Abstract
Utrophin/dystrophin-related protein is the autosomal homologue of the chromosome X-encoded dystrophin protein. In adult skeletal muscle, utrophin is highly enriched at the neuromuscular junction. However, the molecular mechanisms underlying regulation of utrophin gene expression are yet to be defined. Here we demonstrate that the growth factor heregulin increases de novo utrophin transcription in muscle cell cultures. Using mutant reporter constructs of the utrophin promoter, we define the N-box region of the promoter as critical for heregulin-mediated activation. Using this region of the utrophin promoter for DNA affinity purification, immunoblots, in vitro kinase assays, electrophoretic mobility shift assays, and in vitro expression in cultured muscle cells, we demonstrate that ets-related GA-binding protein α/β transcription factors are activators of the utrophin promoter. Taken together, these results suggest that the GA-binding protein α/β complex of transcription factors binds and activates the utrophin promoter in response to heregulin-activated extracellular signal–regulated kinase in muscle cell cultures. These findings suggest methods for achieving utrophin up-regulation in Duchenne’s muscular dystrophy as well as mechanisms by which neurite-derived growth factors such as heregulin may influence the regulation of utrophin gene expression and subsequent enrichment at the neuromuscular junction of skeletal muscle.
INTRODUCTION
Duchenne’s muscular dystrophy (DMD) is the most common X-linked neuromuscular disorder and is caused by genetic mutations leading to quantitative and qualitative disturbances in the expression of dystrophin (Hoffman et al., 1987). Dystrophin belongs to the spectrin superfamily of proteins, which includes the spectrins, the α-actinins, and three close relatives of dystrophin, the chromosome 6–encoded dystrophin-related protein or utrophin (Love et al., 1989; Khurana et al., 1990; Tinsley et al., 1992), the chromosome 18–encoded dystrobrevin (Khurana et al., 1994), and the chromosome-X encoded dystrophin-related protein 2 (Roberts et al., 1996). In muscle, dystrophin is complexed to the membrane bound dystroglycan–sarcoglycan complex, which forms a link with the extracellular matrix via laminin. Mutations in the genes encoding various members of the complex disrupt sarcolemmal integrity and result in a variety of X-linked and limb girdle muscular dystrophies (Campbell, 1996; Brown, 1997).
Utrophin shares extensive sequence homology and organizational motifs with dystrophin and is considered the autosomal homologue of dystrophin (Love et al., 1989; Tinsley et al., 1992). The protein products of these genes are of similar size and abundance in muscle; however, utrophin is more widely distributed than dystrophin (Khurana et al., 1990; Khurana et al., 1991; Love et al., 1991). Additionally, utrophin’s expression continues unabated in DMD muscle, whereas dystrophin is severely reduced or absent (Khurana et al., 1990; Love et al., 1991). The possibility that dystrophin and utrophin share functional properties is reinforced by studies determining that utrophin is up-regulated during the perinatal period and during regeneration, periods during which there is lack of necrosis in dystrophin-deficient muscle (Khurana et al., 1991). Moreover, they have similar affinities for binding F-actin at the amino terminus (Winder et al., 1995). The ability of high levels of utrophin to rescue dystrophin-deficient muscle was recently demonstrated by generating transgenic mice expressing high levels of utrophin and breeding them with dystrophin-deficient mdx mice (Tinsley et al., 1996). Theoretically, the approach of utrophin up-regulation is particularly useful in the case of DMD, because utrophin (being autosomally encoded) is not subject to the chromosome Xp21 mutations that cause DMD. Although transgene-mediated utrophin up-regulation is not directly applicable to DMD patients for obvious reasons, findings of the study have provided a powerful impetus to searching for molecules that can up-regulate the expression of utrophin as a possible means of DMD therapy (Campbell and Crosbie, 1996; Karpati, 1997).
Clues for identification of transcriptional regulators of utrophin come from previous studies characterizing the subcellular distribution of utrophin in the myofiber (Khurana et al., 1991; Nguyen et al., 1991; Ohlendieck et al., 1991). The spatial distribution of utrophin in myofibers parallels that of the nicotinic acetylcholine receptors (nACHRs) to a remarkable degree. Both nACHR and utrophin proteins are codistributed in adult myofiber mainly at the neuromuscular junction (NMJ) with a secondary peak of enrichment at the myotendinous junction (Khurana et al., 1991). Furthermore, in common with the nACHR ε subunit gene promoter, the utrophin promoter contains an N-box motif (Koike et al., 1995; Dennis et al., 1996), which in the case of the nACHR ε subunit gene is critical for mediating synapse-specific transcription via ets-related transcription factors (Koike et al., 1995; Sapru et al., 1998; Schaeffer et al., 1998). Although the functional relevance of this motif in the utrophin promoter is unclear, its existence coupled with highly regulated distribution of gene product in muscle at the NMJ would suggest that nerve-derived growth factors such as heregulin (cf.), which are known to increase the transcription of nACHR subunits would be prime candidates to regulate the activity of the utrophin promoter at this locale as well.
Heregulin is a member of the neuregulin family of polypeptide growth factor homologues, including heregulin, Neu differentiation factor, acetylcholine receptor-inducing activity (ARIA), and glial growth factor (Fischbach and Rosen, 1997). These ligands and their receptors have wide-ranging effects that are considered critical for nervous system development (Meyer and Birchmeier, 1995; Lemke, 1996; Fischbach and Rosen, 1997). The treatment of cultured muscle cells with ARIA, for example, results in dramatic increases in the rate of synthesis and accumulation of acetylcholine receptors and sodium channel gene products in the sarcolemma (Fischbach and Rosen, 1997). The up-regulation occurs via heregulin binding and activating the HER/erbB class of receptor tyrosine kinases followed by activation of the PI3 and MAPK signal pathways, leading to a preferential increase of nACHR subunit gene transcription at the subsynaptic nuclei, which lie juxtaposed to the NMJ (Carraway and Burden, 1995; Tansey et al., 1996; Fischbach and Rosen, 1997).
In this study we have used DNA affinity columns, immunoblots, kinase assays, electrophoretic mobility shift assays (EMSAs), and in vitro expression studies in cultured muscle cells to identify and characterize the mechanism of utrophin regulation at a transcriptional level.
MATERIALS AND METHODS
Constructs
The pGABPα and pGABPβ expression constructs were generated by cloning the human GA-binding protein α (GABPα) and GABPβ1 cDNAs into the mammalian expression vector pCAGGS and have been previously described (Rosmarin et al., 1995). The previously described pPUBF (Dennis et al., 1996) luciferase reporter plasmid contains the entire human utrophin promoter sequence cloned into the pGL2 basic reporter plasmid and was kindly supplied by Drs. Jon Tinsley and Kay Davies (Oxford University, Oxford, England). Additionally, we generated the ΔNBox construct, which has a deletion mutation removing the entire N-box (and core ets-binding site) from the human utrophin promoter and is cloned in the forward orientation in pGL2 Basic vector (Promega, Madison, WI). The ΔNBox reporter was generated by cloning the HindIII–SmaI (1–569 bp) fragment of the human utrophin promoter into pGEM3Zf (Promega), generating the plasmid pGEM-L. Next the PstI–HindIII (659–1242 bp) fragment of the promoter was subcloned into pBluescript SKII+ (Stratagene, La Jolla, CA) yielding pBS-R. The SmaI–PstI fragment (570–658 bp) of the utrophin promoter was used for PCR mutagenesis using the primers NboxlessF (5′-CCCCCCGGGAACGTAGTGGGGCTGATCAACAAAGTTGCTGGGCCGGCGG-3′) and NboxlessR (5′-CCTCCGGCCCGCGCCTCTGCAGCGCTCCGG-3′).
The PCR fragment was cloned into PCR 2.1 vector (Invitrogen, Carlsbad, CA) yielding pI. The SmaI–KpnI fragment of pI was subcloned into pGEM-L yielding pGEM-L+I. Next, the PstI–KpnI fragment of pBS-R was cloned into pGEM-L+I to yield pGEM-L+I+R. The HindIII fragment of pGEM-L+I+R was subcloned into pGL2 Basic to yield the ΔNBox construct. The clone was sequenced to verify orientation and sequence.
Tissue Culture, Transfections, and Luciferase Assays
Primary mouse skeletal muscle cells were generated and propagated essentially as previously described (Rando and Blau, 1994). The L6 rat muscle cell line was obtained and cultured according to standard protocols (American Type Culture Collection, Rockville, MD). For transfections, differentiated muscle cells were plated at 1.2 × 106 cells/35-mm well. Plasmids were transfected using Superfect (Qiagen, Hilden, Germany) in the case of L6 cells and calcium phosphate in the case of primary muscle cells. Cells were transfected with a total of 5 μg of DNA/well in case of heregulin response studies and 7.5 μg of DNA/well in case of GABP response studies. Heregulin (R & D Systems, Minneapolis, MN) in PBS was added to achieve a final concentration 1 nM in cultured cells after transfection. In the case of the pGABPα and pGABPβ experiments, 2.5 μg of each plasmid was used an equivalent amount of either pCAGGS or single-stranded DNA (ssDNA) used as a control. To control for efficiency, we cotransfected the pRL control plasmid at a ratio of 1:250 or 1:400. The pRL plasmid (Promega) is designed to express renilla luciferase driven by a cytomegalovirus promoter and was used as an internal control for efficiency of transfection. Luciferase assays were performed using the dual luciferase reagents on a Turner Designs (Sunnyvale, CA) 20/20 luminometer 24–48 h after transfection according to instructions supplied by the manufacturer (Promega). Promoter activity values were expressed as normalized luciferase activity by dividing the firefly luciferase readings with the renilla luciferase reading for each well. The selective MAPK kinase 1 inhibitor PD98059 was preincubated with muscle cell cultures for inhibiting extracellular signal–regulated kinase (ERK) as suggested by the manufacturer (Calbiochem, Bad Soden, Germany).
Antibodies and Immunoblots
Human GABPα and GABPβ1 cDNAs were cloned into the pGEX vector for generation of GABPα and GABPβ GST fusion proteins (Nuchprayoon et al., 1997). Purified fusion proteins were used as antigens to raise polyvalent antisera in rabbits. Antisera were affinity purified against the appropriate GST fusion protein and negatively selected against GST protein (HTI Bio-Products, Ramona, CA) to yield affinity-purified GABPα and GABPβ antibodies. These reagents were used at 1:20,000 and 1:2000, respectively. Proteins were detected using enhanced chemiluminescence using the Ultra ECL kit (Pierce, Rockford, IL). The anti-phosphotyrosine antibody PY 20 was used for detecting active p185 species of hetrodimerized heregulin receptors (Santa Cruz, Biotechnology, Santa Cruz, CA). The rabbit polyclonal pTEpY antibody (Promega) was used to detect dually phosphorylated, active forms of ERK 1and ERK2.
In Vitro Kinase Assays
In vitro kinase assays were performed as previously described (Tansey et al., 1996). Briefly, cells were treated with heregulin and lysed, and ERK was immunoprecipitated using the goat polyclonal antibody sc-154-G (Santa Cruz) that reacts with both p44 ERK1 and p42 ERK 2. The immunoprecipitated ERK complexes were incubated with 500 ng of GST-GABPα, GST-GABPβ, or myelin basic protein (MBP) as substrates, along with 50 μM ATP and 10 μCi of [γP32]ATP for 1 h at 30°C. The reactions were stopped by addition of protein sample buffer and heating at 98°C for 2 min in the case of GST fusion proteins and orthophosphoric acid in the case of MBP. MBP reaction products (positive control for ERK activation) were spotted onto phosphocellulose paper and counted using a scintillation counter. GST reaction products were resolved on 10% denaturating polyacrylamide gels and detected by autoradiography using a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
EMSAs
Nuclear extracts were prepared from dishes of L6 myotubes cultured to confluence, and total proteins were quantified, as previously described (Ausubel et al., 1995). The double-stranded UtroNBox probe used was 5′-ATCTTCcggaaC-3′ (N-box underlined; ets-domain italicized and lowercase), which was end labeled with [γ-32P]ATP using T4 polynucleotide kinase. Typically, radiolabeled probe (1 ng, ∼10–100,000 cpm) was incubated on ice for 20 min with 10 μg of nuclear extracts or 1 μl of GST fusion proteins or antibodies in a 12.5 μl of reaction buffer containing 20 mM HEPES, pH 7.6, 1 mM MgCl2, 0.1 mM EGTA, 40 mM KCl, 10% glycerol, and 1 μg of ssDNA. Competition with cold probe was performed with preincubation with 1000× excess of unlabeled UtroNBox probe. Complexes were resolved by electrophoresis at 200 V for 2 h on 4% acrylamide gels in 25 mM Tris, 192 mM glycine, and 1 mM EDTA (for nuclear extracts) and 12.5 mM Tris, 96 mM glycine, and 0.5 mM EDTA (for fusion proteins) before autoradiographic analysis using a Storm PhosphorImager.
Quantitative Reverse Transcription-PCR (RT-PCR)
Confluent, differentiated L6 cultures were treated with heregulin (R & D systems) in PBS (final concentration, 1 nM) for 30 min. Controls were treated with an equivalent volume of PBS (typically 1 μl in 10 ml of culture medium). Bioactivity of heregulin was verified by monitoring increased tyrosine phosphorylation of p185 heregulin receptor species in treated cultures (Fischbach and Rosen, 1997). RT-PCR was performed essentially as previously described (Khurana et al., 1991; Ausubel et al. 1995). Briefly, RNA was extracted using Trizol (Life Technologies, Paisly, Scotland) as suggested by the manufacturer. Purified RNA was treated with DNase and repurified to exclude DNA contamination. One microgram of RNA was used as template for oligo-dT–primed RT using Superscript reverse transcriptase enzyme (Life Technologies). Five percent (vol/vol) of the purified cDNA (corresponding to 50 ng of RNA) was used as template for quantitative RT-PCR. Primers used to amplify a 322-bp fragment of rat utrophin (GenBank accession number AJ002967, position 9659–9981) were RUTROF (5′-CAGTATGTGGCCAGAGCACTATGA-3′) and RUTROR (5′-GCAGATTTCTTTGCTCTTCCTCC-3′).
As an internal control for efficiency of RT and quantification, we simultaneously amplified a 194-bp fragment of rat glyceraldehyde 3-phosphate dehydrogenase (GAPDH; accession M17701, position 335–529) using the primers RGAPDHF (5′-CCATGGAGAAGGCTGGGG-3′) and RGAPDHR (5′-CAAAGTTGTCATGGATGACC-3′).
PCR was performed using a 2-min denaturation at 94°C followed by 20 or 25 cycles (for GAPDH and utrophin, respectively) of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by 72°C for 7 min, conditions that had been optimized for exponential phase amplification of both transcripts. Reactions were also performed in parallel, adding 1 μl of [32P]dCTP/100 μl of reaction mixture, for measuring radioactive incorporation. Products were resolved on 2% agarose gels and photographed using ethidium bromide. Photographs were digitized using an Agfa (Mortsel, Belgium) Arcus II scanner at 1600 dots per inch, and bands were quantified using ImageQuant 1.1 software (Molecular Dynamics) for the Macintosh OS 7.5.3 (Apple Computer, Cupertino, CA). Radioactive PCR products were resolved on 5% acrylamide gels, dried and the radioactivity incorporated in bands quantified using a Storm PhosphorImager and ImageQuant 1.1 software. Similar results were obtained in both cases.
DNA Affinity Purification
The utrophin promoter UtroNBox probe described above was ligated using T4 DNA ligase to streptavidin magnetic particles that have previously been coupled to a 16-mer oligonucleotide and used for DNA affinity purification as suggested by the manufacturer (Boehringer Mannheim, Mannheim, Germany). Typically, 50 μg of L6 myotube nuclear extract were incubated with UtroNBox coupled magnetic particles and eluted in 25 μl of high-salt buffer [20 mM HEPES, pH 7.6, 1 mM EDTA, 10 mM (NH4)2 SO4, 1 mM DTT, 0.2% Tween 20 (wt/vol), and 2 M KCl]. The DNA-binding proteins were dialyzed to reduce the salt concentration using a 3500 molecular weight cutoff (Pierce) membrane before analysis.
Statistical Analysis
All data were subjected to Student’s t test for calculation of statistical significance. Where appropriate they were also subjected to an additional parametric test (ANOVA) as well as nonparametric (Wilcoxon’s rank sum) tests for statistical significance. Statistical analysis was performed using Statview 5.0 (SAS Institute, Cary, NC). All data are graphically represented with controls normalized to 100 and increases (or decreases) shown as a percentage of control levels. Error bars are specified in figure legends as SEM or SD.
RESULTS
Heregulin Activates Utrophin Expression
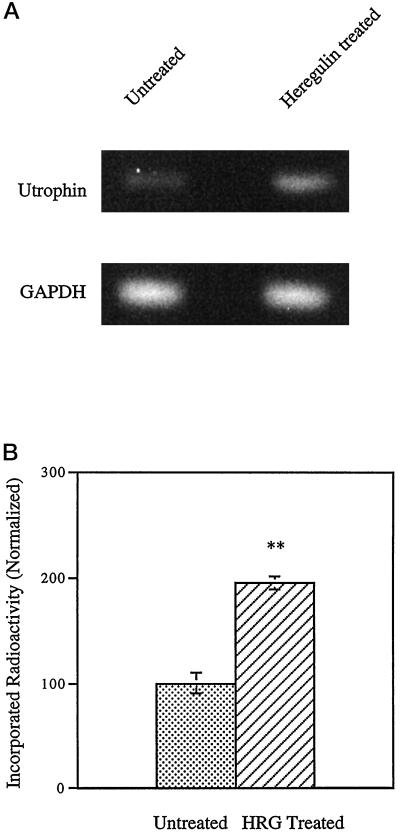
To address whether heregulin regulates utrophin gene expression, we treated rat L6 myotubes with heregulin and processed the cultures for quantitative RT-PCR. As shown in Figure 1, heregulin treatment increased the mRNA level of utrophin to 195% compared with control cultures. Although this technique is both sensitive and specific, it cannot distinguish whether the observed increase of utrophin mRNA is due to increased utrophin gene transcription or changes in mRNA stability.
Figure 1.
Heregulin increases utrophin mRNA in skeletal muscle cultures. Differentiated L6 rat myotubes were incubated with 1 nM heregulin in PBS for 30 min along with controls. RNA was extracted, and quantitative RT-PCR performed. (A) Representative experiment showing the 322-bp utrophin fragment and the 194-bp GAPDH control fragment obtained by RT-PCR. (B) Results of radioactive quantification of four individual experiments taken together. The stippled bars represent utrophin mRNA levels in untreated cells, and cross-hatched bars represent the levels in heregulin (HRG)-treated cultures. Heregulin treatment increases the endogenous utrophin message in muscle cell cultures to 195% of control levels. Error bars indicate SEM; n = 4). Asterisks denote the results were statistically highly significant at p < 0.001.
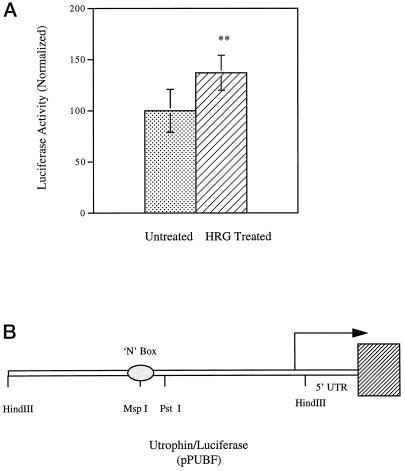
To verify the increase in utrophin expression as well as to determine whether the increases were indeed due to increased transcription of the utrophin promoter, we transfected murine muscle cultures with a reporter construct containing the entire human utrophin promoter driving the expression of firefly luciferase construct pPUBF (Dennis et al., 1996). As shown in Figure 2, heregulin increased the luciferase expression to 138% compared with control cultures. The increased luciferase activity reflects increased de novo transcription of the utrophin promoter in response to heregulin. The data in Figure 2 are a summary of 10 independent experiments, with 5 sets of experiments being done in primary mouse muscle cultures and 5 sets in the rat L6 muscle cell line, with similar increases being noted in each individual experiment.
Figure 2.
Heregulin activates the utrophin promoter in muscle cell cultures. The utrophin promoter–luciferase reporter construct pPUBF was cotransfected into L6 muscle cell lines or mouse primary muscle cultures along with transfection control plasmid pRL and assayed after 24–48 h of incubation with heregulin-containing medium or control-containing medium. (A) pPUBF derived firefly luciferase activity is normalized to pRL-derived renilla luciferase activity (internal control) as 100% in the untreated group and expressed as luciferase activity (normalized). The graph represents the summary of 10 individual experiments, 5 sets of experiments in primary mouse cultures and 5 sets of experiments in L6 rat muscle cell cultures. The stippled bars represent utrophin promoter activity in untreated cells, and cross-hatched bars represent the levels in heregulin (HRG)-treated cultures. (B) Schematic of the pPUBF-luciferase construct (Dennis et al., 1996) and was kind gift from Drs. J. Tinsley and K. Davies. Heregulin increases de novo utrophin transcription in muscle cell cultures to 138% of control levels. Error bars indicate SEM; n = 60. Asterisks denote the results were statistically highly significant at p < 0.001.
Role of N-Box in Transcriptional Activation of Utrophin Expression
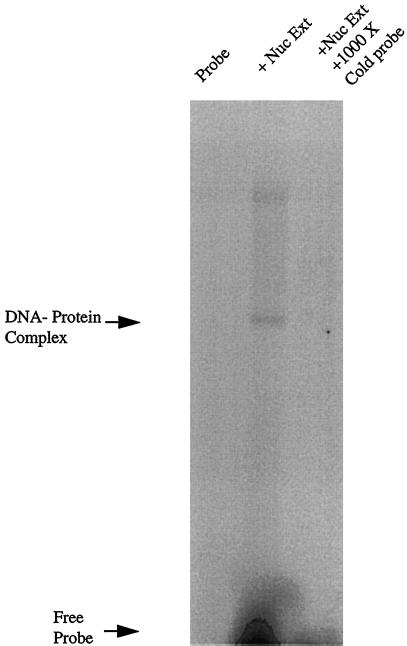
Having determined that heregulin increased utrophin transcription, we addressed whether the region of the utrophin promoter containing the N-box motif was capable of interacting with putative transcription factors. We performed EMSAs to address this issue, using nuclear extracts made from cultured L6 myotubes. Figure 3 demonstrates that the UtroNbox probe (from the region of the utrophin promoter containing the N-box motif) binds a factor(s) present in the nuclear extracts of L6 myotubes. The binding is specific because it could be competed with a 1000× excess of unlabeled probe.
Figure 3.
The utrophin N-Box binds proteins in nuclear extracts of cultured L6 myotubes. EMSA was performed with the radiolabeled UtroNBox probe using 10 μg of nuclear extract made from cultured L6 myotubes. Lane 1, migration of free, unretarded probe control; lane 2, mobility shift of the probe in the presence of a N-box–binding factor present in L6 nuclear extracts; lane 3, control for specificity showing that the mobility shift was competed with a 1000× excess of unlabeled probe.
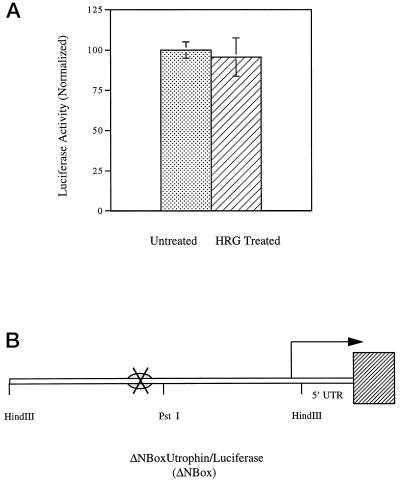
The previous experiments suggested that the N-box specifically bound nuclear factor(s) and may be involved in transcriptional regulation. To ask whether this region of the promoter mediated heregulin-induced activation, we generated a mutant reporter construct of the human utrophin promoter containing a 6-bp deletion of the N-box motif, called the ΔN-box construct. L6 muscle cultures were transfected with the ΔN-box construct and treated with heregulin. As shown in Figure 4, heregulin did not increase the expression of luciferase in L6 muscle cultures that had been transfected with the ΔN-box reporter construct compared with controls. These data suggest that the N-box region is critical for the heregulin-induced transcriptional activation of the utrophin promoter.
Figure 4.
The N-Box motif in the utrophin promoter mediates transcriptional activation by heregulin in cultured muscle cells. The mutant utrophin promoter–luciferase reporter construct ΔNBox (deleted in the N-box) was cotransfected into L6 muscle cell lines along with transfection control plasmid pRL and assayed after 24 h of incubation with either heregulin-containing medium or controls. (A) The ΔNBox-derived firefly luciferase activity is normalized to pRL-derived renilla luciferase activity (internal control) as 100% in the untreated state and expressed as luciferase activity (normalized). The stippled bars represent mutant utrophin promoter activity in untreated cells, and cross-hatched bars indicate the levels in heregulin (HRG)-treated cultures. (B) Schematic of the ΔNBox–luciferase construct; the cross depicts the site of deletion mutation removing the N-box. Heregulin does not activate the mutant utrophin promoter bearing a deletion of the N-Box. Error bars indicate SEM; n = 36. The differences of expression are not statistically significant.
GABPα/β Transcription Factors Are Mediators of Utrophin Activation
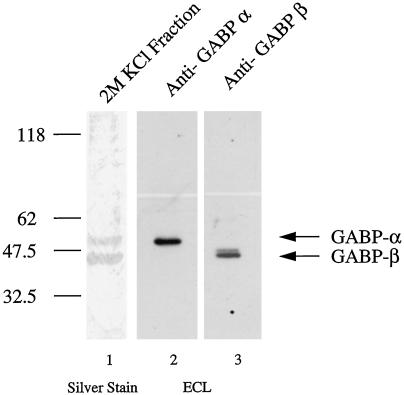
To identify the transcriptional factors that bind and activate the utrophin promoter in response to heregulin, we made a DNA affinity column using the UtroNBox probe coupled to magnetic particles. This column was used to purify promoter-binding proteins from nuclear extracts of cultured L6 myotubes. As shown in Figure 5, lane 1, we purified two proteins of 43 and 58 kDa from these nuclear extracts. Because, the molecular masses exactly matched members of the ets-related GABPα/β transcription factors that have been implicated in case of the nACHR genes (Sapru et al., 1998; Schaeffer et al., 1998), we performed immunoblot analysis using affinity-purified antibodies against GABPα and GABPβ on the purified fractions (Brown and McKnight, 1992; de la Brousse et al., 1994), to determine the molecular identity of the purified protein. As shown in Figure 5, lanes 2 and 3, the 43- and 58-kDa proteins were indeed recognized by antibodies specific for GABPβ and GABPα transcription factors.
Figure 5.
Identification of transcription factors that bind the N-Box of the utrophin promoter. UtroNBox-coupled magnetic particles were used to perform DNA affinity chromatography to purify promoter-binding proteins from nuclear extracts of cultured L6 myotubes. Fifty micrograms of nuclear extracts were used, and binding proteins were eluted in 25 μl of 2 M KCl. A 15-μl aliquot was resolved using 12% SDS-PAGE gels and subjected to silver staining (lane 1), or 5-μl aliquots were immunoblotted with affinity-purified anti-GABPα and -GABPβ antibodies and subjected to enhanced chemiluminescence detection (lanes 2 and 3). Lane 1, silver-stained proteins demonstrating that the molecular masses of purified proteins (43 and 58 kDa) exactly matches the molecular masses of GABPβ and GABPα heterodimeric complex of transcription factors; lane 2, the 58-kDa band in the purified fraction is recognized by affinity-purified anti-GABPα antibodies; lane 3, the 43-kDa band in the purified fraction is recognized by affinity-purified anti-GABPβ antibodies. The additional high-molecular-mass species presumably represents a GABPβ isoform sharing sequence similarity with the GABPβ1 isoform, against which the antibodies were raised. The anti-GABPβ1 antibodies used in this study (see MATERIALS AND METHODS) are predicted to recognize all GABPβ isoforms.
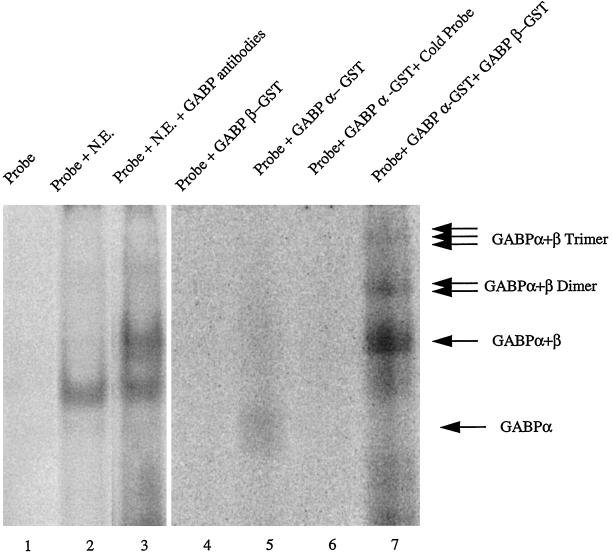
To independently verify the role of GABPα/β transcription factors in heregulin-mediated transcriptional activation, we asked whether the region of utrophin promoter containing the N-box motif was capable of specifically interacting with these transcription factors by performing a supershift assay using antibodies against GABPα/β as well as EMSA with GABPα/β fusion proteins (Rosmarin et al., 1995). Figure 6 demonstrates that the UtroNBox probe specifically bound transcription factors present in nuclear extracts (lane 2) and that the addition of antibodies directed GABPα/β caused a further mobility shift or “supershift” (lane 3), suggesting that the GABP transcription factors were present in the nuclear extracts made from muscle cells. Furthermore, the probe was capable of binding the GABPα fusion protein (Figure 6, lane 5). This binding was specific, because it could be competed with an excess of unlabeled probe (Figure 6, lane 6). As predicted by structural studies of GABP complex (Brown and McKnight, 1992; de la Brousse et al., 1994), the UtroNBox probe did not bind GABPβ alone (Figure 6, lane 4); however, it bound the in vitro reconstituted heterodimeric complex of GABPα/β with enhanced efficiency (Figure 6, lane 7), as evidenced by the formation of multimeric complexes of the UtroNBox probe with GABPα/β.
Figure 6.
The utrophin N-Box binds the heterodimeric GABPα/β transcription factor. EMSA was performed with the radiolabeled oligonucleotide UtroNBox probe, nuclear extracts, and antibodies against GABPα/β (for supershift assays) in lanes 1–3. In lanes 4–7 EMSA was performed using purified GABPα and GABPβ GST fusion proteins and this probe. Lane 1, lack of mobility shift when the probe is electrophoresed alone; lane 2, mobility shift when the probe is incubated with nuclear extracts from cultured muscle cells; lane 3, the addition of antibodies directed against GABPα/β causes a further mobility shift or supershift compared with the mobility shift obtained using nuclear extracts alone; lane 4, lack of mobility shift using the GABPβ protein, suggesting that the UtroNBox probe does not bind GABPβ by itself; lane 5, mobility shift when the probe is incubated with GABPα fusion protein; lane 6, specificity of the interaction with GABPα, because it is competed with a 1000× excess of unlabeled probe; lane 7, mobility shifts with the formation of GABPα/β multimers when the probe is incubated with both GABPα and GABPβ fusion proteins, suggesting enhancement of GABPα binding by reconstitution of the heterodimeric GABPα/β transcription factor complex.
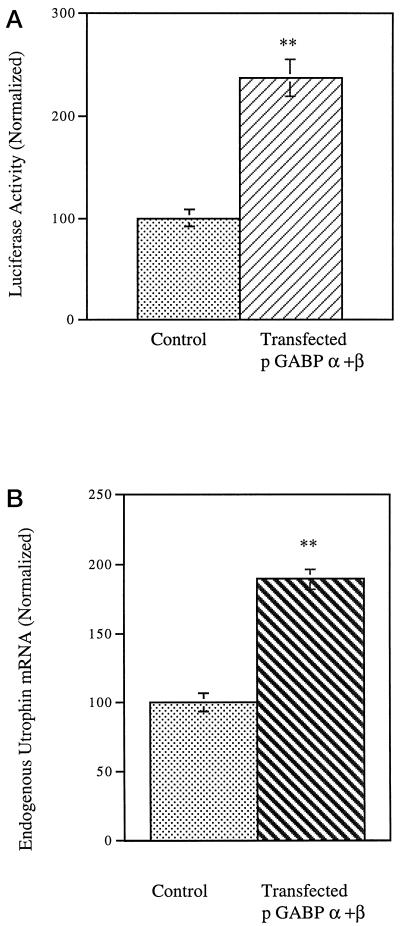
Having determined that the GABPα/β complex was biochemically associated with the N-box region of the utrophin promoter, we asked whether GABPα/β can functionally activate the promoter and increase utrophin transcription. We transfected L6 myotubes with pPUBF along with expression constructs encoding GABPα and GABPβ (Rosmarin et al., 1995; Dennis et al., 1996). Control cultures were transfected with the reporter construct along with either empty vector pCAGGS or an equivalent amount of ssDNA. As shown in Figure 7A, the expression of GABPα and GABPβ in these cultures increased the luciferase expression to 238% compared with control cultures. The increased luciferase activity reflects transcriptional activation of the utrophin promoter by the heterodimeric complex of GABPα and GABPβ in muscle cells. To independently verify the role of GABPα/β transcription factors in regulation of utrophin gene expression, we processed cultures that had been transfected with expression constructs encoding GABPα and GABPβ for quantitative RT-PCR. As shown in Figure 7B, cultures transfected with GABPα/β showed a 189% increase in the utrophin mRNA levels compared with control cultures. These data demonstrate that transfection with GABPα/β increases the de novo transcription of the utrophin promoter and consequently increases the levels of endogenously expressed utrophin mRNA.
Figure 7.
GABPα/β activates the utrophin promoter in muscle cell cultures. (A) The utrophin promoter–luciferase reporter construct PUBF was cotransfected into L6 muscle cell lines along with expression constructs pGABPα, pGABPβ, and pCAGGS (empty vector) along with transfection control pRL and assayed after 24 h of incubation. PUBF-derived firefly luciferase activity is normalized to pRL-derived renilla luciferase activity (internal control) in control transfectants as 100% and expressed as luciferase activity (normalized). The stippled bars represent utrophin promoter activity in cells transfected with empty vector pCAGGS, and cross-hatched bars represent the levels in cultures transfected with pGABPα and pGABPβ. (B) L6 muscle cell lines were transfected with expression constructs pGABPα, pGABPβ, and pCAGGS (empty vector) as control. RNA was extracted, and quantitative RT-PCR was performed for estimating the level of utrophin mRNA. The graph shows the results of quantification of five individual experiments. The stippled bars represent utrophin mRNA levels in control cultures, and cross-hatched bars represent the levels in cultures transfected withpGABPα and pGABPβ. GABPα and GABPβ cotransfection increases de novo utrophin transcription in muscle cell cultures to 238% of control levels (error bars indicate SEM; n = 24) and increases endogenous utrophin mRNA by 189% of control levels (error bars indicate SD; n = 5). Asterisks denote the results were statistically highly significant at p < 0.001.
Intracellular Signaling Pathways for Heregulin-mediated Activation of the Utrophin Promoter
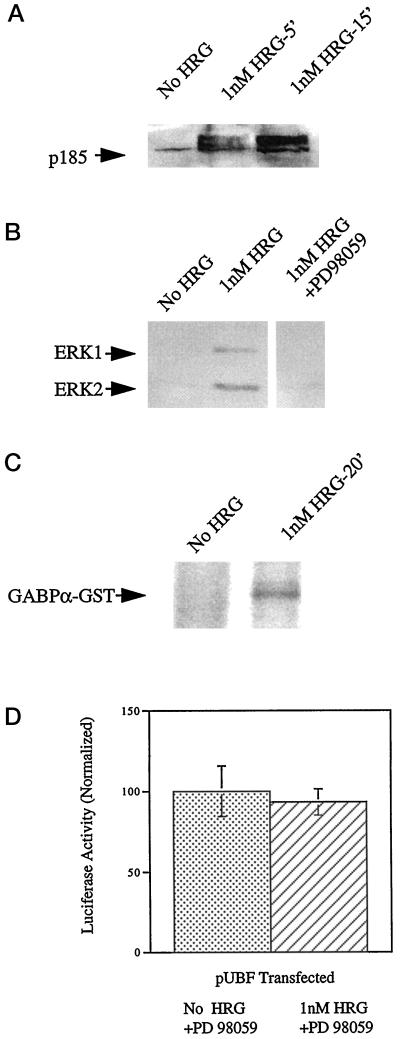
To explore the signaling pathway(s) by which heregulin mediates utrophin promoter activation, we analyzed the ERK pathway, because it has been implicated in regulation of nACHR subunit gene transcription at the subsynaptic nuclei of muscle by heregulin (Tansey et al., 1996; Schaeffer et al., 1998). Additionally, active MAPK (ERK1 and ERK2) has been shown to phosphorylate GABPα (Ouyang et al., 1996; Schaeffer et al., 1998). As shown in Figure 8A, treatment of L6 muscle cell cultures with heregulin led to increased tyrosine phosphorylation and mobility shift of the HER receptors expressed by L6 cells. Furthermore, heregulin treatment led to the activation of MAP kinase family members ERK1 and ERK2. The activation could be completely prevented by preincubation with PD98059 (an inhibitor of MAPK kinase 1, the upstream activator of ERK1 and ERK2), as shown in Figure 8B. Next, we asked whether heregulin-activated ERK was capable of phosphorylating GABP by performing in vitro kinase assays using immunoprecipitated active ERK extracted from heregulin-treated muscle cell cultures. As shown in Figure 8C, heregulin-activated ERK was capable of phosphorylating recombinantly expressed GST-GABPα fusion protein. We could not detect similar phosphorylation of the GST-GABPβ fusion protein (our unpublished results). To address the functional significance of the heregulin-mediated ERK activation, we analyzed whether heregulin was capable of activating the utrophin promoter in muscle cell cultures that had been treated with PD98059. As shown in Figure 8D, heregulin was incapable of activating the utrophin promoter in muscle cell cultures in which ERK activation had been abolished by these pharmacological means, compared with the 138% increase noted when ERK activation was allowed to proceed in a normal manner in cultures (Figure 2) treated with heregulin.
Figure 8.
Heregulin-mediated utrophin promoter activation requires ERK activation. (A) Differentiated L6 rat myotubes were incubated with PBS alone or 1 nM heregulin for 5 and 15 min. Proteins were extracted from cultures and resolved on 10% SDS-polyacrylamide gels and electroblotted onto polyvinylidene difluoride membranes followed by Western blotting with the PY20 antibody to detect phospotyrosine residues. The blot shows a mobility shift and increased tyrosine phosphorylation (activation) of p185 (HER receptor) species and serves as a positive control. (B) Proteins were extracted from control cultures, cultures treated for 20 min with 1 nM HRG, and cultures preincubated with PD98059 before treatment with HRG. Proteins were resolved on 10% SDS-polyacrylamide gels and electroblotted onto polyvinylidene difluoride membranes followed by Western blotting using the pTEpY antibody, which detects dually phosphorylated, active forms of ERK. The blot shows activation of ERK1 and ERK2 by heregulin and prevention of activation by preincubation with PD 98059. (C) In vitro kinase assay was performed using immunoprecipitated ERK from control or muscle cell cultures treated with 1 nM heregulin for 20 min. Proteins were resolved on 10% SDS-polyacrylamide gels, and radioactively labeled proteins were detected by autoradiography. The autoradiograph shows that activated ERK extracted from heregulin-treated cells could phosphorylate the GST-GABPα fusion protein. (D) The utrophin promoter–luciferase reporter construct pPUBF was cotransfected into L6 muscle cell lines along with transfection control plasmid pRL and assayed after 48 h of incubation with PD98059 in heregulin-containing medium or control medium. The stippled bars represent normalized utrophin promoter (luciferase) activity in untreated cells, and cross-hatched bars represent the levels in heregulin-treated cultures. Heregulin does not activate the utrophin promoter in muscle cell cultures treated with PD98059. Error bars indicate SD; n = 6. The differences of expression are not statistically significant.
DISCUSSION
Using a variety of molecular and cell biological techniques we have demonstrated that the growth factor heregulin increases de novo utrophin transcription in skeletal muscle cultures by activating ERK. Using DNA affinity columns, immunoblots, and EMSA, we have identified the ets-related GABPα/β complex as transcriptional mediators that bind and activate the utrophin promoter.
The utrophin promoter is a CpG-rich promoter devoid of TATA or CAAT boxes (Dennis et al., 1996). This organization is typically associated with housekeeping genes; however, the utrophin gene, although ubiquitously expressed, is highly regulated at the level of developmental expression as well as subcellular distribution in both brain and muscle. In the brain utrophin is highly enriched at the astrocytes forming the abluminal aspect of the blood–brain barrier, in close apposition to the extracellular matrix (Khurana et al., 1992; Khurana et al., 1995), whereas in skeletal muscle (an elongated multinucleated cell), utrophin protein is enriched at the NMJ/synapse (Khurana et al., 1991; Nguyen et al., 1991; Ohlendieck et al., 1991), and utrophin transcripts selectively accumulate in the postsynaptic sarcoplasmic compartment, in part because of being preferentially expressed at the subsynaptic nuclei rather than nuclei scattered along the length of the myofiber (Gramolini et al., 1997).
These local control mechanisms are reminiscent of those used by some of the nACHR subunit genes to help control the spatial distribution of ACHR in specific regions of the myofiber. nACHR genes are subject to regulation by contact (at the NMJ) with nerves and nerve-derived growth factors (e.g., ARIA/heregulin, agrin, and calcitonin gene-related peptide) (Duclert and Changeux, 1995; Fischbach and Rosen, 1997). The treatment of cultured muscle cells with heregulin results in dramatic increases in the rate of synthesis and accumulation of nACHR and sodium channels in the sarcolemma. The up-regulation occurs via heregulin binding and activating the HER/erbB class of receptor tyrosine kinases followed by activation of the PI3 and MAPK pathways leading to an increase of nACHR subunit gene transcription. The activation of nACHR subunit gene transcription by neurite-associated heregulin seems to occur preferentially at the nuclei that lie immediately adjacent (and under) the synapse, rather than intramuscular nuclei that lie scattered along the length of the myofiber (Carraway and Burden, 1995; Tansey et al., 1996; Fischbach and Rosen, 1997). Additionally, the nACHR molecules are subject to posttranslational modifications such as increased clustering by activation of the receptor tyrosine kinase MuSK by agrin (Glass et al., 1996). A recent report has suggested the role of agrin in utrophin expression, via a yet to be identified pathway (Gramolini et al., 1998). However, it is unclear whether the effects noted in the study were indeed primarily due to agrin or due to activation of HER receptors (p185) secondary to agrin treatment. Because the authors reported that both the neural and muscle isoforms of agrin increased transcription, it is unlikely that agrin’s effects on utrophin transcription occurred via the physiological (MuSK receptor) pathway for agrin, which is known to discern between these isoforms (Glass et al., 1996). Experiments monitoring the activation of HER and MuSK receptors upon treatment with agrin need to be performed to resolve this issue in a satisfactory manner.
The presence of E-box, N-box, and SP1 motifs in the utrophin promoter suggest that transcriptional factors may contribute to regulation of utrophin transcription (Dennis et al., 1996). The E-box is a binding site for helix-loop-helix proteins of the MyoD family and found in all nACHR subunit gene promoters; however, it does not contribute to synapse-specific expression of the nACHR ε subunit gene (Duclert and Changeux, 1995). The N-box region has previously been shown to be critical for in vivo, synapse-specific expression of the nACHR δ and ε subunits (Duclert and Changeux, 1995). Recently, the heregulin response element of the ACHRε gene was mapped to a region that overlaps the N-box region (Sapru et al., 1998). Additionally, the N-box motif has been shown to be critical for mediating the effect of heregulin on ACHRε transcription via the ets transcription factor GABPα/β (Schaeffer et al., 1998). Taken together, it is evident that the 6-bp N-box (position −60 to −55 in the murine nACHRε gene) is contained within a larger 15-bp heregulin response element (position −69 to −55 in the murine nACHRε gene), and mutations removing the N-box in these experiments also abolished the GABPα/β binding site in the nACHRε gene (Sapru et al., 1998; Schaeffer et al., 1998).
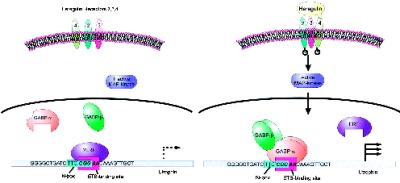
Our results suggest that the heregulin response element in the utrophin gene maps to the N-box region (position −206 to −201 of the utrophin gene, relative to transcription start site). Mutagenesis of the utrophin N-box in our constructs also destroyed the GGA core of the ets-binding site [(C/A)GGA(A/T)] in this region of the utrophin promoter (5′-ATCTTCcggaaC-3′; N-box underlined, ets domain italicized) because of the overlap of these motifs (Figure 9). This mutagenesis was accompanied by a loss of heregulin responsiveness, confirming the suggestion that this region of the utrophin promoter is critical for heregulin responsiveness (Figure 4). The ets transcription factors often cooperate with other factors that bind and activate DNA elements in the vicinity, leading to a myriad of mechanisms capable of achieving tight spatial and developmental control over subcellular expression. In the myeloid lineage SP1 has been shown to cooperate with GABPα/β to activate the CD18 (β2-leukocyte integrin) promoter (Rosmarin et al., 1998). Similar cooperation may be operative for utrophin regulation in muscle as well, because of the coexistence of SP1 and ets-binding sites in the utrophin promoter. Additionally, the utrophin promoter may be subject to transcriptional down-regulation by repressors recognizing the ets site such as ETS2 repressor factor (ERF) or ERF-like molecules (Sgouras et al., 1995). Thus, active repression may be a mechanism involved in the sharp reduction of utrophin that occurs during the perinatal period and leads to the relatively low levels (typically 0.01% of message) of utrophin encountered in adult skeletal muscle (Khurana et al., 1990). Thus, heregulin may influence utrophin expression by changing the relative levels and activity of transcriptional repressors as well as activators such as GABPα/β complex (Figure 9).
Figure 9.
Model for utrophin up-regulation by GABPα/β in muscle cell cultures. In this schematic we propose that in unstimulated muscle cultures (and adult muscle) utrophin is transcribed at low levels, possibly because of transcriptional repression activity at the ets-binding site by repressors such as ERF or ERF-like molecules. Upon stimulation with heregulin, transcription is activated via the MAP kinase pathways by decreasing the repressor activity, as well as increasing the propensity of GABPα/β transcription factors to bind and heterodimerize, leading to an overall increase of utrophin transcription. This sequence is from human utrophin promoter and shows the relative position, content, and overlap of the N-box (turquoise box) and the site bound by the ets transcription factor complex GABPα/β (lilac box) in muscle cell cultures to activate the transcription of utrophin.
In conclusion, we believe that heregulin released from nerve terminals of motor neurons may play a role in controlling the enrichment of utrophin at the NMJ of skeletal muscle during development by transcriptional activation of the utrophin promoter via GABPα/β via activation of ERK. Additionally, our definition of molecules activating transcription of the utrophin gene in this study may provide pharmacological means to achieve utrophin up-regulation in skeletal muscle of DMD patients in the future. However, we think it is important to test these hypotheses in vivo before ascribing these roles to the GABPα/β complex of transcription factors.
ACKNOWLEDGMENTS
We thank Lisbeth Sørensen, Birte Kofoed, Lene Jensen, and Linda Whittaker for superb technical assistance, Drs. Jon Tinsley and Kay Davies for kind gift of the pPUBF plasmid, and Dr. Emanuela Gussoni and Morten Frödin for suggestions and insightful comments. This work was supported in part by National Institutes of Health National Institute of Neurological Diseases and Stroke grant KO8 NS-01858 (to T.S.K.) and National Institute of Diabetes and Digestive and Kidney Diseases grant R29 DK-44728 (to A.G.R.), American Cancer Society grant DHP-11 (to A.G.R.), and grants from the Herbert W. Savit ’49 Fund of Brown University (to A.G.R.), the Lundbeck Foundation, Statens Sundhedsvidenskabelige Forskningsråd, Novo Nordisk, A.P. Møller, and Kong Christian den X Fonds, Denmark (to T.S.K.), The Duchenne Parent Project, The Netherlands (to T.S.K.), and the Copenhagen County Health Services (to S.G.). S.D. is a doctoral fellow of The Howard Hughes Medical Institute.
REFERENCES
- Ausubel F, et al. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- Brown RH. Dystrophin-associated proteins and muscular dystrophies. Annu Rev Med. 1997;48:457–466. doi: 10.1146/annurev.med.48.1.457. [DOI] [PubMed] [Google Scholar]
- Brown TA, McKnight SL. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes & Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- Campbell KP. Three muscular dystrophies: loss of cytoskeletal extracellular matrix linkage. Cell. 1996;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- Campbell KP, Crosbie RH. Muscular dystrophy. Utrophin to the rescue. Nature. 1996;384:308–309. doi: 10.1038/384308a0. [DOI] [PubMed] [Google Scholar]
- Carraway K, Burden SJ. Neuregulins and their receptors. Curr Opin Neurobiol. 1995;5:606–612. doi: 10.1016/0959-4388(95)80065-4. [DOI] [PubMed] [Google Scholar]
- de la Brousse F, Birkenmeier EH, King DS, Rowe LB, McKnight SL. Molecular and genetic characterization of GABP beta. Genes & Dev. 1994;8:1853–1865. doi: 10.1101/gad.8.15.1853. [DOI] [PubMed] [Google Scholar]
- Dennis CL, Tinsley JM, Deconinck AE, Davies KE. Molecular and functional analysis of the utrophin promoter. Nucleic Acids Res. 1996;24:1646–1652. doi: 10.1093/nar/24.9.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclert A, Changeux JP. Acetylcholine receptor gene expression at the developing neuromuscular junction. Physiol Rev. 1995;75:339–368. doi: 10.1152/physrev.1995.75.2.339. [DOI] [PubMed] [Google Scholar]
- Fischbach GD, Rosen KM. ARIA—a neuromuscular junction neuregulin. Annu Rev Neurosci. 1997;20:429–458. doi: 10.1146/annurev.neuro.20.1.429. [DOI] [PubMed] [Google Scholar]
- Glass DJ, et al. Agrin acts via a MuSK receptor complex. Cell. 1996;85:513–523. doi: 10.1016/s0092-8674(00)81252-0. [DOI] [PubMed] [Google Scholar]
- Gramolini AO, et al. Muscle and neural isoforms of agrin increase utrophin expression in cultured myotubes via a transcriptional regulatory mechanism. J Biol Chem. 1998;273:736–743. doi: 10.1074/jbc.273.2.736. [DOI] [PubMed] [Google Scholar]
- Gramolini AO, Dennis CL, Tinsley JM, Robertson GS, Cartaud J, Davies KE, Jasmin BJ. Local transcriptional control of utrophin at the neuromuscular synapse. J Biol Chem. 1997;272:8117–8120. doi: 10.1074/jbc.272.13.8117. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Karpati G. Utrophin muscles in on the action. Nat Med. 1997;3:22–23. doi: 10.1038/nm0197-22. [DOI] [PubMed] [Google Scholar]
- Khurana TS, Engle EC, Bennett RR, Silverman GA, Selig S, Bruns GA, Kunkel LM. (CA) repeat polymorphism in the chromosome 18 encoded dystrophin-like protein. Hum Mol Gen. 1994;3:841–841. doi: 10.1093/hmg/3.5.841-a. [DOI] [PubMed] [Google Scholar]
- Khurana TS, Hoffman EP, Kunkel LM. Identification of a chromosome 6-encoded dystrophin-related protein. J Biol Chem. 1990;265:16717–16720. [PubMed] [Google Scholar]
- Khurana TS, Kunkel LM, Frederickson AD, Carbonetto S, Watkins SC. Interaction of chromosome-6-encoded dystrophin related protein with the extracellular matrix. J Cell Sci. 1995;108:173–185. doi: 10.1242/jcs.108.1.173. [DOI] [PubMed] [Google Scholar]
- Khurana TS, Watkins SC, Chafey P, Chelly J, Tome FM, Fardeau M, Kaplan JC, Kunkel LM. Immunolocalization and developmental expression of dystrophin related protein in skeletal muscle. Neuromuscul Disord. 1991;1:185–194. doi: 10.1016/0960-8966(91)90023-l. [DOI] [PubMed] [Google Scholar]
- Khurana TS, Watkins SC, Kunkel LM. The subcellular distribution of chromosome 6-encoded dystrophin-related protein in the brain. J Cell Biol. 1992;119:357–366. doi: 10.1083/jcb.119.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S, Schaeffer L, Changeux JP. Identification of a DNA element determining synaptic expression of the mouse acetylcholine receptor delta-subunit gene. Proc Natl Acad Sci USA. 1995;92:10624–10628. doi: 10.1073/pnas.92.23.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke G. Neuregulins in development. Mol Cell Neurosci. 1996;7:247–262. doi: 10.1006/mcne.1996.0019. [DOI] [PubMed] [Google Scholar]
- Love DR, et al. An autosomal transcript in skeletal muscle with homology to dystrophin. Nature. 1989;339:55–58. doi: 10.1038/339055a0. [DOI] [PubMed] [Google Scholar]
- Love DR, et al. Tissue distribution of the dystrophin-related gene product and expression in the mdx and dy mouse. Proc Natl Acad Sci USA. 1991;88:3243–3247. doi: 10.1073/pnas.88.8.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- Nguyen TM, Ellis JM, Love DR, Davies KE, Gatter KC, Dickson G, Morris GE. Localization of the DMDL gene-encoded dystrophin-related protein using a panel of nineteen monoclonal antibodies: presence at neuromuscular junctions, in the sarcolemma of dystrophic skeletal muscle, in vascular and other smooth muscles, and in proliferating brain cell lines. J Cell Biol. 1991;115:1695–1700. doi: 10.1083/jcb.115.6.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuchprayoon I, Simkevich CP, Luo M, Friedman AD, Rosmarin AG. GABP cooperates with c-Myb and C/EBP to activate the neutrophil elastase promoter. Blood. 1997;89:4546–4554. [PubMed] [Google Scholar]
- Ohlendieck K, Ervasti JM, Matsumura K, Kahl SD, Leveille CJ, Campbell KP. Dystrophin-related protein is localized to neuromuscular junctions of adult skeletal muscle. Neuron. 1991;7:499–508. doi: 10.1016/0896-6273(91)90301-f. [DOI] [PubMed] [Google Scholar]
- Ouyang L, Jacob KJ, Stanley FM. GABP mediates insulin-increased prolactin gene transcription. J Biol Chem. 1996;271:10425–10428. doi: 10.1074/jbc.271.18.10425. [DOI] [PubMed] [Google Scholar]
- Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol. 1994;125:1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RG, Freeman TC, Kendall E, Vetrie DL, Dixon AK, Shaw SC, Bone Q, Bobrow M. Characterization of DRP2, a novel human dystrophin homologue. Nat Genet. 1996;13:223–226. doi: 10.1038/ng0696-223. [DOI] [PubMed] [Google Scholar]
- Rosmarin AG, Caprio DG, Kirsch DG, Handa H, Simkevich CP. GABP and PU.1 compete for binding, yet cooperate to increase CD18(β2 Integrin) transcription. J Biol Chem. 1995;270:23627–23633. doi: 10.1074/jbc.270.40.23627. [DOI] [PubMed] [Google Scholar]
- Rosmarin AG, Luo M, Caprio DG, Shang J, Simkevich CP. Sp1 cooperates with the ets transcription factor, GABP, to activate CD18(β2 integrin) promoter. J Biol Chem. 1998;273:13097–13103. doi: 10.1074/jbc.273.21.13097. [DOI] [PubMed] [Google Scholar]
- Sapru MK, Florance SK, Kirk C, Goldman D. Identification of a neuregulin and protein-tyrosine phosphatase response element in the nicotinic acetylcholine receptor e subunit gene: regulatory role of an ets transcription factor. Proc Natl Acad Sci USA. 1998;95:1289–1294. doi: 10.1073/pnas.95.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer L, Duclert N, Dymanus MH, Changeux JP. Implication of a multisubunit Ets-related transcription factor in synaptic expression of the nicotinic acetylcholine receptor. EMBO J. 1998;17:3078–3090. doi: 10.1093/emboj/17.11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgouras DN, Athanasiou MA, Beal GJ, Fisher RJ, Blair DG, Mavrothalassitis GJ. ERF: an ets domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J. 1995;14:4781–4793. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey MG, Chu GC, Merlie JP. ARIA/HRG regulates the AChR e subunit gene expression at the neuromuscular synapse via activation of the phosphatidylinositol 3-kinase and RAS/MAPK pathway. J Cell Biol. 1996;134:465–476. doi: 10.1083/jcb.134.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley JM, et al. Primary structure of dystrophin-related protein. Nature. 1992;360:591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- Tinsley JM, Potter AC, Phelps SR, Fisher R, Trickett JI, Davies KE. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature. 1996;384:349–353. doi: 10.1038/384349a0. [DOI] [PubMed] [Google Scholar]
- Winder SJ, et al. Utrophin actin binding domain: analysis of actin binding and cellular targeting. J Cell Sci. 1995;108:63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]