to obtain phosphorus- and nitrogen-specific maps. The image collected at 155 eV is enhanced in phosphorus, whereas the 120-eV image contains predominantly mass information only. Subtracting this 120-eV image from the 155-eV image produces a net phosphorus image. (Structures are represented as gray on a black background.) At this resolution, there are three prominent cellular structures that generate strong phosphorus signals. These are the granular component of the nucleolus (GC; Figure 1, lower left panel), chromatin organized into structures of ≥30 nm (Fig. 1, Chr), and ribosomes (Fig. 1, Rib). The interchromatin space (Figure 1, IS) appears open, with only occasional RNA-containing particles dispersed within this space. Figure 1, middle row, shows a 385-eV mass-dependent image that is used to extract the nitrogen-specific signal (Net N) from a nitrogen enhanced 415-eV energy loss image. In the net nitrogen image, the most prominent structure is the nucleolus. In the region shown, only the granular component of the nucleolus is seen. Although the granular component of the nucleolus is rich in pre-rRNA–containing granules, which are responsible for the generation of a strong phosphorus signal (Figure 1, Net P), it is apparent that there is a much greater mass component provided by protein than, for example, in regions of compact chromatin. Consequently, the nucleolus has a lower nucleic acid density but a similar mass density than comparable regions of condensed chromatin. The most striking difference between the net phosphorus image (Net P), which reflects the density of nucleic acids, and the net nitrogen image (Net N), which reflects the density of protein and nucleic acid, is in the extranucleolar regions of the nucleus. In the net nitrogen image, the interchromatin space does not appear open, as it does in the net phosphorus image. Instead, the interchromatin space is filled with structure, and the regions of chromatin only produce a marginally greater signal than the surrounding interchromatin structure. These results demonstrate that there is an abundance of protein-rich, nucleic acid-depleted structure in the interchromatin space of these paraformaldehyde-fixed cells.
Figure 1.
ESI of cellular structures. The principles of ESI of chromatin, ribonucleoprotein, and protein structures are illustrated. An energy loss series was recorded from an ultrathin section of an Indian muntjac fibroblast cell. The top series shows a reference 120-eV energy loss image, a phosphorus-enhanced 155-eV energy loss image, and a phosphorus distribution map (Net P) calculated from these two images. The bottom panels show analagous pre-edge and element-enriched images used to generate a nitrogen distribution map (Net N). The images in the top two panels show structure as bright on a dark background. Bar, 2.3 μm.
Quantitative Validation of Net Nitrogen and Phosphorus Maps for Identification of Protein- and Nucleic Acid-based Structures
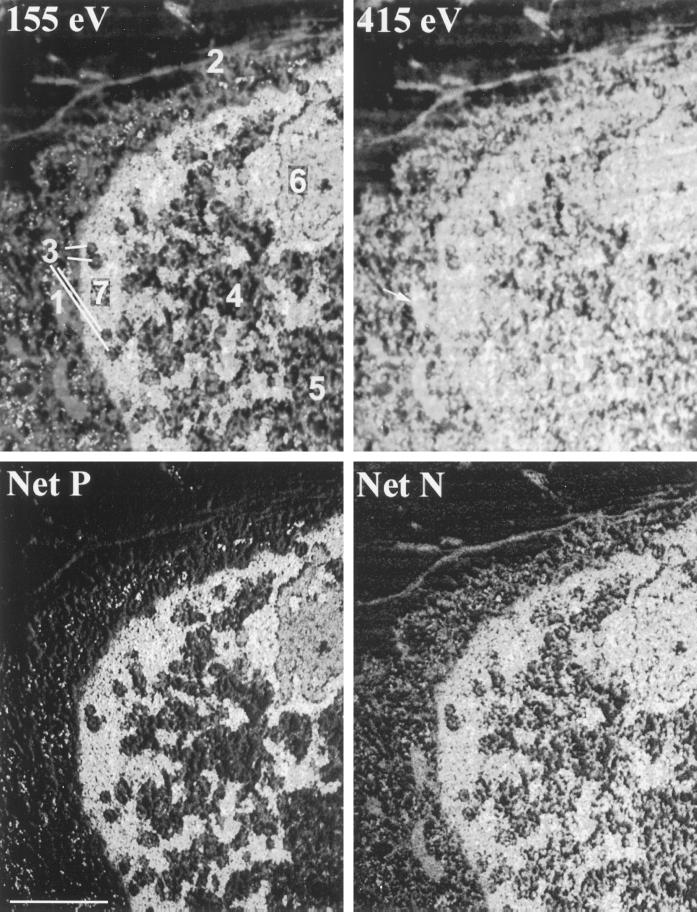
In principle, nucleic acids and proteins can be discriminated through analysis of phosphorus and nitrogen contents in a quantitatively sensitive manner. This principle is qualitatively validated by visual comparisons of structures such as those in Figure 1. We sought to validate this approach further through quantification of phosphorus and nitrogen contents in cellular components of well-characterized composition. The data are presented in Figure 2 and Table 1. Figure 2 shows a 10T½ cell nucleus prepared by paraformaldehyde fixation. Part of the cell nucleus has been cut along its surface, evident by the cross-sectioning of several nuclear pores (3). Cytoskeletal elements (Figure 2, 2) and mitochondria (Figure 2, 1, arrow in 415 eV image) are also represented. These structures serve as references for structures that are predominantly protein in composition. Chromatin (Figure 2, 7) and the granular component of the nucleolus (Figure 2, 6) serve as references for structures that have a high compositional representation of nucleic acid. The IGC (Figure 2, 5) and the interchromatin region (Figure 2, 4) were evaluated for their compositional characteristics relative to these reference structures.
Figure 2.
Relative phosphorus and nitrogen contents of compositionally defined and compositionally undefined cellular structures. An energy loss series was collected on a mouse 10T½ cell. The 155-eV phosphorus-enhanced, 415-eV nitrogen-enhanced, net nitrogen (Net N), and net phosphorus (Net P) images are shown. The regions numbered 1–7 were analyzed quantitatively for both phosphorus and nitrogen content. The arrow in the 415-eV image indicates the position of a mitochondrion (region 1). The results of these measurements are presented in Table 1. Bar, 1 μm.
Table 1.
Relative nitrogen-to-phosphorus ratios of representative cellular structures
| No. | Structure type | N/P ratio |
|---|---|---|
| 1 | Mitochondrion | 5.38 |
| 2 | Ribosome-depleted cytoplasm | 4.44 |
| 3 | Nuclear pores | 3.23 ± 0.30 |
| 4 | RNP-depleted nuclear matrix | 3.37 |
| 5 | Interchromatin granule cluster | 3.04 |
| 6 | Nucleolus (granular component) | 1.42 |
| 7 | Chromatin | 0.97 |
The numbers indicate regions identified in Figure 3. Integrated density for regions containing, with the exception of nuclear pores, at least 1000 pixels were calculated in both the net nitrogen and net phosphorus images. The resulting nitrogen and phosphorus values were then used to determine the net nitrogen-to-phosphorus ratio (N/P ratio).
A quantitative analysis of the relative nitrogen and phosphorus composition is presented in Table 1. It can be seen that an interchromatin region, specifically chosen to avoid nuclear RNP granules (4), and the IGC (5), which contains several small ribonucleoprotein granules within a proportionately larger nonnucleoprotein mass, have a very high nitrogen-to-phosphorus ratio. This reflects the predominance of protein structures within these regions. The granular component of the nucleolus also has a much greater protein contribution to its total mass than the condensed chromatin region. These results indicate that the combined net phosphorus and net nitrogen information can be used to determine the biochemical composition of the individual structures represented in energy filtered electron micrographs.
Visualization of Interchromatin Protein Structure in the Cell Nucleus
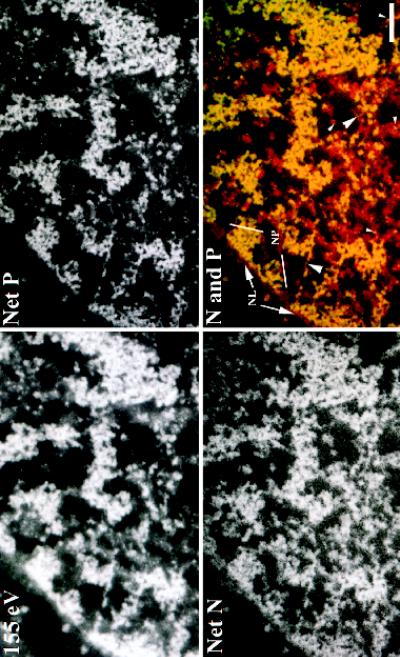
The results of Table 1 validate the comparison of net nitrogen and net phosphorus images for the morphological identification and elemental characterization of cellular complexes. We next addressed whether a protein component could be visualized that had the basic morphological characteristics of a protein matrix. Figure 3 shows a 155-eV energy loss image and net phosphorus and nitrogen images of a subregion of an interphase Indian muntjac fibroblast cell nucleus. In the bottom right panel, the net phosphorus image has been false colored (green) and superimposed on the net nitrogen image (red). As expected, the nucleic acid components of the cell nucleus contain high amounts of both nitrogen and phosphorus, and, consequently, appear yellow in the merged image. Small particles of <5 nm are apparent as small yellow points within the image (Figure 3, small arrowheads in bottom right panel). These are likely RNP particles. Similarly, decondensed chromatin fibers are also observed and appear yellow in the merged image (Figure 3, large arrowheads in bottom right panel; also see corresponding regions of Net P image). Despite the presence of very small structures that contain nucleic acid, most of the nitrogen-containing material outside of the compact regions of chromatin is derived from protein structures, highly depleted in nucleic acid. This appears red in the merged image. There are two nuclear pores that appear as red insertions between the blocks of condensed chromatin on the periphery of this cell nucleus (Figure 3, NP in bottom right panel). These serve as references for structures that are protein based in composition. Additionally, close examination will reveal a narrow band of protein on the cytoplasmic side of the peripheral chromatin (Figure 3, NL in bottom right panel). This reflects the presence of the nuclear lamina. The intervening protein elements within the nucleoplasm have a general filamentous appearance and can be seen to connect adjacent nucleic acid-rich structures. Moreover, the smallest nucleic acid-containing complexes observed in this nuclear section (Figure 3, small arrows in bottom right panel) are associated with this more massive protein architecture.
Figure 3.
(Facing page). Direct visualization of protein architecture within the cell nucleus. An energy loss series of a region of an Indian muntjac fibroblast cell nucleus was collected at high magnification. The 155-eV energy loss, net phosphorus (Net P), and net nitrogen (Net N) images are shown. The bottom right panel shows a superimposition of the net phosphorus image (false-colored green) onto the net nitrogen image (false-colored red). Nucleoproteins, which contain high amounts of both phosphorus and nitrogen, appear yellow in the merged image. Protein structures, which are low in phosphorus, appear red and orange. The positions of nuclear pores (NP) and the nuclear lamina (NL) are indicated. The small arrowheads indicate the positions of small RNP granules, and the large arrowheads indicate the positions of decondensed nucleic acid-containing fibrils that are continuous with more condensed chromatin fibers. Bar, 250 nm.
Visualization of Protein Architecture within the IGC
The IGC represents an excellent example of compartmentalization within the cell nucleus. The best described component of the IGC is a 20- to 25-nm ribonucleoprotein granule (Monneron and Bernhard, 1969; Wassef, 1979). The nature of this granule was recently revealed quantitatively by both ESI (Hendzel et al., 1998) and qualitatively using RNA-specific staining methods (Thiry, 1993; Biggiogera and Fakan, 1998). We have shown that these interchromatin granules each contain between 2000 and 10,000 bases of RNA. There are additional smaller RNPs, ∼5 nm in diameter, that contain amounts of RNA consistent with small nuclear RNPs (Hendzel et al., 1998). The clusters of these granules can exceed diameters of 1 μm. The RNPs within the IGC domain are not so densely clustered that particle–particle associations could be responsible for their compartmentalization. In early investigations, interconnecting fibrils were observed (Monneron and Bernhard, 1969; Puvion and Bernhard, 1975; Wassef, 1979). They were thought to be RNA fibers because they stained by the EDTA-regressive method. However, the specificity of the staining method for RNA is not ensured (Bernhard, 1969). For example, nuclear structures termed promyelocytic leukemia (PML) bodies are well stained using the EDTA-regressive staining procedure (LaMorte et al., 1998) but contain very low amounts of phosphorus (Boisvert and Bazett-Jones, unpublished results). Thus, the method cannot be used to conclude that the fibers are composed of RNA.
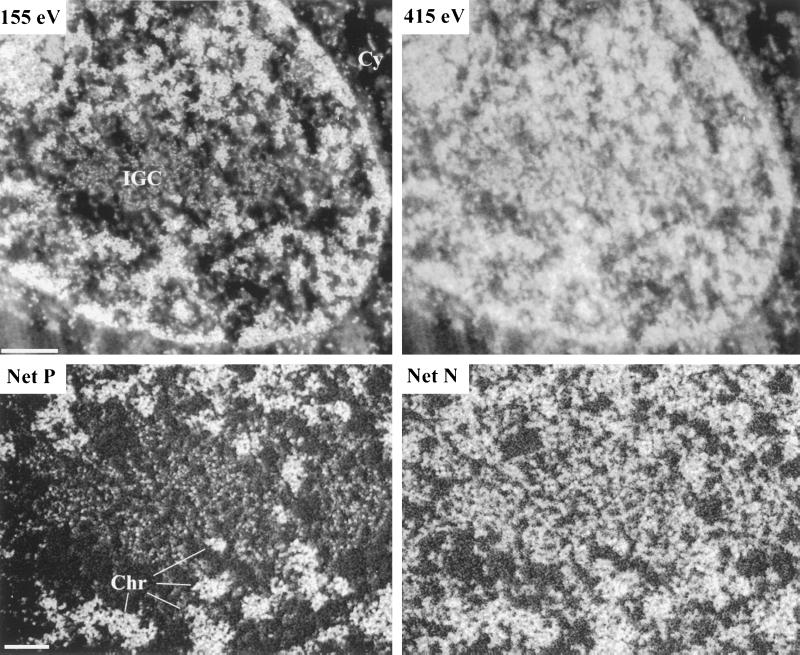
We investigated the organization of IGCs using phosphorus and nitrogen (Figure 4). The top two panels show low-magnification views of a region of an Indian muntjac cell nucleus containing a prominent IGC. In comparing the phosphorus-enhanced 155-eV energy loss image with the nitrogen-enhanced 415-eV image, it can be seen that the IGC stands out in the 155-eV image as a nuclear region relatively depleted in phosphorus. In contrast, the IGC is not a region of obvious signal depletion in the 415-eV image. This reflects the presence of a large amount of protein mass within the IGC, further illustrated in the high-magnification views of the IGC and surrounding nuclear territory presented in the lower two panels. When phosphorus is imaged, the IGC appears as a number of dispersed phosphorus-rich particles (Figure 4, bottom left). This contrasts with the net nitrogen image (Figure 4, bottom right), where the particle nature of the IGC is much less apparent. Instead, the IGC appears as a relatively dense network of protein fibers in which the granules are embedded.
Figure 4.
Phosphorus and Nitrogen distributions within IGCs. An energy loss series was collected for a region of an Indian muntjac fibroblast cell nucleus containing a prominent IGC. The top two panels show the 155 eV phosphorus-enhanced and the 415 eV nitrogen-enhanced images. The bottom two panels show, at higher magnification, the net phosphorus (Net P) and net nitrogen (Net N) maps within and around the IGC. Cy, cytoplasm; Chr, examples of chromatin. Bar, 250 and 200 nm in the top and bottom panels, respectively.
Nuclear Bodies and the Detection of Protein Phosphorylation
The protein components of the cell nucleus can be distinguished from nucleic acid components of the cell nucleus based on phosphorus density. Detection of low levels of phosphorus associated with most cellular components cannot always be detected reliably with only one pre-edge (120-eV) and one post-edge (155-eV) image. Instead, an energy loss spectrum derived from particular objects can be obtained to assess low levels of particular elements (Vazquez-Nin et al., 1996). Nuclear bodies, which are large, protein-based structures in the cell nucleus, and condensed chromatin were imaged at 10-eV energy loss intervals across the phosphorus L2,3 ionization edges of phosphorus and nitrogen. We then plotted the signal intensity at each energy loss to generate an energy loss spectrum for each structure.
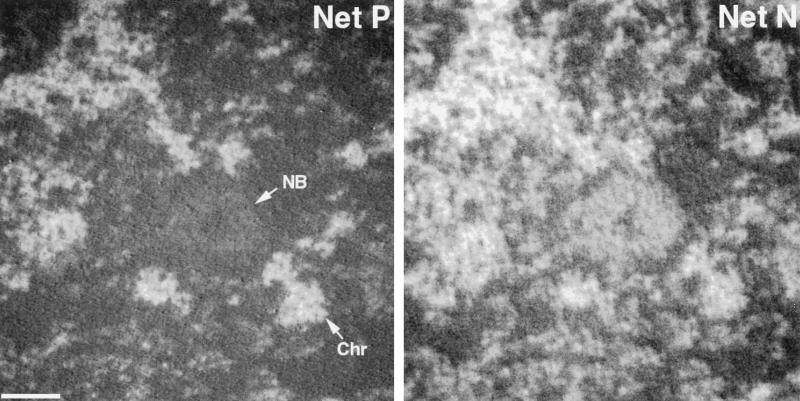
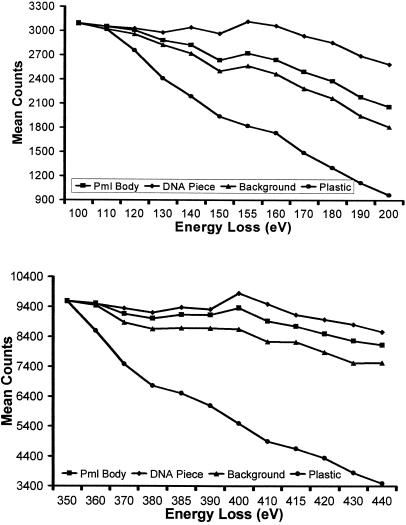
Figure 5 shows a net phosphorus and a net nitrogen map of a region of an SKN cell nucleus that contains a nuclear body. The nuclear body was identified using an anti-CBP antibody and correlative immunofluorescence microscopy (Hendzel et al., 1999). These nuclear bodies are not sites of RNA synthesis (Boisvert and Bazett-Jones, unpublished results). The nuclear body is qualitatively striking in its deficiency in phosphorus. In the energy loss spectrum obtained from the nuclear body (Figure 6), the amount of phosphorus is barely detectable, based on the continuing decline in signal beyond 132 eV (the phosphorus ionization edge). Even less is detected in background regions of the nucleus, represented by regions that do not correspond to structural features. As expected, no phosphorus was detected over regions of the plastic embedding material. Because the phosphorus signal through this body is diffuse and near the nucleoplasmic background, it likely represents the presence of phosphorylated proteins in the nucleoplasm. The nuclear body, on the other hand, is rich in nitrogen (Figure 6, lower panel), contributing to a signal close to that of the chromatin and well above the background nuclear signal.
Figure 5.
Phosphorus and nitrogen mapping of a nuclear body. A nuclear body was identified by fluorescence imaging of an SKN cell nucleus stained with anti-CBP. An energy loss series was then collected on a subregion of the cell nucleus containing the nuclear body. The net phosphorus and net nitrogen maps are shown. NB, nuclear body; Chr, chromatin. Bar, 120 nm.
Figure 6.
Detection of protein phosphorylation. The integrated density of a nuclear body (Pml Body), DNA (Chr in Figure 5), and a background region of the nucleoplasm were calculated across an energy loss series, spanning both the phosphorus ionization edge (upper graph) and the nitrogen ionization edge (lower graph), using a constant exposure time. Under these conditions, the phosphorus and nitrogen signals generated by the plastic decay at the expected rates. In contrast, all three nuclear structures give detectable phosphorus signals between 130- and 160-eV energy losses, characteristic of the presence of a phosphorus L2,3 ionization edge.
DISCUSSION
There are many reasons to believe that the cell nucleus contains a structural component that is capable of organizing and spatially sequestering biochemicals within the cell nucleus. The increased application of confocal microscopy to the understanding of biomolecular organization within the nucleus of fixed biological specimens has provided much of the most compelling evidence. Most biomolecules show some degree of spatial sequestration. These include transcription factors (van Steensel et al., 1995; Htun et al., 1996; Grande et al., 1997; Zeng et al., 1997; Fejes-Toth et al., 1998; Noordmans et al., 1998), RNA-processing machinery (for review, see Spector, 1993), and interphase chromosomes (for review, see Lamond and Earnshaw, 1998). More recently, the study of biomolecular organization in living cells, using fluorescence microscopy, has further indicated the presence of a component of the cell nucleus that is capable of spatially restricting the movement of biomolecular structures within the cell nucleus. The best characterized example is chromatin. Chromatin has been clearly shown not to undergo substantial Brownian motion (Abney et al., 1997; Marshall et al., 1997; Sullivan and Shelby, 1999) despite the fact that transmission electron micrographs demonstrate that the density of chromatin and nonchromatin structures within the cell nucleus is not sufficient to constrain the diffusion of chromatin within the cell nucleus. The absence of substantial Brownian motion of chromatin within the living cell nucleus has led several investigators to suggest that chromatin is confined in situ by a structure analagous to a nuclear matrix (Marshall et al., 1997; Sullivan and Shelby, 1999).
The quantitative resolution of both phosphorus and nitrogen content within the specimen using ESI is sufficient to resolve the compositional differences between many structures within the cell nucleus. For example, the granular component of the nucleolus clearly contains a higher protein-to-nucleic acid stoichiometry than does chromatin. Similarly, the granular component of the nucleolus, which is intermediate in nucleic acid density, is also clearly resolved from protein-based components such as nuclear pores. Using this established method (Hendzel and Bazett-Jones, 1996; Bazett-Jones and Hendzel, 1999; Bazett-Jones et al., 1999), we have addressed the biochemical nature of fine structures within the cell nucleus. This has not been accomplished using heavy atom staining, because it is not possible to visualize all cellular components while also discriminating between protein and nucleic acid components. In particular, decondensed chromatin and RNA fibrils cannot be differentiated from protein filaments of similar dimensions. Moreover, heavy atom contrast agents required in conventional electron microscopy frequently fail to stain some structures or overrepresent others, based on the degree of their chemical reactivity (Abholhassani-Dadras et al., 1996). The resolution of these components by ESI enables us to conclude that the majority of the fine structure found between “condensed” regions of chromatin comprises mainly protein.
Paraformaldehyde fixation is the preferred method of fixation for the preservation of chromatin structure and nuclear volume (Robinett et al., 1996). It has also become a standard for indirect immunofluorescence analysis of the cell nucleus. In such preparations, transcription and splicing factors have been shown to occupy distinct compartments within the cell nucleus. We conclude that there is a protein-based architecture that has the potential to organize factors in the nucleus. Whether the nucleus is similarly organized in a dynamic living state remains to be determined conclusively. It is noteworthy, however, that at least some factors are found in foci (Htun et al., 1996; Becker et al., 1998; Fejes-Toth et al., 1998; Sleeman et al., 1998; Hendzel, Bisgrove, and Godbout, unpublished observations) or speckles (Misteli et al., 1997; Sleeman et al., 1998) within living cells, and that chromatin is restricted from significant diffusion (Abney et al., 1997; Marshall et al., 1997; Sullivan et al., 1999).
The IGC represents the single most striking example of a compartmentalization process within the extranucleolar regions of the cell nucleus. Importantly, this compartment can be visualized as a structurally dynamic domain in which splicing factors enrich in living cells (Misteli et al., 1997). Our results demonstrate that the particle density of ribonucleoprotein granules within the IGC is not sufficient to explain the compartmentalization of these particles through interparticle association. We have found that the IGC contains structures that link the individual granules. Moreover, most of the biological mass that appears to integrate the IGC is contributed by protein. In the case of the IGC, then, there is strong reason to propose that the integration of particles through a protein architecture occurs in living cells. Although our images represent “snapshots in time,” it is clear that nuclear structure is, to some extent, dynamic (Misteli and Spector, 1998; Schul et al., 1998), and, consequently, any underlying architecture must also be capable of dynamic reorganization.
Since the initial characterization of a nuclease and high-salt–resistant matrix-insoluble component of the cell nucleus, the nuclear matrix has been actively studied by a small but persistent group of biochemists and cell biologists. As a means of studying high-affinity interactions between specific nuclear components, the original biochemical fractionation procedure of Berezney and Coffey (1977) and subsequent variations on this protocol (Jackson and Cook, 1985; He et al., 1990) have merit (Stenoien et al., 1998). As a cytological preparation for the investigation of structure–function relationships by transmission electron microscopy, however, the procedure has significant shortcomings. The identification and characterization of the nuclear matrix is absolutely dependent on elution of chromatin before fixation (Jackson and Cook, 1988; He et al., 1990) or after fixation (Nickerson et al., 1997). Because the interaction between a nuclear matrix and chromatin is of fundamental importance for its role in organizing chromatin within the cell nucleus, this shortcoming is very serious. The potential to reorganize components through chromatin elution is the basis for much of the skepticism surrounding the nuclear matrix field. The ideal procedure would leave chromatin components intact and would enable the study of interactions that involve not only protein and RNA but also chromatin, which is the substrate in so many biochemical processes occurring in the cell nucleus. In this study, we have presented, for the first time, compelling evidence of a nuclear protein architecture within a standard cytological preparation, based on the direct discrimination of protein-based and nucleic acid-based structures, visualized in situ.
ACKNOWLEDGMENTS
We thank Manfred Herfort and Maryse Fillion for excellent technical assistance. We also thank Dr. Charlotte Spencer for critical reading of the manuscript. This work was supported by an operating grant provided by the Cancer Research Society.
REFERENCES
- Abholhassani-Dadras S, Vazquez-Nin GJ, Echeverria OM, Fakan S. Image-EELS for in situ estimation of the phosphorus content of RNP granules. J Microsc. 1996;183:215–222. [Google Scholar]
- Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J Cell Biol. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett-Jones DP, Hendzel MJ. Electron spectroscopic imaging of chromatin. Methods. 1999;17:188–200. doi: 10.1006/meth.1998.0729. [DOI] [PubMed] [Google Scholar]
- Bazett-Jones, D.P., Hendzel, M.J., and Kruhlak, M.J. (1999). Stoichiometric analysis of protein and nucleic acid-based structures in the cell nucleus. Micron (in press). [DOI] [PubMed]
- Becker W, Weber Y, Wetzel K, Eirmbter K, Tejedor FJ, Joost H-G. Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J Biol Chem. 1998;273:25893–25902. doi: 10.1074/jbc.273.40.25893. [DOI] [PubMed] [Google Scholar]
- Berezney R, Coffey DS. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60:1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Berezney R, Coffey DS. Nuclear matrix: isolation and characterization of a framework structure from rat liver nuclei. J Cell Biol. 1977;73:616–637. doi: 10.1083/jcb.73.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard W. A new staining procedure for electron microscopical cytology. J Ultrastruct Res. 1969;27:250–265. doi: 10.1016/s0022-5320(69)80016-x. [DOI] [PubMed] [Google Scholar]
- Biggiogera M, Fakan S. Fine structural specific visualization of RNA on ultrathin sections. J Histochem Cytochem. 1998;46:389–395. doi: 10.1177/002215549804600313. [DOI] [PubMed] [Google Scholar]
- Fejes-Toth G, Pearce D, Naray-Fejes-Toth A. Subcellular localization of mineralocorticoid receptors in living cells: effects of receptor agonists and antagonists. Proc Natl Acad Sci USA. 1998;95:2973–2978. doi: 10.1073/pnas.95.6.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande MA, van der Kraan I, de Jong L, van Driel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci. 1997;110:1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- He DC, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Bazett-Jones DP. Probing nuclear ultrastructure by electron spectroscopic imaging. J Microsc. 1996;182:1–14. doi: 10.1046/j.1365-2818.1996.123403.x. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Kruhlak MJ, Bazett-Jones DP. Organization of highly acetylated chromatin around sites of nuclear RNA accumulation. Mol Biol Cell. 1998;9:2491–2507. doi: 10.1091/mbc.9.9.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Kruhlak, M.J., and Bazett-Jones, D.P. (1999). Electron spectroscopic imaging and correlative fluorescent microscopy of the cell nucleus. Scanning (in press).
- Htun J, Barsony J, Renyi I, Gould DL, Hager GL. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc Natl Acad Sci USA. 1996;93:4845–4850. doi: 10.1073/pnas.93.10.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. A general method for preparing chromatin containing intact DNA. EMBO J. 1985;4:913–918. doi: 10.1002/j.1460-2075.1985.tb03718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. Visualization of a filamentous nucleoskeleton with a 23 nm axial repeat. EMBO J. 1988;7:3667–3677. doi: 10.1002/j.1460-2075.1988.tb03248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- LaMorte VJ, Dyck JA, Ochs RL, Evans RM. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Curr Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Cocco L, Riederer BM, Neri LM. The nuclear matrix: a critical appraisal. Histol Histopathol. 1996;11:1035–1048. [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a premRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. The cellar organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian fells. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Nickerson JA, Krockmalnic G, Wan KM, Penman S. The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc Natl Acad Sci USA. 1997;94:4446–4450. doi: 10.1073/pnas.94.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordmans HJ, van der Kraan K, van Driel R, Smeulders AW. Randomness of spatial distributions of two proteins in the cell nucleus involved in mRNA synthesis and their relationship. Cytometry. 1998;33:297–309. doi: 10.1002/(sici)1097-0320(19981101)33:3<297::aid-cyto3>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Puvion E, Bernhard W. Ribonucleoprotein components in liver cell nuclei visualized by cryoultrmicrotomy. J Cell Biol. 1975;67:200–214. doi: 10.1083/jcb.67.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W, de Jong L, van Driel R. Nuclear neighbors: the spatial and functional organization of genes and nuclear domains. J Cell Biochem. 1998;70:159–171. doi: 10.1002/(sici)1097-4644(19980801)70:2<159::aid-jcb2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Sleeman J, Lyon CE, Platani M, Kreivi JP, Lamond AI. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp Cell Res. 1998;243:290–304. doi: 10.1006/excr.1998.4135. [DOI] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Stenoien D, Sharp ZD, Smith CL, Mancini MA. Functional subnuclear partitioning of transcription factors. J Cell Biochem. 1998;70:213–221. [PubMed] [Google Scholar]
- Sullivan KF, Shelby RD. Using time-lapse confocal microscopy for analysis of centromere dynamics in human cells. Methods Cell Biol. 1999;58:183–202. [PubMed] [Google Scholar]
- Thiry M. Differential location of nucleic acids within interchromatin granule clusters. Eur J Cell Biol. 1993;62:259–269. [PubMed] [Google Scholar]
- van Steensel B, Brink M, van der Meulen K, van Binnendijk EP, Wansink DG, de Jong L, de Kloet ER, van Driel R. Localization of the glucocorticoid receptor in discrete clusters in the cell nucleus. J Cell Sci. 1995;108:3003–3011. doi: 10.1242/jcs.108.9.3003. [DOI] [PubMed] [Google Scholar]
- Vazquez-Nin GH, Abolhassani-Dadras S, Echeverria OM, Rouelle-Rossier VB, Fakan S. Phosphorous distribution in perichromatin granules and surrounding neoplasm as visualized by electron spectrosopic imaging. Biol Cell. 1996;87:171–177. [PubMed] [Google Scholar]
- Wassef M. A cytochemical study of interchromatin granules. J Ultrastruct Res. 1979;69:121–133. doi: 10.1016/s0022-5320(79)80047-7. [DOI] [PubMed] [Google Scholar]
- Zeng C, et al. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-α transcription factors. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Cremer T, Saffrich R, Fischer R, Trendelenburg MF, Ansorge W, Stelzer EHK. Structure and dynamics of human interphase chromatin territories in vivo. Hum Genet. 1998;102:241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]