Abstract
We present a new map showing dimeric kinesin bound to microtubules in the presence of ADP that was obtained by electron cryomicroscopy and image reconstruction. The directly bound monomer (first head) shows a different conformation from one in the more tightly bound empty state. This change in the first head is amplified as a movement of the second (tethered) head, which tilts upward. The atomic coordinates of kinesin·ADP dock into our map so that the tethered head associates with the bound head as in the kinesin dimer structure seen by x-ray crystallography. The new docking orientation avoids problems associated with previous predictions; it puts residues implicated by proteolysis-protection and mutagenesis studies near the microtubule but does not lead to steric interference between the coiled-coil tail and the microtubule surface. The observed conformational changes in the tightly bound states would probably bring some important residues closer to tubulin. As expected from the homology with kinesin, the atomic coordinates of nonclaret disjunctional protein (ncd)·ADP dock in the same orientation into the attached head in a map of microtubules decorated with dimeric ncd·ADP. Our results support the idea that the observed direct interaction between the two heads is important at some stages of the mechanism by which kinesin moves processively along microtubules.
INTRODUCTION
Kinesin and nonclaret disjunctional protein (ncd) are microtubule-based molecular motors. They have been shown to function in organelle transport or in the separation of chromosomes (Vale et al., 1985; Endow et al., 1990; Goldstein, 1993; Hirokawa, 1998). Although their directions of movement along microtubules are in opposite directions, the motor domain of ncd is highly homologous to that of kinesin, with 41% amino acid sequence identity and almost identical structural folds (Kull et al., 1996; Sablin et al., 1996). Also, it has been shown that the kinetics of the ATPase of ncd is similar to that of kinesin, especially in that ADP release is the rate-limiting step and that microtubules greatly accelerate this step (Hackney, 1988, 1994; Sadhu and Taylor, 1992; Lockhart and Cross, 1994; Lockhart et al., 1995a; Shimizu et al., 1995; Crevel et al., 1996; Pechatnikova and Taylor, 1997).
The first x-ray structures were obtained for the ∼320 residue “catalytic core” of human kinesin (Kull et al., 1996) and Drosophila ncd (Sablin et al., 1996), but longer constructs have since been solved. Monomeric (Sack et al., 1997) and dimeric (Kozielski et al., 1997) rat kinesin constructs revealed two additional β strands (β9 and β10) at the C terminus, which are thought to be important for determining the directionality, and an α helix (α7) that forms a coiled coil in the dimer structure. In crystals of dimeric kinesin two motor domains are asymmetrically related to each other by an ∼120° rotation. In contrast, the atomic structure of dimeric ncd has shown two identical heads with a 180° relationship, set on either side of a coiled-coil segment (Sablin et al., 1998).
Although all the crystal structures were obtained for molecules with bound ADP, which makes motors bind only weakly to a microtubule, most electron microscopy (EM) studies of motor–microtubule complexes have used 5′-adenylylimidodiphosphate (AMP·PNP), which induces strong binding to the microtubule lattice. The structures of microtubules fully decorated with monomeric kinesin or ncd constructs, as studied by different groups (Hirose et al., 1995, 1996; Hoenger et al., 1995; Hoenger and Milligan, 1997; Kikkawa et al., 1995; Sosa et al., 1997), are mostly in good agreement and show a stoichiometry of one kinesin head per tubulin dimer. At the resolution of these studies (2–3.5 nm), there is no large difference in the structures of the kinesin–tubulin and ncd–tubulin complexes. Studies of microtubules decorated with dimeric constructs have been more controversial.
When we decorated microtubules with dimeric constructs, initially in the presence of AMP·PNP (Hirose et al., 1996), we found a pair of heads associated with each tubulin dimer, with one head bound directly to the microtubule and the other tethered in a relatively fixed position. The position of the tethered heads of kinesin and ncd showed a clear difference; the unattached head of kinesin was closer to the plus end, whereas the unattached head of ncd was pointing toward the minus end of the microtubule. Although subsequent studies gave equivalent results for dimeric ncd (Arnal et al., 1996; Sosa et al., 1997), there has been disagreement over dimeric kinesin. The results of Arnal et al. (1996) were similar to ours, but the structures obtained by Hoenger et al. (1998) showed no additional density compared with the monomeric structure. The latter group have suggested this was because each head of a dimer was bound to separate sites on tubulin. They also reported a dimeric kinesin–tubulin complex in the absence of nucleotides, which again showed only single heads attached to each tubulin dimer. Both groups who saw pairs of heads have found that the second, tethered head of dimeric kinesin (Arnal and Wade, 1998) or ncd (Hirose et al., 1998) changes position relative to the bound head when the nucleotide states are changed.
Biochemical and genetic methods have been used to identify the interaction sites between motor domains and tubulin. A truncated motor domain lacking the first 130 residues of native kinesin was shown to be still able to bind to microtubules (Yang et al., 1989). Alanine scanning of part of the surface of kinesin showed that mutation of residues in regions of the polypeptide chain that form loops L7, L8, and L11 and the L12/α5 complex impaired the interaction between kinesin and tubulin (Woehlke et al., 1997). Of these, L12/α5 seemed to be especially important for microtubule binding. The importance of the loops L8, L11, L12, and, in ncd, L2 in binding to tubulin was also shown by proteolysis experiments; some residues in these loops were protected from proteolysis when bound to microtubules (Alonso et al., 1998).
A complementary approach is to fit the coordinates of the crystal structures into EM density maps of the complexes. Similar predictions were reported by Sosa et al. (1997), in orienting the monomeric ncd·ADP crystal structure into a map of ncd complexed to microtubules in the presence of AMP·PNP, and by Hoenger et al. (1998), in docking the kinesin·ADP crystal structure into a map showing kinesin in the same state. The orientation of the directly bound heads appeared to be in agreement with most of the data obtained by biochemical and mutagenesis studies (Yang et al., 1989; Woehlke et al., 1997; Alonso et al., 1998). This orientation for the bound head was also thought to be reasonably consistent with the atomic structure of dimeric ncd (Sablin et al., 1998).
On the other hand, attempts to relate the crystal structures of dimeric kinesin (Kozielski et al., 1997; Hoenger et al., 1998) to complexes seen by electron microscopy have been unsatisfactory. When one of the heads (“head B” in the crystal structure) of dimeric kinesin is docked in the orientation chosen by Sosa et al. (1997) and Hoenger et al. (1998), the coiled coil that dimerizes the two heads points into the microtubule surface. A similar docking with “head A” as the bound head was not possible, because the second head would then penetrate into the microtubule density. With head B placed in the density of the bound head, the second head should extend from the top right of the bound head, as viewed from the outside of a microtubule with its plus end oriented upward (which we refer to as the “standard” view); however, the three-dimensional (3-D) map of Hoenger et al. (1998) showed very little density here. In contrast, a map of dimeric kinesin complexed with ADP produced by Arnal and Wade (1998) did include extra density. Kozielski et al. (1998) reported that the best fit for the dimer coordinates was obtained with head A as the bound head, rotated by ∼150° compared with the orientation chosen by Hoenger et al. (1998). The coiled-coil segment would then point away from the microtubule surface; however, this arrangement has the drawback of placing the contact that kinesin makes with tubulin on the opposite surface from the one predicted from biochemical studies, leaving loops L11 and L12 on the exposed surface of the motor domain. To explain this, Kozielski et al. (1998) suggested that kinesin might associate with microtubules via different surfaces at different stages in the motility cycle.
Now, however, we show that 3-D images of both dimeric kinesin (newly presented here) and dimeric ncd (Hirose et al., 1998), complexed with ADP and attached to microtubules by one of their two heads, can agree with all the available crystal structures. The best fitting for the directly bound heads in these maps is rotated by ∼60°, compared with the orientation with which Sosa et al. (1997) and Hoenger et al. (1998) fitted the heads into maps of unsuitable nucleotide states, and by ∼90° in the other direction, compared with the orientation selected by Kozielski et al. (1998) to fit into their map showing dimeric kinesin with ADP. Our orientation puts the predicted tubulin-binding loops L8, L11, and L12 toward the microtubule surface and also avoids a clash between the microtubule and the coiled coil of dimeric kinesin. A preliminary comparison of the maps of the ADP-containing and empty states allows us to identify tentatively some atomic structures that move to achieve a strongly bound state.
MATERIALS AND METHODS
Microtubules were assembled from purified pig brain tubulin in a polymerizing solution [80 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8, 1 mM EGTA, 5 mM MgCl2, 1 mM DTT, 0.5 mM GTP, 5% DMSO], stabilized with taxol, centrifuged, and resuspended in a solution without GTP. DMSO was added to increase the proportion of 15-protofilament microtubules (Ray et al., 1993). The motor domain consisting of rat kinesin residues 1–430 was expressed (KΔ430) and purified as described for KΔ401 (Lockhart et al., 1995b). Microtubules diluted in the 2-(N-morpholino)ethanesulfonic acid solution [60 mM 2-(N-morpholino)ethanesulfonic acid solution, 5 mM MgSO4, 1 mM EGTA, 1 mM DTT, 10 μM taxol, pH 6.5] were applied to an EM grid coated with a holey carbon film, and KΔ430 was added to give a final concentration of 10 μM. For freezing in the no-nucleotide state, the microtubule–KΔ430 mixture was incubated on the grids with 2 U/ml apyrase and then rapidly frozen by plunging the grids into an ethane slush. For the ADP state, the microtubule–kinesin mixture was put on to the grid first, and 1 mM ADP, 1 U/ml hexokinase, and 0.01% glucose were added just before freezing. Buffers and chemicals were from Sigma-Aldrich (Dorset, United Kingdom).
The grids were examined using a Gatan cold stage (Pleasanton, CA) in a Philips EM 420 electron microscope (Cheshire, CT) operating at 120 kV and with a defocus of 1300–1600 nm. Images were photographed at a magnification of 36,000 and then scanned in 28-μm steps using a Zeiss Phodis scanner (Thornwood, NY). Images of microtubules with 15 or more protofilaments were selected by measuring their diameters and then by inspecting their Fourier transform patterns, as described (Hirose et al., 1998). Micrographs were recorded with a consistent exposure, and all the images were scanned in the same manner; 796-nm lengths of digitized image were boxed and floated consistently before their Fourier transforms (Figure 1) were calculated. Phases in the transform data from different images were adjusted to match, as well as possible, those of a reference image, by allowing for a phase-origin shift and a rotation about the axis (Figure 2). The final averages included only images whose layer-line data on both sides were completely consistent with the consensus, to be sure of avoiding microtubules with “seams” in the helical lattice (Song and Mandelkow, 1995). Variance maps were calculated by comparing individual real-space maps as described by Trachtenberg and DeRosier (1987). Maps were displayed as described previously (e.g., Hirose et al., 1998). Fitting of the α-carbon backbone of the atomic coordinates into the EM density maps was performed visually using the program MAIN (Turk, 1992). Crystal structures were drawn using Rasmol (Roger Sayle, rasmol@ggr.co.uk) and Molscript (Kraulis, 1991).
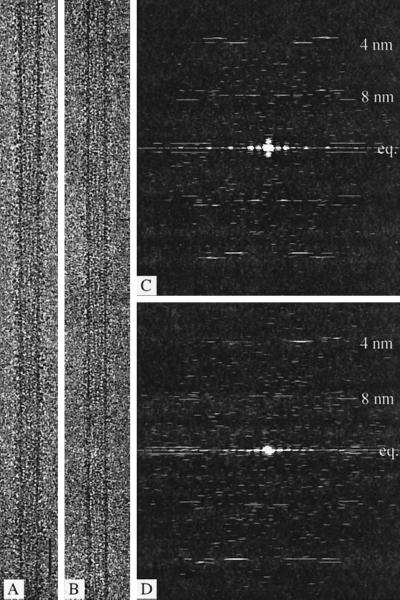
Figure 1.
Electron microscope images and their diffraction patterns. (A and B) Images of 15-protofilament brain microtubules decorated with the double-headed kinesin motor construct (KΔ430) and either treated with apyrase to remove all free nucleotide (A) or in the presence of ADP and hexokinase (B). Bar, 50 nm. (C and D) Computed Fourier transforms from A and B. The equatorial (eq.) and 8-nm layer lines are relatively weaker in patterns from an ADP-containing complex (D) than in those from the nucleotide-free state (C).
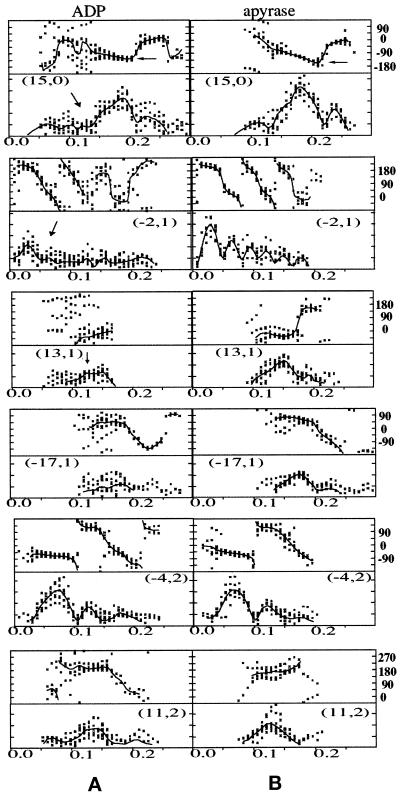
Figure 2.
Amplitudes and phases along the main layer lines of computed Fourier transforms. Phases (in degrees) are plotted in the upper half of each panel; amplitudes (all on the same scale) are shown in the bottom plots. The phases have been corrected for rotations about the microtubule axis and for shifts in the position of the phase origin along the axis to give the best overall agreement among different images. The data points are from individual images of 15-protofilament brain microtubules decorated with KΔ430 in ADP (A) or in the absence of nucleotides (effect of apyrase; B). The plotted lines show the averaged values. The layer-line indexing (n,l) refers to the Bessel order n, or number of equivalent helices in 15-protofilament microtubules, and the layer-line number l, based on a nominal 8-nm axial repeat. Some differences between nucleotide states are indicated by arrows: the phase distribution on the (15,0) layer line does not fall as steeply for ADP as for the tightly bound state; amplitudes on several layer lines are lower for the ADP state.
RESULTS
3-D Images of Kinesin Dimers with ADP or No Nucleotide
Cryoelectron microscope images of reassembled brain microtubules decorated with dimeric kinesin (KΔ430) and frozen after incubation with apyrase to remove traces of ATP and ADP appeared very well ordered (Figure 1A); other images, of specimens to which 1 mM Mg·ADP had been added immediately before freezing, were decorated in a less well-ordered way but still appeared to be saturated with motor domains (Figure 1B). The difference between the strongly bound empty state and the weakly bound ADP state is apparent from the relative intensities of the 4- and 8-nm layer lines in diffraction patterns from the images (Figure 1, C and D). For each type of image, an average data set, calculated from a selected group of helical “sides” that agreed best with one another (Figure 2), was used to calculate the final density maps (Figure 3).
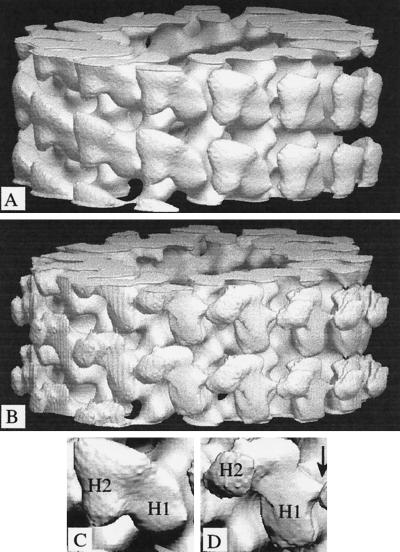
Figure 3.
Surface representations of the 3-D density maps. (A and B) The maps were computed from averaged data (see Figure 2) for 15-protofilament brain microtubules decorated with dimeric kinesin in the absence of free nucleotide (A) or in the presence of ADP (B). They are in the standard orientation, with the microtubule plus end at the top of the page. (C and D) Enlarged views of individual dimeric motors are shown. In each case, H1 is the directly bound head; H2 is the tethered head. An arrow indicates the absence in the nucleotide-free structure (C) of a feature in the ADP-containing structure (D) that can fit loop L12 (see Figure 6c).
In both density maps, one of the two heads in each dimer appears to bind directly to the microtubule surface, whereas the other is tethered to the bound head in a relatively fixed position. The positions of the tethered head relative to that of the bound head vary from what we found for the AMP·PNP state (Hirose et al., 1996), being to the left of the bound head rather than to the right. In the ADP state, as compared with the empty state, the tethered head is shifted more toward the microtubule plus end. The density of the heads in the ADP state is weaker and noisier because the motor domains then bind weakly to tubulin. Small random movements in the weakly attached heads appear to be amplified in the tethered heads; thus the size and shape of the tethered kinesin heads in ADP are not expected to be reliable, although their mean positions should be more so.
Structural differences are also found in the shapes of the bound heads; a protruding feature toward the top right of the bound head in ADP (see Figures 3D, arrow, and 4B, cross section) is missing in the empty state (Figures 3C and 4A). Nor was it seen in our previous maps of kinesin in the AMP·PNP state (Hirose et al., 1995, 1996). To check that images of the two states really are different, variances were calculated from sets of individual 3-D maps, after scaling in real-space (Trachtenberg and DeRosier, 1987). The results (Figure 4) indicate some significant structural differences.
Figure 4.
Contoured cross sections with variance levels. (A and B) Cross sections through the average maps in Figure 3 superimposed on variance values calculated from the individual maps. Arrows point to the projecting feature, sectioned at various levels, present in kinesin·ADP (B) but missing in the empty state (A). (C) Variancesobtained when nucleotide-free and ADP data sets are mixed. All variances are shown on the same scale, with darker density for higher values; there are peaks (arrows) on the exposed surface of the bound heads (H1), close to the point of attachment of the tethered heads (H2) and also close to where the tops of the bound heads contact tubulin, reflecting the changes described in the text.
Fitting the Atomic Coordinates into EM Density Maps
Kinesin Monomer and Dimer.
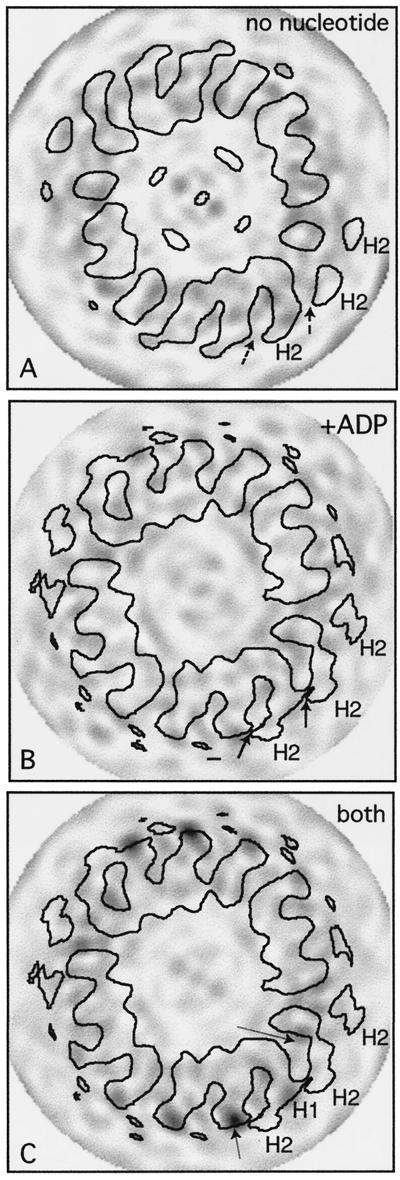
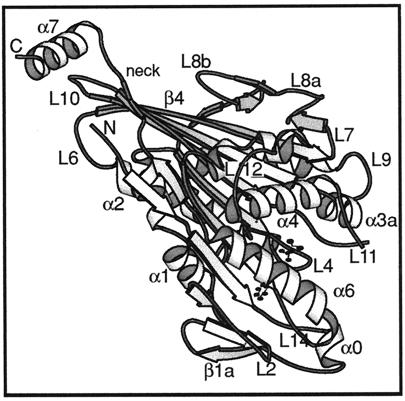
The crystal structures of motor domains of monomeric rat kinesin (Sack et al., 1997) (illustrated in Figure 5) and monomeric human kinesin (Kull et al., 1996) both have bound ADP and are very similar except that the N- and C-terminal segments are missing from the human kinesin structure. Loop L11, which is assumed to be very flexible, is not clearly seen in either structure. Figure 6 shows the best fit, as judged by eye, when we tried docking either set of coordinates into our 3-D EM map of kinesin·ADP. Loop L11 would lie against the right-hand surface of a tubulin protofilament. This orientation seemed to best satisfy the shape of the outer boundaries but also produced a good complementarity between the shapes of the bound kinesin head and the tubulin monomers. In Figure 6, density peaks representing tubulin subunits have been identified by comparing this map with one of an undecorated microtubule (Hirose et al., 1997). The two monomers of a tubulin heterodimer are labeled on the basis of information obtained from images of the very ends of decorated tubulin sheets (reviewed by Amos and Hirose, 1997).
Figure 5.
Atomic structure of monomeric kinesin. A ribbon diagram of the kinesin motor domain (Sack et al., 1997) viewed from an orientation similar to that in a figure below (see Figure 6b). The heart-shaped catalytic domain comes to a point at the top left of the molecule. The neck (including β9 and β10) runs alongside leading to helix α7, part of the rod domain that forms a coiled coil in dimeric kinesin. Bound ADP shown as a ball and stick model. N and C, chain termini.
Figure 6.
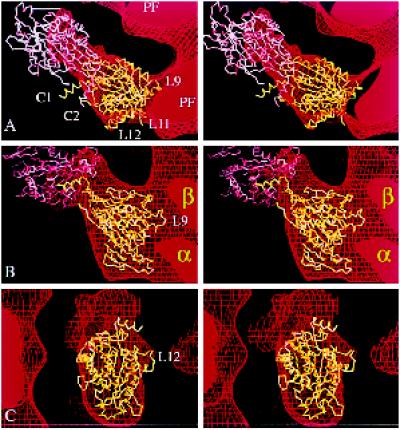
Fitting the atomic structure of kinesin into our kinesin·ADP map. Stereo views of two copies of the α-carbon backbone of the kinesin monomer (Sack et al., 1997) oriented inside part of the EM map (in Figure 3B) that shows tubulin protofilaments (PF) decorated with dimeric kinesin·ADP. (a) A top view, seen from the microtubule plus end. (b) A side view. (c) The view, at 90° to both a and b, showing the attached head from outside the microtubule. The map is represented as a surface net (red lines). One kinesin molecule (shown in yellow) occupies the attached head density; another (in pink) is positioned in the tethered head density. The relative orientation of the pink and yellow molecules mimics, as closely as possible, the asymmetric arrangement of heads in crystals of dimeric kinesin (Kozielski et al., 1997); thus, loop L10 of the attached head B contacts loop L8 of the tethered head A. A small distortion of each C-terminal helix (C1 and C2) would be needed to bring them together to form a coiled coil; this rod would then point almost directly away from the microtubule.
A volume of protein density above the left one-half of the bound head that remains unfilled (Figure 6c) is essentially where the tethered head should be positioned to agree with the dimeric kinesin crystal structure (Kozielski et al., 1997), with the attached head corresponding to head B. Although the map has insufficient density at high radius to accommodate all of head A, probably because of disorder, a second copy of the monomer crystal structure (Figure 6, pink), in an orientation close to that of head A in the crystal structure, readily accounted for the extra density. We saw no sign of density representing a coiled-coil segment (residues 339–370) or of the final C-terminal segment (residues 371–430) of KΔ430, which is predicted to form a random loop followed by another piece of coiled coil; these structures are likely to be poorly ordered and to make no detectable contribution to the EM map. However, in the fitted orientation, the average position of the coiled-coil rod would be expected to point away from the microtubule surface (as can be seen from Figure 6a).
We tried to arrange the monomer structure into our kinesin·ADP map in the same orientation as Hoenger et al. (1998) fitted it into a kinesin·AMP·PNP map, that is, with the bound head rotated by ∼60° (Figure 7, same map shown in Figure 6). The bound head (yellow) lies inside the density, but in the top view (Figure 7a), the shapes do not match as well as in Figure 6a. In the lateral view (Figure 7b), the agreement appears good near the outside surface, but the interface between kinesin and tubulin matches less well. When a second monomer structure (pink) is added, to mimic the dimeric kinesin crystal structure, this molecule is almost completely out of the density of the 3-D map (Figure 7a); it is impossible to move it into the density while keeping an arrangement resembling that in the crystal structure. Nor is it possible to orient the dimer so that the coiled coil fits into the extra density.
Figure 7.
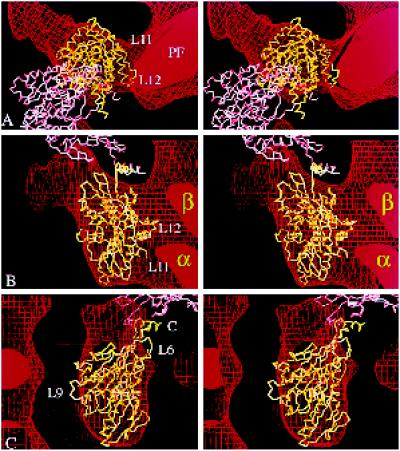
Alternative docking of kinesin into our kinesin·ADP map. (a–c) Stereo views showing two copies of the α-carbon backbone of monomeric kinesin oriented inside the same EM map of kinesin·ADP shown in Figure 6 but docked in the orientation favored by Hoenger et al. (1998). The directly bound head (yellow) fills the outer boundaries almost as well as in Figure 6, but the top (a) and side (c) views fit less accurately, and in the side view (b), the complementarity of kinesin’s inside surface with the tubulin subunits (α and β) is less good. A second molecule (pink) arranged as in the crystallographic structure is completely out of the EM density, as can be seen in a. The coiled coil would point obliquely into the microtubule surface. C, C terminus; PF, protofilament.
ncd Monomer and Dimer.
We have published previously a map of a microtubule decorated with dimeric ncd expressed motor domain (consisting of Drosophila ncd residues 295–700) in the presence of ADP (Hirose et al., 1998) but did not attempt to fit atomic coordinates into it. We have now tried to fit the coordinates of monomeric ncd (Sablin et al., 1996) into density representing each of the two heads (Figure 8). Compared with that in maps of ncd in the AMP·PNP state (Arnal et al., 1996; Hirose et al., 1996, 1998; Sosa et al., 1997), the connection between the bound and tethered heads is shifted away from the top right corner of the bound head to a position more toward the front of the bound head and closer to the minus end of the microtubule. Also, the shape of the top of the attached head is more domed. As in the case of kinesin, the monomeric ncd crystal structure appeared, by eye, to fit best into the bound-head density (crystal structure in yellow) after being rotated by ∼60° from the original alignment (Sosa et al., 1997). The N-terminus, which is thought to be connected to the coiled coil, is inside the density connecting the bound and the tethered heads.
Figure 8.
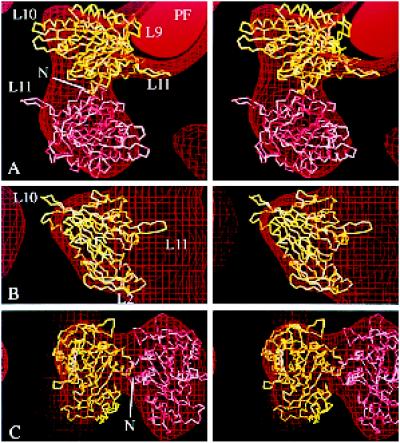
Fitting the atomic structure of ncd into our ncd·ADP map. (a–c) Stereo views of the α-carbon backbone of monomeric ncd (Sablin et al., 1996) oriented inside a map showing tubulin protofilaments (PF) decorated with dimeric ncd with bound ADP (Hirose et al., 1998). As in Figures 6 and 7, the map is represented at a single contour level as a surface net (red lines). One copy of the monomeric ncd crystal structure (yellow) is positioned in the attached head density; another (pink) lies in the tethered head density. The labeled loops (L11, L9, etc.) are equivalent to those in the kinesin structure (see Figure 5). The side view (b) at 90° to a is shown with the tethered head removed. The front view in c is at 90° to both a and b. There is an approximate twofold axis between the heads, running almost vertically in c. N, N terminus.
When a pair of ncd monomers, arranged exactly as in the dimeric ncd crystal structure (Sablin et al., 1998), was docked with the bound head density in the orientation shown in Figure 8, the tethered head fell outside the EM density. Thus, it was necessary to fit the tethered head separately. The shape of the tethered head in the EM map is not sufficiently asymmetrical to fix its orientation unequivocally. However, the shapes of the two heads seem similar, with the top part fatter, and they seem to be related by an approximate twofold symmetry, although the directly attached head is smaller than it should be and the tethered head is larger. The position of the monomer shown in pink in Figure 8 was chosen to maintain an approximate twofold relationship between the heads and to put the two N termini close together in the connection between the heads. There is space in this region that could accommodate the extra residues in the expressed motor domain that are not in the monomer crystal structure (residues 345–667). The orientation chosen by Sosa et al. (1997) for the second head fits almost as well into our map as the one shown in Figure 8. It is related to our docking by an ∼180° rotation around its connection with the first head, which would make both loops L11 point toward the right in Figure 8a.
We also tried docking the ncd dimer with the attached head oriented as described by Sosa et al. (1997) (the orientation of the second head is then quite different from that shown by those authors). In this case, part of the tethered head lies within the EM map density without changing the dimeric arrangement. However, the attached head does not agree with the shape suggested by the density in the EM map; in particular, the point of the heart-shaped monomer, including loop L10, extends well into the density we identify as part of the second head, pushing the coordinates of the latter nearly halfway out of the available density. This problem apparent in the illustration shown by Mandelkow and Hoenger (1999).
Interactions with Tubulin
Monomeric kinesin is represented in Figure 9 as a space-filling model; Figure 9A shows the surface that would face away from the microtubule in the new orientation. The N-terminal 130 residues of kinesin (colored in light-blue), which can be removed without loss of microtubule-binding (Yang et al., 1989), are mostly on this surface. In Figure 9B, the motor has been rotated by 180° to show the surface that would be seen by a tubulin dimer. The loops L7, L8, L9, L11, and L12 and helices α4 and α5 are in the upper one-half of this surface, where they would interact with β-tubulin; α6, L2, L14, α0 are in the lower one-half, where they would contact α-tubulin.
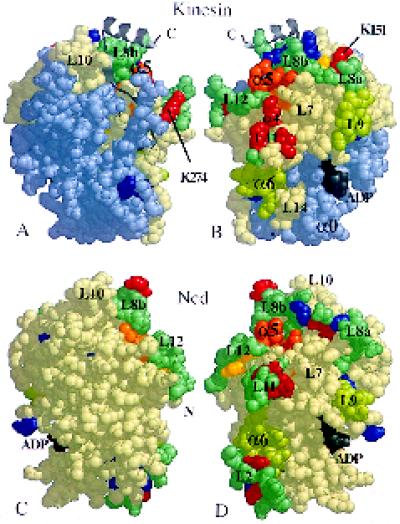
Figure 9.
Exposed and protected surfaces of the bound motor domains. Space-filling images of monomeric kinesin and Ncd, showing the positions of residues that appear to be important. (A and C) The views from directly outside a decorated microtubule for heads bound in the orientation shown in Figure 6; the head (B and D) has been rotated by 180° to show the surface closest to tubulin. Cleavable residues are either protected from proteolysis by microtubules (red), including lysines K151 and K274, or always exposed (blue) (Alonso et al., 1998); yellow residues are cut differentially in ADP versus AMP·PNP, in the absence of microtubules. Alanine scanning (Woehlke et al., 1997) of kinesin suggests that the orange arginine and lysine residues (R280, K283, and R286—corresponding to R278, K281, and R284 in human kinesin, respectively) in the L12/α5 region are important for the binding of microtubules. Some predicted tubulin-binding loops (L7, L8, L11, and L12 [Kull et al., 1996]) are shown in blue-green. Helices α4 and α6 and loops L7 and L9 also appear to be exposed to tubulin. The light-blue part of kinesin represents residues 1–130 (including α0), which can be removed without loss of microtubule binding (Yang et al., 1989). N and C, chain termini.
Most of the proteolysis-susceptible residues in kinesin that are protected by binding the motors to microtubules (Figure 9, residues colored in red [Alonso et al., 1998]) are in the flexible L11 loops or in the region containing loops L12 and L8 and helix α5. Tubulin protects similar sites on ncd from proteolysis, except that there are additional residues in loop L2 (lysines K388 and K390) (Alonso et al., 1998). The residues whose mutation is found to reduce microtubule binding are also in the overlapping area, including the “hot spot” in L12/α5 (colored in orange) (Woehlke et al., 1997). Most of these residues are on the surface facing the microtubule, either in the orientation we have fitted or in that proposed by Sosa et al. (1997) and Hoenger et al. (1998). The important loop L12 is further from the microtubule surface in our orientation. Its position in our docking coincides with a protrusion in the 3-D map of the ADP state, which is absent in the empty state (Figure 3, C and D). An equivalent difference was also seen in our original maps of microtubules decorated with kinesin monomer and imaged in negative stain (Hirose et al. [1995]; see also Hirose et al. [1998], their Figure 9).
DISCUSSION
Several groups have fitted the atomic structures of kinesin or ncd into 3-D EM maps of kinesin or ncd complexed to microtubules (Sosa et al., 1997; Hoenger et al., 1998; Kozielski et al., 1998). Everyone agrees that the point of the heart-shaped monomer in the directly attached heads should point toward the microtubule plus end, but there is disagreement over the angle of rotation about the vertical axis (summarized in Table 1). Our results differ from those published previously in a number of ways that will be discussed individually. In summary, our fitting of kinesin into our map of microtubules decorated in the ADP-filled state not only satisfies the consensus view as to which motor domain loops are involved in microtubule-binding but also suggests the same head-to-head relationship as in the crystal structure of dimeric kinesin·ADP (Kozielski et al., 1997) and avoids steric interference between the stalk domain of dimeric kinesin and the microtubule. Our results also conform with the idea that both kinesin and ncd bind to tubulin in the same way, as predicted by a range of other techniques, and are compatible with the absence of any direct interaction between ncd heads, as shown by the crystal structure of dimeric ncd·ADP (Sablin et al., 1998).
Table 1.
Alternative dockings of kinesin·ADP or Ncd·ADP into EM maps
| Authors | State of motors bound | Heads seen | Binding orientation | Agreement with crystallographic dimer structures? | Agreement with proteolysis and mutagenesis |
|---|---|---|---|---|---|
| Sosa et al., 1997 | Dimeric ncd + AMP·PNP | 2 | −60° | Not good, lacks 2-fold | Yes |
| Hoenger et al., 1998 | Dimeric kinesin + AMP·PNP | 1 | −60° | No, 2nd head or tail would interfere with microtubule | Yes |
| Kozielski et al., 1998 | Dimeric kinesin + ADP | 2 | +90° | Yes | No |
| This paper | Dimeric kinesin + ADP | 2 | 0° | Yes | Yes |
| This paper | Dimeric ncd + ADP | 2 | 0° | OK, 2-fold relationship retained | Yes |
Stoichiometry of Kinesin Heads Bound to Microtubules
All of our maps of dimeric kinesin bound to microtubules have shown the density for two heads associated with each tubulin dimer: one head attached directly to tubulin and a somewhat smaller region of density that we interpret as the second head tethered to the first. The reduced density of the tethered head probably results from random movement of this feature, which seems to be most flexibly attached in the AMP·PNP state (Hirose et al., 1996). Hoenger et al. (1998) detected a slight density increase in their map of dimeric kinesin bound to microtubules with AMP·PNP, compared with monomeric kinesin, but considered it to be insufficient to assign to tethered heads. They, therefore, suggested that interference between the coiled-coil stalk and the microtubule surface causes the two heads to separate and bind to different tubulin dimers. They also suggested that the density we interpreted as the tethered head in our AMP·PNP map (Hirose et al., 1996) might represent the extra piece of polypeptide chain at the C terminus of a dimeric construct. But this idea could certainly not explain the extra density in our ADP map, which points in approximately the opposite direction to the predicted position of the coiled coil for their orientation (Figure 7a) and at ∼90° for our orientation (Figure 6a).
Our results (Lockhart et al., 1995b; Hirose et al., 1996; the maps shown here) suggest that few kinesin dimers are bound by both heads under the conditions used in our experiments. In particular, for EM, microtubules are spread first on the carbon film before being decorated in situ, when we rapidly add a saturating concentration of kinesin dimers so that all the binding sites on tubulin should be occupied by first heads. It is, however, not yet clear what is important in producing the varying conformations seen in different laboratories.
Docking of Kinesin and ncd in Similar Orientations
Sosa et al. (1997) docked the monomeric ncd·ADP crystal structure into an EM map of motor domains bound to microtubules in the AMP·PNP state. In this state, the main contact between the bound and tethered heads is at the very top of the right-hand side of the bound head (Arnal et al., 1996; Hirose et al., 1996; Sosa et al., 1997); thus, the coordinates were put into the bound head density so that the point of the heart would make the connection to the tethered head. The resulting orientation was in agreement with most of the mutagenesis and proteolysis results, and Hoenger et al. (1998) were able to duplicate it when fitting kinesin monomer coordinates into the bound head density in a map of kinesin-decorated microtubules. However, Hoenger et al. (1998) also showed that an arrangement similar to that in the dimeric kinesin crystal structure would be sterically impossible with the bound head in this orientation.
The way we have docked the bound heads of kinesin and ncd is rotated by ∼60° compared with that of Sosa et al. (1997) and Hoenger et al. (1998). In the original orientation, it was predicted that the flexible loop L11 of the bound head contacts tubulin along the left-hand side of a protofilament in the standard view, reaching far into the interprotofilament groove (see Figure 7a). In our orientation, it makes contact with the right-hand side of the ridged surface of the protofilament (Figure 6a).
Most of the proteolysis-susceptible residues in kinesin and ncd that are protected by binding the motors to microtubules (Alonso et al., 1998) are on the surface facing the microtubule, in either orientation. Protection of K151 on loop L8a is more easily accounted for by our result, although it still appears fairly exposed (Figure 9). It may be closer to the microtubule surface in the strongly bound states as a result of conformational changes at the top of the head, because it is known that the adjacent residue Leu153 (Figure 9, yellow) is protected by adding AMP·PNP to the motor domains, even in the absence of microtubules. Also, the tops of kinesin and ncd heads both appear to make closer contact with β-tubulin in the strongly bound states than in the ADP state (see Hirose et al. [1998] for ncd; results for kinesin [our unpublished observations] are similar). These results indicate that some parts of the molecule move toward the microtubule during the transition from weakly to strongly bound states. It seems likely that the changes also bring the important loop L12 closer to the microtubule surface, which would account for the absence of density in this region in the empty state (see Figure 3).
In the ncd·ADP map, the point of the heart now occupies the domed top of the bound head. We found no difficulty in orienting the tethered head, with the two N termini close to one another, to give an approximate twofold relationship, although the position of the twofold axis is not the same as in the crystal structure (Sablin et al., 1998). This can be compared with the arrangement originally predicted by Sosa et al. (1997), in which the heads are not even close to a twofold relationship. However, their choice for the orientation of the tethered head turns out to be related to ours by an ∼180° rotation. In fact, putting the attached head in our orientation and the tethered head in theirs provides a reasonable alternative to that shown in Figure 8, again producing an approximate twofold relationship in the ncd dimer, with the twofold axis rotated to a new position.
Whereas the crystallographic structure found for the kinesin dimer shows a direct interaction between the two heads, the structure of dimeric ncd (Sablin et al., 1998) shows two heads interacting with either side of a coiled coil formed by the neck domains. Sablin et al. (1998) have predicted that this arrangement is unstable and suggest that a small conformational change caused by one head binding to a microtubule might easily cause the neck to lose its coiled-coil structure and no longer hold the two heads closely together. Our results support this idea because we see no density that would account for a coiled coil sandwiched between the two heads. Each head of the dimer in the crystallographic structure would need to twist freely, by ∼75° around the point of connection, to produce the ncd dimer configuration shown in Figure 8. Indeed, unzipping of the coiled coil might cause the heads to twist in this way.
Alternative Conformations of Dimeric Kinesin·ADP
Our map of dimeric kinesin bound to microtubules in the absence of nucleotide is in good agreement with that of Arnal and Wade (1998). However, there is a marked difference between their map of the ADP-containing state and ours. Their map shows a conformation similar to that of dimeric kinesin in the AMP·PNP (ATP-like) state, with the tethered head attached to the right-hand side of the attached head (Arnal et al., 1996; Hirose et al., 1996) but extending more in the direction of the microtubule plus end than does that in the AMP·PNP state. In our map of the ADP state, the tethered head is toward the left-hand side, resembling the empty state but tilting more toward the microtubule plus end. Kozielski et al. (1998) have docked the kinesin dimer coordinates, with head A as the attached head, in an orientation rotated by ∼90° from ours. This docking also avoids interference between the microtubule and the coiled-coil stalk but is unlikely to be correct because it does not agree with the data from proteolysis protection and mutagenesis.
We do need to understand, however, why Arnal and Wade (1998) saw dimeric kinesin·ADP in a different conformation from ours. One possibility is that bound kinesin heads can be trapped in two nonequivalent ADP-containing states. Kinesin and ncd remain bound to tubulin while ATP is hydrolyzed and the freed γ-phosphate may also be released quickly, leading to a “final” weakly bound ADP-containing state before the head detaches (reviewed by Cross, 1997). The head then attaches to a new site on the protofilament (the “initial” weakly binding state), which catalyzes the release of ADP, followed by the strongly binding empty state. In our experiments, microtubules were first decorated with motor domains in the empty state, and ADP was added immediately before freezing. Arnal and Wade (1998), on the other hand, observed decorated microtubules after incubating them for 5 min with motors and ADP in a buffer that included 10 mM phosphate. It is possible that differences in the experimental conditions have driven the motors into two nonequivalent ADP-containing states. The map shown here may, for example, represent the initial ADP state, whereas that of Arnal and Wade may represent a posthydrolysis state. We have some preliminary results that support this idea and suggest that it will be possible to dock head A of the dimer structure into maps of the second ADP state in the same orientation that we have docked head B into the map shown in Figure 6.
In conclusion, our docking of motor domains attached to microtubules gives the only orientation that is consistent with proteolysis and mutagenesis data, while also supporting the kinesin dimer conformation seen by crystallography (see Table 1). It seems likely that the asymmetric interaction between the two heads of kinesin could be strong and specific enough to be preserved under both crystallizing and EM specimen-preparation conditions. We suggest that this structure is probably close to a conformation that occurs during the hydrolysis cycle and that the asymmetry reflects the known functional asymmetry of dimeric kinesin (Hackney, 1994; Gilbert et al., 1995; Jiang et al., 1997).
ACKNOWLEDGMENTS
We are very grateful to Juan Fan for providing purified tubulin. We thank R. Fletterick and colleagues for the atomic coordinates of monomeric kinesin and ncd. The coordinates for 2KIN and 3KIN were obtained from the Brookhaven database. J.L. was the recipient of a long-term European Molecular Biology fellowship during this project.
Abbreviations used:
- AMP·PNP
5′-adenylylimidodiphosphate
- EM
electron microscopy
- KΔ430
expressed motor domain consisting of rat kinesin residues 1–430
- ncd
nonclaret disjunctional protein
- 3-D
three-dimensional
REFERENCES
- Alonso MC, Vanderkerckhove J, Cross RA. Proteolytic mapping of kinesin/ncd-microtubule interface: nucleotide-dependent conformational changes in the loops L8 and L12. EMBO J. 1998;17:945–951. doi: 10.1093/emboj/17.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos LA, Hirose K. The structure of microtubule-motor complexes. Curr Opin Cell Biol. 1997;9:4–11. doi: 10.1016/s0955-0674(97)80145-7. [DOI] [PubMed] [Google Scholar]
- Arnal I, Metoz F, DeBonis S, Wade RH. Three-dimensional structure of functional motor proteins on microtubules. Curr Biol. 1996;6:1265–1270. doi: 10.1016/s0960-9822(02)70712-4. [DOI] [PubMed] [Google Scholar]
- Arnal I, Wade RH. Nucleotide-dependent conformations of the kinesin dimer interacting with microtubules. Structure. 1998;6:33–38. doi: 10.1016/s0969-2126(98)00005-7. [DOI] [PubMed] [Google Scholar]
- Crevel IM-T, Lockhart A, Cross RA. Weak and strong states of kinesin and ncd. J Mol Biol. 1996;257:66–76. doi: 10.1006/jmbi.1996.0147. [DOI] [PubMed] [Google Scholar]
- Cross RA. Molecular motors: the natural economy of kinesin. Curr Biol. 1997;7:R631–R633. doi: 10.1016/s0960-9822(06)00320-4. [DOI] [PubMed] [Google Scholar]
- Endow SA, Henikoff S, Soler Niedziela L. Mediation of meiotic and early mitotic chromosome segregation in Drosophila by a protein related to kinesin. Nature. 1990;345:81–83. doi: 10.1038/345081a0. [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Webb MR, Brune M, Johnson KA. Pathway of processive ATP hydrolysis by kinesin. Nature. 1995;373:671–676. doi: 10.1038/373671a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LSB. With apologies to Scheherazade—tails of 1001 kinesin motors. Annu Rev Genet. 1993;27:319–351. doi: 10.1146/annurev.ge.27.120193.001535. [DOI] [PubMed] [Google Scholar]
- Hackney DD. Kinesin ATPase: rate-limiting ADP release. Proc Natl Acad Sci USA. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. The rate-limiting step in microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains occurs while bound to the microtubule. J Biol Chem. 1994;269:16508–16511. [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Hirose K, Amos WB, Lockhart A, Cross RA, Amos LA. Three-dimensional cryoelectron microscopy of 16-protofilament microtubules: structure, polarity and interaction with motor protein. J Struct Biol. 1997;118:140–148. doi: 10.1006/jsbi.1997.3840. [DOI] [PubMed] [Google Scholar]
- Hirose K, Cross RA, Amos LA. Nucleotide-dependent structural changes in dimeric ncd molecules complexed to microtubules. J Mol Biol. 1998;278:389–400. doi: 10.1006/jmbi.1998.1709. [DOI] [PubMed] [Google Scholar]
- Hirose K, Lockhart A, Cross RA, Amos LA. Nucleotide-dependent angular change in kinesin motor domain bound to tubulin. Nature. 1995;376:277–279. doi: 10.1038/376277a0. [DOI] [PubMed] [Google Scholar]
- Hirose K, Lockhart A, Cross RA, Amos LA. Three-dimensional cryoelectron microscopy of dimeric kinesin and ncd motor domains on microtubules. Proc Natl Acad Sci USA. 1996;93:9539–9544. doi: 10.1073/pnas.93.18.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A, Milligan RA. Motor domains of kinesin and ncd interact with microtubule protofilaments with the same binding geometry. J Mol Biol. 1997;265:553–564. doi: 10.1006/jmbi.1996.0757. [DOI] [PubMed] [Google Scholar]
- Hoenger A, Sablin EP, Vale RD, Fletterick RJ, Milligan RA. 3-dimensional structure of a tubulin-motor-protein complex. Nature. 1995;376:271–274. doi: 10.1038/376271a0. [DOI] [PubMed] [Google Scholar]
- Hoenger A, Sack S, Thormählen M, Marx A, Müller J, Gross H, Mandelkow E. Image reconstructions of microtubules decorated with monomeric and dimeric kinesins: comparison with x-ray structure and implications for motility. J Cell Biol. 1998;141:419–430. doi: 10.1083/jcb.141.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Stock MF, Li X, Hackney DD. Influence of the kinesin neck domain on dimerization and ATPase kinetics. J Biol Chem. 1997;272:7626–7632. doi: 10.1074/jbc.272.12.7626. [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Ishikawa T, Wakabayashi T, Hirokawa N. 3-dimensional structure of the kinesin head-microtubule complex. Nature. 1995;376:274–279. doi: 10.1038/376274a0. [DOI] [PubMed] [Google Scholar]
- Kozielski F, Arnal I, Wade RH. A model of the microtubule–kinesin complex based on electron cryomicroscopy and x-ray crystallography. Curr Biol. 1998;8:191–198. doi: 10.1016/s0960-9822(98)70083-1. [DOI] [PubMed] [Google Scholar]
- Kozielski F, Sack S, Marx A, Thormählen M, Schönbrunn E, Biou V, Thompson A, Mandelkow EM, Mandelkow E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–941. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript—a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart A, Crevel IM-TC, Cross RA. Kinesin and ncd bind through a single head to microtubules and compete for a shared MT binding site. J Mol Biol. 1995a;249:763–771. doi: 10.1006/jmbi.1995.0335. [DOI] [PubMed] [Google Scholar]
- Lockhart A, Cross RA. Origins of reversed directionality in the ncd molecular motor. EMBO J. 1994;13:751–757. doi: 10.1002/j.1460-2075.1994.tb06317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart A, Cross RA, McKillop DFA. ADP release is the rate-limiting step of the MT activated ATPase of nonclaret disjunctional and kinesin. FEBS Lett. 1995b;368:531–535. doi: 10.1016/0014-5793(95)00723-m. [DOI] [PubMed] [Google Scholar]
- Mandelkow E, Hoenger A. Structures of kinase and kinesin-microtubule interactions. Curr Opin Cell Biol. 1999;11:34–44. doi: 10.1016/s0955-0674(99)80005-2. [DOI] [PubMed] [Google Scholar]
- Pechatnikova E, Taylor EW. Kinetic mechanism of monomeric nonclaret disjunctional protein (Ncd) ATPase. J Biol Chem. 1997;272:30735–30740. doi: 10.1074/jbc.272.49.30735. [DOI] [PubMed] [Google Scholar]
- Ray S, Meyhöfer E, Milligan RA, Howard J. Kinesin follows the microtubule’s protofilament axis. J Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablin EP, Case RB, Dai SC, Hart CL, Ruby A, Fletterick RJ, Vale RD. Direction determination in the minus-end-directed kinesin motor ncd. Nature. 1998;395:813–816. doi: 10.1038/27463. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- Sack S, Müller J, Marx A, Thormählen M, Mandelkow EM, Brady ST, Mandelkow E. X-ray structure of motor and neck domains from rat brain kinesin. Biochemistry. 1997;36:16155–16165. doi: 10.1021/bi9722498. [DOI] [PubMed] [Google Scholar]
- Sadhu A, Taylor EW. A kinetic study of the kinesin ATPase. J Biol Chem. 1992;267:11352–11359. [PubMed] [Google Scholar]
- Shimizu T, Sablin E, Vale RD, Fletterick R, Pechatnikova E, Taylor EW. Expression, purification, ATPase properties, and microtubule-binding properties of the ncd motor domain. Biochemistry. 1995;34:13259–13266. doi: 10.1021/bi00040a042. [DOI] [PubMed] [Google Scholar]
- Song Y-H, Mandelkow E. The anatomy of flagellar microtubules: polarity, seam, junctions, and lattice. J Cell Biol. 1995;128:81–94. doi: 10.1083/jcb.128.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa H, Dias DP, Hoenger A, Whittaker M, Wilson-Kubalek E, Sablin E, Fletterick RJ, Vale RD, Milligan RA. A model for the microtubule-ncd motor protein complex obtained by cryo-electron microscopy and image analysis. Cell. 1997;90:217–224. doi: 10.1016/s0092-8674(00)80330-x. [DOI] [PubMed] [Google Scholar]
- Trachtenberg S, DeRosier DJ. Three-dimensional structure of the frozen-hydrated flagellar filament. The left-handed filament of Salmonella typhimurium. J Mol Biol. 1987;195:581–601. doi: 10.1016/0022-2836(87)90184-7. [DOI] [PubMed] [Google Scholar]
- Turk D. Weiterentwicklung eines Programms für Molekülgraphik und Elektrondichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklärungen. Ph.D. Thesis. München. Germany: Technische Universität; 1992. [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- Yang JT, Laymon RA, Goldstein LSB. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analyses. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]