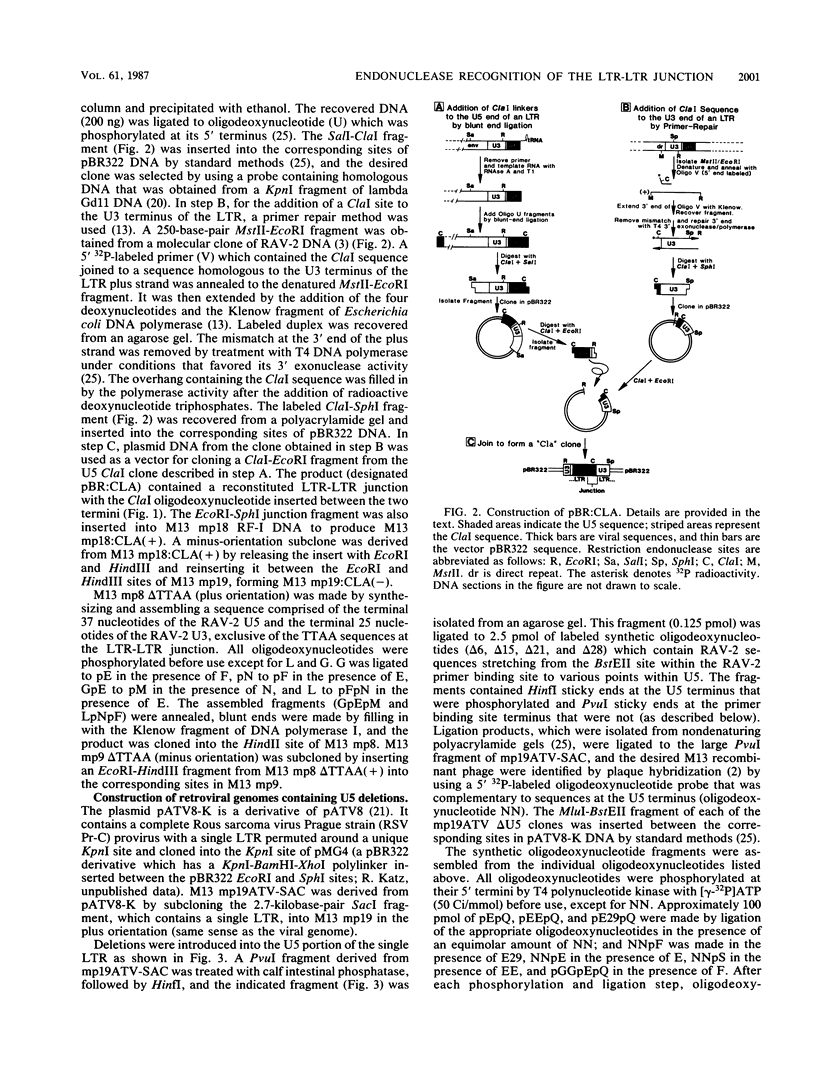
Abstract
Integration of retroviral DNA is a site-specific reaction involving an endonuclease encoded by the viral pol gene (pol-endo). In vitro the pol-endo from avian sarcoma and leukosis viruses (ASLVs) cleaves both DNA strands near the U5-U3 junction of tandem long terminal repeats (LTR-LTR junction) in single-stranded and replicative form (RF)-I substrates. We have reported previously that the sequences that are required for cleavage of single-stranded substrates by the alpha beta form of the pol-endo differ for the plus and minus strands (G. Duyk, M. Longiaru, D. Cobrinik, R. Kowal, P. deHaseth, A. M. Skalka, and J. Leis, J. Virol. 56:589-599, 1985). This is not the case with RF-I substrates, in which a maximum of 22 base pairs of U5 and 8 base pairs of U3 were required for alpha beta pol-endo cleavage in each strand. Insertion of a palindromic octanucleotide (CATCGATG) at the LTR-LTR junction abolished cleavage in RF-I but not in single-stranded DNA substrates. Deletion of the four nucleotides (TTAA) at the junction prevented cleavage in the plus strand of RF-I DNA, but did not affect cleavage of single-stranded DNA. Furthermore, the alpha beta form of ASLV pol-endo did not recognize heterologous LTR-LTR junction sequences from the reticuloendotheliosis virus or Moloney murine leukemia virus in either substrate form, despite their sequence and structural similarities to the ASLV junction. These results support a role for a sequence-specific interaction between the ASLV pol-endo and the LTR-LTR junction domains that are required for cleavage. By using the infectious Rous sarcoma virus clone pATV8-K, we introduced a set of deletions into the U5 region that would be incorporated into the LTR-LTR junction on viral replication. In the unintegrated provirus, the deletions started 43 base pairs from the LTR-LTR junction and extended various lengths toward the junction. Results of transfection studies with these clones indicated that the U5 sequences that are required for virus production in vivo correspond to those that are required for cleavage of RF-I DNA in vitro.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander F., Leis J., Soltis D. A., Crowl R. M., Danho W., Poonian M. S., Pan Y. C., Skalka A. M. Proteolytic processing of avian sarcoma and leukosis viruses pol-endo recombinant proteins reveals another pol gene domain. J Virol. 1987 Feb;61(2):534–542. doi: 10.1128/jvi.61.2.534-542.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bizub D., Katz R. A., Skalka A. M. Nucleotide sequence of noncoding regions in Rous-associated virus-2: comparisons delineate conserved regions important in replication and oncogenesis. J Virol. 1984 Feb;49(2):557–565. doi: 10.1128/jvi.49.2.557-565.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone L. R., Skalka A. Two species of full-length cDNA are synthesized in high yield by melittin-treated avian retrovirus particles. Proc Natl Acad Sci U S A. 1980 Feb;77(2):847–851. doi: 10.1073/pnas.77.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers M. H., Beaucage S. L., Efcavitch J. W., Fisher E. F., Goldman R. A., deHaseth P. L., Mandecki W., Matteucci M. D., Rosendahl M. S., Stabinsky Y. Chemical synthesis and biological studies on mutated gene-control regions. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):411–418. doi: 10.1101/sqb.1983.047.01.048. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985 Sep;42(2):573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- Duyk G., Leis J., Longiaru M., Skalka A. M. Selective cleavage in the avian retroviral long terminal repeat sequence by the endonuclease associated with the alpha beta form of avian reverse transcriptase. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6745–6749. doi: 10.1073/pnas.80.22.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyk G., Longiaru M., Cobrinik D., Kowal R., deHaseth P., Skalka A. M., Leis J. Circles with two tandem long terminal repeats are specifically cleaved by pol gene-associated endonuclease from avian sarcoma and leukosis viruses: nucleotide sequences required for site-specific cleavage. J Virol. 1985 Nov;56(2):589–599. doi: 10.1128/jvi.56.2.589-599.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W., Verma I. M. Studies on the reverse transcriptase of RNA tumor viruses. Structural relatedness of two subunits of avian RNA tumor viruses. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4991–4994. doi: 10.1073/pnas.71.12.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeddel D. V., Shepard H. M., Yelverton E., Leung D., Crea R., Sloma A., Pestka S. Synthesis of human fibroblast interferon by E. coli. Nucleic Acids Res. 1980 Sep 25;8(18):4057–4074. doi: 10.1093/nar/8.18.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S., Traktman P., Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981 Apr;38(1):239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb M., Grandgenett D. P. Endonuclease activity of purified RNA-directed DNA polymerase from avian myeloblastosis virus. J Biol Chem. 1979 Mar 10;254(5):1606–1613. [PubMed] [Google Scholar]
- Grandgenett D. P., Knaus R. J., Hippenmeyer P. J. Antibodies against a synthetic peptide of the avian retrovirus pp32 protein and the beta DNA polymerase subunit. Virology. 1983 Oct 15;130(1):257–262. doi: 10.1016/0042-6822(83)90137-x. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Schiff R. D. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978 Aug;89(1):119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C. Site-specific nicking at the avian retrovirus LTR circle junction by the viral pp32 DNA endonuclease. Nucleic Acids Res. 1985 Sep 11;13(17):6205–6221. doi: 10.1093/nar/13.17.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Swanstrom R., Olsen J. C. Nuclease mechanism of the avian retrovirus pp32 endonuclease. J Virol. 1986 Jun;58(3):970–974. doi: 10.1128/jvi.58.3.970-974.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishinuma F., DeBona P. J., Astrin S., Skalka A. M. Nucleotide sequence of acceptor site and termini of integrated avian endogenous provirus ev1: integration creates a 6 bp repeat of host DNA. Cell. 1981 Jan;23(1):155–164. doi: 10.1016/0092-8674(81)90280-4. [DOI] [PubMed] [Google Scholar]
- Katz R. A., Omer C. A., Weis J. H., Mitsialis S. A., Faras A. J., Guntaka R. V. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus DNA: structure of large terminal repeats in circle junctions. J Virol. 1982 Apr;42(1):346–351. doi: 10.1128/jvi.42.1.346-351.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopchick J. J., Ju G., Skalka A. M., Stacey D. W. Biological activity of cloned retroviral DNA in microinjected cells. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4383–4387. doi: 10.1073/pnas.78.7.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis J., Duyk G., Johnson S., Longiaru M., Skalka A. Mechanism of action of the endonuclease associated with the alpha beta and beta beta forms of avian RNA tumor virus reverse transcriptase. J Virol. 1983 Feb;45(2):727–739. doi: 10.1128/jvi.45.2.727-739.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. C., Swanstrom R. A new pathway in the generation of defective retrovirus DNA. J Virol. 1985 Dec;56(3):779–789. doi: 10.1128/jvi.56.3.779-789.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. Circles with two tandem LTRs are precursors to integrated retrovirus DNA. Cell. 1984 Mar;36(3):673–679. doi: 10.1016/0092-8674(84)90347-7. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature. 1983 Nov 10;306(5939):155–160. doi: 10.1038/306155a0. [DOI] [PubMed] [Google Scholar]
- Rho H. M., Grandgenett D. P., Green M. Sequence relatedness between the subunits of avian myeloblastosis virus reverse transcriptase. J Biol Chem. 1975 Jul 10;250(13):5278–5280. [PubMed] [Google Scholar]
- Schiff R. D., Grandgenett D. P. Partial phosphorylation in vivo of the avian retrovirus pp32 DNA endonuclease. J Virol. 1980 Dec;36(3):889–893. doi: 10.1128/jvi.36.3.889-893.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984 Jul;37(3):1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka A. M., Duyk G., Longiaru M., DeHaseth P., Terry R., Leis J. Integrative recombination--a role for the retroviral reverse transcriptase. Cold Spring Harb Symp Quant Biol. 1984;49:651–659. doi: 10.1101/sqb.1984.049.01.073. [DOI] [PubMed] [Google Scholar]
- Tanese N., Roth M. J., Goff S. P. Analysis of retroviral pol gene products with antisera raised against fusion proteins produced in Escherichia coli. J Virol. 1986 Aug;59(2):328–340. doi: 10.1128/jvi.59.2.328-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder N. D., Boeke J. D. The filamentous phage (Ff) as vectors for recombinant DNA--a review. Gene. 1982 Jul-Aug;19(1):1–10. doi: 10.1016/0378-1119(82)90183-4. [DOI] [PubMed] [Google Scholar]
- deHaseth P. L., Goldman R. A., Cech C. L., Caruthers M. H. Chemical synthesis and biochemical reactivity of bacteriophage lambda PR promoter. Nucleic Acids Res. 1983 Feb 11;11(3):773–787. doi: 10.1093/nar/11.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]