Abstract
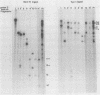
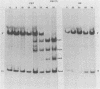
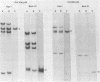
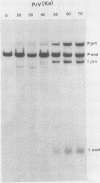
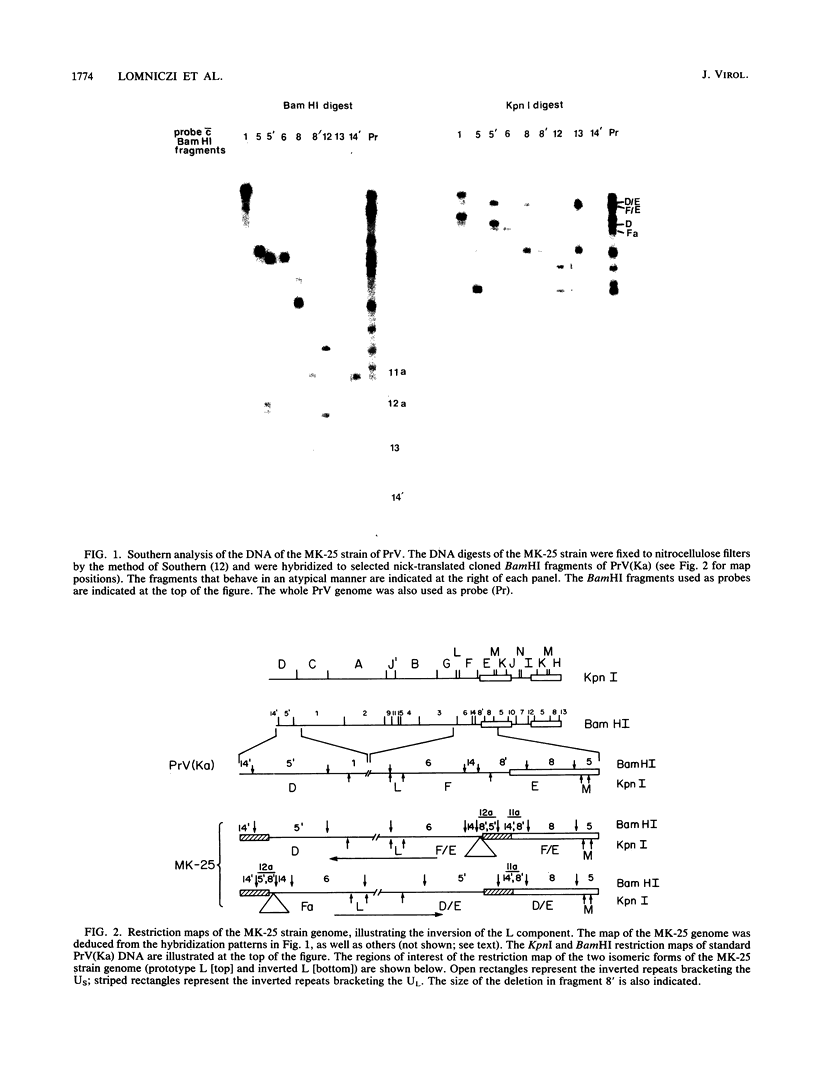
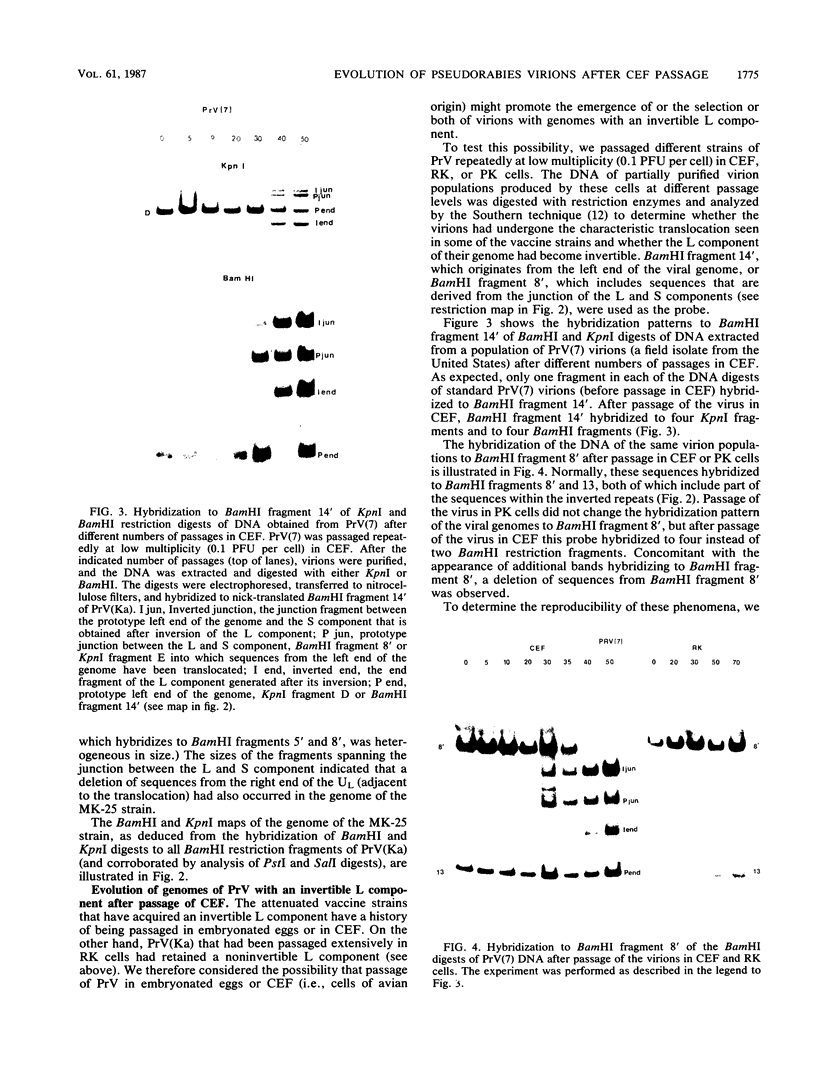
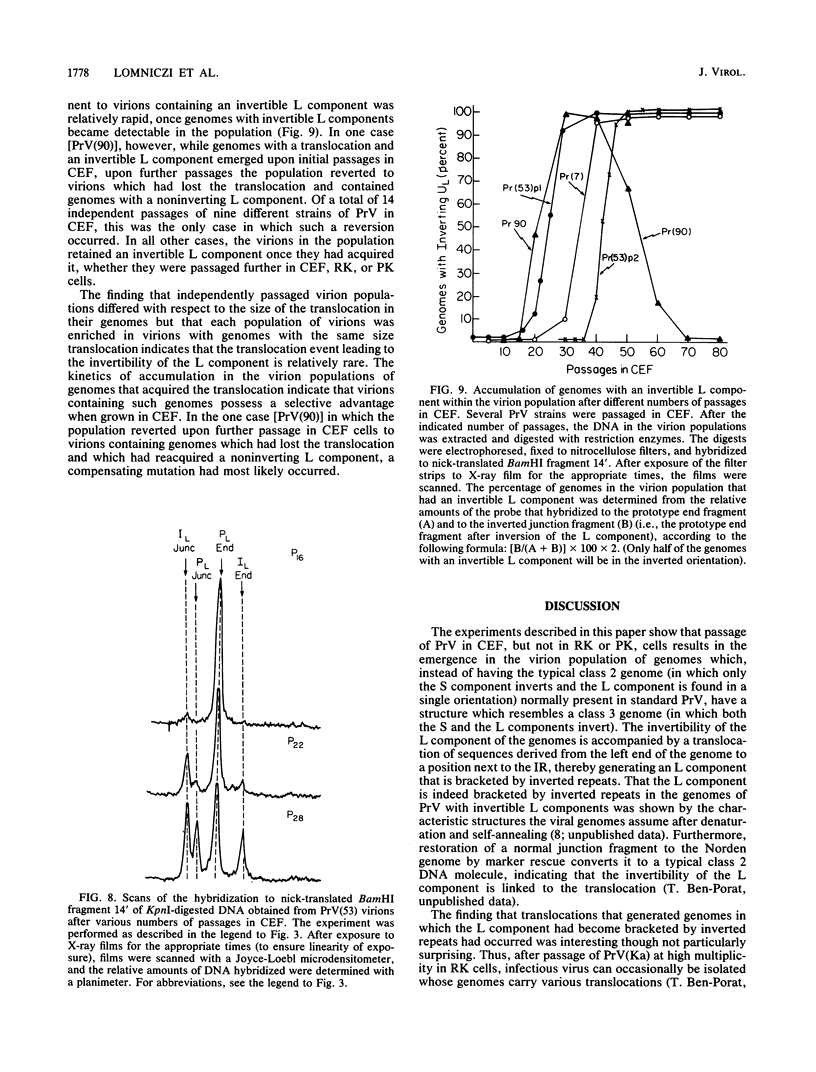
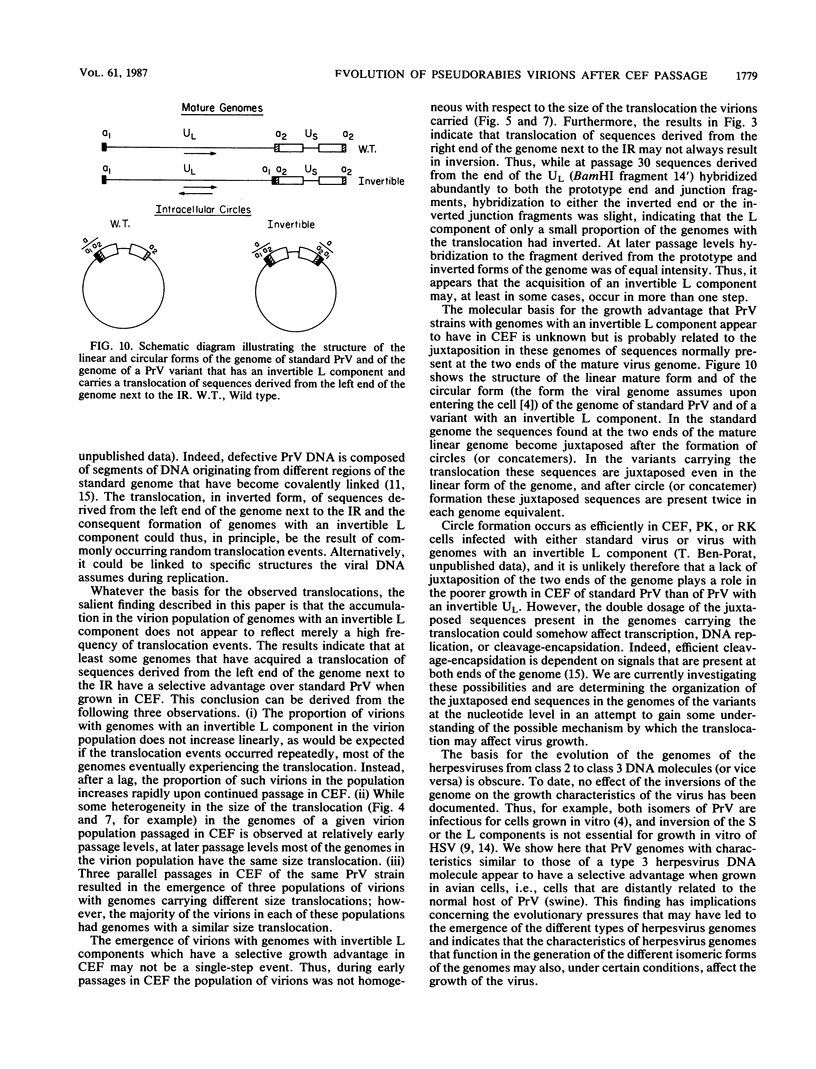
The genome of pseudorabies virus consists of two components, short (S) and long (L). Only the S component is bracketed by inverted repeats, and only the S component inverts itself relative to the L component, giving rise to two isomeric forms of the genome. An attenuated vaccine strain of pseudorabies virus (Norden), however, has a genome which is found in four isomeric forms (B. Lomniczi, M. L. Blankenship, and T. Ben-Porat, J. Virol. 49:970-979, 1984). To determine the basis for the atypical structure of the genome of the Norden strain, we examined more than 40 field isolates of pseudorabies virus; all contained genomes in which the L component was fixed in only one orientation relative to the S component. Several independently generated vaccine strains which have been passaged extensively in chicken embryos and chicken embryo fibroblast (CEF) cell cultures were also analyzed; they possessed an invertible L component. Furthermore, emergence of pseudorabies virus variants with an invertible L component was observed after passage of the virus in CEF, but not in rabbit kidney or pig kidney, cells. The invertibility of the L component was associated consistently with a translocation of sequences from the left end of the genome to a position next to the inverted repeat sequence of the S component. Three observations indicate that genomes with an invertible L component (and the translocation) have a selective growth advantage over standard pseudorabies virus when grown in CEF. The proportion of virions with such genomes does not increase linearly as would be expected if the translocation events occurred repeatedly, most genomes eventually experiencing the translocation. Instead, after a lag, the proportion of such virions in the population increases relatively rapidly. The genome structures that are generated upon independent passage in CEF of each virion population were relatively homogeneous. Some heterogeneity was observed at relatively early stages of the emergence of the genomes carrying the translocation; at later stages, virions with genomes with a specific size translocation predominated in the virus population. Parallel passages in CEF of the same pseudorabies virus strain resulted in the emergence of populations of virions with genomes with different size translocations. However, in each of the passaged populations of virions the majority of virions had genomes with the same size translocation. The most likely interpretation of these results is that virions with genomes carrying the translocations that emerge upon passage of the virus in CEF have a selective advantage when grown in these cells.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Porat T., Demarchi J. M., Kaplan A. S. Characterization of defective interfering viral particles present in a population of pseudorabies virions. Virology. 1974 Sep;61(1):29–37. doi: 10.1016/0042-6822(74)90239-6. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Rixon F. J., Blankenship M. L. Analysis of the structure of the genome of pseudorabies virus. Virology. 1979 Jun;95(2):285–294. doi: 10.1016/0042-6822(79)90484-7. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Rixon F. J. Replication of herpesvirus DNA. IV: analysis of concatemers. Virology. 1979 Apr 15;94(1):61–70. doi: 10.1016/0042-6822(79)90438-0. [DOI] [PubMed] [Google Scholar]
- EAGLE H. Amino acid metabolism in mammalian cell cultures. Science. 1959 Aug 21;130(3373):432–437. doi: 10.1126/science.130.3373.432. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Watson D. H. Unity and diversity in the herpesviruses. J Gen Virol. 1977 Oct;37(1):15–37. doi: 10.1099/0022-1317-37-1-15. [DOI] [PubMed] [Google Scholar]
- Ladin B. F., Ihara S., Hampl H., Ben-Porat T. Pathway of assembly of herpesvirus capsids: an analysis using DNA+ temperature-sensitive mutants of pseudorabies virus. Virology. 1982 Jan 30;116(2):544–561. doi: 10.1016/0042-6822(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Lomniczi B., Blankenship M. L., Ben-Porat T. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the genomes. J Virol. 1984 Mar;49(3):970–979. doi: 10.1128/jvi.49.3.970-979.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poffenberger K. L., Tabares E., Roizman B. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proc Natl Acad Sci U S A. 1983 May;80(9):2690–2694. doi: 10.1073/pnas.80.9.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rixon F. J., Ben-Porat T. Structural evolution of the DNA of pseudorabies-defective viral particles. Virology. 1979 Aug;97(1):151–163. doi: 10.1016/0042-6822(79)90381-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tatarov G. Apathogener Mutant des Aujeszky-Virus, induziert von 5-Jodo-2-Deoxyuridin (JUDR) Zentralbl Veterinarmed B. 1968 Oct;15(8):847–853. [PubMed] [Google Scholar]
- Wilkie N. M., Davison A., Chartrand P., Stow N. D., Preston V. G., Timbury M. C. Recombination in herpes simplex virus: mapping of mutations and analysis of intertypic recombinants. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):827–840. doi: 10.1101/sqb.1979.043.01.089. [DOI] [PubMed] [Google Scholar]
- Wu C. A., Harper L., Ben-Porat T. cis Functions involved in replication and cleavage-encapsidation of pseudorabies virus. J Virol. 1986 Aug;59(2):318–327. doi: 10.1128/jvi.59.2.318-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]