Abstract
We have screened for temperature-sensitive (ts) fission yeast mutants with altered polarity (alp1–15). Genetic analysis indicates that alp2 is allelic to atb2 (one of two α-tubulin genes) and alp12 to nda3 (the single β-tubulin gene). atb2+ is nonessential, and the ts atb2 mutations we have isolated are dominant as expected. We sequenced two alleles of ts atb2 and one allele of ts nda3. In the ts atb2 mutants, the mutated residues (G246D and C356Y) are found at the longitudinal interface between α/β-heterodimers, whereas in ts nda3 the mutated residue (Y422H) is situated in the domain located on the outer surface of the microtubule. The ts nda3 mutant is highly sensitive to altered gene dosage of atb2+; overexpression of atb2+ lowers the restrictive temperature, and, conversely, deletion rescues ts. Phenotypic analysis shows that contrary to undergoing mitotic arrest with high viability via the spindle assembly checkpoint as expected, ts nda3 mutants execute cytokinesis and septation and lose viability. Therefore, it appears that the ts nda3 mutant becomes temperature lethal because of irreversible progression through the cell cycle in the absence of activating the spindle assembly checkpoint pathway.
INTRODUCTION
Microtubules are important and ubiquitous structures that play essential roles in various cellular processes, including motility, mitosis, transport of proteins and mRNAs, and cell morphogenesis (reviewed in Mitchison and Kirschner, 1986; Lehmann, 1995; Hyman and Karsenti, 1996). They assemble from heterodimers composed of α- and β-tubulin subunits, which are both evolutionarily highly conserved. Additional members of tubulin subfamilies including γ-tubulin exist in various species (Oakley, 1992; Burns, 1995). The crystallographic structure of the α/β-heterodimer has been solved recently (Nogales et al., 1998); however, many fundamental aspects of microtubule function and dynamics are still not fully understood.
Genetically amenable organisms such as yeasts and fungi have proven to be ideal systems with which to investigate the cellular function of microtubules (Oakley and Morris, 1981; Neff et al., 1983; Hiraoka et al., 1984; Toda et al., 1984; Schatz et al., 1986a). Saccharomyces cerevisiae (budding yeast) and Schizosaccharomyces pombe (fission yeast), although very divergent in evolution, have analogous genomic organization of tubulin-encoding genes (i.e., two α-tubulin genes and one β-tubulin gene) and show similar genetic properties in those genes (Neff et al., 1983; Hiraoka et al., 1984; Toda et al., 1984; Adachi et al., 1986; Schatz et al., 1986a,b). In both yeasts, a single β-tubulin–encoding gene is essential for cell viability (nda3+ in fission yeast and TUB2 in budding yeast) (Neff et al., 1983; Hiraoka et al., 1984). In contrast, two homologous α-tubulin–encoding genes (nda2+ and atb2+ in fission yeast, and TUB1 and TUB3 in budding yeast) show different genetic behavior. One of the two homologues is essential for cell viability (nda2+ and TUB1), whereas the other (atb2+ and TUB3) is not; however, the nonessential α-tubulin can compensate for loss of the essential α-tubulin gene when overexpressed, indicating that these two pairs of α-tubulin molecules are functionally interchangeable (Adachi et al., 1986; Schatz et al., 1986b).
Determining the molecular basis for the temporal and spatial definition of cell polarity is a fundamental problem in cell biology. The shape of every eukaryotic cell is believed to be maintained by the cytoskeleton, which consists of three filamentous systems: actin-based microfilaments, tubulin-based microtubules, and intermediate filaments. The cytoskeleton does not act as a static structural scaffold of the cell. Instead, in virtually every system characterized to date, the cytoskeleton is highly dynamic, frequently changing its structure during cell cycle progression and developmental differentiation. This dynamic behavior is intimately related to growth control (Drubin and Nelson, 1996; Gönczy and Hyman, 1996).
Fission yeast cells are rod shaped and have defined growth polarity during both the vegetative cycle and developmental states (Mitchison and Nurse, 1985; Snell and Nurse, 1993). Previous work from our laboratory and other laboratories has clearly shown the importance of microtubule integrity in determining growth polarity. Mutations in the tubulin genes (Toda et al., 1983; Umesono et al., 1983b) or their regulators (Mata and Nurse, 1997; Hirata et al., 1998) result in altered cell shapes such as bent or branched cells. The cortical actin-dependent pathway is also important for the maintenance of growth polarity as well as cytokinesis (Gould and Simanis, 1997). As in other organisms, the localization of these cytoskeletal molecules changes dramatically during cell cycle progression (Marks and Hyams, 1985; Marks et al., 1986; Tanaka and Kanbe, 1986; Hagan and Hyams, 1988), and these molecules play key roles in specific cell cycle events, as demonstrated by the fact that genes encoding cytoskeletal components and regulators were first identified as cell division cycle (cdc) mutants (Nurse et al., 1976; Gould and Simanis, 1997).
To understand in more detail the molecular pathways that regulate cell polarity, we have undertaken a large-scale screen of temperature-sensitive (ts) mutants to identify those with altered growth polarity (alp) (Hirata et al., 1998). We have isolated a class of alp mutants that become bent or branched and lose microtubules after incubation at the restrictive temperature. Genetic as well as molecular analyses indicate that two of these alp loci are allelic to atb2 and nda3, encoding α2-tubulin and β-tubulin, respectively. The identification of mutations in the atb2 gene as ts polarity mutants is intriguing because previous studies have shown that cells in which atb2+ is deleted are viable, with few defects (Adachi et al., 1986), which suggests that the alp2 mutants we have isolated here must be dominant in nature. The identification of nda3 as a ts mutant is also interesting because despite extensive and systematic mutational analysis of the β-tubulin gene in the past, very few ts mutants have been isolated (Yamamoto, 1980; Oakley and Morris, 1981; Umesono et al., 1983a,b; Thomas et al., 1985; Huffaker et al., 1988; Matsuzaki et al., 1988; Stearns and Botstein, 1988; Reijo et al., 1994; Sage et al., 1995). We have determined, therefore, the mutation sites of the tubulin genes in these ts mutants. There is a single point mutation in each mutant, affecting codons that correspond to amino acids that are highly conserved through evolution. Furthermore, in contrast to previously isolated tubulin mutants, detailed phenotypic analyses suggest that the spindle assembly checkpoint control might not be operational in these ts tubulin mutants.
MATERIALS AND METHODS
Strains, Media, and Chemicals
All mutant strains were derived from HM123 (h−leu1) (Table 1). JY6 (h+leu1his2), and TP108-3D (h+leu1 ura4his2) were used for backcrossing mutants. Complete medium, YPD (1% yeast extract, 2% polypeptone, and 2% dextrose), which contains 10 μg/ml Phloxine B (Sigma, St. Louis, MO) (called YPDP), YES (0.5% yeast extract, 3% dextrose, and 75 μg/ml adenine, histidine, leucine, and uracil), modified synthetic EMM2, and MES (3% malt extract, and 75 μg/ml adenine, histidine, leucine, and uracil) have been described previously (Moreno et al., 1991). Plates contained 1.6% agar.
Table 1.
Strain list
| Strains | Genotypes | Derivations |
|---|---|---|
| HM123 | h−leu1 | Our stock |
| JY6 | h+leu1his2 | Our stock |
| TP108-3D | h+leu1ura4his2 | Our stock |
| TP71-7C | h−arg3ura2 | Our stock |
| Δatb2 | h−leu1atb2::LEU2 | Adachi et al. (1986) |
| KM52-110 | h−leu1nda2-KM52 | Toda et al. (1984) |
| DH1-7C | h−leu1atb2-996 | This study |
| DH1-2D | h−leu1ura4atb2-996 | This study |
| DH1-2B | h+leu1ura4his2atb2-996 | This study |
| 1212 | h−leu1atb2-1212 | This study |
| DH2-8D | h−leu1alp1-1315 | Hirata et al. (1998) |
| DH12 | h−leu1nda3-1828 | This study |
| PR7 | h−leu1ura4nda3-1828 | This study |
| PR11 | h−leu1nda3-1828atb2::LEU2 | This study |
| TPR19A | mei1-B102leu1 arg1atb2-996 | This study |
Genetic Techniques and Nomenclatures
Standard procedures for S. pombe genetics were followed as described (Moreno et al., 1991). Cell number was measured using Sysmex F-800 (TOA Medical Electronics, Tokyo, Japan). S. pombe cells were transformed using the lithium method (Ito et al., 1983). A temperature-sensitive phenotype is abbreviated to the lowercase letters ts, e.g., ts atb2. Proteins are designated by an uppercase first letter, e.g., Atb2. Gene disruptions are abbreviated as the gene preceded by Δ such as Δatb2.
Isolation of ts Mutants with Polarity Defects
Wild-type HM123 cells were mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine as described previously (Uemura and Yanagida, 1984). Approximately 300 viable cells were spread per one YPDP plate. Plates were incubated at 29°C for 4 d, and colonies were then replica plated on YPDP and incubated at 36°C for 1 d. Strains that did not form colonies or that formed sick dark-red colonies were picked, and the morphology of these cells was examined by Calcofluor staining (Streiblová and Wolf, 1972). Approximately 1 of 50 colonies showed either no growth or retarded growth at the restrictive temperature. In total, 200,960 colonies were screened in this way, and 2822 ts mutants were isolated. Mutants that showed altered cell shape (bent or branched) at 36°C were selected for further analysis. These mutants were grown in liquid YPD and shifted to 36°C for 8 h, and the cell morphology was examined once again by Calcofluor staining. In total, 22 ts mutants that showed bent, curved, or branched morphologies were isolated and designated alp (Table 2).
Table 2.
Complementation groups and defective phenotypes of the alp mutants
| Loci | Alleles | Morphologya | Microtubules | Genes/product |
|---|---|---|---|---|
| alp 1 | 1315, 1356, 1584 | Bent/branched | No | Cofactor Db |
| 2 | 996, 1212, 1377 | Bent/branched | No | atb2+/α2-tubulin |
| 3 | 149, 601 | Curved/cut | ||
| 4 | 225, 1891 | Bent/cut | Few | |
| 5 | 1134 | Bent | ||
| 6 | 719 | Bent/cut | ||
| 7 | 738 | Bent/cut | ||
| 8 | 1146c | Bent/branched | tea1+ | |
| 9 | 1437 | Bent | ||
| 10 | 103 | Curved | ||
| 11 | 924 | Bent/branched | No | Cofactor Bd |
| 12 | 1828 | Bent/branched | No | nda3+/β-tubulin |
| 13 | 1837 | Bent/cut | ||
| 14 | 1270 | Bent/branched | Few | |
| 15 | 1218 | Bent/cut |
Mutants that showed >1% of cells with branched morphology or 10% of “cut” at 36°C for 8 h are indicated as “branched” or “cut” (Hirano et al., 1986), respectively.
The original isolate (1146) contained two mutations; one in tea1 (Mata and Nurse, 1997), which leads to branched morphology at 36°C, and the other in an unknown locus, which causes temperature-sensitive growth.
Radcliffe and Toda, unpublished observations.
Complementation Analysis
h+ mutant strains were isolated after mating of the mutant strains to JY6 (h+leu1his2). Crisscross mating was performed, and free spores were plated on two YPDP plates, one of which was incubated at 29°C, the other at 36°C. If the difference in the number of colonies formed between these two plates was >104-fold, the two mutants were assigned as allelic. Strain 1146 (alp8) (Table 2) contained two mutations: one is responsible for the morphological defects, which is allelic to tea1 (Mata and Nurse, 1997), and the other is responsible for the ts growth phenotype.
Dominance–Recessive Test
A stable prototrophic diploid heterozygous for the atb2 locus was constructed between the ts mei1-B102atb2-996 strain (TPR19A) (Table 1) and a mater strain (TP71-7C), and temperature sensitivity was examined.
DNA Preparation and Manipulation
Standard molecular biology techniques were followed as described (Sambrook et al., 1989). Enzymes were used as recommended by the suppliers (New England Biolabs, Beverly, MA, and Boehringer Mannheim, Indianapolis, IN). Nucleotide sequencing was performed by the dideoxy method (Sanger et al., 1977).
Identification of the alp2 and alp12 Loci as atb2 and nda3, Respectively
S. pombe genomic libraries constructed in the vectors pDB248 (Beach and Nurse, 1981; Hirano et al., 1988) and pUR19 (Barbet et al., 1992) were used for the isolation of plasmids that complemented the ts alp2 or alp12 mutant (DH1-7C: h−leu1alp2 [atb2]-996; DH1-2D: h−leu1 ura4alp2 [atb2]-996; DH1-2B: h+leu1ura4his2alp2 [atb2]-996; and PR7: h−leu1ura4alp12 [nda3]-1828) (Table 1). For the atb2 mutant, 4 of 36,000 colonies transformed with the pDB248-based library were capable of growing at 36°C, whereas for alp12, 5 of 20,000 colonies transformed with the pUR19-based library grew at 36°C. Segregation analysis indicated that the Ts+ phenotype was plasmid dependent. Plasmid DNAs were recovered from these transformants. Four different plasmids (pALA200, pALB200, pALC200, and pALD200) and three different plasmids (pCR8 [isolated three times], pCR9, and pCR10) were recovered from the alp2 and alp12 transformants, respectively. Restriction mapping indicated that three (pALA200, pALC200, and pALD200) contained overlapping inserts and carried the atb2+ gene, whereas pALB200 contained the nda2+ gene. pCR8, 9, and 10 all contained overlapping inserts and carried the nda3+ gene.
Identification of the alp2 loci as atb2 has been described previously (Yaffe et al., 1996). Furthermore, tetrad analysis between ts atb2-996 and an atb2 disruptant (Δatb2) (Table 1) showed a tight linkage; 23 tetrads were dissected, and all showed parental ditypes. Allelism between alp12 and nda3 was determined as follows. pCR10 was transformed into the ts alp12 (nda3)-1828 mutant. At 36°C, a number of Ts+Ura+ colonies appeared that stained less intensely on YPDP (at an approximate frequency of 10−2). Segregation analysis indicated that the Ura+ phenotype was stable, showing that pCR10 had integrated stably in the genome via homologous recombination. Free spore analysis between these integrants and a mater strain (TP108-3D) (Table 1) showed that no ts segregant appeared from 103 colonies, indicating that alp12 is allelic to nda3.
Cloning of the ts atb2 and nda3 Genes and Determination of the Mutation Sites
To determine the mutation sites in the ts atb2 mutants, 0.6-kb (corresponding to the first 166 residues of Atb2) and 1.3-kb (residues 167–449) HindIII fragments that comprise the entire atb2+ gene (Toda et al., 1984) were cloned into an integration vector and used to transform ts atb2-996 and -1212 strains (Yaffe et al., 1996). Both of the ts mutants were suppressed by the plasmid containing the 1.3-kb fragment but not by that containing the 0.6-kb fragment, indicating that the mutation sites of atb2-996 and -1212 are located in the 1.3-kb HindIII fragment. A DNA fragment corresponding to amino acids 167–449 was amplified from ts atb2-996 and -1212 strains by PCR using the following oligonucleotides as primers: Alp2-N1, AAAAAAGCTTCAATTTTCTATGTATCC; and Alp2-C1, AAAAGGATCCTTAGTACTCTTCTTCCA (underlined are HindIII and BamHI sites, respectively). The 858-bp amplified fragment was subcloned into pUC19 (Vieira and Messing, 1982). In each case, two independent PCRs were performed, amplified fragments were cloned separately, and the nucleotide sequences were determined. The same sequence was obtained from the independent clones and contained a single point mutation (see RESULTS), indicating that the base changes were not due to errors during PCR.
To determine the mutation site of the ts nda3 mutant, the entire ORF was cloned into pUC19 from the ts nda3-1828 mutant by PCR using the following two oligonucleotides: Nda3-N3P, TATGCTGCAGCTAACGAAACTCACCTAC; and Nda3-C1B, TATGGATCCAACGTAGATAAACACT (underlined are PstI and BamHI sites, respectively). Sequencing of the two clones derived from independent PCRs showed nine putative deviations (leading to amino acid substitutions at six places) in the ts nda3-1828 strain from the published nucleotide sequence of the wild-type nda3+ gene (Hiraoka et al., 1984). To determine which site is mutated in ts nda3-1828, the wild-type nda3+ gene was cloned in a similar manner, and its nucleotide sequence was determined. Comparison of the nucleotide sequences of ts nda3-1828 and wild-type nda3+ revealed that the published data contain sequencing errors in eight positions. In ts nda3-1828, nucleotide 1566 (A of initiator methionine is denoted as +1) is mutated from T to C, which results in substitution of Tyr by His at residue 422 (see RESULTS). These sequence data are available from EMBL/GenBank/DDBJ under accession numbers AF042827 (nda3+) and AF042828 (nda3–1828).
Immunochemical Assays
For indirect immunofluorescence microscopy, the methanol fixation method was used (Alfa et al., 1993). TAT-1 antibody (provided by Dr. Keith Gull, University of Manchester, Manchester, United Kingdom) and Cy3-conjugated sheep anti-mouse immunoglobulin G (Sigma) were used to visualize microtubules, and DAPI was used for chromosomal DNA.
Cell extracts were prepared as described (Matsusaka et al., 1995), except that HB buffer (Moreno et al., 1991) was used in the disruption of cells. Standard procedures for immunoblotting were followed (Harlow and Lane, 1988). Monoclonal anti-β-tubulin antibody (Sigma), anti-α-tubulin (TAT-1, gift from Dr. Keith Gull), and anti-Cdc2 (Y100, gift from Dr. Hiroyuki Yamano, ICRF) were used as primary antibodies. Horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Bio-Rad, Richmond, CA) and a chemiluminescence system (ECL, Amersham, Arlington Heights, IL) were used to detect bound antibody.
RESULTS
Isolation of Mutants That Are Defective in Growth Polarity
We performed a large-scale visual screen for mutants with defects in growth polarity. We first isolated ts mutants and then examined the cell morphology of these mutants after incubation at the restrictive temperature using Calcofluor, which stains septa and growing ends of the cell (Streiblová and Wolf, 1972; Mitchison and Nurse, 1985). Mutants with bent, curved, or branched morphology were selected. In total, 22 different mutants were obtained, and complementation tests indicated that these represent 15 loci, designated alp1–15 (Table 2). Some examples of the cell morphology and DAPI staining of alp2 and 12 mutants are shown in Figure 1.
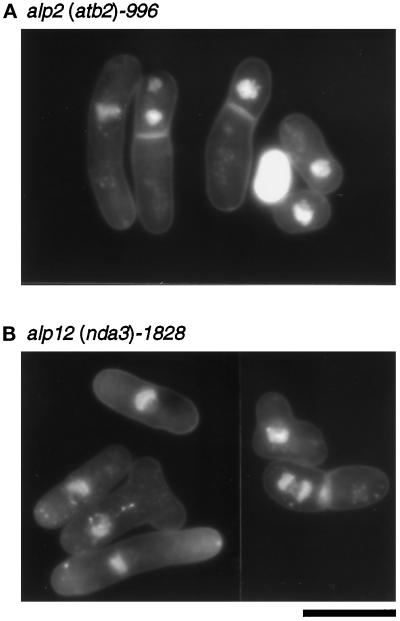
Figure 1.
Nuclear staining of ts alp2 (atb2) and alp12 (nda3) cells. alp2 (atb2)-996 (A, DH1–7C; Table 1) or alp12 (nda3)-1828 cells (B, DH12) were grown exponentially at 26°C, shifted to 35.5°C, and incubated for 6 h. Cells were fixed and stained with DAPI. Bar, 10 μm.
Classification of Polarity Mutants with Microtubule Staining
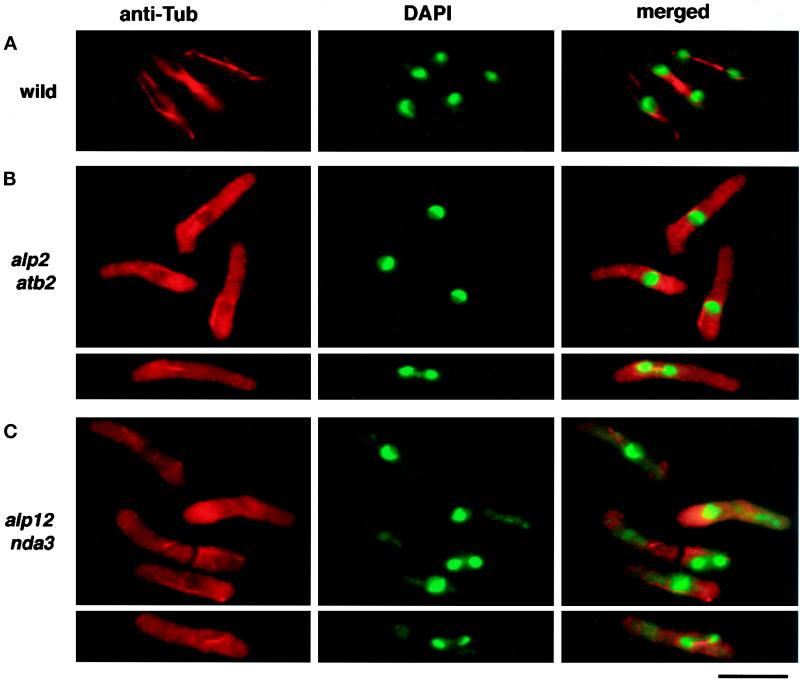
It has become clear that microtubules are important for the determination of cell shape and for growth polarity of fission yeast (Toda et al., 1983; Yaffe et al., 1996; Mata and Nurse, 1997; Hirata et al., 1998). We therefore used indirect immunofluorescence microscopy by using anti-tubulin antibody (TAT-1, kindly provided by Dr. Keith Gull) to examine the microtubule cytoskeleton of the alp mutants. It was clear from these analyses that at least four (alp1, 2, 11, and 12) of the genes identified in our screen were required for the maintenance of microtubule structures, because the microtubules in these mutants became fragile and disappeared when the temperature shifted. In this study, we have characterized alp2 and alp12 at the molecular level. Three alleles of alp2 (-996, -1212, -1377) (Table 2) and one allele of alp12 (-1828) were identified. Microtubule staining of alp2 and alp12 mutants is shown in Figure 2. Cytoplasmic microtubules became unstable and disappeared after 6 h incubation at 35.5°C, although some short nuclear spindle microtubules still remained (Figure 2, B and C, bottom panels).
Figure 2.
Microtubule staining in ts alp2 (atb2) and alp12 (nda3) cells. Mutant and wild-type cells were prepared as in Figure 1, fixed in methanol, and stained with anti-tubulin antibody (TAT-1, left panel) or DAPI (middle panel). Merged figures are shown in the right panel. Representative figures are shown for wild-type (top row), ts alp2 (atb2) (second and third rows), and alp12 (nda3) (fourth and fifth rows).
In addition to the loss of microtubule structures, alp2 and alp12 mutants showed various defective phenotypes in cell cycle and cell shape control. These include displacement of the nucleus (24% for alp2-996 and 17% for alp12-1828 after 8 h at 35.5°C) (Figure 1) and accumulation of septated cells. Also, asymmetrical rather than medial septation was often observed in many cells (∼50% of septated cells), resulting in anucleate cells following cytokinesis (8% for alp2-996 and 2% for alp12-1828).
Identification of the alp2 and alp12 Mutations as Novel Alleles in Mutant Tubulin Genes
We cloned the alp2+ and alp12+ genes by complementation using a fission yeast genomic library constructed with a multicopy vector. Four plasmids were isolated that suppressed ts alp2-996, and three suppressed alp12-1828. Restriction mapping of these plasmids indicated that the four plasmids suppressing the alp2 mutation consist of two previously identified genes, nda2+ and atb2+, which encode the two α-tubulins (α1- and α2-tubulin, respectively) (Toda et al., 1984; Adachi et al., 1986), and all three alp12-complementing plasmids contained the nda3+ gene, which encodes β-tubulin (Hiraoka et al., 1984). Genetic analysis indicated that alp2 was allelic to atb2, and alp12 was allelic to nda3 (see MATERIALS AND METHODS). Therefore, we shall hereafter use atb2 for alp2 and nda3 for alp12 preceded by ts, such as ts atb2.
Mutations in tubulin genes, or genes involved in microtubular pathways, often result in supersensitivity to antimicrotubule drugs such as members of the benzimidazole family of compounds (Umesono et al., 1983b; Adachi et al., 1986; Hirata et al., 1998). Consistent with this, ts atb2 and nda3 strains were supersensitive to the benzimidazole compound thiabendazole. They could not form colonies on rich media plates containing 10 μg/ml at 20°C, whereas wild-type cells could (Table 3).
Table 3.
Sensitivity of the atb2 and nda3 mutants to thiabendazole (TBZ)
| Strainsa | TBZ
(μg/ml)b
|
|
|---|---|---|
| 28°C | 20°C | |
| ts atb2 | + | − |
| ts nda3 | ++ | − |
| alp1 | + | − |
| Δatb2 | + | + |
| cs nda2 | − | NAc |
| Wild type | ++ | ++ |
Strains used are as follows: ts atb2 (DH1-7C; see Table 1); ts nda3 (DH12); alp1 (DH2-8D; Hirata et al., 1998), and Δatb2 (atb2::LEU2; Adachi et al., 1986); cs nda2 (h−leu1nda2-KM52; Toda et al., 1984); and wild type (HM123).
Mutant cells were streaked on rich YPD plates containing 10 μg/ml of TBZ and incubated at 28° or 20°C for 7 d. ++, Colony size was indistinguishable from that on plates without TBZ: +, tiny colonies were formed. −, no colonies were formed.
NA, Not applicable.
Genetic Interaction of ts atb2 and nda3 Mutations with Other Tubulin Genes
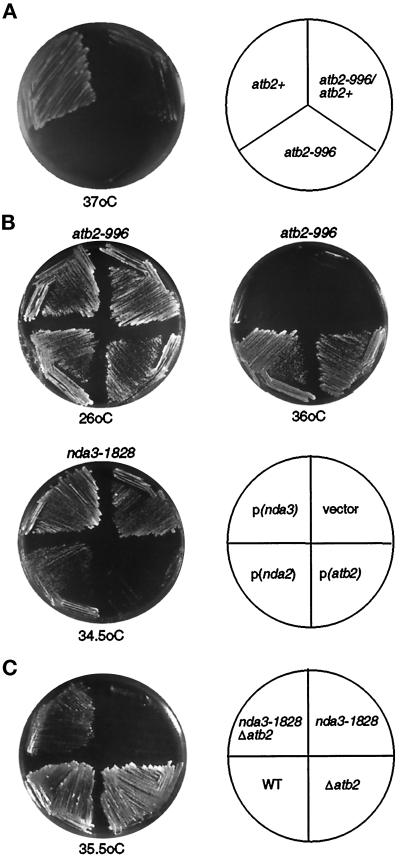
It was curious that the nonessential atb2 gene was identified in this study as a ts mutation. The mutant Atb2 proteins (α2-tubulin) must have a dominantly harmful effect on the maintenance of microtubules because deletion of atb2+ does not show a lethal or ts phenotype (Adachi et al., 1986). In line with this notion, genetic analysis demonstrated that the ts atb2 mutant is dominant because a heterozygous diploid between atb2-996 and atb2+ showed the ts phenotype, although some weak growth was observed compared with a haploid ts alp2-996 strain (Figure 3A). If the mutant Atb2 protein were to inhibit the function of β-tubulin by abortive binding, effectively taking the β-tubulin out of the pool of available subunits, it might be expected that an increased dosage of β-tubulin gene would suppress ts atb2 mutations; however, a multicopy plasmid containing nda3+ was incapable of suppressing ts atb2 (Figure 3B, top right plate). This result suggests that the phenotypic dominance of ts atb2 (α2-tubulin) over nda2+ (α1-tubulin) is not ascribable to a loss of β-tubulin function.
Figure 3.
Genetic interaction between ts tubulin mutants and other tubulins. (A) Dominance–recessive test of ts atb2 mutation. Haploid wild-type (atb2+, TP71–7C; Table 1), ts atb2 (atb2-996, TPR19A), and a heterozygous diploid constructed by crossing these two strains (atb2-996/atb2+) were streaked on rich YPD plates and incubated at 37°C for 2 d. (B) atb2-996 mutant cells (top two plates) or nda3-1828 (bottom left plate) were transformed with an empty vector (vector), a multicopy plasmid containing atb2+ [pALA200, p(atb2)], nda2+ [pALB200, p(nda2)], or nda3+ [pCR9, p(nda3)]. Transformants were incubated at either 26°C (B, top left), 36°C (B, top right) or 34.5°C (bottom left) for 3 d. (C) Four strains (top left, nda3-1828Δatb2; top right, nda3-1828; bottom right, Δatb2; bottom left, wild type) were streaked on YPD plates and incubated at 35.5°C for 2 d.
Crossing ts atb2-996 and cs nda2-KM52 (Umesono et al., 1983b) indicates that the double mutants are synthetically lethal. Among 12 tetrads dissected, no double mutants (14 spores predicted) were obtained, whereas 14 wild-type and 10 of each single mutant strain formed colonies at 29°C, a temperature permissive for both mutants. Thus for both ts atb2 and cs nda2 mutants, functional wild-type proteins of the other α-tubulin homologue are required for viability at the permissive temperature.
In the case of ts nda3, a different effect from increased dosage of α-tubulin-encoding genes was observed. Overexpression of atb2+ led to enhanced temperature sensitivity in the ts nda3 mutant (Figure 3B, bottom left plate). Incubation of ts nda3 on minimal plates at 34.5°C allowed colony formation in the ts nda3 mutant containing an empty vector, whereas mutant cells containing a multicopy plasmid carrying atb2+ were incapable of forming colonies at this temperature. This suggested that the ts nda3 mutant is highly sensitive to altered gene dosage of atb2+. To examine the effect of atb2+ gene dosage in more detail, the atb2+ gene was disrupted in the ts nda3 strain. In reverse parallel with the toxic effect of overexpression, the deletion of atb2+ partially rescued the temperature sensitivity of nda3-1828 (Figure 3C). This result indicates that the defective phenotypes observed in the ts nda3 mutant can be ascribed, at least in part, to the existence of the Atb2 protein. It is noteworthy that the toxic effect is α2-tubulin specific, because ts nda3 cells carrying the nda2+ gene encoding α1-tubulin on an equivalent plasmid were capable of forming colonies, although of a slightly smaller size compared with those containing vector alone (Figure 3B, bottom left plate). Multicopy plasmids containing γ-tubulin (Horio et al., 1991) do not suppress either ts atb2 or nda3, nor do they enhance the lethality conferred by these mutations (Hirata and Toda, unpublished observations), suggesting that the interactions we report are specific.
Determination of Mutation Sites in the ts atb2 and nda3 Mutants
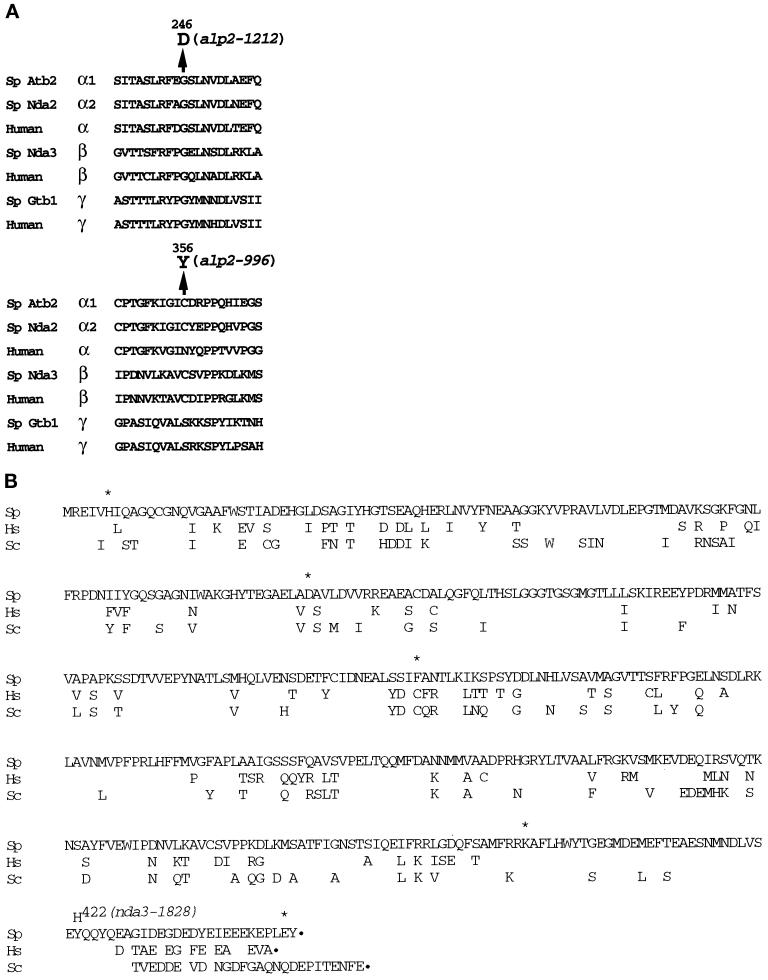
We determined the mutation sites of the ts atb2 and nda3 mutants (atb2-996 and -1212 and nda3-1828; see MATERIALS AND METHODS). In each case, a single point mutation was found, which resulted in alteration of an amino acid residue in Atb2 and Nda3. In ts atb2-996, nucleotide 1065, guanine (A of the initiator methionine is denoted as +1), was mutated to adenine, which resulted in the substitution of cysteine 356 (TGT) with tyrosine (TAT; mutated nucleotides are underlined). In the case of ts atb2-1212, nucleotide 737, guanine, was mutated to adenine, which resulted in the substitution of glycine 246 (GGC) with aspartate (GAC). A comparison of amino acid sequences around the mutated residues is shown in Figure 4A. Both of the mutated residues have been highly conserved throughout evolution. In particular, glycine 246 is of interest because this residue is invariant not only in fission yeast α1-tubulin (Nda2) and α-tubulins from other species, but also in β- and γ-tubulins in virtually every species (Figure 4A).
Figure 4.
Determination of the mutation sites in the ts atb2 (alp2) and nda3 (alp12) mutants. Amino acid comparisons of α-tubulin (A) and β-tubulin (B) together with the amino acid substitution caused by a point mutation in each ts mutant are shown. Sp stands for fission yeast (S. pombe), Sc for budding yeast (S. cerevisiae), and Hs for human (H. sapiens). atb2-996 contains a single base change at nucleotide number 1065 (G to A, A of the initiator methionine is denoted as +1), resulting in an amino acid substitution of cysteine 356 with tyrosine. atb2-1212 contains a single base change at nucleotide 737 (G to A), which results in an amino acid substitution of glycine 246 with aspartate. nda3-1828 contains a single base change at position 1566 (from T to C), resulting in substitution of tyrosine 422 with histidine. Only nonconserved amino acid residues are shown in B for Hs and Sc. Asterisks show amino acids where the previously published data (Hiraoka et al., 1984) are incorrect because of sequencing errors.
The mutation site of ts nda3-1828 is located near the C terminus of the Nda3 protein. Nucleotide 1566 was mutated from thymine to cytosine, which results in the substitution of tyrosine 422 (TAT) with histidine (CAT) (Figure 4B). Like glycine 246, this tyrosine is also conserved in all members of tubulins. It is noteworthy that the region around tyrosine 422 is rich in acidic amino acid residues, which are believed to be important for interactions with other proteins. Extensive mutational analysis of the budding yeast β-tubulin gene TUB2 has shown the C terminus of β-tubulin to be essential for the normal function of microtubules (Reijo et al., 1994).
ts atb2 Alters the Cellular Ratio of α1- to α2-Tubulin
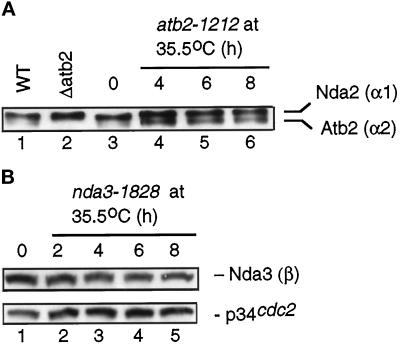
Why do the atb2 mutations that we isolated show a dominant ts growth defect? As shown above, it is not due to the absorption of β-tubulin. One possible explanation might be an altered cellular ratio of α1- and α2-tubulins. To examine this possibility, immunoblotting using anti-α-tubulin antibody was performed with protein extracts from a ts atb2 mutant. As described previously (Adachi et al., 1986), the two closely migrating bands of 55 and 57 kDa in wild-type extracts correspond to Atb2 and Nda2, respectively (Figure 5A, lane 1) (note that p57nda2 is more abundant than p55atb2). In an atb2-deleted strain, the p55atb2 band is missing (Δatb2; Figure 5A, lane 2). In atb2-1212 cells grown at the permissive temperature (Figure 5A, lane 3), the ratio of Atb2 to Nda2 is similar to that seen in wild-type extracts, because the level of p57nda2 is much higher than that of p55atb2. After shifting to the restrictive temperature of 35.5°C, the relative level of p55atb2 increased (Figure 5A, lanes 4–6). Although the increase was modest, it was reproducible. Thus it is possible that the dominance of atb2-1212 is ascribable to the higher relative abundance of mutant Atb2 protein over Nda2. Suppression by high dosage of the nda2+ gene supports this notion, although how ts Atb2 protein interferes with microtubule function still remains to be determined (see DISCUSSION).
Figure 5.
The level of tubulin proteins in ts atb2 and nda3 mutants. (A) Total cell extracts were prepared from wild type (HM123, lane 1; Table 1), deleted atb2 (Δatb2, lane 2), or ts atb2-1212 (1212, lanes 3–6). Cells were cultured at 26°C in lanes 1 and 2. atb2-1212 cells were first grown at 26°C (lane 3) and shifted to 35.5°C. Samples were collected at 4 (lane 4), 6 (lane 5), and 8 (lane 6) h. Protein (2 μg) from cell extracts was run in SDS-PAGE, and immunoblotting was performed with anti-α-tubulin antibody (TAT-1). (B) ts nda3-1828 cells (DH12) were grown as described in A, and samples were taken every 2 h after the shift. Cell extracts (20 μg) were run in SDS-PAGE, and immunoblotting was performed with mouse anti-β-tubulin antibody (Sigma) (top) or anti-Cdc2 antibody as a loading control (bottom).
It should be noted that despite the altered ratio of α1- to α2-tubulin, the total amount of α-tubulin proteins remained almost constant, suggesting that regulatory mechanisms maintain a steady-state level of α-tubulin protein, as has been reported in other organisms (May et al., 1990; Gonzalez-Garay and Cabral, 1996). This supports previous data that were suggestive of this form of regulation (Adachi et al., 1986). In contrast to the increase of Atb2 protein in the ts atb2 mutant, the level of β-tubulin remained unchanged in the ts nda3 mutant (Figure 5B). Thus mutation in the C-terminal amino acid (Y422H, Figure 4B) did not affect the stability of the β-tubulin molecule.
The Spindle Assembly Checkpoint Is Not Operational in ts Tubulin Mutants
The temporal order within the cell cycle is maintained by surveillance mechanisms called checkpoints (Hartwell and Weinert, 1989). The spindle assembly checkpoint prevents cells from proceeding into mitosis when the bipolar spindle function is compromised (Murray, 1995; Wells, 1996). In fission yeast, it has been shown that a cs nda3 mutation (nda3-KM311) activates the spindle assembly checkpoint at the restrictive temperature so that cells arrest in midmitosis with condensed chromosomes and maintain high viability (Hiraoka et al., 1984; Moreno et al., 1989). Upon release from the mitotic block, nda3-KM311 cells reinitiate anaphase in a highly synchronized manner (Hiraoka et al., 1984). We therefore were interested in determining whether the spindle assembly checkpoint is also intact in the newly isolated ts nda3 mutant.
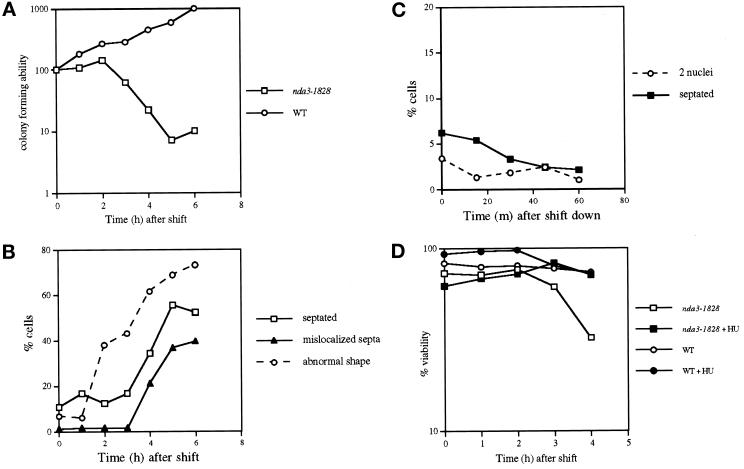
Contrary to our initial expectations, the spindle assembly checkpoint appeared not to be operational in the ts nda3 mutant cells. As shown in Figure 6, A and B, the septation index increased 3 h after temperature shift-up and was accompanied by a sharp drop in viability. More than 60% of the septated cells had mislocalized septa (Figure 6B). Also, nuclear displacement was evident, and in some cells partial chromosome segregation occurred (Figure 1B). Polarity defects such as bent or branched cell morphology were apparent at 2 h. Consistent with continued cell cycle progression in these mutants, interphase-like hemispherical chromosomes, rather than condensed chromosomes, were seen after 6 h at 35.5°C (Figure 1B). This contrasts with the cs allele of nda3 (-KM311), which results in accumulation of increasing numbers of cells with condensed chromosomes with prolonged incubation at the restrictive temperature (Umesono et al., 1983a; Hiraoka et al., 1984).
Figure 6.
ts nda3 mutants proceed through the cell cycle and lose viability at the restrictive temperature. (A and B) ts nda3 mutant (DH12; Table 1) and wild-type (HM123) cells were grown at 26°C and shifted to 35.5°C, and aliquots were collected hourly. After appropriate dilution (10−4), cells were plated on rich YES plates to examine colony-forming ability per culture (A, the value at time 0 was taken as 100%; ○, wild type; □, ts nda3). Vertical axis (%) is shown logarithmically. Septation index and percentage of cells with abnormal cell morphology in the ts nda3 mutant (B, □ and ○, respectively) were observed with Calcofluor staining, and the percentage of cells that showed septa displaced from the center of the cell is also shown (▴). In wild-type cells, these abnormal cells were never observed (<0.1%). (C) For the reversibility experiments, ts nda3 cells incubated at 35.5°C for 2 h (note that viability is still high at this time point; see A) were shifted down to 26°C, and septation index (using Calcofluor, ▪) and the percentage of cells with two nuclei (using DAPI, ○) were scored at 15-min intervals for 1 h. (D) The same strains as in A (wild type shown as circles and ts nda3 as squares) were grown in the presence (closed symbols) or absence (open symbols) of 10 mM hydroxyurea in rich YPD for 3 h at 27°C and shifted to 35.5°C. Cell number was measured at each time point, and viability was examined by plating cells (10−4 dilution) on rich YES plates. After plates were incubated at 27°C, the number of colonies was counted, and viability at each time point was calculated by dividing the number of viable cells by the cell number. Vertical axis (%) is shown logarithmically.
To examine the reversibility of the ts nda3 mutant incubated at the restrictive temperature, the mutant cells were incubated for various periods (2, 4, and 6 h) at 35.5°C before being returned to 26°C, and the percentage of mitotic or septated cells was measured. No synchronous anaphase or septation was observed (Figure 6C). These results show that contrary to the cell cycle arrest phenotype of cs nda3 mutants, in the ts nda3 mutant the spindle assembly checkpoint is not functional, and mutant cells proceed into mitosis and subsequent events such as septation and cytokinesis at the restrictive temperature. A similar result was obtained with a ts atb2 mutant, namely mitosis, and septation occurred at the restrictive temperature, although viability remained high (Figure 1A) (Radcliffe and Toda, unpublished observations).
If the ts nda3 mutant becomes lethal because of cell cycle progression in the absence of activation of the spindle checkpoint, it would be expected that the lethality of the mutant would be rescued by blockage of the cell cycle before entry into mitosis. That is indeed the case. The ts nda3 mutant arrested in early S phase by hydroxyurea (HU) treatment did not lose viability for up to 4 h incubation at the restrictive temperature, whereas the control culture in the absence of the drug lost viability sharply (Figure 6D). Further incubation at the restrictive temperature in the presence of HU (>4 h) led to a loss of viability of the ts nda3 mutant, probably because of progression of the cell cycle by prolonged exposure to HU (21% after 6 h) (Sazer and Nurse, 1994). We therefore conclude that the loss of viability in the ts nda3 mutant is ascribable to irreversible progression of the cell cycle without normal function of spindles, in which the spindle assembly checkpoint is not activated.
DISCUSSION
Cell Polarity and Microtubules
Microtubules have been shown to be indispensable in the execution of diverse cellular processes in many systems, including motility, mitosis, protein and mRNA transport, and cell morphogenesis. In fission yeast, microtubules also execute several distinct functions, including chromosome segregation (Umesono et al., 1983a,b), distribution of organelles, in particular mitochondria and Golgi (Ayscough et al., 1993; Yaffe et al., 1996), and cell polarity and morphogenesis (Toda et al., 1983; Mata and Nurse, 1997; Hirata et al., 1998). Fission yeast tubulin mutants were originally isolated on the basis of cell cycle defective phenotypes in mitosis (nuclear division arrest [nda]) (Toda et al., 1983). In this study we have screened for ts mutants with altered polarity of cell growth (alp mutant) and have identified new mutant alleles in tubulin genes. This clearly demonstrates that, as in other eukaryotes, microtubules are crucial elements in the determination of cell polarity in fission yeast. How do microtubules regulate cell polarity? A recent study has identified a “marker” molecule that translocates into the cell tips via microtubules and determines the orientation of cell tip growth (Mata and Nurse, 1997). There may be several molecules that mark the growing tips in a microtubule-dependent manner, and other alp+ genes may encode such molecules.
Work from budding yeast, on the other hand, suggests that in this organism the main role of microtubules is in mitosis and meiosis, namely nuclear migration and chromosome segregation (Huffaker et al., 1988; Jacobs et al., 1988). This suggests that despite the similarity in genomic organization and genetic properties of tubulin genes in these two yeasts (Neff et al., 1983; Hiraoka et al., 1984; Toda et al., 1984; Adachi et al., 1986; Schatz et al., 1986a,b), their biological roles have diverged considerably during evolution. This may be reflected in the different distribution of cytoplasmic microtubules. Fission yeast has long cytoplasmic arrays that run along the cell axis, whereas budding yeast lacks this kind of network structure; instead, cytoplasmic microtubules emanate from the spindle pole body and form bundles (Adams and Pringle, 1984; Kilmartin and Adams, 1984; Hagan and Hyams, 1988).
Why Do the atb2 Mutants That Are Isolated Become Temperature Sensitive?
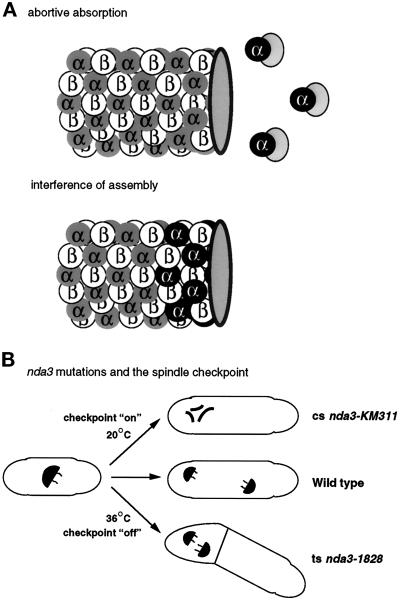
How does ts α2-tubulin (Atb2) protein act in a dominant manner over α1-tubulin (Nda2) at the restrictive temperature? There are several possibilities. The first is that ts Atb2 protein forms an abortive complex with some other protein(s), which usually execute a function that is essential for microtubule biogenesis and cell viability (Figure 7A). This scenario is not unique: a similar mutant has been reported and analyzed previously. Fission yeast contains two genes encoding type I protein phosphatases, dis2+ and sds21+ (Ohkura et al., 1989). Unlike nda2+ and atb2+, neither dis2+ nor sds21+ is essential by itself, but simultaneous disruption leads to lethality. The dis2-11 mutation, however, shows a dominant cs phenotype. In this case, the reason that dis2-11 shows cs appears to be due to the absorption, by cs Dis2, of a protein called Sds22, which is a regulatory subunit of both type I protein phosphatases and is essential for cell viability (Ohkura and Yanagida, 1991; Stone et al., 1993). Multicopy plasmids containing dis2+, sds21+, or sds22+ genes are capable of suppressing cs dis2-11. These interactions are strikingly similar to the interactions between the tubulin genes that we present here. The observation that in the ts atb2 mutant the relative ratio of Nda2 and ts Atb2 protein alters in a way that ts atb2 cells appear to contain more ts Atb2 protein is intriguing. It has been reported that free α-tubulin molecules become highly unstable (Tian et al., 1997). It is possible that in the ts atb2 mutant interaction between Nda2 and other essential proteins is compromised, which may lead to an apparent reduction of the relative ratio of Nda2 and ts Atb2 protein. The rescue of ts atb2 by high-dosage nda2+ gene supports this assumption.
Figure 7.
Possible models for novel ts mutations in tubulin genes. (A) ts Atb2 protein irreversibly absorbs proteins essential for microtubule biogenesis and assembly, which results in the disappearance of microtubules at the restrictive temperature (abortive absorption). ts Atb2 protein (α2-tubulin) is incorporated into microtubule structures, but interferes with the assembly of microtubules, which also results in microtubule disassembly (interference of assembly). ts Atb2 protein is shown as black circles with α in white letters, Nda2 (α1-tubulin) as gray circles with α in black letters. Hypothetical proteins that are essential for microtubule structures are shown in dotted circles bound to ts Atb2 proteins. (B) Differences in the defective phenotypes of cs nda3-KM311 and ts nda3-1828 mutants are shown. nda3 mutants arrest at mitosis with condensed chromosomes in which the spindle assembly checkpoint is activated (on), whereas ts nda3 mutants proceed through mitosis without activating the checkpoint (off), form septa, and eventually partial chromosome segregation occurs.
β-Tubulin is a strong candidate for a protein that is absorbed by the ts Atb2 protein. The data, however, show that this is probably not the case, because an increased dosage of the β-tubulin gene did not suppress the ts atb2 mutant. Other candidates for interacting proteins are the cofactors that are required for the correct folding of tubulin molecules (Tian et al., 1996). Cofactors B, D, and E are proposed to interact with α-tubulin to produce assembly-competent α/β-tubulin heterodimers (Tian et al., 1997). All of these molecules are essential for cell viability in fission yeast (cofactor D/Alp1 [Hirata et al., 1998; Radcliffe and Toda, unpublished observations]). Preliminary analysis from our laboratory, however, makes this possibility also unlikely because none of the fission yeast homologues of these cofactors suppress the ts atb2 mutant when introduced on multicopy plasmids (Radcliffe and Toda, unpublished observations). There may, of course, be other α-tubulin binding proteins, such as microtubule-associated proteins (MAPs), that await identification.
The Implications of the Mutation Sites in ts Atb2 Protein
The second possibility is that ts Atb2 protein dominantly interferes with the assembly of newly synthesized α/β-tubulin heterodimers (Figure 7A). The electron crystallographic structure of the α/β heterodimer was recently solved (Nogales et al., 1998). The tubulin molecule is divided into three functional domains, namely the N-terminal domain (1–205), which is responsible for GTP binding, the intermediate domain (206–381), which is required for heterodimer and/or intradimer formation, and the remaining C-terminal domain, which is thought to be important for interactions with various MAPs and motors. Assignment of the mutation sites of ts atb2 (-996 and -1212), which reside in the central domain, to the three-dimensional structure of α/β-heterodimers has proved illuminating. Cysteine 356 (mutated to tyrosine in atb2-996) is located at the longitudinal interface between the α- and β-tubulin monomers (Nogales et al., 1998). It is also the residue whose analogous position in β-tubulin is the binding site for the tubulin-depolymerizing drug colchicine (Bai et al., 1996). Interestingly, glycine 246 (mutated to aspartate in atb2-1212) is spatially adjacent to the β-sheet in which cysteine 356 is included, and furthermore, both of these amino acid residues are in close contact with the GTP/GDP exchangeable site of β-tubulin (Nogales et al., 1998). It is also noteworthy that the region adjacent to glycine 246 (242LRFEG246) shows homology to regions that possess ribose-binding activity within various ATPases (Burns and Farrell, 1996). All of these facts raise the interesting possibility that the ts atb2 mutation alters the physical interaction between α- and β-tubulin, or between α/β-heterodimers, and is accompanied by an alteration in the level of GTP/GDP exchange or hydrolysis of β-tubulin.
In the worm Caenorhabditis elegans, the mutation analogous to G246D was isolated in β-tubulin–encoding mec-7 (G244S, called u129) (Savage et al., 1994). Consistent with the fission yeast atb2-1212 mutant, the u129 mutant exhibits a dominant phenotype. Thus it appears that for either α- or β- tubulin, this conserved glycine has an essential role in microtubule biogenesis and/or assembly, and that mutation of this residue results in a dominant phenotype. Given the high degree of conservation in both sequence (invariant glycine) and function (dominant mutations) in higher eukaryotic systems in which mutants are not readily available, inducible expression of this mutant form of tubulin might be a useful approach for conditionally disrupting microtubules. This hypothesis is currently being tested.
ts Mutation in the β-Tubulin–encoding nda3+ Gene
The toxicity of the high-dosage atb2+ gene and suppression by its deletion in the ts nda3 mutant suggest an altered affinity of ts Nda3 toward the Atb2 protein. It is possible that heterodimers of Atb2/ts Nda3 might specifically interfere with microtubule assembly and/or maintenance in this mutant. However, because nda2+ is an essential gene, we are unable to similarly test whether its deletion rescues ts of nda3-1828. Further analysis will be required to establish any distinction between the two α-tubulin molecules in terms of their specific interaction with ts Nda3.
The mutation site in nda3-1828 resides in the C-terminal region of the molecule Y422H. In animal cells, the C-terminal region is responsible for interactions with MAPs (MAP-1 and MAP-2) (Rivas et al., 1988; Cross et al., 1991), and the crystallographic analysis shows that the C-terminal region resides on the outer surface of the microtubule (Nogales et al., 1998). Systematic mutational analysis of budding yeast β-tubulin also suggests that regions near the C terminus are essential for microtubular function (Reijo et al., 1994), and a truncation from glutamate 431 results in a ts growth defective phenotype (Matsuzaki et al., 1988). Recently an essential protein that shows properties similar to mammalian MAPs has been identified in budding yeast (Irminger-Finger et al., 1996). It is therefore possible that the C-terminal region is also required for an interaction with MAPs in fission yeast and that this interaction is perturbed in the ts nda3 mutant.
Despite extensive mutational analysis of β-tubulin genes, only a few ts mutants have been successfully isolated to date. In contrast, many cs mutations have been isolated (Oakley and Morris, 1981; Thomas et al., 1985; Huffaker et al., 1988; Matsuzaki et al., 1988; Stearns and Botstein, 1988; Davis et al., 1994; Reijo et al., 1994; Savage et al., 1994; Sage et al., 1995). In view of this, the nda3-1828 mutant is of interest and may be a useful tool with which to obtain further insight into the structure and function of microtubules.
Mitotic Spindle Assembly Checkpoint and ts Tubulin Mutants
A surprising observation arising from this study is that in both ts atb2 and nda3 mutants, it appears that the spindle assembly checkpoint is not operational (Figure 7B). In these mutants, cell cycle events such as septation, which usually occur after bipolar spindle function, continue to take place at the restrictive temperature despite defects in mitosis. In contrast, previously identified cs nda3-KM311 mutant cells arrested uniformly in midmitosis with condensed chromosomes and no septa (Umesono et al., 1983b; Hiraoka et al., 1984). This is the typical terminal phenotype when the spindle assembly checkpoint is functional. One possible explanation of this phenotypic difference is that in the ts nda3 mutant, microtubular function, especially spindle function, is insufficiently defective to activate the spindle assembly checkpoint. As shown in Figure 2, we sometimes observe short residual spindle microtubules in ts nda3 cells incubated at the restrictive temperature. A similar phenotype has been observed in ts atb2 and cs nda2 mutants (Umesono et al., 1983b; this study). Because fission yeast has two α-tubulin–encoding genes, a single mutation may be unable to completely abrogate the function of the other gene, and as a result partial microtubular function remains. It is therefore possible that the temperature lethality of nda3-1828 mutant cells arises from abortive cell cycle progression attributable to residual spindle function, preventing activation of the checkpoint machinery rather than cell cycle arrest caused by the loss of microtubules.
It is worth pointing out that the phenotype of the ts nda3 mutant, which loses viability at the restrictive temperature, is similar to that of spindle assembly checkpoint mutants such as cs nda3-KM311 mad2, nda3-KM311cdc16, and nda3-KM311dma1 double mutants in which components of the spindle assembly checkpoints are missing (Fankhauser et al., 1993; Murone and Simanis, 1996; He et al., 1997). The nda3-1828 mutant could prove useful in the genetic dissection of the transduction mechanisms that monitor spindle defects via the checkpoint machinery.
ACKNOWLEDGMENTS
We thank Drs. Keith Gull for the TAT-1 antibody, Hiroyuki Yamano for anti-Cdc2 antibody, Anthony Carr for the pUR19-based fission yeast genomic library, Yasuhisa Adachi for strains, Mikiko Fukui and Mark Eddison for help with characterization of the alp mutants, and Juan Mata for help with allelism tests between alp8 and tea1. We thank Drs. Mitsuhiro Yanagida for stimulative discussion and Paul Nurse and Iain Hagan for critical reading of this manuscript and useful suggestions. The initial part of this work is supported by a research grant from Kyowa Hakko.
REFERENCES
- Adachi Y, Toda T, Yanagida M. Differential expression of essential and nonessential α-tubulin genes in Schizosaccharomyces pombe. Mol Cell Biol. 1986;6:2168–2178. doi: 10.1128/mcb.6.6.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AEM, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. [Google Scholar]
- Ayscough K, Hajibagheri NM, Watson R, Warren G. Stacking of Golgi cisternae in Schizosaccharomyces pombe requires intact microtubules. J Cell Sci. 1993;106:1227–1237. doi: 10.1242/jcs.106.4.1227. [DOI] [PubMed] [Google Scholar]
- Bai R, Pei X-F, Boyé O, Getahun Z, Grover S, Bekisz J, Nguyen NY, Brossi A, Hamel E. Identification of cysteine 354 of β-tubulin as part of the binding site for the A ring of colchicine. J Biol Chem. 1996;271:12639–12645. doi: 10.1074/jbc.271.21.12639. [DOI] [PubMed] [Google Scholar]
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Beach D, Nurse P. High frequency transformation of the fission yeast Schizosaccharomyces pombe. Nature. 1981;290:140–142. doi: 10.1038/290140a0. [DOI] [PubMed] [Google Scholar]
- Burns RG. Identification of two new members of the tubulin family. Cell Motil Cytoskeleton. 1995;31:255–258. doi: 10.1002/cm.970310402. [DOI] [PubMed] [Google Scholar]
- Burns RG, Farrell KW. Getting to the heart of β-tubulin. Trends Cell Biol. 1996;6:297–303. doi: 10.1016/0962-8924(96)10024-6. [DOI] [PubMed] [Google Scholar]
- Cross D, Domenguez J, Maccioni RB, Avila J. MAP1 and MAP2 binding sites at the C terminus of β-tubulin. Studies with synthetic tubulin peptides. Biochemistry. 1991;30:5372–5376. doi: 10.1021/bi00231a036. [DOI] [PubMed] [Google Scholar]
- Davis A, Sage CR, Dougherty CA, Farrell KW. Microtubule dynamics modulated by guanosine triphosphate hydrolysis activity of β-tubulin. Science. 1994;264:839–842. doi: 10.1126/science.8171338. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Marks J, Reymond A, Simanis V. The S. pombe cdc16 gene is required for maintenance of p34cdc2 kinase activity and regulation of septum formation: a link between mitosis and cytokinesis. EMBO J. 1993;12:2697–2704. doi: 10.1002/j.1460-2075.1993.tb05931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, Hyman AA. Cortical domains and the mechanisms of asymmetric cell division. Trends Cell Biol. 1996;6:382–387. doi: 10.1016/0962-8924(96)10035-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Garay ML, Cabral F. alpha-Tubulin limits its own synthesis: evidence for a mechanism involving translational repression. J Cell Biol. 1996;135:1525–1534. doi: 10.1083/jcb.135.6.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Funahashi S, Uemura T, Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cut mutants that block nuclear division but not cytokinesis. EMBO J. 1986;5:2973–2979. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Hiraoka Y, Yanagida M. A temperature-sensitive mutation of the S. pombe gene nuc2+ that encodes a nuclear scaffold-like protein blocks spindle elongation in mitotic anaphase. J Cell Biol. 1988;108:243–253. doi: 10.1083/jcb.106.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes β-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J. 1998;17:658–666. doi: 10.1093/emboj/17.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. The fission yeast γ-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- Huffaker TC, Thomas JH, Botstein D. Diverse effects of β-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A, Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Irminger-Finger I, Hurt E, Roebuck A, Collart MA, Edelstein SJ. MHP1, an essential gene in Saccharomyces cerevisiae required for microtubule function. J Cell Biol. 1996;135:1323–1339. doi: 10.1083/jcb.135.5.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs CW, Adams AE, Szaniszlo PJ, Pringle JR. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R. Cell-cell signaling, microtubules, and the loss of symmetry in the Drosophila oocyte. Cell. 1995;83:353–356. doi: 10.1016/0092-8674(95)90111-6. [DOI] [PubMed] [Google Scholar]
- Marks J, Hagan IM, Hyams JS. Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J Cell Sci Suppl. 1986;5:229–241. doi: 10.1242/jcs.1986.supplement_5.15. [DOI] [PubMed] [Google Scholar]
- Marks M, Hyams JS. Localization of F-actin through the cell division cycle of Schizosaccharomyces pombe. Eur J Cell Biol. 1985;39:27–32. [Google Scholar]
- Mata F, Nurse P. tea1p and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Matsusaka T, Hirata D, Yanagida M, Toda T. A novel protein kinase gene ssp1+ is required for alteration of growth polarity and actin localization in fission yeast. EMBO J. 1995;14:3325–3338. doi: 10.1002/j.1460-2075.1995.tb07339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F, Matsumoto S, Yahara I. Truncation of the carboxy-terminal domain of yeast β-tubulin causes temperature-sensitive growth and hypersensitivity to antimitotic drugs. J Cell Biol. 1988;107:1427–1435. doi: 10.1083/jcb.107.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May GS, Waring RB, Morris NR. Increasing tubC beta-tubulin synthesis by placing it under the control of a benA beta-tubulin upstream sequence causes a reduction in benA beta-tubulin level but has no effect on microtubule function. Cell Motil Cytoskeleton. 1990;16:214–220. doi: 10.1002/cm.970160308. [DOI] [PubMed] [Google Scholar]
- Mitchison JM, Nurse P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1985;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner MW. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analyses of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:773–782. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murone M, Simanis V. The fission yeast dma1 gene is a component of the spindle assembly checkpoint, required to prevent septum formation and premature exit from mitosis if spindle function is compromised. EMBO J. 1996;15:6605–6616. [PMC free article] [PubMed] [Google Scholar]
- Murray AW. The genetics of cell cycle checkpoints. Curr Opin Genet. 1995;5:5–11. doi: 10.1016/s0959-437x(95)90046-2. [DOI] [PubMed] [Google Scholar]
- Neff N, Thomas JH, Grisafi P, Botstein D. Isolation of the β-tubulin gene from yeast and demonstration of its essential function in vivo. Cell. 1983;33:211–219. doi: 10.1016/0092-8674(83)90350-1. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Oakley BR. Gamma-tubulin: the microtubule organizer? Trends Cell Biol. 1992;2:1–5. doi: 10.1016/0962-8924(92)90125-7. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Morris NR. A β-tubulin mutation in Aspergillus nidulans that blocks microtubule function without blocking assembly. Cell. 1981;24:837–845. doi: 10.1016/0092-8674(81)90109-4. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Adachi Y, Kinoshita N, Niwa O, Toda T, Yanagida M. Cold-sensitive and caffeine supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 1988;7:1465–1473. doi: 10.1002/j.1460-2075.1988.tb02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H, Kinoshita N, Miyatani S, Toda T, Yanagida M. The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type I protein phosphatases. Cell. 1989;57:997–1007. doi: 10.1016/0092-8674(89)90338-3. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Yanagida M. S. pombe gene sds22+ essential for a mid-mitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-I. Cell. 1991;64:149–157. doi: 10.1016/0092-8674(91)90216-l. [DOI] [PubMed] [Google Scholar]
- Reijo RA, Cooper EM, Beagle GJ, Huffaker TC. Systematic mutational analysis of the yeast β-tubulin gene. Mol Biol Cell. 1994;5:29–43. doi: 10.1091/mbc.5.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas CI, Vera JC, Maccioni RB. Anti-idiotypic antibodies that react with microtubule-associated proteins are present in the sera of rabbits immunized with synthetic peptides from tubulin’s regulatory domain. Proc Natl Acad Sci USA. 1988;85:6092–6096. doi: 10.1073/pnas.85.16.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage CR, Dougherty CA, Davis AS, Burns RG, Wilson L, Farrell KW. Site-directed mutagenesis of putative GTP-binding of yeast β-tubulin: evidence that α-, β-, and γ-tubulins are atypical GTPases. Biochemistry. 1995;34:7409–7419. doi: 10.1021/bi00022a014. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage C, Xue Y, Mitani S, Hall DRZ, Chalfie M. Mutations in the Caenorhabditis elegans beta-tubulin gene mec-7: effects on microtubule assembly and stability and on tubulin autoregulation. J Cell Sci. 1994;107:2165–2175. doi: 10.1242/jcs.107.8.2165. [DOI] [PubMed] [Google Scholar]
- Sazer S, Nurse P. A fission yeast RCC1-related protein is required for the mitosis to interphase transition. EMBO J. 1994;13:606–615. doi: 10.1002/j.1460-2075.1994.tb06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ, Pillus L, Grisafi P, Solomon F, Botstein D. Two functional α-tubulin genes of the yeast Saccharomyces cerevisiae encode divergent proteins. Mol Cell Biol. 1986a;6:3711–3721. doi: 10.1128/mcb.6.11.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ, Solomon F, Botstein D. Genetically essential and nonessential α-tubulin genes specify functionally interchangeable proteins. Mol Cell Biol. 1986b;6:3722–2733. doi: 10.1128/mcb.6.11.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell V, Nurse P. Investigation into the control of cell form and polarity: the use of morphological mutants in fission yeast. J Cell Sci Dev (Suppl) 1993;1993:289–299. [PubMed] [Google Scholar]
- Stearns T, Botstein D. Unlinked noncomplementation: isolation of new conditional-lethal mutations in each of the tubulin genes of Saccharomyces cerevisiae. Genetics. 1988;119:249–260. doi: 10.1093/genetics/119.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Yamano H, Kinoshita N, Yanagida M. Mitotic regulation of protein phosphatases by the fission yeast sds22 protein. Curr Biol. 1993;3:13–26. doi: 10.1016/0960-9822(93)90140-j. [DOI] [PubMed] [Google Scholar]
- Streiblová E, Wolf A. Cell wall growth during the cell cycle of Schizosaccharomyces pombe. Z Allg Mikrobiol. 1972;12:673–684. [PubMed] [Google Scholar]
- Tanaka K, Kanbe T. Mitosis in the fission yeast Schizosaccharomyces pombe as revealed by freeze-substitution electron microscopy. J Cell Sci. 1986;80:253–268. doi: 10.1242/jcs.80.1.253. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Neff NF, Botstein D. Isolation and characterization of mutations in the β-tubulin gene of Saccharomyces cerevisiae. Genetics. 1985;112:715–734. doi: 10.1093/genetics/111.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded β-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- Tian G, Lewis SA, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan NJ. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Adachi Y, Hiraoka Y, Yanagida M. Identification of the pleiotropic cell cycle gene NDA2 as one of two different α-tubulin genes in Schizosaccharomyces pombe. Cell. 1984;37:233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]
- Toda T, Umesono K, Hirata A, Yanagida M. Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J Mol Biol. 1983;168:251–270. doi: 10.1016/s0022-2836(83)80017-5. [DOI] [PubMed] [Google Scholar]
- Uemura T, Yanagida M. Isolation of type I and II topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K, Hiraoka Y, Toda T, Yanagida M. Visualization of chromosomes in mitotically arrested cells of the fission yeast Schizosaccharomyces pombe. Curr Genet. 1983a;7:123–128. doi: 10.1007/BF00365637. [DOI] [PubMed] [Google Scholar]
- Umesono K, Toda T, Hayashi S, Yanagida M. Two cell division cycle genes NDA2 and NDA3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity of antimitotic benzimidazole compounds. J Mol Biol. 1983b;168:271–284. doi: 10.1016/s0022-2836(83)80018-7. [DOI] [PubMed] [Google Scholar]
- Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wells WAE. The spindle assembly checkpoint: aiming for a perfect mitosis, every time. Trends Cell Biol. 1996;6:228–234. doi: 10.1016/0962-8924(96)10018-0. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Hirata D, Verde F, Eddison M, Toda T, Nurse P. Microtubules mediate mitochondrial distribution in fission yeast. Proc Natl Acad Sci USA. 1996;93:11664–11668. doi: 10.1073/pnas.93.21.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M. Genetic analysis of resistant mutants to antimitotic benzimidazole compounds in Schizosaccharomyces pombe. Mol Gen Genet. 1980;180:231–234. doi: 10.1007/BF00267375. [DOI] [PubMed] [Google Scholar]