Abstract
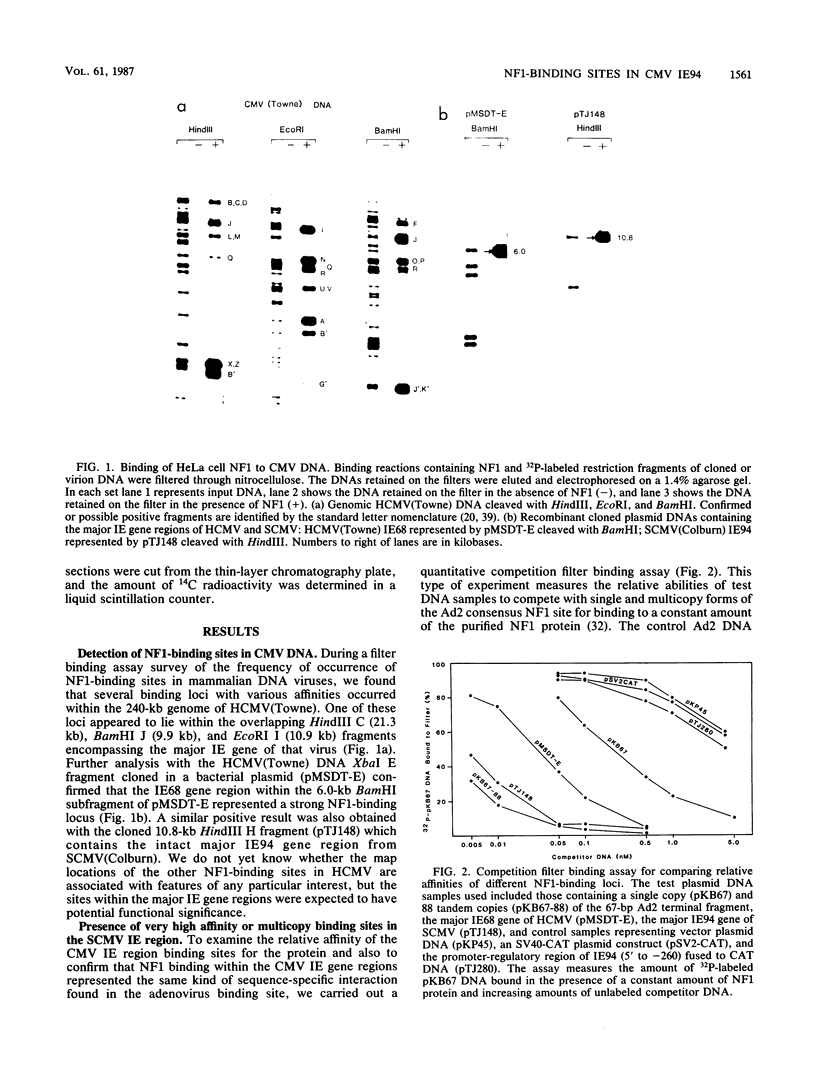
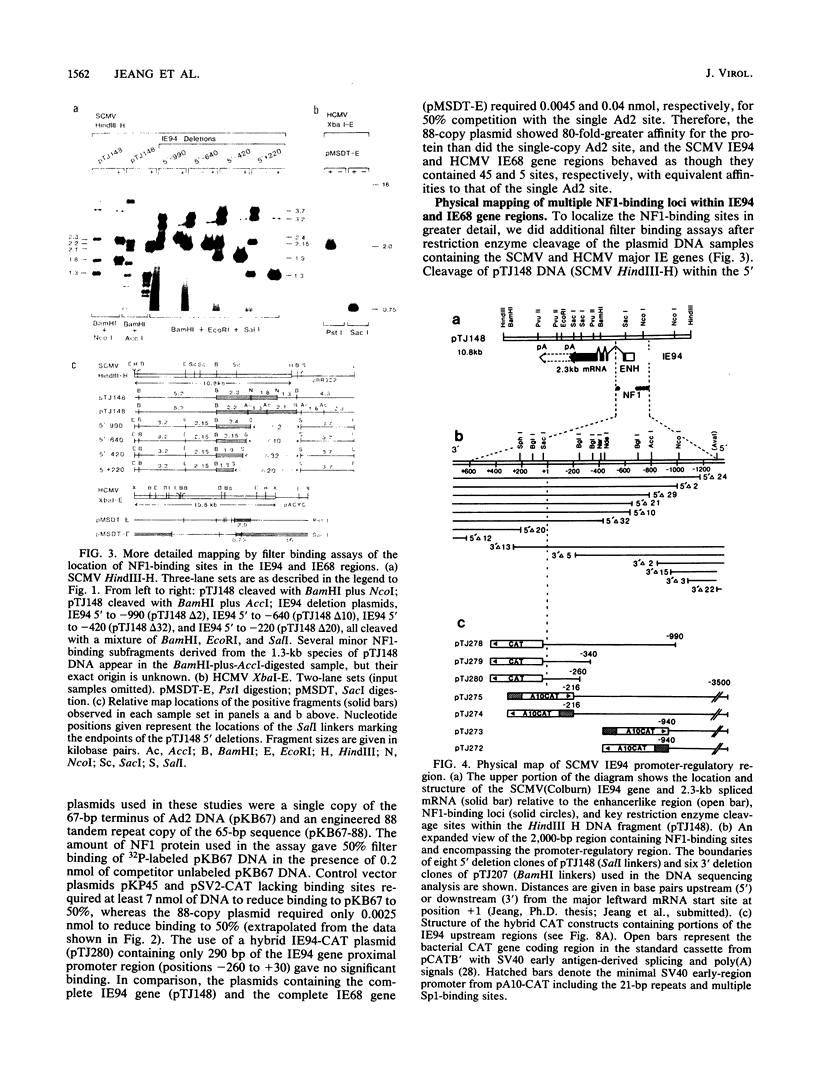
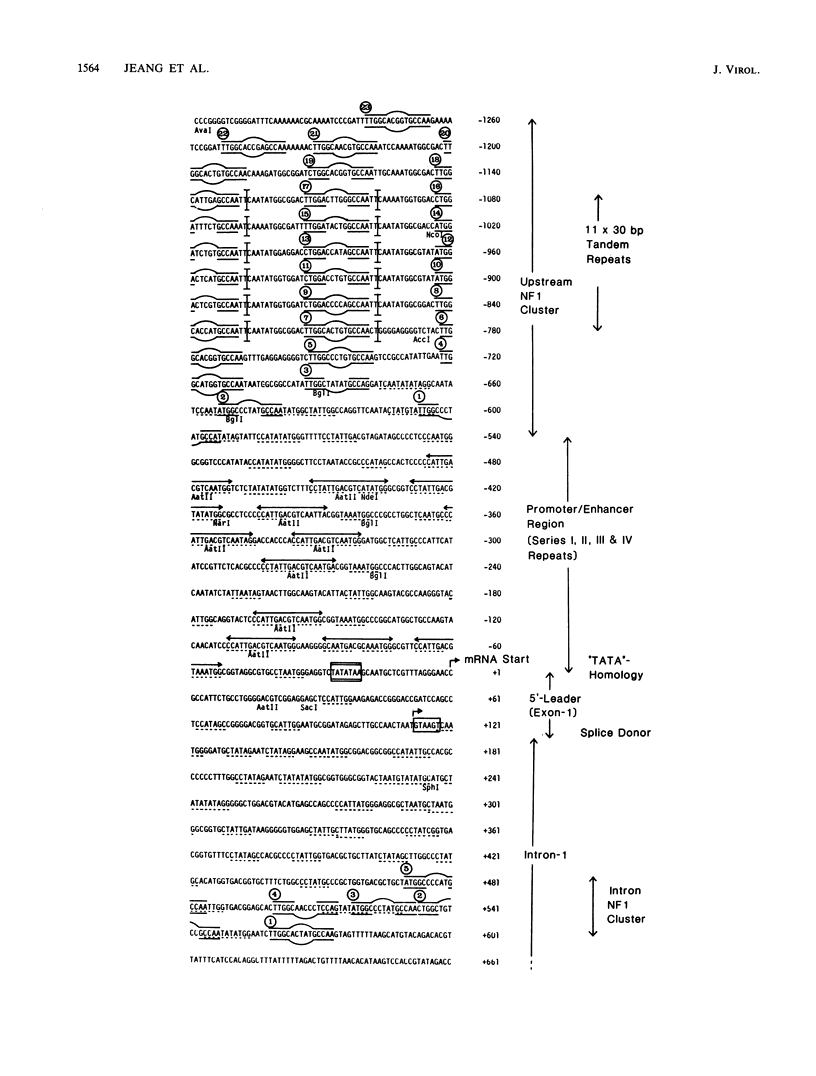
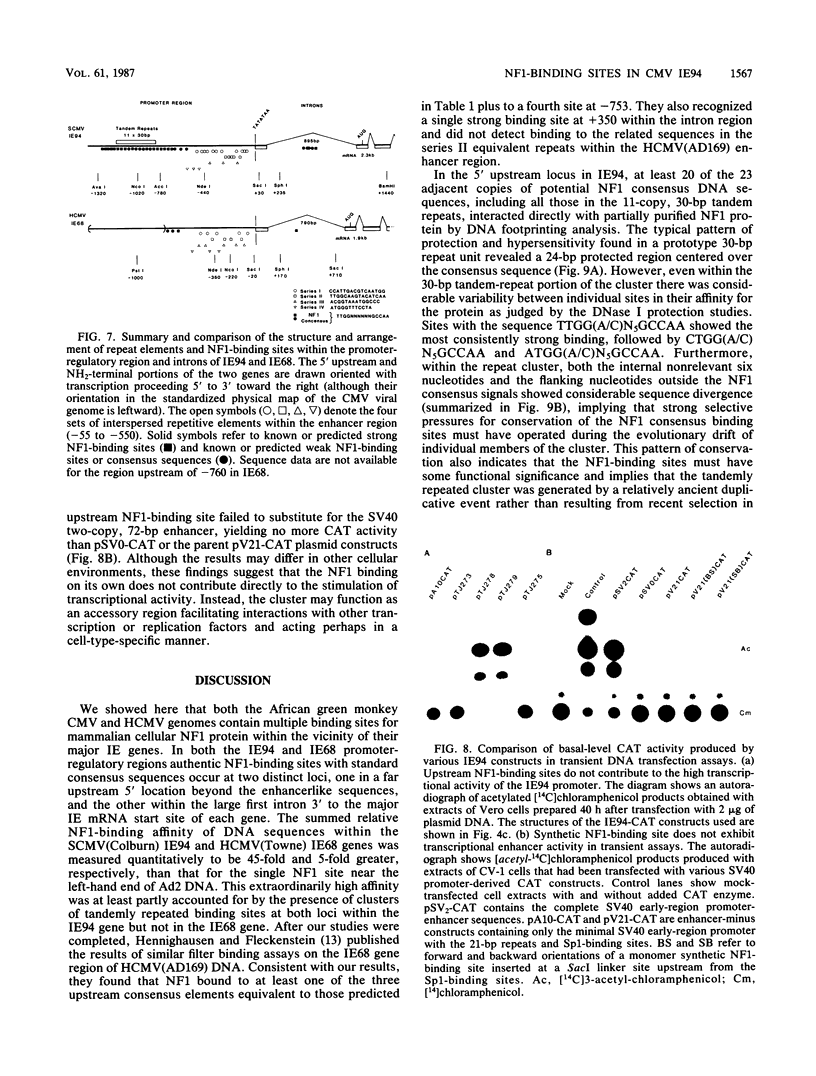
We show that the large DNA genomes of human and simian cytomegaloviruses (HCMV and SCMV, respectively) each contain multiple binding sites for purified cellular nuclear factor 1 (NF1) protein. Examination of the major immediate-early (IE) gene region in the HindIII H fragment of SCMV (Colburn) by filter binding assays showed that it competed 45-fold better than the single adenovirus type 2 binding site for NF1 protein and that it contained at least two distinct binding loci. Direct DNase I footprinting analyses of the 5' upstream locus detected at least 20 adjacent NF1-binding sites located between positions -600 and -1300 relative to the IE94 mRNA start site. DNA sequence analysis of the region revealed a conserved consensus NF1 recognition element (T)TGG(C/A)N5GCCAA embedded within each of 23 highly diverged 30-base-pair tandem repeats, together with a second downstream cluster of five consensus NF1-binding sites between positions +470 and +570 in the large first intron. Two separate NF1-binding loci were also found in the equivalent IE68 gene of HCMV(Towne) DNA, but in this case the DNA sequence and competition filter binding experiments indicated a maximum of only four to five consensus binding sites encompassing the promoter-enhancer region. In transient expression assays, neither the isolated upstream IE94 tandem repeats nor a synthetic single-copy consensus NF1-binding site acted as transcriptional cis activators or enhancers when placed adjacent to the simian virus 40 minimal early region promoter. We conclude that the large and complex 5' upstream promoter-regulatory region for the SCMV IE94 gene comprises two distinct domains. The previously described four sets of 13- to 18-base-pair interspersed repeat elements between -55 and -580 provide most of the high basal transcriptional strength, whereas the arrangement of further upstream tandemly repeated NF1-binding sites may contribute significantly to the expanded biological host range for expression of SCMV IE94 compared with HCMV IE68.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akrigg A., Wilkinson G. W., Oram J. D. The structure of the major immediate early gene of human cytomegalovirus strain AD169. Virus Res. 1985 Mar;2(2):107–121. doi: 10.1016/0168-1702(85)90242-4. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgmeyer U., Nowock J., Sippel A. E. The TGGCA-binding protein: a eukaryotic nuclear protein recognizing a symmetrical sequence on double-stranded linear DNA. Nucleic Acids Res. 1984 May 25;12(10):4295–4311. doi: 10.1093/nar/12.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M., Weber F., Jahn G., Dorsch-Häsler K., Fleckenstein B., Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985 Jun;41(2):521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J., Jr Adenovirus DNA replication in vitro. Proc Natl Acad Sci U S A. 1979 Feb;76(2):655–659. doi: 10.1073/pnas.76.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Rawlins D. R. Template requirements for the initiation of adenovirus DNA replication. Proc Natl Acad Sci U S A. 1984 Jan;81(1):100–104. doi: 10.1073/pnas.81.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski R. M., Adhya S., Nagata K., Guggenheimer R. A., Hurwitz J. Site-specific DNA binding of nuclear factor I: analyses of cellular binding sites. Mol Cell Biol. 1985 May;5(5):964–971. doi: 10.1128/mcb.5.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggenheimer R. A., Stillman B. W., Nagata K., Tamanoi F., Hurwitz J. DNA sequences required for the in vitro replication of adenovirus DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3069–3073. doi: 10.1073/pnas.81.10.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczöl E., Andrews P. W., Plotkin S. A. Cytomegalovirus replicates in differentiated but not in undifferentiated human embryonal carcinoma cells. Science. 1984 Apr 13;224(4645):159–161. doi: 10.1126/science.6322309. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Ambinder R., Ciufo D., Hayward S. D., LaFemina R. L. Structural organization of human herpesvirus DNA molecules. J Invest Dermatol. 1984 Jul;83(1 Suppl):29s–41s. doi: 10.1111/1523-1747.ep12281149. [DOI] [PubMed] [Google Scholar]
- Hennighausen L., Fleckenstein B. Nuclear factor 1 interacts with five DNA elements in the promoter region of the human cytomegalovirus major immediate early gene. EMBO J. 1986 Jun;5(6):1367–1371. doi: 10.1002/j.1460-2075.1986.tb04368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L., Siebenlist U., Danner D., Leder P., Rawlins D., Rosenfeld P., Kelly T., Jr High-affinity binding site for a specific nuclear protein in the human IgM gene. Nature. 1985 Mar 21;314(6008):289–292. doi: 10.1038/314289a0. [DOI] [PubMed] [Google Scholar]
- Huang E. S., Pagano J. S. Human cytomegalovirus. II. Lack of relatedness to DNA of herpes simples I and II, Epstein-Barr virus, and nonhuman strains of cytomegalovirus. J Virol. 1974 Mar;13(3):642–645. doi: 10.1128/jvi.13.3.642-645.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang K. T., Chin G., Hayward G. S. Characterization of cytomegalovirus immediate-early genes. I. Nonpermissive rodent cells overproduce the IE94K protein form CMV (Colburn). Virology. 1982 Sep;121(2):393–403. doi: 10.1016/0042-6822(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Jeang K. T., Cho M. S., Hayward G. S. Abundant constitutive expression of the immediate-early 94K protein from cytomegalovirus (Colburn) in a DNA-transfected mouse cell line. Mol Cell Biol. 1984 Oct;4(10):2214–2223. doi: 10.1128/mcb.4.10.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang K. T., Gibson W. A cycloheximide-enhanced protein in cytomegalovirus-infected cells. Virology. 1980 Dec;107(2):362–374. doi: 10.1016/0042-6822(80)90304-9. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Yamamoto K. R., Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985 Sep;42(2):559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- LaFemina R. L., Hayward G. S. Replicative forms of human cytomegalovirus DNA with joined termini are found in permissively infected human cells but not in non-permissive Balb/c-3T3 mouse cells. J Gen Virol. 1983 Feb;64(Pt 2):373–389. doi: 10.1099/0022-1317-64-2-373. [DOI] [PubMed] [Google Scholar]
- LaFemina R., Hayward G. S. Constitutive and retinoic acid-inducible expression of cytomegalovirus immediate-early genes in human teratocarcinoma cells. J Virol. 1986 May;58(2):434–440. doi: 10.1128/jvi.58.2.434-440.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Enomoto T., Lichy J. H., Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Hurwitz J. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6177–6181. doi: 10.1073/pnas.80.20.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. A., Groudine M. Transcriptional regulation of the human cytomegalovirus major immediate-early gene is associated with induction of DNase I-hypersensitive sites. Mol Cell Biol. 1986 Feb;6(2):452–461. doi: 10.1128/mcb.6.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins D. R., Milman G., Hayward S. D., Hayward G. S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985 Oct;42(3):859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- Rawlins D. R., Rosenfeld P. J., Wides R. J., Challberg M. D., Kelly T. J., Jr Structure and function of the adenovirus origin of replication. Cell. 1984 May;37(1):309–319. doi: 10.1016/0092-8674(84)90327-1. [DOI] [PubMed] [Google Scholar]
- Reisman D., Yates J., Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985 Aug;5(8):1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld P. J., Kelly T. J. Purification of nuclear factor I by DNA recognition site affinity chromatography. J Biol Chem. 1986 Jan 25;261(3):1398–1408. [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Stenberg R. M., Thomsen D. R., Stinski M. F. Structural analysis of the major immediate early gene of human cytomegalovirus. J Virol. 1984 Jan;49(1):190–199. doi: 10.1128/jvi.49.1.190-199.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski M. F., Roehr T. J. Activation of the major immediate early gene of human cytomegalovirus by cis-acting elements in the promoter-regulatory sequence and by virus-specific trans-acting components. J Virol. 1985 Aug;55(2):431–441. doi: 10.1128/jvi.55.2.431-441.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski M. F., Thomsen D. R., Stenberg R. M., Goldstein L. C. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol. 1983 Apr;46(1):1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D. Localization of an origin of DNA replication within the TRS/IRS repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1(7):863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen D. R., Stenberg R. M., Goins W. F., Stinski M. F. Promoter-regulatory region of the major immediate early gene of human cytomegalovirus. Proc Natl Acad Sci U S A. 1984 Feb;81(3):659–663. doi: 10.1073/pnas.81.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen D. R., Stinski M. F. Cloning of the human cytomegalovirus genome as endonuclease XbaI fragments. Gene. 1981 Dec;16(1-3):207–216. doi: 10.1016/0378-1119(81)90077-9. [DOI] [PubMed] [Google Scholar]
- Yates J., Warren N., Reisman D., Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]