Abstract
In Caenorhabditis elegans, the EGF receptor (encoded by let-23) is localized to the basolateral membrane domain of the epithelial vulval precursor cells, where it acts through a conserved Ras/MAP kinase signaling pathway to induce vulval differentiation. lin-10 acts in LET-23 receptor tyrosine kinase basolateral localization, because lin-10 mutations result in mislocalization of LET-23 to the apical membrane domain and cause a signaling defective (vulvaless) phenotype. We demonstrate that the previous molecular identification of lin-10 was incorrect, and we identify a new gene corresponding to the lin-10 genetic locus. lin-10 encodes a protein with regions of similarity to mammalian X11/mint proteins, containing a phosphotyrosine-binding and two PDZ domains. A nonsense lin-10 allele that truncates both PDZ domains only partially reduces lin-10 gene activity, suggesting that these protein interaction domains are not essential for LIN-10 function in vulval induction. Immunocytochemical experiments show that LIN-10 is expressed in vulval epithelial cells and in neurons. LIN-10 is present at low levels in the cytoplasm and at the plasma membrane and at high levels at or near the Golgi. LIN-10 may function in secretion of LET-23 to the basolateral membrane domain, or it may be involved in tethering LET-23 at the basolateral plasma membrane once it is secreted.
INTRODUCTION
A key aspect of epithelial cell polarity is that the apical and basolateral membrane compartments express different classes of transmembrane proteins, allowing these domains to be structurally and functionally distinct (for reviews, see Rodriguez and Nelson, 1989; Rodriguez and Powell, 1992; Drubin and Nelson, 1996). One example of the difference between these membrane domains is that signaling from mesodermal tissues to ectodermal epithelia is inherently polarized, because ligands are presented predominantly to the basolateral face of epithelia. Signaling receptors, such as the hepatocyte growth factor receptor and the epidermal growth factor receptor, can be asymmetrically expressed on the basolateral membrane domains of epithelia (Maratos et al., 1987; Mullin and McGinn, 1988; Crepaldi et al., 1994) and are thus responsive to ligands present in the basal extracellular space.
A key issue is to understand the mechanism of asymmetric expression of transmembrane proteins on the surface of polarized cells. cis-acting amino acid sequences have been identified that are required for basolateral sorting and retention of the EGF receptor and other basolateral transmembrane proteins (Mostov et al., 1986; Casanova et al., 1991; Hunziker et al., 1991; Matter et al., 1992; Hobert and Carlin, 1995; Hobert et al., 1997). However, very little is known about the trans-acting proteins (or protein complexes) that bind to cis-acting sorting sequences and mediate basolateral localization.
We are studying basolateral localization of the EGF receptor tyrosine kinase (encoded by the gene let-23) in the Caenorhabditis elegans vulval precursor cells. The six vulval precursor cells (P3.p to P8.p) are epithelial cells that lie in an anterior–posterior row along the ventral midline of hermaphrodite worms. The anchor cell is located on the basal side of the vulval precursor cell P6.p, and it secretes an EGF-like signal (encoded by lin-3) that activates LET-23 receptor tyrosine kinase (RTK) in P6.p (for review, see Kornfeld, 1997). Activation of LET-23 directly induces P6.p to divide in a stereotypic pattern, giving rise to the inner portion of the adult vulval structure (termed the 1° cell fate), and also causes P6.p to secrete a lateral signal (Aroian et al., 1990). The lateral signal and possibly the anchor cell signal acting at a distance cause adjacent vulval precursor cells (P5.p and P7.p) to divide in a different pattern, giving rise to the outer portion of the vulva (2° cell fate) (Katz et al., 1995; Koga and Ohshima, 1995; Simske and Kim, 1995; for review, see Simske and Kim, 1998). The remaining vulval precursor cells (P3.p, P4.p, and P8.p) are not induced by either the anchor cell signal or the lateral signal and adopt a nonvulval epithelial cell fate (3° cell fate).
lin-2 and lin-7 are required for basolateral localization of LET-23 RTK in vulval precursor cells (Simske et al., 1996). In lin-2 and lin-7 mutants, LET-23 is mislocalized from the basolateral membrane domain to the apical domain of vulval precursor cells, and this mislocalization causes loss of LET-23 signaling and a vulvaless phenotype (Simske et al., 1996). LIN-7 binds to the cytoplasmic tail of LET-23, indicating that it plays a direct role in LET-23 localization (Simske et al., 1996; Kaech et al., 1998).
There are at least two important differences between the cellular phenotypes caused by mutations in genes that act in LET-23 receptor localization (lin-2 and lin-7) and genes that act directly in the LET-23 RTK/LET-60 Ras/MPK-1 MAP kinase signaling pathway. Loss-of-function mutations in lin-2 or lin-7 result in an incompletely penetrant (95%) vulvaless phenotype and do not appear to affect other cells that respond to LET-23 signaling (Ferguson and Horvitz, 1985). In contrast, loss-of-function mutations in let-23 result in a completely penetrant vulvaless phenotype and cause defects in at least five cell types other than the vulval precursor cells (Kornfeld, 1997).
lin-10 null mutations cause a specific, incompletely penetrant vulvaless phenotype, similar to lin-2 and lin-7. Because lin-2 and lin-7 function in basolateral localization of LET-23 RTK, this genetic result suggests that lin-10 may also function in receptor localization in the vulval precursor cells. In this paper, we investigate the role of lin-10 in LET-23 RTK localization.
MATERIALS AND METHODS
Genetics and Strains
Standard methods were used to handle and maintain C. elegans (variety Bristol, strain N2) at 20°C (Wood, 1988). Alleles used in this work were described (Ferguson and Horvitz, 1985; Kim and Horvitz, 1990), except lin-10(sy217) (Sternberg, personal communication) and let-23(sy5) (Aroian et al., 1990). Homozygous let-23(sy5) rol-6(e187); lin-1(sy254) animals are sterile and non-Let; these animals were obtained for anti-LET-23 staining by selecting F1 Rol progeny from let-23(sy5) rol-6(e187)/dpy-10(e128); lin-1(sy254) (Kaech, personal communication).
Genetic complementation of lin-10 by nDf23 was determined by scoring egg-laying ability of progeny from a cross between lin-10(n1508); him-5(e1490) males and nDf23/unc-13(e1091) lin-11(n566) hermaphrodites. Approximately half of the progeny were males (indicating that most or all of the F1 were cross-progeny), and all of the hermaphrodites were wild-type for egg laying and vulval morphology (n > 400). To verify that lin-10(n1508)/nDf23 animals were present in the F1 progeny, 20 non-Egl F1 progeny were isolated, allowed to self-fertilize, and scored for phenotypic segregation of F2 progeny. Eight F1 produced F2 progeny that were all non-Unc, indicating that these F1 were of genotype lin-10(n1508)/nDf23; him-5(e1490)/+. The remaining 12 F1 produced F2 progeny that were approximately one-fourth Unc, indicating that these F1 were of genotype lin-10(n1508)/unc-13(e1091) lin-11(n566); him-5(e1490)/+ or were self-progeny of genotype nDf23/unc-13(e1091) lin-11(n566).
Homozygous nDf23 embryos were picked as self-progeny of nDf23/unc-13(e1091) lin-11(n566) hermaphrodites that fail to develop into L1 larva within 24 h of egg laying. PCR was performed on single unhatched embryos as described (Williams et al., 1992) using listed primers (Table 1).
Table 1.
Oligonucleotide primers used
| nDf23 mappinga | ||
| unc-15 | TTTCTCAACTCGAGAAGGCC | TCGGCAAGAGCCTCCTTGGC |
| ceh-8 | TGACTTTTCTGTAAATGTCC | GCTGGAGGAAGCATTGGCGG |
| T01G9.2 | GCAATACGAGATTCGTACGG | GGAAGAGATTTAAATGCGGG |
| mei-1 | TTCTGGTACAAATGGACGGG | TTAATTTCATCGGATTTCGG |
| lin-10 cDNA constructionb | ||
| GCAGGTCGACATGTCATCTGAAGCAGTAGCACAAGCC | ||
| lin-10 SSCP analysis and sequencing | ||
| Exon 1 | ATGTCCTTTTCCCGCTTCCTCTC | CTGATTTGATTTTGTCTTGCTTTTTC |
| Exon 2 | CCGCCGCCTGACCACAAAAATAG | TTATAAAACAAACCTGAGAAGAAG |
| Exon 3 | ATGCTTATTGGATGTTTGATGATGC | CGAACCACATTTATTTATTTTAGAG |
| Exon 4 | GGCAGTGTTGTGTCCTCTAATGTGTC | CTCTTCTTCTCACCAAAATGTTCTC |
| Exon 5 | GGACGGAACTTAGAGACCACACG | CGAAATAAAACAACAGCCAAAAATG |
| Exons 6, 7 | GGATTCTATTGATGTTGTTGTGAGC | GAAGAGATTATGTTGATGTGGTTGC |
| Exons 7, 8 | GCAACCACATCAACATAATCTCTTC | GTTCCATTTTCCTTGTTCTCTTCAG |
| Exon 8 | CTGAAGAGAACAAGGAAAATGGAAC | TTTAGCTTTGAAACACTGAAAACAT |
| Exon 9 | CACTTCAATTTCCTGCTCATTTTCC | TTCACAACTTACTTTCTTTGCTGTT |
| Exons 10, 11 | CATTCTTTTCAACCCACTTCTCC | TAAAAACGAACCCATCAGAACAAG |
| Exons 11, 12 | GTTATATTGCGGACATTGGAGATTTAG | GTTTCTAGTTAGTTAACATGTCAATTTCC |
| Exon 13 | CTTCAATAGGTACTTCAGCATCAAA | GTTTCTAGTTAGTTAACATGTCAATTTCC |
| Exons 14, 15 | CTCCCATCAGAACACTATAATTTAAT | TGAAATGAAATGGATGTGAAGAGG |
| Inverse PCR | ||
| n299 5′ | GGCTGTGGCTTGTGCTACTGCTTC | GGCAGTGTTGTGTCCTCTAATGTGTC |
| n299 3′ | CTATGAGATTGGGAGAAGTGGG | CTCCCATCAGAACACTATAATTTAAT |
| nl299 | TTTAGCTTTGAAACACTGAAAACAT | CGGGTGCATCAACAAAAGGTG |
Primer pair used for the C09H6.2 locus is the same as that used in lin-10 SSCP analysis and sequencing (exon 4).
Underlined nucleotides were added to YK114C6 to create a full-length cDNA.
Molecular Genetics
Transgenic lines were established by injecting indicated DNA along with the cotransformation marker unc-29(+) (cosmid F35D3) into gonads of indicated strains and selecting non-Unc progeny (Mello et al., 1991). All DNA clones were injected at concentrations of 50–100 μg/ml, except Y42H1 (4 μg/ml) and Y55A5 (15 μg/ml). The YK114C6 cDNA (provided by Y. Kohara, National Institute of Genetics, Shizuoka, Japan) is a lin-10 cDNA that is nearly full length, because it contains the entire predicted coding region for C09H6.2 except nine nucleotide bases at the 5′ end (including the predicted start codon). These nine nucleotide bases were added to the 5′ end of YK114C6 by PCR using the indicated primer (Table 1) and the M13 reverse primer, and the resulting full-length cDNA was subcloned into the lin-31 expression vector pB255 (Tan, personal communication).
The lin-10(n299) polymorphism was characterized using inverse PCR as follows. PCR and Southern blotting experiments showed that lin-10(n299) contains a DNA rearrangement, and that the 5′ break point of this rearrangement was between a primer binding site (at 1.8 kb in Figure 3C) and an EcoRI site (at 5.9 kb in Figure 3C). DNA upstream (relative to the direction of lin-10 transcription) of the primer binding site is present in lin-10(n299) DNA, and DNA downstream of the primer binding site (including the EcoRI site) is absent. We used inverse PCR to amplify a portion of the altered EcoRI fragment in lin-10(n299) DNA. Genomic DNA from lin-10(n299) and wild-type animals was digested with EcoRI and then circularized by self-ligation. Because the EcoRI fragment is circular, PCR primer pairs designed against the lin-10 DNA sequence (Table 1) could be used to amplify the novel sequence present in the altered EcoRI fragment from lin-10(n299). Products of inverse PCR were different in size using wild-type and lin-10(n299) DNA, indicating that the amplified DNA contained the lin-10(n299) rearrangement break point. Sequencing of this PCR product revealed that lin-10(n299) results in the fusion of lin-10 DNA from the intron 3′ to exon 3 from the central region of chromosome I with DNA corresponding to cosmid DY3 located on the right arm of chromosome I. A similar inverse PCR experiment was used to analyze the 3′ rearrangement break point in lin-10(n299), showing that lin-10 DNA 5′ to exon 10 was fused to DNA corresponding to cosmid YK858 located on the right arm of chromosome I. The simplest explanation for these results is that the genomic region corresponding to exons 4–9 of lin-10 is deleted in lin-10(n299), and that there is an insertion of ∼300 kb of DNA (spanning cosmids YK858–DY3) in an inverted orientation.
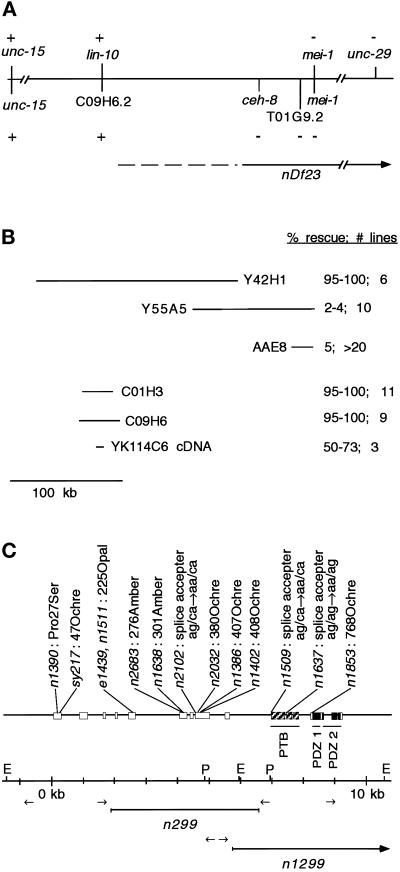
Figure 3.
Molecular identification of lin-10. (A) Genetic and physical maps of the lin-10 region. Genetic and physical markers are shown above and below the line, respectively. (+ and −) Presence and absence of genetic markers (shown by complementation with nDf23) or physical markers (shown using PCR experiments in nDf23 homozygous embryos), respectively. A continuous line, no line, or a dashed line next to nDf23 indicates the region known to be deleted by nDf23, the region known to be unaffected by nDf23, and the region that may or may not be deleted by nDf23, respectively. (B) Genomic and cDNA clones used in transformation rescue experiments. Results of transformation rescue experiments using lin-10(n1390) are shown. Only 0.3% of lin-10(n1390) animals lay eggs.Shown are the percent of transgenic animals that lay eggs and the number of independent transgenic lines. The lin-10 cDNA (YK114C6) was expressed from the vulval-specific lin-31 promoter. Genomic clones Y55A5 and AAE8 also cause a protruding vulval phenotype. Rescue data for cosmid AAE8 was reported by Kim and Horvitz (1990). (C) lin-10 genomic structure was determined by aligning YK114C6 cDNA sequence with the genomic sequence. Exons are indicated by boxes. Regions predicted to encode PTB and PDZ domains are indicated. Positions and molecular lesions associated with lin-10 mutant alleles are indicated. Restriction sites EcoRI (E) and PstI (P) and primer pairs (arrows) used for inverse PCR experiments are indicated.
PCR and Southern blotting experiments showed that lin-10(n1299) contained a deletion or rearrangement that did not include a primer binding site (at 5.5 kb in Figure 3C) or sequences upstream of this site but did include a PstI site (at 6.9 kb in Figure 3C) and sequences downstream of this site. Similar to lin-10(n299), we used inverse PCR to amplify the rearranged PstI fragment from lin-10(n1299) animals. DNA sequencing revealed that lin-10(n1299) is an 11.6-kb deletion that removes the entire 3′ end of lin-10 including exons 10–15 and also deletes an adjacent predicted gene (C09H6.1).
Single-strand conformation polymorphism (SSCP) analysis was performed by PCR amplification of lin-10 mutant and wild-type genomic DNA in the presence of 1 μCi of [α-33P]dATP using the indicated primers (Table 1). Radiolabeled PCR products were separated using an MDE gel (FMC Bioproducts, Rockland, ME; 0.4× concentrate plus 10% glycerol) and analyzed by autoradiography. Point mutations were determined by sequencing both DNA strands using the indicated primers (Table 1) or by sequencing one DNA strand in at least two independent experiments.
Antibodies and Western Blotting
Recombinant LET-23 (residues 1127–1323) and LIN-10 (residues 1–721) were expressed in bacteria as 6-His fusion proteins, and purified on an Ni2+ column. These recombinant proteins were used to raise rabbit polyclonal antibodies against LET-23 and LIN-10. These rabbit polyclonal antibodies were affinity purified using 6-His LIN-10 or 6-His LET-23 recombinant proteins on a Ni2+ column. For Western blot analysis, 20 μl of mixed stage worms were heated in loading buffer at 95°C for 10 min; total protein was separated using an 8% SDS-PAGE gel and detected using anti-LIN-10 antisera (1:1000) and chemiluminescence (SuperSignal; Pierce, Rockford, IL).
Fluorescence Microscopy and Imaging
LET-23 staining was performed essentially as described (Finney and Ruvkun, 1990), except Triton X-100 was used at 0.1%. Anti-LET-23 antibody was used at 1:1000, and MH27 mAb (a gift from Bob Waterston, Washington University, St. Louis, MO) at 1:1500. For LIN-10 staining, N2 worms were first fixed in 33% saturated picric acid, 11% formalin, 2.2% acetic acid, 46% methanol for 1 h at 23° (including three quick freeze–thaw cycles after 30 min), then washed extensively in 20 mM H3BO3, 10 mM NaOH, 2% β-mercaptoethanol, 0.1% Triton X-100, and finally stained as described (Finney and Ruvkun, 1990). Anti-LIN-10 antibody was used at 1:200. Sialyltransferase-green fluorescent protein (ST-GFP; provided by Ian Trowbridge, Salk Institute for Biological Studies, La Jolla, CA) was subcloned into the heat shock expression vector pPD49.83 (Mello and Fire, 1995), and transgenic worms were generated as above. ST-GFP transgenic worms were heat shocked at 30°C for 30 min, allowed to recover at 20°C for 4 h, fixed (as described for LIN-10), and stained with anti-LIN-10 antibodies (1:400) and anti-GFP mAb (1:400; 3E6; Quantum Biotechnology, Montreal, Canada).
Immunofluorescence micrographs were obtained using a wide-field fluorescence microscope. Where indicated, three-dimensional images were recorded with a charge-coupled device camera using a DeltaVision wide-field fluorescence microscope (Applied Precision, Issaquah, WA). Optical sections were collected by moving the samples through the focal plane of the lens at 0.15- or 0.20-μm increments, and data stacks were deconvolved with an empirically measured optical transfer function using a constrained iterative method (Agard et al., 1989). Image display and analysis were performed using the Image Visualization Environment software package (Chen et al., 1996).
RESULTS
Localization of LET-23 RTK in Vulval Epithelial Cells
Previous studies using a LET-23:GFP reporter protein indicated that LET-23 RTK is concentrated at the basal side of the cell junctions in wild-type vulval precursor cells (Simske et al., 1996). In this report, we have confirmed and extended these studies by raising antibodies against LET-23 and using them in immunocytochemical experiments to examine subcellular localization of endogenous LET-23 RTK in the vulval precursor cells. Anti-LET-23 antibodies stain LET-23 specifically, because only background staining is observed in a let-23 null mutant (Figure 1A).
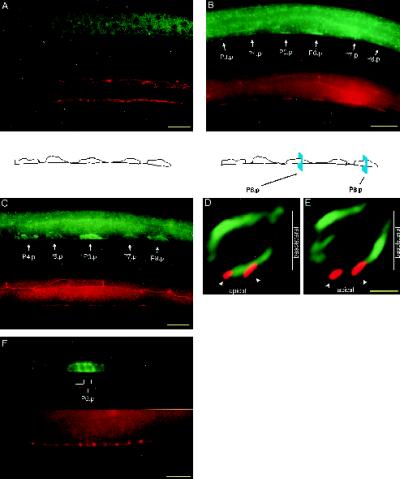
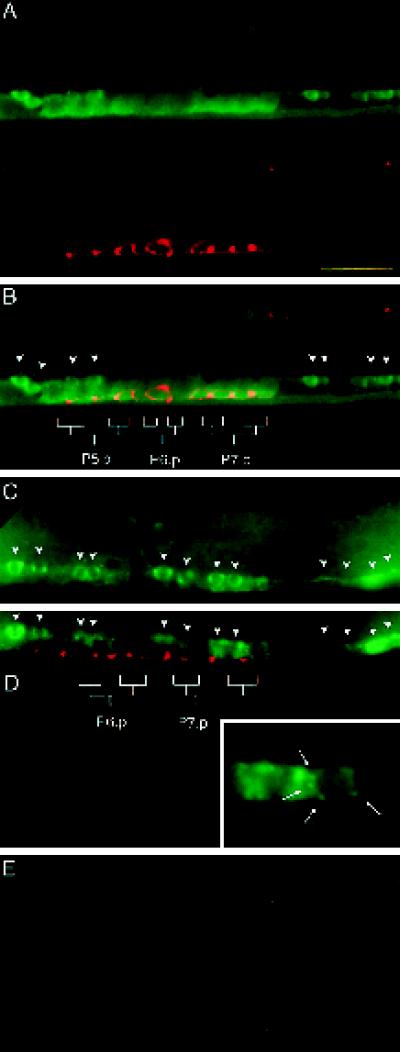
Figure 1.
Localization of LET-23 RTK in vulval epithelial cells. Vulval precursor cells are stained with anti-LET-23 antibodies (green) and a cell junction mAb marker (MH27, red). (A) let-23(sy5) rol-6(e187); lin-1(sy254) mutant shows no specific LET-23 staining. (B) Lateral view of an early L2 wild-type hermaphrodite. Strong LET-23 staining is observed in the apical membrane domain, which appears as a thin line along the ventral surface. Weak LET-23 staining is observed in the basolateral membrane domain, located dorsal and anterior to the cell junctions. At this stage, the cell junctions are small, separate rings that appear as lines when seen on edge. (C) Lateral view of a late L2–early L3 wild-type hermaphrodite. LET-23 staining appears in the basolateral membrane domains of the vulval precursor cells. Cell junctions have elongated along the anterior–posterior axis, such that the junction from one vulval precursor cell connects with the junctions from adjacent vulval precursor cells. (D and E) Optical cross-sections of vulval precursor cells P6.p (D) and P8.p (E) from a late L2–early L3 wild-type hermaphrodite. The basolateral membrane domain is above the cell junctions, and the apical membrane domain is between or slightly below the cell junctions. LET-23 appears predominantly basolateral in P8.p and both basolateral and apical in P6.p. Outlines of the vulval precursor cells depicted in C and of the cross-sectional areas depicted in D and E are shown. Cross-sections were generated by rotating three-dimensional images 90° about the dorsal–ventral axis and projecting 1 μm along the anterior–posterior axis. (F) Lateral view of an L3 wild-type hermaphrodite, showing granddaughter cells of P6.p. LET-23 staining appears in both the apical and basolateral membrane domains. Dorsal is up and anterior is left, except for D and E. Bars, 10 μm (A-C and F); 2 μm (D and E).
In wild-type larvae, LET-23 RTK is first expressed in the vulval precursor cells in the early L2 stage and appears on both the basolateral and apical membrane domains, with higher levels in the apical membrane domain (Figure 1B). At the late L2 to early L3 stage (which cell ablation studies have indicated is the time that the 1° cell fate becomes determined [Kimble, 1981]), the vulval precursor cells are small (∼5 μm in cross-section), and the apical membrane domain is small relative to the basolateral membrane domain. To obtain high-resolution images of the distribution of LET-23 on the surface of the vulval precursor cells, we obtained three-dimensional images by deconvolving serial sections taken with a DeltaVision wide-field microscope (see MATERIALS AND METHODS). Cross-sectional images of the vulval precursor cells were generated by rotating computer-generated images using three-dimensional imaging software, so that the basolateral membrane domain, the cell junctions (stained with mAb MH27), and the apical membrane domain could be clearly resolved (Figure 1, D and E). LET-23 staining is observed throughout the basolateral membrane of all six vulval precursor cells (Figure 1, C–E). In five of the vulval precursor cells (P3.p, P4.p, P5.p, P7.p, and P8.p), little or no LET-23 is present in the apical membrane domain. In the remaining vulval precursor cell (P6.p), LET-23 is present in both the basolateral and apical membrane domains (Figure 1, D and E). As P6.p divides in early L3, LET-23 staining continues to be present in both the basolateral and apical membrane domains (Figure 1F).
lin-10 Is Required for Basolateral Localization of LET-23 RTK in the Vulval Precursor Cells
Genetic results suggest that lin-10 may be involved in a common cellular function with lin-2 and lin-7, indicating a possible role for lin-10 in basolateral localization of LET-23. To directly test this possibility, we examined LET-23 localization in lin-10 mutants. In the vulval precursor cells of lin-10 mutants, strong LET-23 staining was observed in the apical membrane domains, and no LET-23 staining was observed in the basolateral membrane domains (Figure 2). We observed LET-23 staining exclusively on the apical membrane domains of the vulval precursor cells and their descendants, from the onset of LET-23 expression in early L2 through the beginning of vulval cell divisions in L3. These results show that lin-10 is required for basolateral localization of LET-23 RTK in the vulval precursor cells.
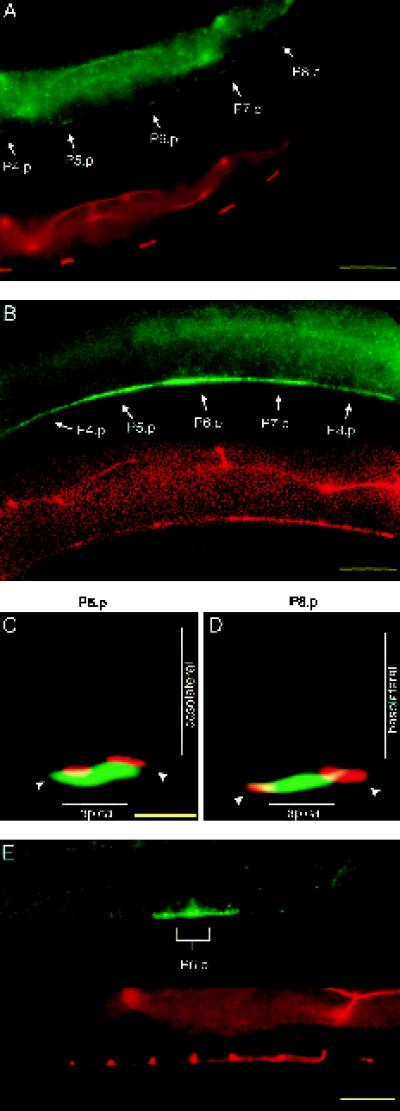
Figure 2.
Basolateral localization of LET-23 requires lin-10. Immunofluorescent micrographs of vulval precursor cells stained with anti-LET-23 antibodies (green) and a cell junction mAb marker (MH27, red) in lin-10(sy217) (A–D) and lin-10(n1509) (E) mutants are shown. (A) Lateral view of an early L2 hermaphrodite. (B) Lateral view of a late L2–early L3 hermaphrodite. Optical cross-sections of P6.p (C) and P8.p (D) in a late L2–early L3 hermaphrodite were generated as in Figure 1, D and E. (E) Lateral view of the two daughter cells from P6.p. Bars, 10 μm (A, B, and E); 2 μm (C and D).
The loss of basolateral expression of LET-23 in lin-10 mutants is not the result of a general loss of cell polarity. First, the vulval precursor cells secrete components of the basal lamina from their basal sides and components of the cuticle from their apical sides. The general morphology of both the basal lamina and the cuticle surrounding the vulval precursor cells appears normal in lin-10 mutants, as observed using Nomarski microscopy. Second, the cell junctions (comprising both the adherens and tight junctions) form the physical barrier between the apical and basolateral membrane domains of vulval precursor cells and are necessary for maintaining epithelial cell polarity. The cell junctions appear normal in lin-10 mutants, as indicated by staining with the cell junction mAb MH27 (Figure 2).
lin-10 Encodes a Protein with Regions of Similarity to Mammalian X11/mint Proteins
During mapping experiments involving a gene closely linked to lin-10 on linkage group I, we obtained data indicating that the previous molecular identification of lin-10 (Kim and Horvitz, 1990) was incorrect. nDf23 is a chromosomal deficiency that does not delete lin-10, because it genetically complements lin-10 loss-of-function mutations (Ferguson and Horvitz, 1985). We verified this result by mating lin-10(n1508); him-5(e1490) males with nDf23/unc-13(e1091) lin-11(n566) hermaphrodites and found that all of the F1 cross-progeny (n > 400, half of which should be of genotype lin-10(n1508)/nDf23) could lay eggs and had wild-type vulval morphology. Next, we mapped the end of the nDf23 deficiency by determining which physical map loci are deleted in nDf23 using a PCR-based strategy. First, homozygous nDf23 embryos were obtained by allowing heterozygous nDf23/unc-13(e1091) lin-11(n566) hermaphrodites to self-fertilize and then selecting dead embryos 24 h later (these progeny are presumably homozygous nDf23 mutants). Primers designed to amplify DNA from physical map loci (Table 1) were used in PCR experiments to determine whether specific physical map loci were deleted in individual homozygous nDf23 embryos. All PCR reactions included primers that amplify DNA from unc-15 (not deleted by nDf23) and mei-1 (deleted by nDf23) as internal positive and negative controls. PCR products corresponding to unc-15 and C09H6.2 were consistently amplified from nDf23 homozygotes, whereas PCR products corresponding to ceh-8, T01G9.2 (previously identified as lin-10) and mei-1 were not amplified (Figure 3A). These results indicate that nDf23 does not delete the lin-10 genetic locus (because it complements lin-10 mutations), but it does delete the gene previously reported to encode lin-10 (T01G9.2) and flanking genes ceh-8 and mei-1. Thus, T01G9.2 cannot correspond to the lin-10 genetic locus, as was previously reported by Kim and Horvitz (1990).
We identified an alternate genomic region, located ∼180 kb to the left of T01G9.2, that is not deleted by nDf23 and that can rescue the lin-10 vulvaless phenotype in transgene experiments. Yeast artificial chromosome Y42H1, cosmid C01H3, or cosmid C09H6 can strongly rescue the lin-10 mutant phenotype, generating rescued animals that lay eggs and have an essentially wild-type vulval morphology (Figure 3B). In contrast, lin-10 animals that are transgenic for genomic clones that contain T01G9.2 show weak rescue of the vulvaless phenotype and exhibit a protruding vulval phenotype (Kim and Horvitz, 1990; Figure 3B). It is likely that the weak rescue provided by genomic clones that span T01G9.2 represents extragenic suppression of the lin-10 mutant phenotype.
The DNA sequence of cosmid C09H6 has been determined by the C. elegans genome sequencing consortium, and analysis of this sequence revealed a gene (C09H6.2) that is predicted to encode a protein with PDZ domains. Because lin-2 and lin-7 also contain PDZ domains, we specifically investigated whether C09H6.2 corresponds to lin-10. We obtained six cDNA clones originating from C09H6.2 from a random collection of expressed sequence tags (kindly provided by Y. Kohara), and one of these cDNA clones (YK114C6) appears to be nearly full length, because it begins three codons from the start codon and ends after the stop codon predicted from the genomic sequence. We used PCR to insert the first three codons of C09H6.2 into the YK114C6 cDNA and then showed that expression of the full-length coding sequence under control of the vulval-specific lin-31 promoter (Tan et al., 1998) can rescue the lin-10 mutant phenotype (Figure 3B). This result indicates that the PDZ-containing C09H6.2 gene is likely to correspond to lin-10.
To conclusively show that the candidate gene C09H6.2 corresponds to the genetic lin-10 locus, we examined C09H6.2 for molecular lesions in 18 lin-10 mutant alleles. Primers were designed to amplify each of the candidate gene exons and exon–intron boundaries in PCR experiments (Table 1). In two lin-10 mutant strains, primer pairs failed to amplify large portions of C09H6.2. Specifically, exons 4–10 did not amplify in lin-10(n299), and exons 11–15 did not amplify in lin-10(n1299), suggesting that lin-10(n299) and lin-10(n1299) cause significant deletions of the C09H6.2 coding sequence. To determine the exact nature of these alterations, we used inverse PCR to amplify DNA spanning the lin-10(n299) and lin-10(n1299) rearrangement break points (see MATERIALS AND METHODS). We determined the DNA sequence of the inverse PCR products and compared them to the wild-type DNA sequence. lin-10(n1299) has an 11.6-kb deletion that removes five exons from the 3′ end of C09H6.2 and also deletes the entire coding region of a neighboring gene (C09H6.1) (Figure 3C). lin-10(n299) has a complex rearrangement in which 4.7 kb of C09H6.2 (including six exons) is replaced by an ∼300-kb insertion from another region of chromosome I (spanning cosmids DY3 to ZK858 on the physical map) (Figure 3C).
For the remaining 16 lin-10 alleles, we searched for point mutations in all exon and exon–intron boundaries in C09H6.2 by using single-strand conformational polymorphism analysis. For three alleles (n1508, n1541, and n1636), no polymorphisms were identified; for 13 alleles, we identified exactly one single-strand conformational polymorphism that was associated with each allele. PCR fragments containing each of the polymorphisms were sequenced. The identified polymorphisms include one missense mutation, three RNA splice acceptor mutations, and nine nonsense mutations (Figures 3 and 4). One nonsense mutation in particular provides strong evidence that C09H6.2 corresponds to the lin-10 genetic locus. The vulvaless phenotype of lin-10(n1638) is suppressed by the amber suppresser tRNA mutation sup-5(e1464) (Kim and Horvitz, 1990), and we found that C09H6.2 con-tains an amber stop mutation at amino acid 301 in lin-10(n1638) mutants.
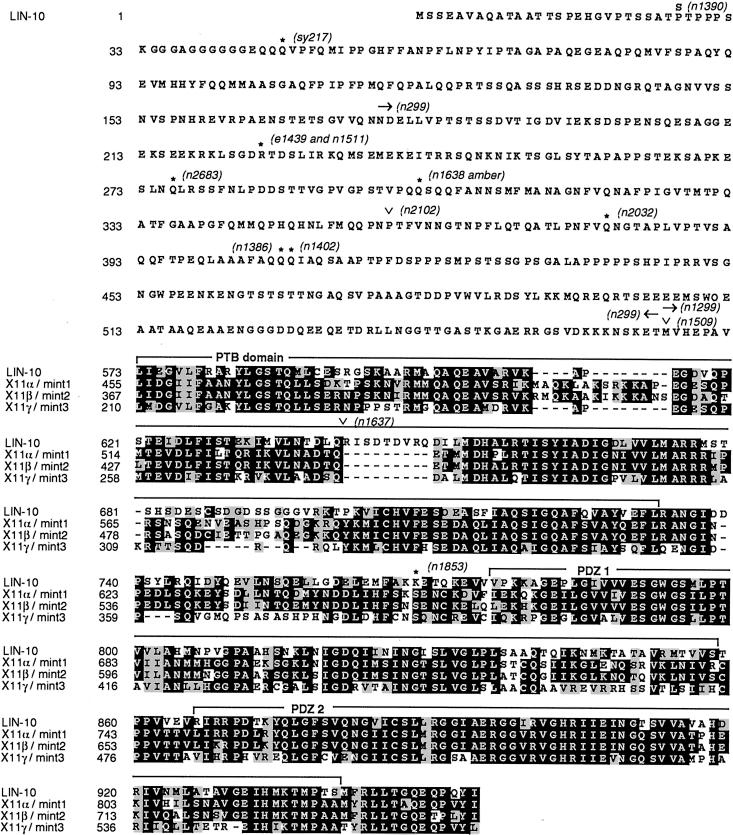
Figure 4.
LIN-10 amino acid sequence, shown aligned with the C terminus of mammalian X11/mint proteins. Identical amino acids are boxed in black, and similar amino acids are boxed in gray. No significant sequence similarity was detected in pair-wise alignment of N-terminal LIN-10 (amino acids 1–572) with each of the X11/mint proteins. PTB, PDZ 1, and PDZ 2 domains are indicated. Positions and molecular lesions associated with lin-10 mutant alleles are indicated: *, Stop codons; ∨, RNA splice acceptor mutations; arrows, codons deleted by chromosomal deletions. These sequence data are available from GenBank.
The previously reported lin-10 (T01G9.2) contains an insertion of the Tc1 transposable element into an intron in a strain carrying lin-10(n1299) and produces an altered mRNA transcript in a strain containing lin-10(n299) (Kim and Horvitz, 1990). Although highly unlikely, strains carrying these two lin-10 mutations both have polymorphisms in two tightly linked genes (C09H6.2 and T01G9.2). Deficiency-mapping results, transformation rescue experiments, DNA sequence analysis of 15 lin-10 alleles, and identification of an amber-suppressible amber stop mutation provide compelling evidence that C09H6.2 corresponds to lin-10 rather than the gene previously identified (T01G9.2). Thus, it is likely that the DNA changes in T01G9.2 in lin-10(n299) and lin-10(n1299) strains correspond to linked polymorphisms rather than lin-10 mutations.
The C09H6.2 gene corresponding to lin-10 is predicted to encode a 954–amino acid protein with regions of similarity to mammalian X11/mint proteins (Duclos et al., 1993; Borg et al., 1996, 1998b; Okamoto and Sudhof, 1997; Butz et al., 1998) (Figure 4). The 382 amino acids in the C-terminal region of LIN-10 are similar to the C-terminal region of X11α/mint1 (55% identical), X11β/mint2 (56% identical), and X11γ/mint3 (51% identical). The N-terminal 572 amino acids of LIN-10 show little or no sequence similarity with X11/mint proteins or other known proteins.
Vulval Induction Mediated by Mutant LIN-10 Lacking PDZ Domains
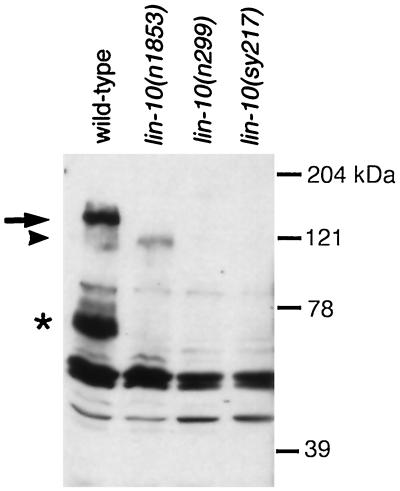
We prepared affinity-purified antibodies against bacterially expressed LIN-10 and used them to characterize the expressed LIN-10 protein. Western blotting experiments using these anti-LIN-10 antibodies show one band corresponding to full-length LIN-10 at 140 kDa and a smaller band migrating at 70 kDa (Figure 5). The structure of the smaller protein has not been determined but may result from differential RNA splicing or protein degradation. Both 140- and 70-kDa bands are absent in protein extracts from lin-10(n299) and lin-10(sy217) mutants.
Figure 5.
LIN-10 is truncated in lin-10(n1853) and absent in two other mutants. LIN-10 was detected by analyzing protein extracts from hermaphrodites using Western blots and anti-LIN-10 antibodies. Full-length LIN-10 is indicated by an arrow. A smaller LIN-10 product (*) may correspond to a LIN-10 degradation product or an alternate LIN-10 isoform. Truncated LIN-10 product in lin-10(n1853) is indicated by an arrowhead. The lin-10(n1853) truncation does not affect the epitope used to generate anti-LIN-10 antibodies (see MATERIALS AND METHODS). No LIN-10 expression is observed in a deletion mutant, lin-10(n299), or in a mutant carrying an early stop mutation, lin-10(sy217). Molecular size markers are shown.
Characterization of the lin-10(n1853) allele indicates that the PDZ domains may not be essential for LIN-10 function. lin-10(n1853) is a stop mutation that is predicted to truncate the C-terminal 186 amino acids of LIN-10, including both PDZ domains (Figure 4). We investigated expression of the mutant LIN-10 protein in lin-10(n1853) animals using Western blotting with anti-LIN-10 antibodies (Figure 5). In protein extracts from lin-10(n1853) animals, anti-LIN-10 antibodies detect a new band at 120 kDa but do not detect either the 140- or 70-kDa band observed in protein extracts from wild-type animals. The level of the 120-kDa product in lin-10(n1853) animals is significantly lower than the level of the 140-kDa, full-length LIN-10 product in wild-type animals (Figure 5). These results suggest that lin-10(n1853) results in truncation of 186 amino acids from full-length LIN-10 (from 140 to 120 kDa) and also reduces expression of the truncated protein. Furthermore, lin-10(n1853) eliminates expression of the smaller LIN-10 protein. lin-10(n1853) results in a weak vulvaless phenotype (15% of homozygous mutants are egg laying defective; n = 166), whereas mutations that eliminate lin-10 activity result in a strong vulvaless phenotype (95% are egg laying defective) (Ferguson and Horvitz, 1985; Kim and Horvitz, 1990). The weak vulvaless phenotype in lin-10(n1853) animals could result from reduced LIN-10 protein levels or reduced activity of the mutant LIN-10 proteins. In either case, the presence of significant levels of vulval induction in lin-10(n1853) mutants indicates that truncated LIN-10 protein lacking both PDZ domains retains significant genetic activity in vulval induction.
LIN-10 Expression and Subcellular Distribution
Next, we used the anti-LIN-10 antibodies in immunocytochemical experiments to examine the LIN-10 expression pattern and subcellular distribution. The anti-LIN-10 antibodies specifically detect LIN-10, because we did not observe any staining in lin-10(n299) mutants (Figure 6E). In late L3 larvae, LIN-10 is present in descendants of the vulval precursor cells (Figure 6). In these cells, LIN-10 staining is concentrated in punctate spots around the nucleus, and staining is diffuse in the cytoplasm and at the plasma membrane. We have not detected LIN-10 staining in the vulval precursor cells in L2 or early L3 larvae, possibly because strong LIN-10 staining in adjacent ventral cord neurons obscures low-level LIN-10 expression in the vulval precursor cells at this stage.
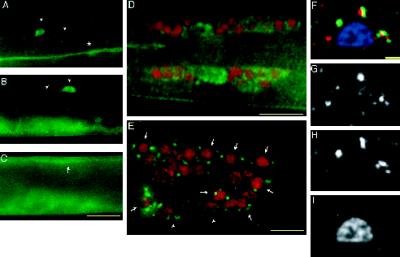
Figure 6.
LIN-10 is expressed in the developing vulva. (A–D) Wild-type, late L3 hermaphrodites. (A) Lateral view of the developing vulva. The upper panel shows staining with anti-LIN-10 antibodies (green), and the lower panel shows staining with the celljunction mAb marker MH27 (red). (B) Panels in A are merged. Positions of vulval epithelia (granddaughter cells of P5.p, P6.p, and P7.p) are indicated. Arrowheads indicate cell bodies of ventral cord neurons. Vulval epithelial cells can be distinguished from ventral cord neurons by their larger size and the presence of stained cell junctions (red). LIN-10 is present in the cytoplasm and at the plasma membrane. (C and D) Two focal planes showing the developing vulva and ventral cord neurons. Positions of ventral cord neurons are indicated by arrows. Some staining in D is from ventral cord neurons out of the plane of focus. Punctate, perinuclear LIN-10 staining can be seen in some vulval epithelial cells (D, inset, arrows). (E) lin-10(n299) late L3 hermaphrodite stained with anti-LIN-10 antibodies (green) as above. The region of developing vulva and ventral cord neurons is shown as in A–D. Bar, 10 μm.
LIN-10 is expressed at high levels in neurons, including ring neurons, ventral cord neurons, dorsal cord neurons, lateral neurons, and neurons in the tail (Figures 6C and 7, A–E). This result supports previous results showing that LIN-10 functions to localize the glutamate receptor GLR-1 to the postsynaptic elements in neurons (Rongo et al., 1998). LIN-10 appears in neuronal processes and cell bodies. In neural cell bodies, a small amount of LIN-10 appears diffusely throughout the cytoplasm, whereas the majority of LIN-10 is concentrated in discrete perinuclear structures (Figure 7, D and E), similar to perinuclear structures observed in vulval epithelial cells. To determine whether these perinuclear structures correspond to Golgi, we used ST-GFP as a marker for the trans-cisterna of the Golgi (Jamora et al., 1997). We expressed ST-GFP in transgenic worms using a heat shock promoter and examined the subcellular localization of LIN-10 and ST-GFP using anti-LIN-10 and anti-GFP antibodies. In single neurons expressing both endogenous LIN-10 and transgenic ST-GFP, the subcellular pattern of LIN-10 staining is similar to that of ST-GFP staining. Deconvolution of images obtained in double-staining experiments revealed that LIN-10 staining is closely associated with ST-GFP staining (Figure 7, F–I), but LIN-10 staining is consistently offset (by 0.2–0.5 μm) from ST-GFP staining. These results indicate that LIN-10 is localized in the trans-cisterna of the Golgi or is localized in a compartment closely associated with the trans-cisterna, such as the trans-Golgi network.
Figure 7.
LIN-10 is expressed in neurons. (A–C) Wild-type, late L3 hermaphrodite stained with anti-LIN-10 antibodies (green). LIN-10 is present in ventral cord processes (A, *), lateral neural cell bodies and processes (A and B, arrowheads), and dorsal cord processes (C, arrow). Ventral staining in B and C is from the developing vulva out of the plane of focus. (D and E) Wild-type worms were stained with anti-LIN-10 antibodies (green) and DAPI (pseudocolored red), and images were taken with a DeltaVision wide-field microscope and deconvolved (see MATERIALS AND METHODS). (D) Lateral view of the nerve ring of an L4 hermaphrodite showing LIN-10 staining in nerve ring cell bodies and nerve ring neuropil. (E) Lateral view of an adult hermaphrodite tail showing punctate, perinuclear LIN-10 staining around neural nuclei (arrows). Some LIN-10 staining may also be associated with nonneural nuclei (arrowheads). (F) Merged image showing a neural cell body expressing transgenic ST-GFP and stained with anti-LIN-10 antibodies (red), anti-GFP antibodies (green), and DAPI (blue). (G) LIN-10 staining; (H) ST-GFP staining; (I) DAPI staining. Bars, 10 μm (A–E); 1 μm (F–I).
DISCUSSION
Basolateral Expression of LET-23 RTK in the Vulval Precursor Cells
We have confirmed and extended previous experiments aimed at determining the expression pattern of LET-23 RTK on the surface of vulval precursor cells. Here, we used anti-LET-23 antibodies to stain worms in immunocytochemical experiments and observed the stained vulval precursor cells using a DeltaVision deconvolution microscope. These experiments indicate that the relative distribution of LET-23 RTK on the apical and basolateral membrane domains is dynamic. LET-23 is initially expressed in early L2 on both the apical and basolateral membrane domains of the vulval precursor cells, with higher expression on the apical surface. In late L2 to early L3, at the time of vulval cell fate determination (Kimble, 1981), LET-23 is expressed predominantly on the basolateral membrane domain of the vulval precursor cells (with the exception of P6.p). In P6.p, LET-23 is expressed on both the apical and basolateral membrane domains and remains distributed on both of the membrane domains as P6.p divides after vulval induction.
These staining experiments suggest that there is temporal and positional regulation of LET-23 RTK localization on the surface of the vulval precursor cells. A temporal shift occurs in middle L2, when LET-23 becomes predominantly basolateral rather than apical. One interesting possibility is that basolateral targeting pathways are developmentally regulated, such that they are weakly expressed during early L2 (and consequently a significant fraction of LET-23 is apical) but begin to be strongly expressed in late L2 (and consequently most of LET-23 is basolateral).
In late L2 to early L3, the six vulval precursor cells have different positions relative to the anchor cell, such that P6.p is closest to the anchor cell and is induced to express the 1° cell fate. At this stage, LET-23 is predominantly basolateral in five vulval precursor cells (P3.p, P4.p, P5.p, P7.p, and P8.p) but is both basolateral and apical in P6.p. Several explanations could account for the difference in LET-23 subcellular localization between P6.p and the other vulval precursor cells. One possibility is that activation of LET-23 tyrosine kinase activity in P6.p could directly affect LET-23 localization. Another possibility is that apical accumulation of LET-23 in P6.p is a consequence of increased LET-23 expression, because there is a dramatic increase in LET-23 expression in P6.p at the beginning of vulval induction (Simske et al., 1996; Candia, personal communication).
lin-10 Acts in Basolateral Localization of LET-23 RTK and Encodes a Protein with Regions of Similarity to Mammalian X11/mint Proteins
We show that lin-10 is required for basolateral localization of LET-23 RTK in the vulval precursor cells. In lin-10 mutant animals, LET-23 is mislocalized from the basolateral membrane domain to the apical membrane domain. Presumably, apical mislocalization of LET-23 in lin-10 mutants prevents LET-23 receptor from binding to the anchor cell signal in the basal extracellular space, so that the LET-23 signaling pathway is not activated and the vulva is not induced.
Our results suggest that lin-10 acts cell autonomously in the vulval precursor cells to localize LET-23 RTK, because expression of the lin-10 cDNA from a promoter specific for the Pn.p cells (the lin-31 promoter) rescues the lin-10 vulvaless phenotype (Figure 3B), but expression from a neuronal-specific promoter (the glr-1 promoter) does not (Rongo et al., 1998). The pattern of LIN-10 expression is consistent with a cell-autonomous function, because LIN-10 is expressed in many cells, including vulval epithelia.
lin-10 encodes a protein with a C-terminal region similar to mammalian X11/mint proteins. The X11/mint protein family is characterized by a highly similar C-terminal region, including a phosphotyrosine-binding (PTB) domain similar to the Shc PTB domain and two PDZ (PSD-95/discs-large/ZO-1) domains. The N-terminal region of mammalian X11/mint proteins shows reduced sequence similarity, and protein interaction studies indicate that this region may be functionally divergent (Duclos et al., 1993; Borg et al., 1996, 1998a; Okamoto and Sudhof, 1997; Butz et al., 1998). Two members of this protein family (X11α/mint1 and X11β/mint2) are expressed in neurons and have been identified in a protein complex with Munc18-1, an important regulator of vesicle docking and exocytosis (Okamoto and Sudhof, 1997). X11α/mint1 and X11β/mint2 have also been implicated in trafficking or metabolism of the β-amyloid precursor protein (Borg et al., 1998b). A third member of this protein family (X11γ/mint3) is expressed ubiquitously, and its function is not known (Borg et al., 1998a; Butz et al., 1998). The PTB domain of X11/mint and related PTB domains can bind a specific tyrosine peptide motif (YENPTY) independently of tyrosine phosphorylation (Borg et al., 1996; Zambrano et al., 1997). The PTB domain of X11/mint has also been shown to bind specifically to phosphatidylinositol-4,5-bisphosphate and phosphatidylinositol-4-phosphate (Okamoto and Sudhof, 1997). PDZ domains typically bind to the C termini of transmembrane proteins, and PDZ-containing proteins are known to be involved in receptor localization, receptor clustering, or linking together multiple components of signaling pathways (for review, see Kim, 1997).
Common Elements in Receptor Localization in Epithelia and Neurons
Like epithelia, neurons are polarized cells in which specialized membrane domains are structurally and functionally distinct, and it has been proposed that epithelia and neurons share common mechanisms for regulated protein trafficking to distinct membrane domains (Dotti and Simons, 1990). Our results together with recent work from others provide direct evidence for such a common mechanism. In addition to its role in LET-23 RTK localization at the basolateral membrane domain of epithelia, LIN-10 acts in localization of the glutamate receptor GLR-1 at postsynaptic elements in neurons (Rongo et al., 1998).
How might LIN-10 function in correctly localizing LET-23 RTK and GLR-1 glutamate receptor to their respective target membranes? LIN-10 is at the plasma membrane in vulval epithelia and in neural processes, and this localization is consistent with a role in regulation of exocytosis of receptor-containing secretory vesicles at the target membrane domain or tethering receptors at the target membrane domain once they are secreted. However, the majority of LIN-10 protein appears to be intracellular. In neurons this pool of LIN-10 is in close association with a marker for the trans-cisterna of the Golgi, suggesting that LIN-10 may be concentrated in the Golgi cisterna or the trans-Golgi network. This localization is consistent with a role in sorting receptors such as LET-23 and GLR-1 into vesicles destined for their respective target membrane domains. The subcellular distribution of C. elegans LIN-10 closely resembles that of one of its mammalian homologues (X11a/mint1), which was recently shown to be largely cytosolic and concentrated in the Golgi apparatus in mammalian neurons (Borg et al., 1999).
A Conserved Protein Complex Mediates Basolateral Localization of LET-23 RTK in Vulval Epithelia
In addition to lin-10, two other genes (lin-2 and lin-7) function in basolateral localization of LET-23 (Simske et al., 1996). Furthermore, recent evidence indicates that LIN-10 is part of a protein complex with LIN-2 and LIN-7, and that LIN-7 directly binds to the cytoplasmic C terminus of LET-23 (Kaech et al., 1998). These results suggest that the mechanism of LET-23 basolateral localization involves direct protein interactions between trans-acting factors (LIN-2, LIN-7, and LIN-10) and a cytoplasmic, cis-acting element in the LET-23 C terminus.
LIN-2, LIN-7 and LIN-10 are evolutionarily conserved (Kaech et al., 1998; Borg et al., 1998a; Butz et al., 1998; this report). C. elegans LIN-2 is highly similar to mammalian CASK and contains a Ca2+/calmodulin-dependent kinase II domain, a conserved LIN-7 binding region, a PDZ domain, an Src homlogy region 3 domain, and a guanylate kinase domain (Hoskins et al., 1996; Kaech et al., 1998). C. elegans LIN-7 is highly similar to three mammalian homologues (termed mLin-7A, mLin-7B, and mLin-7C) and contains a PDZ domain and a conserved LIN-2 binding region (Simske et al., 1996; Kaech et al., 1998). As discussed above, C. elegans LIN-10 has regions of similarity with mammalian X11/mint proteins. Recently, these mammalian homologues have been found to form a ternary complex similar to the C. elegans LIN-2/LIN-7/LIN-10 proteins (Kaech et al., 1998; Borg et al., 1998a, 1999; Butz et al., 1998). Specifically, mammalian LIN-7 binds to mammalian LIN-2/CASK, and mammalian LIN-2/CASK binds to mammalian LIN-10/X11/mint proteins.
ACKNOWLEDGMENTS
We thank John Laudenslager and Ian Trowbridge for sialyltransferase-GFP. We thank Alex Hajnal, Bob Coffey, and the rest of the Kim laboratory for helpful discussions and critical comments on the manuscript. We thank Y. Kohara for providing the lin-10 cDNA clones, Catherine Ying for antibodies, and Abby Dernberg for technical assistance with three-dimensional imaging. Some strains were provided by the Caenorhabditis Genetics Center. This work was supported by grants from National Institutes of Health and the National Cancer Institute.
REFERENCES
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Aroian RV, Koga M, Mendel JE, Ohshima Y, Sternberg PW. The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature. 1990;348:693–699. doi: 10.1038/348693a0. [DOI] [PubMed] [Google Scholar]
- Borg JP, Lopez-Figueroa MO, de Taddeo-Borg M, Kroon DE, Turner RS, Watson SJ, Margolis B. Molecular analysis of the X11-mLin-2/CASK complex in brain. J Neurosci. 1999;19:1307–1316. doi: 10.1523/JNEUROSCI.19-04-01307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg JP, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg JP, Straight SW, Kaech SM, de Taddeo-Borg M, Kroon DE, Karnak D, Turner RS, Kim SK, Margolis B. Identification of an evolutionarily conserved heterotrimeric protein complex involved in protein targeting. J Biol Chem. 1998a;273:31633–31636. doi: 10.1074/jbc.273.48.31633. [DOI] [PubMed] [Google Scholar]
- Borg JP, Yang Y, de Taddeo-Borg M, Margolis B, Turner RS. The x11alpha protein slows cellular amyloid precursor protein processing and reduces abeta40 and abeta42 secretion. J Biol Chem. 1998b;273:14761–14766. doi: 10.1074/jbc.273.24.14761. [DOI] [PubMed] [Google Scholar]
- Butz S, Okamoto M, Sudhof TC. Molecular architecture of a novel junctional complex coupling vesicular traffic to cell adhesion. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- Casanova JE, Apodaca G, Mostov KE. An autonomous signal for basolateral sorting in the cytoplasmic domain of the polymeric immunoglobulin receptor. Cell. 1991;66:65–75. doi: 10.1016/0092-8674(91)90139-p. [DOI] [PubMed] [Google Scholar]
- Chen H, Hughes DD, Chan TA, Sedat JW, Agard DA. IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J Struct Biol. 1996;116:56–60. doi: 10.1006/jsbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Crepaldi T, Pollack AL, Prat M, Zborek A, Mostov K, Comoglio PM. Targeting of the SF/HGF receptor to the basolateral domain of polarized epithelial cells. J Cell Biol. 1994;125:313–320. doi: 10.1083/jcb.125.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Simons K. Polarized sorting of viral glycoproteins to the axon and dendrites of hippocampal neurons in culture. Cell. 1990;62:63–72. doi: 10.1016/0092-8674(90)90240-f. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Duclos F, Boschert U, Sirugo G, Mandel JL, Hen R, Koenig M. Gene in the region of the Friedreich ataxia locus encodes a putative transmembrane protein expressed in the nervous system. Proc Natl Acad Sci USA. 1993;90:109–113. doi: 10.1073/pnas.90.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Horvitz HR. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985;110:17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Hobert M, Carlin C. Cytoplasmic juxtamembrane domain of the human EGF receptor is required for basolateral localization in MDCK cells. J Cell Physiol. 1995;162:434–446. doi: 10.1002/jcp.1041620316. [DOI] [PubMed] [Google Scholar]
- Hobert ME, Kil SJ, Medof ME, Carlin CR. The cytoplasmic juxtamembrane domain of the epidermal growth factor receptor contains a novel autonomous basolateral sorting determinant. J Biol Chem. 1997;272:32901–32909. doi: 10.1074/jbc.272.52.32901. [DOI] [PubMed] [Google Scholar]
- Hoskins R, Hajnal AF, Harp SA, Kim SK. The C. elegans vulval induction gene lin-2 encodes a member of the MAGUK family of cell junction proteins. Development. 1996;122:97–111. doi: 10.1242/dev.122.1.97. [DOI] [PubMed] [Google Scholar]
- Hunziker W, Harter C, Matter K, Mellman I. Basolateral sorting in MDCK cells requires a distinct cytoplasmic domain determinant. Cell. 1991;66:907–920. doi: 10.1016/0092-8674(91)90437-4. [DOI] [PubMed] [Google Scholar]
- Jamora C, Takizawa PA, Zaarour RF, Denesvre C, Faulkner DJ, Malhotra V. Regulation of Golgi structure through heterotrimeric G proteins. Cell. 1997;91:617–626. doi: 10.1016/s0092-8674(00)80449-3. [DOI] [PubMed] [Google Scholar]
- Kaech SM, Whitfield CW, Kim SK. Basolateral membrane localization of the receptor tyrosine kinase LET-23 is mediated by the LIN-2/LIN-7/LIN-10 protein complex in the vulval precursor cells of C. elegans. Cell. 1998;94:761–771. doi: 10.1016/s0092-8674(00)81735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz WS, Hill RJ, Clandinin TR, Sternberg PW. Different levels of the C. elegans growth factor LIN-3 promote distinct vulval precursor fates. Cell. 1995;82:297–307. doi: 10.1016/0092-8674(95)90317-8. [DOI] [PubMed] [Google Scholar]
- Kim SK. Polarized signaling: basolateral receptor localization in epithelial cells by PDZ-containing proteins. Curr Opin Cell Biol. 1997;9:853–859. doi: 10.1016/s0955-0674(97)80088-9. [DOI] [PubMed] [Google Scholar]
- Kim SK, Horvitz HR. The Caenorhabditis elegans gene lin-10 is broadly expressed while required specifically for the determination of vulval cell fates. Genes & Dev. 1990;4:357–371. doi: 10.1101/gad.4.3.357. [DOI] [PubMed] [Google Scholar]
- Kimble J. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol. 1981;87:286–300. doi: 10.1016/0012-1606(81)90152-4. [DOI] [PubMed] [Google Scholar]
- Koga M, Ohshima Y. Mosaic analysis of the let-23 gene function in vulval induction of Caenorhabditis elegans. Development. 1995;121:2655–2666. doi: 10.1242/dev.121.8.2655. [DOI] [PubMed] [Google Scholar]
- Kornfeld K. Vulval development in Caenorhabditis elegans. Trends Genet. 1997;13:55–61. doi: 10.1016/s0168-9525(97)01005-6. [DOI] [PubMed] [Google Scholar]
- Maratos FE, Kao CY, Verdin EM, King GL. Receptor-mediated vectorial transcytosis of epidermal growth factor by Madin-Darby canine kidney cells. J Cell Biol. 1987;105:1595–1601. doi: 10.1083/jcb.105.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter K, Hunziker W, Mellman I. Basolateral sorting of LDL receptor in MDCK cells: the cytoplasmic domain contains two tyrosine-dependent targeting determinants. Cell. 1992;71:741–753. doi: 10.1016/0092-8674(92)90551-m. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. In: Shakes D, Epstein H, editors. Methods in Cell Biology. San Diego: Academic Press; 1995. pp. 451–482. [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostov KE, de Bruyn Kops A, Deitcher DL. Deletion of the cytoplasmic domain of the polymeric immunoglobulin receptor prevents basolateral localization and endocytosis. Cell. 1986;47:359–364. doi: 10.1016/0092-8674(86)90592-1. [DOI] [PubMed] [Google Scholar]
- Mullin JM, McGinn MT. Epidermal growth factor-induced mitogenesis in kidney epithelial cells (LLC-PK1) Cancer Res. 1988;48:4886–4891. [PubMed] [Google Scholar]
- Okamoto M, Sudhof TC. Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J Biol Chem. 1997;272:31459–31464. doi: 10.1074/jbc.272.50.31459. [DOI] [PubMed] [Google Scholar]
- Rodriguez BE, Nelson WJ. Morphogenesis of the polarized epithelial cell phenotype. Science. 1989;245:718–725. doi: 10.1126/science.2672330. [DOI] [PubMed] [Google Scholar]
- Rodriguez BE, Powell SK. Polarity of epithelial and neuronal cells. Annu Rev Cell Biol. 1992;8:395–427. doi: 10.1146/annurev.cb.08.110192.002143. [DOI] [PubMed] [Google Scholar]
- Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM. LIN-10 is a shared component of the polarized protein trafficking pathway in neurons and epithelial cells. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- Simske JS, Kaech SM, Harp SA, Kim SK. LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell. 1996;85:195–204. doi: 10.1016/s0092-8674(00)81096-x. [DOI] [PubMed] [Google Scholar]
- Simske JS, Kim SK. Sequential signaling during Caenorhabditis elegans vulval induction. Nature. 1995;375:142–146. doi: 10.1038/375142a0. [DOI] [PubMed] [Google Scholar]
- Simske JS, Kim SK. Hormons and Growth Factors in Development and Neoplasia. R.B. Dickson and D.S. Salomon, New York: Wiley-Liss; 1998. Pattern formation by sequential signaling during C. elegans vulval Induction; pp. 3–17. [Google Scholar]
- Tan PB, Lackner MR, Kim SK. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell. 1998;93:569–580. doi: 10.1016/s0092-8674(00)81186-1. [DOI] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB. The Nematode Caenorhabditis elegans. New York: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Zambrano N, et al. Interaction of the phosphotyrosine interaction/phosphotyrosine binding-related domains of Fe65 with wild-type and mutant Alzheimer’s beta-amyloid precursor proteins. J Biol Chem. 1997;272:6399–6405. doi: 10.1074/jbc.272.10.6399. [DOI] [PubMed] [Google Scholar]