Abstract
In an attempt to correlate the ability to activate transcription with affinity for single-stranded DNA, both wild-type and temperature-sensitive pseudorabies virus immediate-early proteins were tested for the ability to bind to single-stranded DNA columns. Wild-type and temperature-sensitive immediate-early proteins bound to nonspecific single-stranded DNA columns with similar affinities at both 0 and 40 degrees C. There did not seem to be a direct correlation between the ability to activate transcription and the ability to bind to single-stranded DNA. To study further the interactions that are involved in binding to single-stranded DNA, we expressed the immediate-early protein in an Escherichia coli expression vector. In this system the expressed immediate-early protein was not phosphorylated, nor could it be complexed with mammalian cell factors. The first trp construct did not express a soluble form of the immediate-early protein, presumably due to the insoluble nature of the trp leader. We deleted a large segment of the trpE gene and found that the immediate-early fusion protein was soluble. We tested this protein for its affinity for single-stranded DNA by passage over single-stranded DNA cellulose columns. The bacterially expressed immediate-early protein bound single-stranded DNA at least as well as did the wild-type protein. Affinity for single-stranded DNA did not appear to be dependent on the phosphorylation state nor on the presence of mammalian cell factors.
Full text
PDF
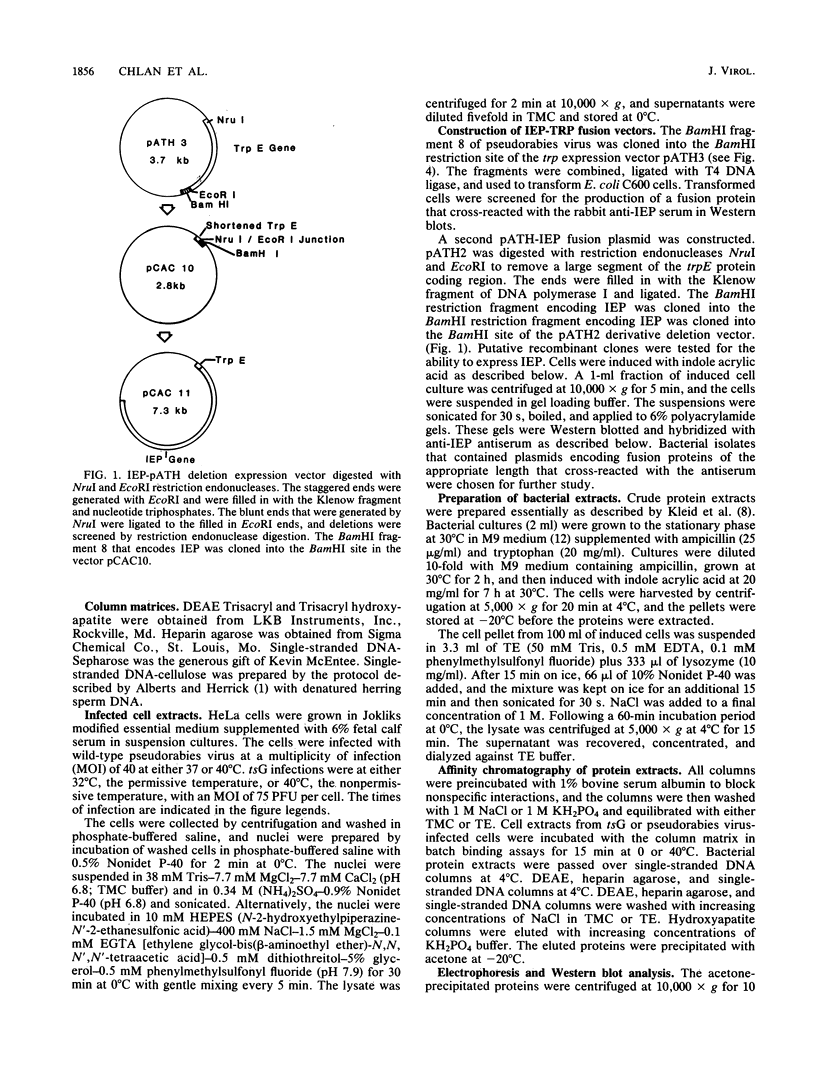




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C. Transient gene expression control: effects of transfected DNA stability and trans-activation by viral early proteins. Mol Cell Biol. 1985 May;5(5):1034–1042. doi: 10.1128/mcb.5.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard P., Faber S., Wilcox K. W., Pizer L. I. Herpes simplex virus immediate early infected-cell polypeptide 4 binds to DNA and promotes transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(11):4016–4020. doi: 10.1073/pnas.83.11.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. S., Cann A. J., Shah N. P., Gaynor R. B. Functional relation between HTLV-II x and adenovirus E1A proteins in transcriptional activation. Science. 1985 Nov 1;230(4725):570–573. doi: 10.1126/science.2996140. [DOI] [PubMed] [Google Scholar]
- Feldman L. T., Imperiale M. J., Nevins J. R. Activation of early adenovirus transcription by the herpesvirus immediate early gene: evidence for a common cellular control factor. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4952–4956. doi: 10.1073/pnas.79.16.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. J., Powell K. L. DNA-binding properties of a herpes simplex virus immediate early protein. J Virol. 1982 Dec;44(3):1084–1087. doi: 10.1128/jvi.44.3.1084-1087.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Treisman R., Maniatis T. Transcriptional activation of cloned human beta-globin genes by viral immediate-early gene products. Cell. 1983 Nov;35(1):137–148. doi: 10.1016/0092-8674(83)90216-7. [DOI] [PubMed] [Google Scholar]
- Kleid D. G., Yansura D., Small B., Dowbenko D., Moore D. M., Grubman M. J., McKercher P. D., Morgan D. O., Robertson B. H., Bachrach H. L. Cloned viral protein vaccine for foot-and-mouth disease: responses in cattle and swine. Science. 1981 Dec 4;214(4525):1125–1129. doi: 10.1126/science.6272395. [DOI] [PubMed] [Google Scholar]
- Kristie T. M., Roizman B. Alpha 4, the major regulatory protein of herpes simplex virus type 1, is stably and specifically associated with promoter-regulatory domains of alpha genes and of selected other viral genes. Proc Natl Acad Sci U S A. 1986 May;83(10):3218–3222. doi: 10.1073/pnas.83.10.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristie T. M., Roizman B. DNA-binding site of major regulatory protein alpha 4 specifically associated with promoter-regulatory domains of alpha genes of herpes simplex virus type 1. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4700–4704. doi: 10.1073/pnas.83.13.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- O'Hare P., Hayward G. S. Three trans-acting regulatory proteins of herpes simplex virus modulate immediate-early gene expression in a pathway involving positive and negative feedback regulation. J Virol. 1985 Dec;56(3):723–733. doi: 10.1128/jvi.56.3.723-733.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. J., Tevethia M. J. Identification of a human cytomegalovirus virus DNA segment that complements an adenovirus 5 immediate early mutant. Virology. 1986 Jun;151(2):329–338. doi: 10.1016/0042-6822(86)90053-x. [DOI] [PubMed] [Google Scholar]
- Tanese N., Roth M., Goff S. P. Expression of enzymatically active reverse transcriptase in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4944–4948. doi: 10.1073/pnas.82.15.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R. J., Clements J. B. Characterization of transcription-deficient temperature-sensitive mutants of herpes simplex virus type 1. Virology. 1978 Dec;91(2):364–379. doi: 10.1016/0042-6822(78)90384-7. [DOI] [PubMed] [Google Scholar]
- Wilcox K. W., Kohn A., Sklyanskaya E., Roizman B. Herpes simplex virus phosphoproteins. I. Phosphate cycles on and off some viral polypeptides and can alter their affinity for DNA. J Virol. 1980 Jan;33(1):167–182. doi: 10.1128/jvi.33.1.167-182.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]






