Abstract
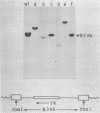
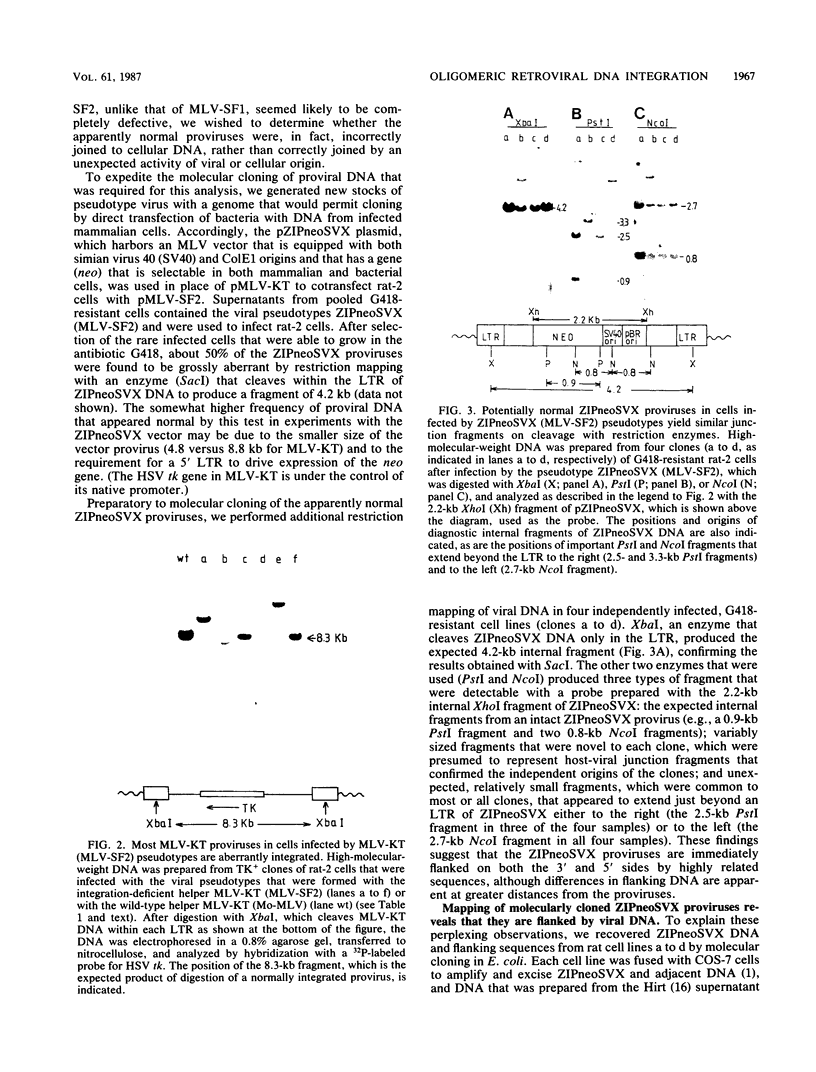
Mutants with lesions engineered into the 3' region of the pol gene of murine leukemia virus (MLV), the region that encodes p40pol, have allowed us to define a function that is required for the normal integration of retroviral DNA. Two such mutants, MLV-SF1, which bears a single missense mutation (L. A. Donehower and H. E. Varmus, Proc. Natl. Acad. Sci. USA 81:6461-6465, 1984), and MLV-SF2, which has a frameshift mutation at the same site, have been used to explore the consequences of integration deficiencies. Pseudotypes that were formed with viral proteins supplied by these mutants and a replication-defective genome that carried a selectable genetic marker induced colony formation at least 100-fold less efficiently than did pseudotypes that were formed with proteins encoded by wild-type helper virus. Most of the proviruses from the mutant pseudotypes were manifestly aberrant when they were analyzed by mapping with restriction enzymes. Rare proviruses that appeared to be normal by restriction enzyme mapping were molecularly cloned and subjected to more detailed study. Two proviruses resulting from infection with the MLV-SF1 pseudotype were identical or nearly identical to wild-type MI V proviruses. Two base pairs were missing from the ends of the long terminal repeats at the host-viral junctions, and 4- or 5-base-pair duplications of host DNA flanked the proviruses. Thus, the missense mutant appears to retain a low level of normal integration activity. Four proviruses from cells that were infected with the MLV-SF2 pseudotype were flanked on both sides by viral DNA of either helper or vector origin, with a single long terminal repeat at each end of the intact vector DNA and with irregular junctions of viral and host DNAs. These results are most simply explained by the nonspecific integration of dimeric or trimeric forms of viral DNA, which may arise during infection by replicative or recombinational mechanisms.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cepko C. L., Roberts B. E., Mulligan R. C. Construction and applications of a highly transmissible murine retrovirus shuttle vector. Cell. 1984 Jul;37(3):1053–1062. doi: 10.1016/0092-8674(84)90440-9. [DOI] [PubMed] [Google Scholar]
- Coffin J. M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979 Jan;42(1):1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- Colbère-Garapin F., Horodniceanu F., Kourilsky P., Garapin A. C. A new dominant hybrid selective marker for higher eukaryotic cells. J Mol Biol. 1981 Jul 25;150(1):1–14. doi: 10.1016/0022-2836(81)90321-1. [DOI] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985 Sep;42(2):573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Varmus H. E. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyk G., Leis J., Longiaru M., Skalka A. M. Selective cleavage in the avian retroviral long terminal repeat sequence by the endonuclease associated with the alpha beta form of avian reverse transcriptase. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6745–6749. doi: 10.1073/pnas.80.22.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyk G., Longiaru M., Cobrinik D., Kowal R., deHaseth P., Skalka A. M., Leis J. Circles with two tandem long terminal repeats are specifically cleaved by pol gene-associated endonuclease from avian sarcoma and leukosis viruses: nucleotide sequences required for site-specific cleavage. J Virol. 1985 Nov;56(2):589–599. doi: 10.1128/jvi.56.2.589-599.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger K. R., Wong E. A., Wahl G., Capecchi M. R. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol. 1982 Nov;2(11):1372–1387. doi: 10.1128/mcb.2.11.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Goubin G., Hill M. Monomer and multimer covalently closed circular forms of Rous sarcoma virus DNA. J Virol. 1979 Feb;29(2):799–804. doi: 10.1128/jvi.29.2.799-804.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Schiff R. D. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978 Aug;89(1):119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C. Site-specific nicking at the avian retrovirus LTR circle junction by the viral pp32 DNA endonuclease. Nucleic Acids Res. 1985 Sep 11;13(17):6205–6221. doi: 10.1093/nar/13.17.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kopchick J. J., Harless J., Geisser B. S., Killam R., Hewitt R. R., Arlinghaus R. B. Endodeoxyribonuclease activity associated with Rauscher murine leukemia virus. J Virol. 1981 Jan;37(1):274–283. doi: 10.1128/jvi.37.1.274-283.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung H. J., Shank P. R., Bishop J. M., Varmus H. E. Identification and characterization of dimeric and trimeric circular forms of avian sarcoma virus-specific DNA. Virology. 1980 Jun;103(2):425–433. doi: 10.1016/0042-6822(80)90201-9. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nissen-Meyer J., Nes I. F. Purification and properties of DNA endonucleases associated with Friend leukemia virus. Nucleic Acids Res. 1980 Nov 11;8(21):5043–5055. doi: 10.1093/nar/8.21.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. Circles with two tandem LTRs are precursors to integrated retrovirus DNA. Cell. 1984 Mar;36(3):673–679. doi: 10.1016/0092-8674(84)90347-7. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature. 1983 Nov 10;306(5939):155–160. doi: 10.1038/306155a0. [DOI] [PubMed] [Google Scholar]
- Pellicer A., Wigler M., Axel R., Silverstein S. The transfer and stable integration of the HSV thymidine kinase gene into mouse cells. Cell. 1978 May;14(1):133–141. doi: 10.1016/0092-8674(78)90308-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984 Jul;37(3):1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O., Gregg R. G., Boggs S. S., Koralewski M. A., Kucherlapati R. S. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985 Sep 19;317(6034):230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- Stacey D. W. Messenger activity of virion RNA for avian leukosis viral envelope glycoprotein. J Virol. 1979 Mar;29(3):949–956. doi: 10.1128/jvi.29.3.949-956.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabin C. J., Hoffmann J. W., Goff S. P., Weinberg R. A. Adaptation of a retrovirus as a eucaryotic vector transmitting the herpes simplex virus thymidine kinase gene. Mol Cell Biol. 1982 Apr;2(4):426–436. doi: 10.1128/mcb.2.4.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K. R., Folger K. R., Capecchi M. R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986 Feb 14;44(3):419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- Topp W. C. Normal rat cell lines deficient in nuclear thymidine kinase. Virology. 1981 Aug;113(1):408–411. doi: 10.1016/0042-6822(81)90168-9. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., Rands E., Chattopadhyay S. K., Lowy D. R., Verma I. M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982 Feb;41(2):542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]