Abstract
The Drosophila melanogaster HSC3 and HSC4 genes encode Hsc70 proteins homologous to the mammalian endoplasmic reticulum (ER) protein BiP and the cytoplasmic clathrin uncoating ATPase, respectively. These proteins possess ATP binding/hydrolysis activities that mediate their ability to aid in protein folding by coordinating the sequential binding and release of misfolded proteins. To investigate the roles of HSC3 (Hsc3p) and HSC4 (Hsc4p) proteins during development, GAL4-targeted gene expression was used to analyze the effects of producing dominant negatively acting Hsc3p (D231S, K97S) and Hsc4p (D206S, K71S) proteins, containing single amino acid substitutions in their ATP-binding domains, in specific tissues of Drosophila throughout development. We show that the production of each mutant protein results in lethality over a range of developmental stages, depending on the levels of protein produced and which tissues are targeted. We demonstrate that the functions of both Hsc3p and Hsc4p are required for proper tissue establishment and maintenance. Production of mutant Hsc4p, but not Hsc3p, results in induction of the stress-inducible Hsp70 at normal temperatures. Evidence is presented that lethality is caused by tissue-specific defects that result from a global accumulation of misfolded protein caused by lack of functional Hsc70. We show that both mutant Hsc3ps are defective in ATP-induced substrate release, although Hsc3p(D231S) does undergo an ATP-induced conformational change. We believe that the amino acid substitutions in Hsc3p interfere with the structural coupling of ATP binding to substrate release, and this defect is the basis for the mutant proteins’ dominant negative effects in vivo.
INTRODUCTION
The Hsp70 protein family belongs to a class of molecular chaperones that are involved in a variety of essential cellular processes including de novo protein folding, oligomeric protein assembly, protein translocation across intracellular membranes, and protein degradation (Gething and Sambrook, 1992; Hartl, 1996). Hsp70 proteins bind and release unfolded polypeptides to prevent off-pathway folding reactions.
Hsp70 proteins have been shown to be structurally subdivided into an N-terminal ATPase domain and a C-terminal substrate-binding domain. The ATP binding/hydrolysis activity of Hsp70 allows it to cycle between conformations that have either high or low affinity for protein substrates (Hendrick and Hartl, 1993; Hartl, 1996). For example, studies have demonstrated that ATP binding promotes the release of bound substrate from Hsp70 (Palleros et al., 1993; Schmid et al., 1994) and peptide binding by Hsp70 stimulates its ATPase activity (Flynn et al., 1989; Sadis and Hightower, 1992), indicating the structural coupling between the ATPase- and peptide-binding domains. A variety of Hsp70 cofactors also help regulate the Hsp70 reaction cycle (Hartl, 1996).
The HSP70 family of Drosophila contains two heat-inducible members (HSP68 and HSP70) as well as five constitutively expressed members (HSC1–HSC5) (Rubin et al., 1993). The HSC4 protein (Hsc4p) is the most abundantly produced of the multiple cytoplasmic Hsc70 members, and the HSC3 protein (Hsc3p) is the sole endoplasmic reticulum (ER) Hsc70 family member. Hsc3p and Hsc4p are homologous to the mammalian ER protein BiP and the cytoplasmic clathrin-uncoating ATPase, respectively. We wished to determine whether Hsc3p and Hsc4p were essential for Drosophila viability or whether functionally related Hsc70 or Hsp70 proteins could compensate for their loss. It had been shown that yeast requires at least one cytoplasmic Hsp70 belonging to the Ssap family (Werner-Washburne et al., 1987) as well as the single ER Hsp70, Kar2p (Normington et al., 1989), for viability. Additionally, it was not known whether either cytoplasmic Hsc70 or ER Hsc70 would be required in all tissues and stages of development in a multicellular organism such as Drosophila.
Because there were no known null mutations for either of these genes at the time this work began, we created dominant negatively acting Hsc3p and Hsc4p that interfered with wild-type Hsc3p and Hsc4p activity, respectively. The structural information obtained from the x-ray crystallographic analysis of the ATPase domain of bovine Hsc70 (Flaherty et al., 1990) was used to select highly conserved residues that were thought to be essential for chaperone activity, and these residues were substituted in Drosophila Hsc3p and Hsc4p. Previous work by Rubin (1993) demonstrated that specific amino acid substitutions in the ATPase domain of both cytoplasmic Hsc4p and ER Hsc3p produced dominant negatively acting proteins in vitro and in vivo, respectively. The amino acid substitutions (D10S, K71S, E175S, D206S) in Hsc4p resulted in loss of function in an in vitro clathrin-uncoating assay, and mutant Hsc4p (D206S) was shown to dominantly inhibit uncoating by wild-type Hsc4p in this assay. The corresponding amino acid substitutions in Hsc3p (D35S, K97S, E201S, D231S) were produced in Drosophila using a heat-inducible HSP70 promoter in a wild-type background. Whereas Hsc3p (D35S) had little effect on viability, Hsc3p (E201S, K97S, and D231S) caused a dominant loss of viability, with K97S and D231S displaying the strongest effects.
Other workers have also demonstrated mutations that map to the ATP-binding domain of the Escherichia coli, Hsp70, DnaK, or eukaryotic ER Hsc70, produced dominant negatively acting proteins in vivo. In a genetic screen conducted to isolate dnak mutations that affected regulation of the E. coli heat shock response (Wild et al., 1992), the dominant negative class of dnak mutations all mapped to the ATP-binding site. Furthermore, production of a dominant negative hamster BiP ATPase mutant (T37G) in mammalian cells resulted in vesiculation of the ER membranes (Hendershot et al., 1995). A biochemical characterization of various ATP-binding site mutations in hamster BiP (Wei et al., 1995), bovine Hsc70 (Wilbanks et al., 1994), and E. coli DnaK (Buchberger et al., 1994; Kamath-Loeb et al., 1995) revealed that certain amino acid substitutions specifically interfered with either ATP binding, ATP hydrolysis, or an ATP-induced conformational change in the protein.
In this study, we use GAL4-targeted gene expression (Brand and Perrimon, 1993) to analyze the effects of producing dominant negatively acting cytoplasmic Hsc4p (D206S, K71S) and ER Hsc3p (D231S, K97S) in specific tissues of Drosophila throughout their development. We show that the activity of both the ER and cytoplasmic Hsc70 is required throughout Drosophila development for both formation and maintenance of the specific tissues examined. Additionally, we present evidence suggesting that the lethality in flies is caused by tissue-specific defects that resulted from a global accumulation of misfolded protein resulting from lack of functional Hsc70. Finally, evidence is presented that the amino acid substitutions D231S and K97S in Hsc3p interfere with the structural coupling of ATP binding to substrate release, and this defect is the basis for the mutant HSC3 proteins’ dominant negative effects in vivo.
MATERIALS AND METHODS
Mutagenesis, Cloning, and DNA-sequencing Procedures
pUAST Constructs.
The HSC3-coding region carrying the wild-type sequence or mutations D231S or K97S were shuttled as a 2.1-kilobase (kb) EcoRI fragment from the pHS-Casper vector (Rubin, 1993) to the UAS P-element vector (pUAST) (Brand and Perrimon, 1993). The resulting constructs were called 3/WT/UAS, 3/231/UAS, and 3/97/UAS. The HSC4-coding region carrying the wild-type sequence in pUC18 was excised as a 2.2-kb EcoRI fragment and inserted into the pUAST vector to create 4/WT/UAS. To create 4/206/UAS, a BamHI/XhoI fragment carrying the mutation D206S from plasmid pT7HSC4-(D206S) (Rubin, 1993) was cloned into the HSC4-coding region in place of the corresponding wild-type DNA. The complete mutant HSC4-coding region was cloned into the pUAST vector as a 2.2-kb EcoRI fragment. To create 4/71/UAS, the HSC4 wild-type coding region was inserted into the pALTER phagemid vector. Single-stranded mutagenesis procedures (altered site mutagenesis system, Promega, Madison, WI) were used to create the K71S mutation, and the complete mutant HSC4-coding region was cloned as a 2.2-kb EcoRI fragment into the pUAST vector. DNA sequencing (Sequenase Version 2.0 Kit, United States Biochemical, Cleveland, OH) of the regions surrounding each mutation in all final pUAST constructs was performed to confirm the presence of each mutation.
Bacterial Expression Vector pET-15b Constructs.
Construction of pET-15b constructs, carrying the mutant HSC3 (D231S, K97S)-coding regions, were initiated by performing PCR reactions using P-element vector constructs 3/231/UAS and 3/97/UAS as templates. Primers were selected so that the coding region was begun at the start site of the mature protein, directly after the leader sequence cleavage site. The PCR-generated 2.0-kb NdeI/BamHI mutant HSC3 (D231S, K97S) fragments were cloned into pET-15b, downstream of the T7 RNA polymerase promoter, prokaryotic ribosomal binding site, and N-terminal 6X-histidine tag. The resulting pET vector constructs were called 3/97/pET and 3/231/pET. DNA sequencing of the regions surrounding each mutation in all final pET vector constructs was performed to confirm the presence of each mutation.
Creation of P-Element-transformed Fly Lines Containing Mutant or Wild-Type HSC3 and HSC4 Genes
P-element germ-line transformations with the pUAST constructs were performed as described by Robertson et al. (1988) to create fly lines containing wild-type or mutant HSC3 and HSC4 genes under the control of the UAS promoter. To determine on which chromosome the P-element inserted, lines heterozygous for the TM3 or TM6 balancers were mated to w126 flies and segregation of the w+ marker was scored: if segregation of w+ was neither with a third chromosome balancer or a sex chromosome, it was inferred to segregate with the second chromosome. Balancer chromosomes were subsequently crossed away by successive matings to w126.
Ab Staining of Embryos
Embryos were dechorionated in 50% Clorox and fixed 5 min in a 1:1 mix of heptane/4% paraformaldhyde in PBS, and the vitelline membranes were removed by shaking in heptane/methanol. Eggs were immediately washed with two 5-min changes of 1 ml of PT (PBS, 0.1% Tween 20). The embryos were incubated overnight at 4°C in a 1:500 dilution of rabbit anti-β-galactosidase Ab (Cappel, West Chester, PA) or a 1:1000 dilution of rabbit anti-HRP Ab (Cappel). After two 30-min washes in 1 ml PT, embryos were incubated overnight at 4°C in a 1:500 dilution of biotinylated anti-rabbit secondary Ab (Vector Laboratories, Burlingame, CA). The next morning, embryos were incubated for 1 h in avidin and biotinylated HRP (Vectastain Elite ABC kit, Vector Laboratories). Peroxidase was detected using diaminobenzidine (0.3 mg/ml in PT) as a substrate. Embryos were dehydrated in ethanol and cleared and mounted in xylene (Sigma Chemical, St. Louis, MO) and viewed with a Zeiss Axioplane microscope (Carl Zeiss, Thornwood, NY). Photographs were taken with Kodak Technical Pan film at ASA 100 (Eastman Kodak, Rochester, NY).
Protein Sample Preparation, Western Blots, and One-dimensional (1D) and Two-dimensional (2D)-PAGE
Protein samples from heat-shocked flies were prepared for 2D-PAGE analysis by placing flies in a preheated 37°C standard media vial containing a moistened paper towel. Flies were heat shocked for 2.5 h at 37°C. Three heat-shocked adult female flies were placed in 50 μl of sonication buffer (10 mM Tris, pH 7.4, 5 mM MgCl2, 50 μg/ml Rnase A, 10 μg/ml chymostatin, 1 mM PMSF, and 10 μg/ml leupeptin). The samples were homogenized, and then sonicated for 1 min at 4°C (Ultra Systems Sonicator). Dnase I was added to 20 μg/ml and samples were incubated for 15 min. Solid urea (46 mg) was added to each sample followed by 77 μl of lysis buffer (9.5 M urea, 2% NP-40, 1.6% ampholines, pH 5–7, 0.4% ampholines, pH 3–10, 5% mercaptoethanol). Samples were microfuged to pellet any insoluble material before the equivalent of one fly was loaded per gel. Larval protein samples were prepared for 2D-PAGE as described above. The samples were analyzed by 2D-PAGE as described previously (O’Farell, 1975). Isoelectric focusing was performed using a pH gradient between 5 and 7. Second-dimension SDS-PAGE analysis was performed as described by Laemmli (1970) using 10% polyacrylamide gels. Western blot analysis was performed as described previously (Palter et al., 1986).
Antibodies
Antibodies employed are as follows: rat mAb to Hsc3p (clone 5/80.5, kindly provided by Belinda Bullard), rat mAb 7.FB, specific for heat-inducible hsp70 and rat mAb 7.10, specific for the C terminus of Hsc70 (both antibodies kindly provided by Susan Lindquist). The rabbit polyclonal Ab specific for Hsc4p was produced as follows: the HSC4-specific peptide CLDEDNLKTKISDSDRTT was synthesized and coupled to a carrier protein (keyhole lympet hemocyanin) (synthesized by Quality Controlled Biochemicals, Hopkinton, MA) for use as an immunogen in rabbits. The resulting polyclonal Hsc4p antibodies were purified using an HSC4 peptide affinity column as described (Sulfolink coupling gel, Pierce Chemical, West Chester, PA).
Purification of Wild-Type and Mutant Hsc3p
BL21(D3) bacterial strains containing either 3/WT/pET, 3/97/pET, or 3/231/pET were grown at 37°C in 1 l of LB containing 50 μg/ml carbenicillin to an OD600 of 0.7 before being induced by 1 mM isopropyl-β-d-thiogalactopyranoside. After growing an additional 2 h, induced cells were pelleted at 4000 rpm and resuspended in 50 ml cold Buffer A (0.2 M NaCl, 5 mM MgCl2, 20 mM Tris, pH 7.0, 1 mM DTT, 10 μg/ml chymostatin and leupeptin, 1 mM PMSF). Cells were lysed by the addition of 1 mg/ml lysozyme, incubated on ice for 1 h, and then sonicated for three 30-s bursts at 4°C at setting 4 (Ultra Systems Sonicator) to shear bacterial DNA. The protein extract was centrifuged for 1 h at 100,000 × g to remove insoluble material before half the extract was loaded onto a 2-ml ATP-agarose affinity column (ATP-agarose; Sigma Chemical) preequilibrated with Buffer A. After loading, the column was run as described by Mehta (1993). Fractions containing ATP-eluted Hsc3p were pooled together, and proteins were precipitated using 70% saturated (NH4)2SO2 to remove free ATP. Proteins were resuspended in 10 ml Buffer B (5 mM imidizol, 0.5 M NaCl, 20 mM Tris, pH 7.9), dialyzed against three changes of Buffer B, and then loaded onto a 0.5-ml Ni-NTA column (Qiagen, Chatsworth, CA) preequilibrated with Buffer B. After loading, the column was run as described by the manufacturer (Qiagen). Hsc3ps were eluted from Ni-NTA with Buffer B containing 400 mM imidizole, and then dialyzed against three changes of Buffer C (40 mM Tris-HCl, pH 7.6, 10 mM Mg (Oac)2, 20 mM NaCl, 20 mM KCl, 0.3 mM EDTA, 2 mM DTT).
Trypsin Digestion of Wild-Type or Mutant Hsc3p
Proteolytic cleavage of Hsc3p was carried out essentially as described previously (Buchberger et al., 1994) by preincubating 10 μg of Hsc3p in Buffer C (total volume of 20 μl) for 30 min at 30°C in the presence of either 2 mM ATP, ADP, or no nucleotide. After starting the proteolytic digestion of Hsc3p by the addition of 0.15 μg trypsin, aliquots were taken at 15- and 45-min time points and added directly to 3× SDS sample buffer (Laemmli, 1970). Samples were boiled immediately to stop the reaction and analyzed by 1D SDS-PAGE using 12% polyacrylamide gels. The proteolytic digestion fragments were detected using either Coomassie blue stain or Silver Stain Plus (Bio-Rad Laboratories, Hercules, CA).
Protein Labeling and Immune Complex Precipitation
Ten salivary glands from third instar larvae were detached in Ringer’s solution and immediately placed into a droplet of 30 μl Robb’s medium minus methionine containing 35S-labeled l-methionine (1.25 μCi/μl) in a small Petri dish. The glands were incubated at 25°C for 1 h in a humid chamber before being transferred to an Eppendorf tube containing 1 ml Buffer D (0.5% NP-40, 50 mM Tris, pH 8.0, 8 mM EDTA, or 10 U apyrase, 10 μg/ml leupeptin and chymostatin, 1 mM PMSF). Glands were washed in 1 ml Buffer D, centrifuged 30 s, and resuspended in 50 μl Buffer D. Protein extracts were prepared by lysing glands by sonicating at setting 4 (Ultra Systems Sonicator) for 1 min on ice. Radioactive incorporation was determined by trichoroacetic acid precipitation on glass fiber filters.
Coimmunoprecipitations were performed essentially as described previously (Hendershot et al., 1995). All steps were performed at 4°C. For each coimmunoprecipitation sample, 650,000 cpm of labeled clarified protein lysate was adjusted to 100 μl with Buffer D. Rat mAb to Hsc3p (50 μl) was added to each sample (or no primary Ab for a background control), and the samples were incubated at 4°C for 1 h on a rotator. A 1:500 dilution of secondary rabbit anti-rat Ab (Cappel) was added to each sample (including the background control) and slowly rotated for an additional 30 min. Protein A-Sepharose beads (100 μl, 10% beads in Buffer D) were added to each sample. After rotating at 4°C for 1 h, the beads were washed with three 1-ml changes of Buffer D and then two additional changes of 1 ml ATPase buffer (20 mM HEPES, pH 7.0, 75 mM KCl, 2 mM MgCl2). The immune-complexed beads were resuspended in 1 ml ATPase buffer and divided equally into two tubes. ATPase buffer (0.5 ml), with or without 2 mM ATP, was added and samples were slowly rotated for 30 min at room temperature. The beads were washed with three 1-ml changes of Buffer D, and then resuspended in 15 μl 1× SDS-sample buffer. Protein bands were eluted from the beads by heating to 85°C for 10 min and the proteins were analyzed by SDS-PAGE using 10% polyacrylamide gels. Proteins were visualized by fluorography using EN3HANCE (Dupont, Boston, MA) and were quantitated by scanning densitometry using a Personal Densitometer Scanner (model 3554) and Image Quantitator 3.3 program (Molecular Dynamics, Sunnyvale, CA)
RESULTS
The Establishment of Independent Fly Lines Expressing Different Levels of Mutant Hsc3p and Hsc4p
It was not known whether the production of mutant Hsc3p and Hsc4p in specific tissues of Drosophila in a wild-type background would cause observable phenotypes. To increase the likelihood of producing mutant phenotypes, we wanted to select flies that produced a sufficiently high ratio of mutant to wild-type protein in specific tissues. When the GAL4-targeted gene expression system is used as a method of gene induction, there are three factors that influence the amount of protein produced in a specific tissue: 1) the level of the GAL4 transcription factor induced in a given specific tissue, 2) the stage in development when GAL4-mediated expression begins and 3) the variable levels of HSC3 or HSC4 gene expression in different transformed fly lines, which depends on the integration site of the UAS-driven P-element–transposed gene. Therefore, multiple independent fly lines were created for each mutant and wild-type HSC3 and HSC4 construct. It is inferred that when different GAL4 lines are used to induce gene expression in a certain tissue, the absolute level of mutant protein produced (and therefore the exact ratio of mutant to wild-type protein) would change with each GAL4 line used. However, the relative level of mutant protein produced between each independently transformed fly line (high, medium, or low; Table 1) would remain the same regardless of the GAL4 lines used.
Table 1.
Levels of expression in independently derived HSC3 and HSC4 fly lines
| Transgenic fly linea | Levels of expressionb |
|---|---|
| 4/WT/A | + |
| 4/WT/B | ++ |
| 4/206/D | + |
| 4/206/E | ++ |
| 4/71/I | + |
| 4/71/G | ++ |
| 3/WT/A | + |
| 3/WT/B | ++ |
| 3/231/A | + |
| 3/231/D | +++ |
| 3/97/B | + |
| 3/97/D | +++ |
4/WT, HSC4 wild-type gene; 4/206, HSC4 with D206 substitution; 4/71, HSC4 with K71S substitution; 3/WT, HSC3 wild-type gene; 3/231, HSC3 with D231 substitution; 3/97, HSC3 with K97 substitution. Amino acid substitutions in HSC4(D206S, K71S) correspond to those substitutions in HSC3(D231S, K97S).
Fly strains producing the highest and lowest levels of accumulated mutant or wild-type proteins where +++ is ∼75–85% the level of wild-type proteins, ++ is ∼35–55% the level of wild-type protein, and + is ∼20–30% the level of wild-type protein.
In order to accurately assess the levels of mutant protein produced by each fly strain, it was important that the mutant proteins be expressed in a wide range of cell types so that they could be easily detected in total fly protein extracts. Therefore, enhancer trap line 337 (Manseau et al., 1997), a strain that ubiquitously expresses high levels of the GAL4 transcription factor, was chosen to quantitate the relative levels of mutant and wild-type Hsc3p and Hsc4p. When flies from GAL4 line 337 were mated to flies from lines containing either wild-type HSC3 or HSC4 gene constructs, no adverse effects were observed in the resulting progeny. However, when flies from GAL4 line 337 were mated to flies from each of the mutant fly lines, all of the resulting progeny died during either the first- or second-instar larval stage. Therefore, this analysis was performed on staged early first-instar larvae from each independently transformed fly line.
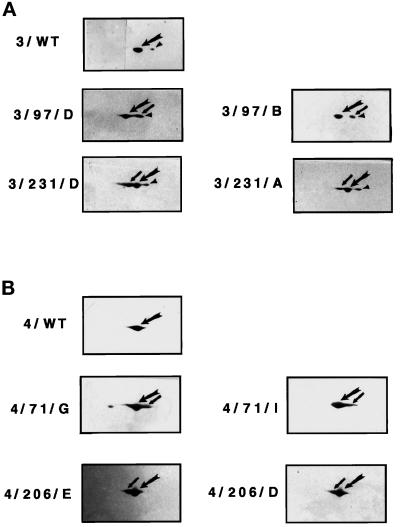
The amino acid substitutions in mutant Hsc3p and Hsc4p alter their pIs, changing their electrophoretic mobility, and thus enable them to be distinguished from wild-type proteins on 2D gels. Hsc3p is normally present in two distinct forms: the unmodified protein and the more acidic ADP-ribosylated modified protein (Rubin et al., 1993). The relative levels of mutant to wild-type protein for each fly line were determined by 2D-PAGE/Western blot analysis (Figure 1, A and B; note that the mutant Hsc3p does not run as a modified form). A minimum of four independent fly lines for each construct were used. For fly strains producing mutant Hsc3p (D231S, K97S), the level of accumulated mutant protein ranged from ∼30 to 85% of the wild-type Hsc3p. For fly strains producing mutant Hsc4p (D206S, K71S), the level of accumulated mutant protein ranged from ∼20 to 55% of the level of wild-type Hsc4p (Table 1; Figure 1, A and B).
Figure 1.
Level of mutant Hsc3p and Hsc4p produced in independent transformed fly lines. Flies homozygous for the GAL4 337 (ubiquitous) transgene were crossed to flies homozygous for either wild-type or mutant HSC3 or HSC4. 2D-PAGE/Western blot analysis using antibodies specific for either Hsc3p or Hsc4p was performed on equivalent amounts of total protein isolated from 60 staged, first-instar larvae progeny from each of the above crosses. Panel A represents HSC3 expression; panel B represents HSC4 expression. Small arrow points to mutant protein; large arrow points to wild-type protein; arrowhead points to modified wild-type Hsc3p. The acidic portion of the gel is to the right and the basic portion is to the left.
Expression of Varying Levels of Mutant HSC3 and HSC4 Produce a Range of Dominant Negative Effects
The level of mutant HSC3 and HSC4 gene expression varied between different transformants (Table 1). These variable levels of gene expression allowed us to assay the phenotypic effects of producing increasing levels of mutant protein in specific tissues of the flies. We wished to determine whether there would be a threshold level of mutant protein necessary to produce an observable mutant phenotype and if so, whether this mutant phenotype would be observed only at a particular stage of development. Also, would different levels of mutant protein produce different phenotypes?
To answer these questions, each of the highest and lowest expressing independent fly lines containing either wild-type or mutant HSC3 (D231S, K97S) and HSC4 (D206S, K71S) were mated to the muscle/mesoderm GAL4 line 24-B (Brand and Perrimon, 1993), and the resulting phenotypes were observed. The GAL4 line 24-B was chosen for use in this study because of its relatively high level of tissue-specific GAL4-mediated expression and its production of GAL4 early in embryonic development (beginning ∼3.5 h after fertilization).
We found that fly lines expressing different levels of wild-type Hsc3p and Hsc4p in the mesoderm/muscles exhibited a normal phenotype. However, fly lines expressing different levels of mutant Hsc3p and Hsc4p in the mesoderm/muscles died at different stages of development, ranging from embryogenesis to late pupation (Table 2). For each construct, flies producing higher levels of mutant protein exhibited lower viability and died earlier in development. Specifically, we observed that higher levels of protein accumulation for each mutant HSC3 and HSC4 construct correlated with flies dying earlier in development (Tables 1 and 2; 3/231/D (+++)>3/231/A(+); 3/97/D(+++)>3/97/B(+); 4/206/E(++)>4/206/D(+); 4/71/G(++)>4/71/I(+)). The expression of mutant HSC4 (D206S) in the mesoderm/muscles produced the strongest dominant negative phenotype. This effect was observed even when protein levels of HSC4 (D206S) were equivalent to or less than protein levels of mutant HSC3 (D206S, K97S). Flies expressing HSC4 (D206S) died as early as late embryogenesis (Tables 1 and 2; 4/206/E(++)>3/231/D(+++) and 3/97/D(+++)).
Table 2.
Expression of variable levels of mutant HSC3 and HSC4 produce a range of dominant negative effects
| Fly line | Total eggs | No. of eggs that hatch | % of eggs that die as embryosa | Outcome of surviving embryosb |
|---|---|---|---|---|
| 4/WT/A | 349 | 303 | 13% | Eclose as adult flies |
| 4/WT/B | 324 | 290 | 10.5% | Eclose as adult flies |
| 4/206/D | 298 | 155 | 48% | First-instar larvae |
| 4/206/E | 327 | 104 | 68% | First-instar larvae |
| 4/71/I | 295 | 265 | 11% | Late pupae |
| 4/71/G | 306 | 270 | 12% | Second-instar larvae |
| 3/WT/A | 329 | 276 | 16% | Eclose as adult flies |
| 3/WT/B | 332 | 302 | 10% | Eclose as adult flies |
| 3/231/A | 325 | 287 | 12% | 85% die as late pupae; 15% eclose as adult flies |
| 3/231/D | 286 | 237 | 17% | Second-instar larvae |
| 3/97/B | 340 | 305 | 11% | Late pupae |
| 3/97/D | 312 | 286 | 8% | Second-instar larvae |
Flies, homozygous for either mutant or wild-type HSC3 or HSC4 were mated to flies homozygous for the mesoderm/muscle GAL4 line 24-B. The progeny embryos were allowed to develop for 32 h at 25°C before the hatched egg casings were counted to determine the percentage of eggs that hatched. We found that ∼10–15% of eggs normally die as embryos using the standard fly line w126. Developing larvae were observed either on apple juice agar plates or transferred to standard media vials to determine when in development the flies died.
Developmental times (in hours) of Drosophila at 25°C: 22 h, first instar larvae hatches from egg; 70 h, second larval molt; 120 h, pupation; 220 h, eclosion (emergence of adult flies).
To determine whether additional wild-type Hsc3p would rescue flies producing mutant Hsc3p (D231S) in a mesoderm/muscle pattern, fly strains were produced that were both homozygous for mutant HSC3 (D231S) (line 3/231/A; Table 1) and carried an extra P-element–transposed copy of the wild-type HSC3 gene on a third chromosome balancer (TM6+) in addition to the endogenous wild-type HSC3 gene on the X chromosome. Such flies were crossed to the mesoderm/muscle GAL4 line, and the viability of the progeny was scored. In this genetic background, without additional wild-type Hsc3p, 1–2% of the flies eclosed whereas when an extra copy of the wild-type HSC3 transgene was present in flies expressing mutant Hsc3p, viability was 16% (our unpublished results). These results demonstrated that the observed lethality was caused by the presence of mutant Hsc3p(D231S) since it could be alleviated by increasing the level of wild-type Hsc3p.
Larvae Expressing Mutant HSC4, but Not HSC3, Undergo a Stress Response Indicated by the Presence of the Heat-inducible Hsp70 Protein
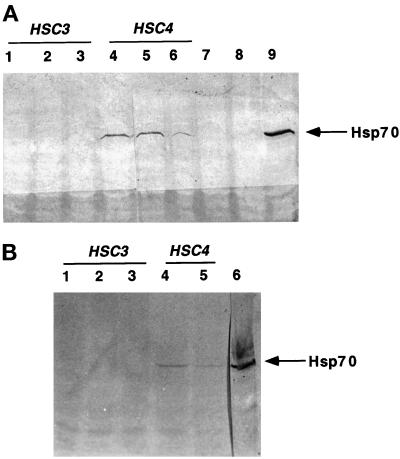
In response to cellular accumulation of misfolded proteins, Drosophila undergo a stress response that is characterized by the presence of the heat-inducible Hsp70 and Hsp68 proteins (Craig and Gross, 1991). If production of mutant Hsp70 resulted in an accumulation of misfolded proteins, one might expect to observe induction of the heat-inducible Hsp70 and Hsp68. Therefore, we wished to determine whether production of mutant cytoplasmic Hsc4p or mutant ER Hsc3p would cause a stress response at normal temperatures. Western blot analysis using an Ab specific to the heat-inducible Hsp70 was performed on equivalent amounts of total protein isolated from 120 staged first-instar larvae expressing either wild-type or mutant HSC3 or HSC4 in the mesoderm/muscles (GAL4 line 24-B) or a ubiquitous (GAL4 line 337) cell type pattern (Figure 2). Although expression of either the highest levels of mutant HSC4 (D206S, K97S) (lines 4/206/E, 4/71/G) and mutant HSC3 (D231S, K97S) (lines 3/231/D, 3/97/D) resulted in loss of viability for both GAL4 lines tested, the stress-inducible Hsp70 protein was only induced in response to production of mutant Hsc4p. Protein samples were analyzed from stages before developmental arrest, and, therefore, heat-inducible Hsp70 was not induced as a consequence of cell death.
Figure 2.
Larvae producing mutant Hsc4p undergo a stress response at normal temperatures, indicated by the presence of the heat-inducible Hsp70. Flies homozygous for either transgenes GAL4 24-B (mesoderm/muscle) or 337 (ubiquitous) were crossed to flies homozygous for either HSC4/WT, HSC3/WT, HSC3 (D231S, K97S), or HSC4 (D206S, K71S). Western blot analysis using Ab 7.FB (specific to heat-inducible Hsp70) was performed on equivalent amounts of total protein isolated from 60 staged, first-instar larvae progeny from each of the above crosses. Panel A shows GAL4 line 24-B mesoderm/muscle expression: lane 1, line 3/WT/B; lane 2, 3/97/D; lane 3, 3/231/D; lane 4, 4/71/G; lane 5, 4/206/E; lane 6, 4/206/D; lane 7, 4/WT/B; lane 8, w126 (negative control); lane 9, w126 (flies heat shocked for 1.5 h at 37°C; positive control). Panel B shows GAL4 line 337 ubiquitous expression: lane 1, line 3/231/D; lane 2, 3/97/D; lane 3, 4/WT/B; lane 4, 4/71/G; lane 5, 4/206/E; lane 6, w126 (flies heat shocked for 1.5 h at 37°C; positive control). Thick arrow denotes Hsp70 protein.
Specific Expression of Mutant HSC4 in Mesoderm/Muscle Causes Muscle Malformation
We observed that when the highest levels of mutant Hsc3p (line 3/231D) and Hsc4p (line 4/206/E) were produced in the mesoderm/muscle cells of embryos, the phenotype of the unhatched eggs and dying larvae resembled those of known muscle mutants as described by Fyrberg et al. (1994). Prelarvae that died were unable to puncture the egg membrane and hatch, and those larvae that did hatch were unable to crawl. Therefore, we wished to determine whether mutant Hsc3p and Hsc4p induced malformation of the muscles in Drosophila embryos, causing the observed muscle mutant phenotypes.
To readily visualize the muscle pattern in whole-mount embryos, we used a myosin heavy chain promoter-β-galactosidase (MHC-β-gal) transgene that confers lacZ expression in the developing muscles. A fly line was created containing both GAL4 24-B (mesoderm/muscle) and MHC-β-gal transgenes. These flies were mated to flies producing the highest levels of either wild-type Hsc4p (line 4/WT/B), mutant Hsc3p (line 3/231D), or mutant Hsc4p (line 4/206/E) to induce mutant protein production in the mesoderm/muscles. Wild-type and mutant embryos were stained with anti-β-galactosidase antibodies to examine their muscle pattern. A minimum of 100 mutant or wild-type embryos at 13–14.5 h of development were examined in detail, as this is when the mature muscle pattern has fully formed (Drysdale et al., 1993). Mutant embryos from each of the crosses were allowed to develop and were shown to die at the previously observed times in development (Table 2). In addition, the mutant’s cuticle and denticle belts, which reflect the pattern of the underlying epidermis, were found to be identical in mutant and wild-type embryos. Because the muscles insert into the epidermis (Drysdale et al., 1993), any muscle defects observed could not be caused by irregularities in the epidermal pattern and therefore could be attributed to the muscles themselves.
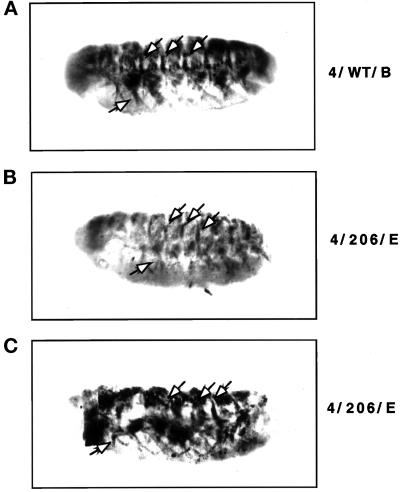
We found that embryos that produce high levels of either wild-type Hsc4p or mutant Hsc3p (D231S) in the mesoderm/muscle cells formed a characteristic wild-type muscle pattern, as described by Drysdale et al. (1993) (Figure 3A). However, embryos producing high levels of mutant Hsc4p in the mesoderm/muscle cells displayed a range of severe muscle pattern defects. First, the overall intensity of lacZ expression in mutant HSC4 embryos was significantly weaker than in wild-type embryos, most likely due to loss of muscle cells expressing lacZ (note: to visualize the weakly stained muscles, embryos are overstained and photographs are overexposed; Figure 3, B and C). Furthermore, the mutant HSC4 embryos also exhibited gaps in the muscle pattern (Drysdale et al., 1993), indicating the loss of muscle fibers, again suggesting muscle-specific cell death. Finally, the mutant embryos displayed an irregular muscle pattern when compared with that of the wild-type embryos.
Figure 3.
The production of mutant Hsc4p in a mesoderm/muscle-specific pattern causes muscle malformation in developing Drosophila embryos. Flies carrying both the MHC-β-gal and GAL24-B (mesoderm/muscle) transgenes were crossed to flies homozygous for either HSC4/WT (line 4/WT/B), HSC3 (D231S) (line 3/231/D), or HSC4 (D206S) (line 4/206/E) and the progeny embryos were stained with anti-β-galactosidase antibodies. Embryos are shown at ∼14.5 h of development (stage 16). Embryos are aligned with their anterior end to the left and their ventral side down. (A) HSC4/WT expression. HSC3 (D231S) embryos exhibit a wild-type muscle pattern and are therefore not shown. (B and C) Mutant HSC4 (D206S) expression. Mutant embryos (B) exhibited significantly weaker β-galactosidase staining when compared with wild-type embryos under identical conditions. To visualize the weakly stained muscles, it was necessary to overexpose the photographs and overstain the mutant embryos, thus increasing background staining. The embryo in panel B is presented in a focal plane to show its malformed segmentally repeated muscle fibers. All mutant embryos (B and C) exhibit gaps in their muscle patterns as well as malformation of the muscle fiber pattern when compared (see arrows).
Interestingly, the production of mutant Hsc4p in the mesoderm/muscle tissue caused a more severe dominant negative effect than that of mutant Hsc3p. For example, although production of either mutant Hsc3p or Hsc4p in the mesoderm/muscle tissues of flies both produced a dominant lethal phenotype, only mutant Hsc4p production caused death as early as embryogenesis (Table 2) and malformation of the muscles in the developing embryo. Because flies producing mutant Hsc3p formed a normal muscle pattern during embryogenesis and yet showed evidence of paralysis later in development, the ER Hsc3p is likely required to maintain muscle function throughout development.
Specific Expression of Mutant HSC3 and HSC4 in the Nervous System Causes Defects in Developing Larvae
Directly after hatching, wild-type larvae normally burrow into the media of a standard food vial before crawling up the vial walls, ∼5 d later, as third-instar larvae. However, we observed that larvae producing the highest levels of mutant Hsc3p or Hsc4p in the CNS, peripheral nervous system (PNS), and brain (using GAL4 lines 266Y, 156Y; Manseau et al., 1997) crawled up the walls of the media vials almost immediately after hatching, where they died 2–3 d later.
To further study this defect, a behavioral assay was performed in which embryos producing either wild-type Hsc4p (line 4/WT/B), Hsc3p (D231S) (line 3/231/D), or Hsc4p(D206S)(line 4/206/E) in the PNS, CNS, and brain were allowed to hatch on an apple juice agar plate. A yeast paste food source was added to the middle of the plate, and the first-instar larvae were scored after a time interval to determine whether they could locate the food. We found that the majority of newly hatched first instar wild-type and mutant HSC3 (D231S) larvae were able to sense, locate, and burrow into the yeast paste food source within ∼20 min. The majority of the mutant HSC4 (D206S) larvae were unable to locate the food source and subsequently died scattered randomly on the agar plate (Table 3). To determine whether the mutant HSC3 (D231S) (line 3/231/D) larvae would exhibit this abnormal behavior later in development, staged 2-d-old mutant and wild-type larvae were transferred from a standard media vial to the edges of an apple juice agar plate, and the behavioral assay was repeated. We found that whereas the wild-type larvae could locate the food source, the mutant HSC3(D231S) larvae could not and died scattered on the plate (Table 4). Mutant HSC3 and HSC4 larvae that accidentally did encounter the food source were unable to burrow into it and also died. The crawling of the mutant HSC3 and HSC4 larvae was significantly slower than that of the wild-type larvae, which may also have contributed to their inability to locate and burrow into the food source.
Table 3.
Newly hatched larvae producing mutant Hsc4p in the CNS, PNS, and brain exhibit a behavioral abnormality
| Fly line | Total larvae tested | No. of larvae in yeast after 20 min | % larvae that find yeast |
|---|---|---|---|
| 4/WT/B | 144 | 120 | 83 |
| 4/206/E | 120 | 20 | 16 |
| 3/231/D | 133 | 118 | 88 |
Embryos expressing either wild-type Hsc4p, mutant Hsc4p (D206S), or mutant Hsc3p (D231S) in the CNS, PNS, and brain (GAL4 line 156Y) were allowed to hatch on an apple juice agar plate. A yeast paste food source was added to the middle of the plate 5 h after hatching, and the first-instar larvae were scored after a 20-min time interval to determine whether they could locate the food.
Table 4.
Two-day-old larvae producing mutant Hsc3p in the CNS, PNS, and brain exhibit a behavioral abnormality
| Fly line | Total larvae tested | No. of larvae in yeast after 20 min | % larvae that find yeast |
|---|---|---|---|
| 4/WT/B | 74 | 63 | 85 |
| 3/231/D | 68 | 12 | 18 |
Two-day-old larvae producing either wild-type Hsc4p or mutant Hsc3p (D231S) in the CNS, PNS, and brain were transferred from standard media vials to an apple juice agar plate, and the behavioral assay (see Table 3) was repeated.
To detect any abnormalities in the neuronal pattern of the mutant HSC3 or HSC4 embryos, antibodies specific for HRP that bind to neuronal membranes in Drosophila (Jan and Jan, 1982) were used as a specific neuronal marker. However, both mutant HSC3 and HSC4 embryos exhibited no obvious defects in their neuronal Ab-staining pattern (our unpublished data). As larvae producing either mutant Hsc3p or Hsc4p in the PNS, CNS, and brain exhibited behavioral defects as well as larval lethality, it is unlikely that these proteins have no function in the nervous system. Therefore, these results suggest that because GAL4-mediated expression in the nervous system came on relatively late during embryonic development for all the nervous system GAL4 lines tested (stage 13/14), a normal embryonic neuronal pattern was established. However, later expression of mutant HSC3 and HSC4 caused the nervous system to function abnormally during larval development.
Amino Acid Substitutions D231S and K97S in Hsc3p Do Not Directly Interfere with ATP Binding
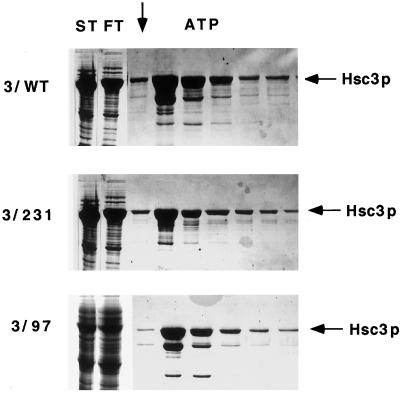
We next wished to investigate the mechanism by which mutant Hsc3p or Hsc4p interfered with wild-type protein function that, in turn, resulted in a dominant negative phenotype. To obtain sufficient purified wild-type and mutant protein to perform a structural analysis, the proteins were expressed in E. coli using Histidine-tagged vectors. A structural analysis of the mutant proteins was performed using Hsc3p since wild-type Hsc3p was previously shown to be correctly folded when expressed in E. coli whereas Hsc4p was not (Mehta, 1993). Both wild-type and mutant Hsc3p (D231S, K97S) were engineered so that a 6X-His tag was inserted in place of the ER leader sequence, allowing the proteins to be synthesized in E. coli as cleaved, mature ER Hsc3p. The wild-type and mutant Hsc3p (D231S, K97S) were first fractionated on an ATP agarose affinity column so that the proteins could be purified free of nonfunctional, misfolded proteins. Retention on the ATP-agarose was also used to determine if the HSC3 mutations (D231S, K97S) interfered with ATP binding. Both wild-type and mutant Hsc3p (D231S, K97S) were found to bind ATP-agarose affinity columns, indicating that the amino acid substitutions did not directly interfere with ATP binding (Figure 4).
Figure 4.
ATP binding of Hsc3p with amino acid substitutions. Wild-type and mutant Hsc3p (D231S, K97S) were applied to an ATP agarose affinity column and washed with low- and high-salt washes before Hsc3p was eluted from the column with 5 mM ATP. Each fraction was analyzed by 1D-PAGE/Coomassie blue staining using 10% polyacrylamide gels. The arrow denotes the start of the ATP-eluted fractions. ST is the starting material and FT is the column flowthrough. The major band eluted is the size expected for full-length Hsc3p, whereas lower-molecular-weight bands are Hsc3p degradation products.
The ATP eluted wild-type and mutant (D231S, K97S) Hsc3p fractions were next purified free of both the ATP binding bacterial Hsp70, DnaK, and most Hsc3p degradation products by using an Ni-NTA agarose affinity column. Unfortunately, Hsc3p (K97S) was not retained on the nickel column, because of loss of residues at the N terminus that contained the 6X-histidine tag. Thus, the partially truncated Hsc3p (K97S) was not used in further studies.
Wild-Type and Mutant Hsc3p (D231S) Exhibit Different Conformational States When Bound by ATP or ADP
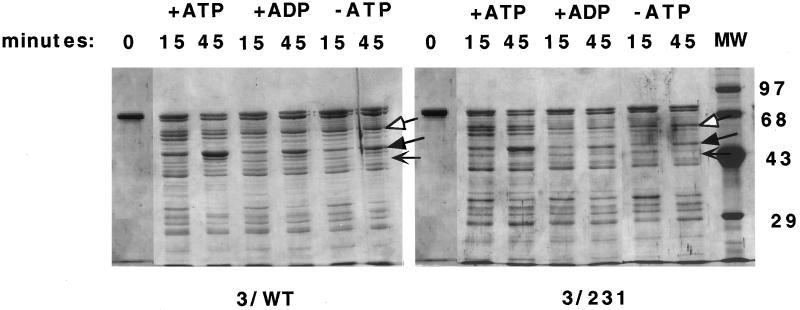
It is thought that when Hsc70 binds ATP, a conformational change is relayed from the ATP-binding domain to the peptide-binding domain such that bound substrate is released in an altered form (Hartl, 1996). This model is supported by experiments demonstrating that the ATP, ADP, and no nucleotide forms of DnaK can be distinguished by limited trypsin digestion. The different conformations of DnaK produced different sized proteolytic fragments (Liberek et al., 1991; Buchberger et al., 1995; Kamath-Loeb et al., 1995). A similar analysis was performed using the purified Drosophila wild-type Hsc3p to determine whether this eukaryotic Hsc70 also displayed different conformations in the presence of ATP, ADP, or no nucleotide. Additionally, limited trypsin digestion was performed on mutant Hsc3p(D231S) to determine whether this mutation interfered with an ATP-induced conformational change in the protein.
Limited proteolysis of both wild-type and mutant Hsc3p (D231S) revealed that the proteins underwent similar ATP-induced conformational changes (Figure 5). In the presence of ATP and trypsin, both mutant and wild-type Hsc3p were converted into an array of proteolytic fragments, including a prominent triplet consisting of 62-, 61-, and 60-kDa fragments and an intense 45-kDa fragment. However, in the presence of ADP or no nucleotide and trypsin, the 62- and 60-kDa fragments virtually disappeared and the intense 45-kDa fragment was found in significantly smaller amounts. These results demonstrated that both the Drosophila wild-type and mutant Hsc3p (D231S) underwent similar conformational changes upon binding ATP. A mixture of the purified wild-type and mutant Hsc3p (D231) was analyzed by 2D-PAGE, confirming the presence of the D231S mutation. Additionally, the ADP, ATP, and no nucleotide proteolytic digestion patterns of mutant Hsc3p (D231S) were all similar to that of wild-type Hsc3p, indicating that the D231S substitution did not drastically alter the structure of the protein.
Figure 5.
Wild-type and mutant Hsc3p exhibit distinct ATP and ADP conformational states. Wild-type and mutant (D231S) Hsc3p were incubated with trypsin in the presence of either ATP, ADP, or no nucleotide (−ATP lane). At the indicated time points, aliquots were removed from the digest and subjected to 1D-PAGE using 12% polyacrylamide gels, followed by Coomassie blue staining. Molecular mass standards (in kDa) are indicated. The arrows are as follows: white arrow, triplet proteolytic fragments (60–62 kDa) present with ATP and absent with ADP or no nucleotide for both wild-type and mutant Hsc3p; black arrow, the 45-kDa proteolytic fragment present in large amounts with ATP and in small amounts with ADP or no nucleotide for both wild-type and mutant Hsc3p; and thin black arrow, the difference in the proteolytic digestion pattern (40–45 kDa) between wild-type and mutant Hsc3p (D231S), probably caused by the mutation slightly altering the mutant protein structure and therefore its susceptibility to trypsin.
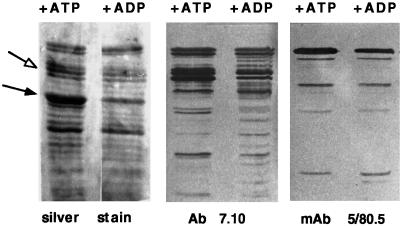
Mapping of ATP-affected Tryptic Cleavage Fragments
The mutant Hsc3p (D231S) was shown to undergo ATP-induced conformational changes similar to that of wild-type Hsc3p. Therefore, we wished to determine whether the ATP-induced conformational changes that were detected occurred in the N-terminal ATP-binding domain, the C-terminal substrate-binding domain, or both. Immunoblot analysis was used to map tryptic cleavage fragments containing C- or N-terminal epitopes. We used two C terminus-specific antibodies (Abs) that recognized either an epitope located at the end of the C terminus (mAb 5/80.5) or a region near or possibly included within the proposed C-terminal substrate-binding region (within 153 amino acids immediately following the ATPase domain in DnaK) (mAb 7.10). Unfortunately, the only available N-terminal Ab (Ab 358) did not recognize Hsc3p and therefore could not be used in this experiment.
We found that the 60- to 62-kDa triplet proteolytic digestion fragments observed with ATP, which become a doublet in the presence of ADP or no nucleotide, were recognized by the C-terminal mAb 7.10 but not mAb 5/80.5 (Figure 6). This result indicated that either these ATP-affected tryptic cleavage fragments reside in or near the C-terminal substrate-binding domain or that they span both the N-terminal ATPase and C-terminal substrate- binding domains. The 45-kDa fragment, which increases in abundance in the presence of ATP, was not recognized by either C-terminal Ab used (Figure 6). This result suggests that this major ATP-induced structural change was restricted to the N-terminal ATP-binding domain.
Figure 6.
Identification of ATP-affected tryptic fragments. Wild-type Hsc3p was incubated with trypsin in the presence of either ATP or ADP. Aliquots were removed from each digest after 45 min and separated by 1D-PAGE using 12% polyacrylamide gels. Tryptic fragments were analyzed by either silver stain or Western blot analysis using C-terminal Abs 5/80.5 and 7.10. White arrow denotes triplet proteolytic fragments present with ATP and absent with ADP or no nucleotide. Black arrow denotes 45-kDa fragment present in large amounts with ATP and in small amounts with ADP or no nucleotide.
The Amino Acid Substitutions D231S and K97S in Hsc3p Interfere with ATP-induced Substrate Release
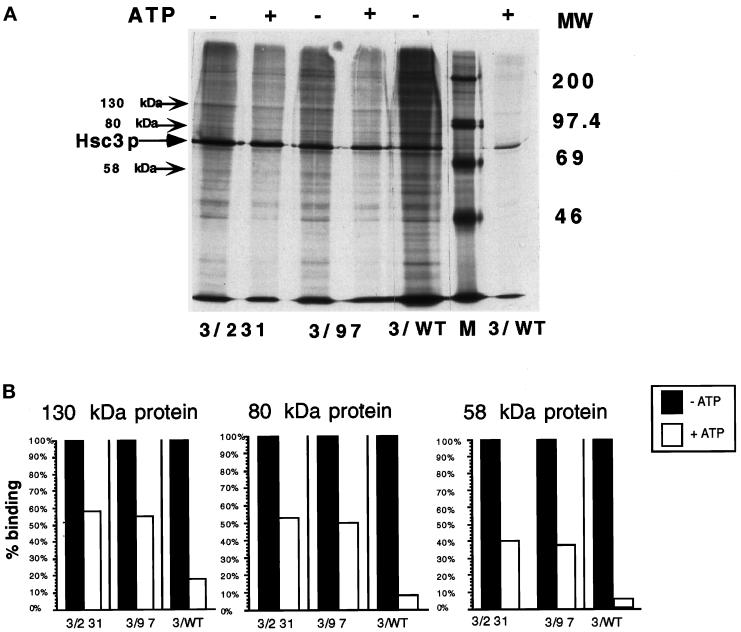
We wished to determine whether the amino acid substitutions D231S and K97S interfered with the structural coupling of nucleotide binding to substrate release. Therefore, Hsc3p coimmunoprecipitation experiments were carried out with labeled mutant and wild-type fly extracts to determine whether mutant Hsc3p (D231S, K97S) would remain bound to substrate in the presence of ATP, as ATP has been shown to promote substrate release (Palleros et al., 1993).
Coimmunoprecipitation experiments were performed using labeled protein extracts from salivary glands producing exogenous mutant or wild-type Hsc3p from a salivary gland-specific GAL4 line. This line produces mutant Hsc3p (D231S, K97S) at ∼25% the level of wild-type Hsc3p found in the salivary glands (our unpublished data). The coimmunoprecipitation experiments could not be performed on flies producing mutant Hsc4p (D206S, K71S), as the resulting malformed salivary glands were extremely small or absent.
Stable Hsc3p–substrate immune complexes from mutant and wild-type salivary gland extracts were incubated with or without ATP. The resulting coprecipitated proteins were compared in the +ATP and −ATP lanes, and three representative proteins for each were quantitated by scanning densitometry (Figure 7, A and B). We observed that upon addition of ATP, 5–18% of coimmunoprecipitated Hsc3p substrate (depending upon the substrate quantitated) remained bound to Hsc3p–substrate immune complexes from wild-type Hsc3p extracts. However, upon addition of ATP, 40–60% of coimmunoprecipitated Hsc3p substrate remained bound to Hsc3p–substrate immune complexes from mutant Hsc3p (D231S, K97S) extracts. The observation that significantly more substrate remained in an ATP-stable immune complex from mutant D231S and K97S Hsc3p extracts that contain both wild-type and mutant protein indicates that the mutant proteins failed to release substrate in the presence of ATP. These results indicate that Asp 231 and Lys 97 play a role in ATP-induced substrate release.
Figure 7.
The detection of ATP-induced substrate release from wild-type or mutant (D231S, K97S) Hsc3p–substrate complexes. (A) [35S]methionine-labeled salivary gland protein extracts from larvae producing either wild-type Hsc3p or mutant Hsc3p (D231S, K97S) in a wild-type background were coimmunoprecipitated with mAb 5/80.5 (specific for Hsc3p) in the presence of EDTA, which inhibits substrate release. The resulting mutant or wild-type Hsc3p–substrate immune complexes were divided equally, incubated with buffer containing 2 mM ATP or no ATP, and analyzed by 1D-SDS/PAGE using 10% polyacrylamide gels and fluorography. The three arrows represent 130-, 80-, and 58-kDa protein bands that were selected for quantitation by scanning densitometry (shown in panel B). (B) Three coimmunoprecipitated substrate proteins randomly selected for and denoted by arrows in panel A were quantitated by scanning densitometry. The amount of bound protein without ATP represented 100% binding. The amount of protein remaining after the ATP treatment was calculated as a percent of the ATP (−) bands.
DISCUSSION
Both Hsc3p and Hsc4p Function Are Required for Tissue Establishment and Maintenance
We began this study to determine whether both cytoplasmic Hsc4p and ER Hsc3p function was required in specific tissues and stages of development in Drosophila. HSC4 is expressed at a level 30–70 times greater than HSC1 and HSC2, which also encode cytoplasmic Hsc70 proteins (Craig et al., 1983). In addition, we did not know whether expression of mutant Hsc4p would elicit the expression of the heat-inducible HSP68 and HSP70 genes encoding proteins that like Hsc4p, can be both cytoplasmic and nuclear. Potentially, any of these cytoplasmic Hsc70 could compensate for loss of Hsc4p function. Hsc3p is the sole ER Hsc70 and therefore could not be compensated by other Hsc70s. Since null mutations for either gene were not available, we used mutant Hsc3p (D231S, K97S) and Hsc4p (D206S, K71S), which had previously been shown to be dominant negatively acting proteins (Rubin, 1993), to interfere with the activity of the wild-type Hsc70.
Several lines of evidence in the present investigation indicate that both Hsc4p and Hsc3p are essential proteins that are required throughout Drosophila development for both tissue establishment and maintenance. We show that when GAL4 targeted gene expression is used to produce dominant negatively acting Hsc4p and Hsc3p in either a mesoderm/muscle, ubiquitous, or nervous system pattern of cells, lethality is observed over a range of developmental stages (Table 2) depending on the level of production of the mutant protein (Tables 1 and 2; Figure 1, A and B) and the specific tissue and time in development that the GAL4 line activated mutant protein production. We demonstrate that lethality is caused by tissue-specific defects that result from mutant Hsc70 production. For example, embryos expressing high levels of mutant Hsc4p in muscles displayed a range of muscle pattern defects similar to those previously described (Figure 3) (Drysdale et al., 1993; Fyrberg et al., 1994), whereas embryos expressing mutant Hsc3p, while having a normal muscle pattern, died later in development and displayed evidence of paralysis. We observe that for the same GAL4 lines and comparable levels of mutant protein expression, the phenotypes produced by mutant Hsc4p are always more severe than those produced by mutant Hsc3p (Table 2). Additionally, we show that when either mutant Hsc3p or Hsc4p is produced in the PNS, CNS, or brain, the larvae exhibited behavioral defects before dying as first- or second-instar larvae (Tables 3 and 4). Such larvae were defective in their ability to locate a food source, and their crawling and burrowing ability were impaired. These particular GAL4 lines all activate mutant protein expression relatively late during embryonic development, allowing a normal neuronal pattern to be established during embryogenesis. However, the nervous system subsequently ceased to function, indicating that Hsc3p and Hsc4p function is required both to establish correct tissue patterns (muscles) and also to maintain proper tissue function (muscles and nervous system). Mutant expression in other tissues (such as a ubiquitous pattern and salivary glands) always resulted in eventual tissue destruction. Preliminary experiments, using acridine orange staining, did not indicate that cell death induced by mutant Hsc70 resulted from apoptosis as opposed to necrosis (our unpublished results). In conclusion, we believe that both Hsc3p and Hsc4p function is required for general cell viability throughout development.
Other cytoplasmic Hsc70s were not able to compensate for the loss of Hsc4p function. Either the other Hsc70s were not present at sufficient levels to compensate for Hsc4p loss, or our dominant acting mutant Hsc4p interfered with the activity of those Hsc70s as well. Since we began this study, null or strong hypomorphic mutations for HSC4 have been obtained. A P-element insertion maps near the start of the 5′-noncoding region of HSC4 (l(3)03550, A. Spradling, BDGP), and small deficiencies of the region were obtained by excision of this element using either transposase (Df(3)PG4 from 88E3,4–88E8,9; Hing, Sun, and Artavanis-Tsakonas, personal communication) or x-rays (Schmucker et al., 1997). Flies homozygous for l(3)03550 die as third-instar larvae or pupae (Perrimon et al., 1996; Braun et al., 1997), whereas we show that flies expressing high levels of dominant negatively acting mutant Hsc4p die during late embryogenesis probably by “poisoning” the activity of the maternal Hsc4p already present in the embryo. A similar poisoning model for dominant acting DnaK (Liberek et al., 1991; Wild et al., 1992; Buchberger et al., 1994) and ER BiP (Hendershot et al., 1995) has also been proposed. Perrimon et al. (1996) have shown that l(3)03550 has a lethal maternal effect phenotype resulting in embryonic head and abdominal cuticular defects. In addition, l(3)03550 and its alleles were named the scattered locus (scd) because they cause defects in the larval visual system, affecting both Bolwig’s organ and proper projection of Bolwig’s nerve (Schmucker et al. 1997). The P-element insertion, l(3)03550, is also an enhancer trap line, and lacZ expression has been detected in the glia cells of the embryonic optic stalk and neurons of the optic lobe anlagen (Schmucker et al., 1997). Additionally, lacZ expression from l(3)03550 has been shown to be concentrated in the larval lymph glands and hemocytes (Braun et al., 1997). Larvae homozygous for this insertion exhibited melanotic tumors in the hemolymph. HSC4 transcripts are most likely present in all cells during embryonic development, but are enriched in cells active in endocytosis or those undergoing rapid growth, such as neuroblasts (Perkins et al., 1990). The larval behavioral defects we observe when dominant mutant Hsc4p is targeted to the PNS, CNS, or brain may be caused by a failure to establish and maintain proper axonal connections, as is observed in HSC4 loss-of-function mutants (Schmucker et al. 1997).
Lethality Induced by Production of Dominant Negatively Acting Hsc3p and Hsc4p Is Likely Caused by a Global Accumulation of Misfolded Proteins in the Cell
When cells are exposed to a variety of metabolic stresses, they induce specific Hsp70 proteins. The primary inducer of this response is believed to be the presence of an excess of misfolded proteins (Craig and Gross, 1991; Hendrick and Hartl, 1993). We show that expression of dominant negatively acting Hsc4p, but not Hsc3p, resulted in induction of the stress-inducible Hsp70 proteins at normal temperatures (Figure 2). We interpret this result to indicate that dominant negatively acting Hsc4p interfered with the function of wild-type Hsc4p and thus allowed misfolded proteins to accumulate in the cytoplasm. Our results are consistent with studies in yeast showing that in response to an accumulation of misfolded proteins in the ER, Kar2p (yeast ER Hsc70) synthesis is specifically enhanced while nuclear/cytosolic Hsp70 synthesis is not (Normington et al., 1989). Therefore, we believe that expression of the dominant negative acting Hsc3p caused an accumulation of misfolded proteins specifically in the ER, which resulted in lethality but would not have been expected to induce Hsp70. Our results support the view that the cell senses and responds differently to an accumulation of misfolded proteins in different intracellular compartments (Shamu et al., 1994). However, we cannot formally exclude a model in which mutant Hsc4p directly induces synthesis of Hsp70 protein, such as by regulating the trimeric state of the HSF transcription factor that regulates the HSP70 genes (Lis and Wu, 1993; Morimoto, 1993). Of note, no effect on HSF by Hsc4p has been observed in vitro thus far (Wu, personal communication).
We believe our data support a model in which a threshold level of accumulated misfolded proteins determines when in development lethality occurs. Flies producing different levels of mutant Hsc4p and Hsc3p in specific tissues died at different stages of development, ranging from late embryogenesis to late pupae (Table 2). The earliest developmental lethality occurred in fly lines expressing the highest levels of mutant Hsc3p and Hsc4p for any given GAL4 line. Furthermore, even the lowest levels of mutant Hsc70 could induce lethality, albeit at later stages of development. We additionally observed that there was always a delay between induction of mutant protein and lethality, consistent with the idea that lethality occurred only when a threshold level of misfolded protein was reached. Thus, the mutant Hsc70-induced lethality is similar to the slow and degenerative human amyloid diseases such as Alzheimer’s disease (Holtzman and Mobley, 1991).
BiP (ER Hsc70) has been shown to be involved in cellular secretory processes by transiently interacting with newly translocated ER proteins to promote their proper folding and/or assembly, by tightly binding to permanently misfolded proteins to block their secretion (Gething and Sambrook, 1992), and it is also required for protein translocation across the ER membrane (Vogel et al., 1990). Cytosolic Hsc70s promote proper folding and assembly of nascent cytosolic proteins (Gething and Sambrook, 1992), uncoat clathrin-coated vesicles (Chappell et al., 1986), target denatured proteins for lysosomal degradation (Gething and Sambrook, 1992), and are required for the translocation of proteins into both the ER and mitochondria (Chirico et al., 1988; Deshaies et al., 1988). Dominant negative acting Hsc3p and Hsc4p that disrupted these processes would likely lead to a global accumulation of misfolded proteins, resulting in detrimental pleiotropic effects. We observed that at comparable levels of expression, mutant Hsc4p consistently caused more severe defects than mutant Hsc3p (Table 2). Loss of function of cytoplasmic Hsc70 would be expected to be more detrimental to cells than loss of ER Hsc70, as it would lead to misfolded proteins in all cellular compartments, in contrast to ER Hsc70, which is needed only for folding and translocation of ER proteins.
Amino Acid Substitutions, D231S and K97S, in the ATP-binding Domain of Hsc3p Interfere with the Coupling of ATP Binding and Substrate Release
When Hsc70 binds ATP, a conformational change is relayed from the ATP-binding domain to the peptide-binding domain such that bound substrate is released in an altered conformation (Hartl, 1996). We demonstrated by limited trypsin digestion that wild-type Hsc3p displays distinct ATP-bound and ADP-bound conformational states (Figure 5) and undergoes ATP-induced substrate release (Figure 7). Therefore, Drosophila Hsc3p, similar to bacterial DnaK (Liberek et al., 1991; Buchberger et al., 1995), hamster BiP (Wei et al., 1995), and bovine Hsc70 (Ha and McKay, 1995), undergoes an ATP-induced conformational change that occurs concomitantly with release of bound substrate (Buchberger et al., 1994). However, unlike the three distinct ATP, ADP, and no nucleotide conformational states shown for DnaK (Buchberger et al., 1995), we found only two conformational states: one for ATP and one for ADP and no nucleotide. Wei and Hendershot (1995) similarly observe only two conformational states for mammalian BiP. However, we cannot exclude the possibility that our “nucleotide-free” Hsc3p may still have had ADP bound, as Gao et al. (1994) have reported ADP can be difficult to remove.
We show that although both mutant Hsc3p(D231S and K97S) bind ATP (Figure 4), they both failed to undergo ATP-induced substrate release in extracts of salivary glands producing mutant Hsc3p (Figure 7, A and B). We were able to examine only the ATP-induced conformational changes of Hsc3p (D231S), as the bacterially expressed Hsc3p(K97S) was truncated. We observed similar ATP-induced conformational changes in Hsc3p(D231S) and wild-type Hsc3p, as assayed by limited trypsin digestion (Figure 5). Our mapping of the ATP-affected tryptic cleavage fragments suggested that the major ATP-induced structural change we observed (the appearance of an intense 45-kDa band) was restricted to the ATP-binding domain (Figure 6). Previous work examining dominant negative DnaK (Liberek et al., 1991; Buchberger et al., 1995; Kamath-Loeb et al., 1995) and BiP (Hendershot et al., 1995; Wei et al., 1995; Morris et al., 1997) have shown that mutants unable to undergo ATP-induced substrate release were defective for ATP-induced conformational changes, as assayed by limited proteolytic digestion. In contrast to our result, Kamath-Loeb et al. (1995) found that DnaK (D201N) (corresponding to D231 in Hsc3p) did not show any ATP-induced conformational changes. This difference may reflect a difference in the Hsc70 proteins, the different residue that was used for substitution, or different domains detected by trypsin cleavage. Our work, therefore, is the first to directly show that upon binding ATP, a dominant negative Hsc70 undergoes a conformational change in the ATP-binding domain, but fails to release substrate, presumably because the conformational change relayed to the peptide-binding domain is absent or aberrant.
The structure of the ATPase domain of bovine Hsc70, as determined by x-ray crystallography, consists of two large globular domains separated by a deep cleft (Flaherty et al., 1990). Two crossed α-helices connect these two subdomains, separating the cleft into an upper cleft (at the bottom of which nucleotide and the required Mg2+ and K+ ions bind) and a lower cleft. The α-helices contain the hinge residues that, upon ATP binding, allow for the movement of the two subdomains. The proposed role of hinge residue E171 (E175 in bovine Hsc70) is to position a critical Mg2+ ion (via water molecules) that serves as a bridge between the two ATPase subdomains (Buchberger et al., 1994). Mutations affecting E171 of DnaK have been shown to interfere with the coupling of the ATPase- and substrate-binding domains, resulting in dominant negative DnaK that is defective for both ATP-induced conformational changes and subsequent substrate release (Wild et al., 1992; Buchberger et al., 1995). In the structure of the ATPase domain, both Asp206 (D231 in Hsc3p) and Lys71 (K97 in Hsc3p) are located in close proximity to Glu175, and both were proposed to contribute to its environment (Buchberger et al., 1995). Therefore, it is plausible that these residues also affect the coupling of the ATPase- and substrate-binding domains. Mutant DnaK (D201N) (D206 in bovine Hsc70 and Hsc4p; D231 in Hsc3p) binds peptides normally, but its ATPase cannot be stimulated by peptides (Kamath-Loeb et al., 1995). The DnaK (D201N) ATPase cannot be stimulated by the cochaperones DnaJ or GrpE either, and it cannot be released from GrpE by ATP. These data again suggest that D201 mutants are defective in the coupling of the ATPase- and substrate-binding domains.
Residues Asp206 and Lys71 (D231 and K97 in Hsc3p) were also substituted with Ser in the 44-kDa ATPase domain of bovine Hsc70 (Flaherty et al., 1994; O’Brien et al., 1996). These substitutions either abolished (Lys71) or significantly reduced (Asp206) the rate of ATP hydrolysis. However, it is unlikely that a defect in the ATPase activity of mutant Hsc3p (D231S, K97S) is the sole cause of the inability of mutant Hsc3p to undergo ATP-induced substrate release. Both mutant DnaK (T199A) (Palleros et al., 1993) and mutant BiP (T229G) (Wei et al., 1995) that are defective only for ATP hydrolysis have been shown to still undergo an ATP-induced conformational change and, consequently, substrate release. In addition, we have previously shown (Rubin et al., 1993) that D35S in Hsc3p does not cause dominant lethality in flies, even though the same mutation in bovine Hsc70 (Asp10) reduces the ATPase activity by 99% (Wilbanks et al., 1994). Rubin (1993) has shown that both D206S and K71S in Hsc4p abolished the ability of the proteins to uncoat clathrin-coated vesicles, and Ha et al. (1997) have shown that bovine Hsc70s defective for ATP hydrolysis are completely deficient for chaperone activities. Therefore, we believe that whereas ATP hydrolysis mutants may be nonfunctional, only those that impede the transduction of the conformational change from the ATP-binding domain to the substrate-binding domain, will be dominant negative mutations.
Possible Mechanisms for Mutant Hsc70-induced Dominant Lethality
Thus far, all dominant negative mutant Hsc70s, including our mutant Hsc3p, are defective in ATP-induced substrate release (Figure 7, A and B). However, why are these mutant Hsc70 dominant over the wild-type? The classical explanation for dominant negative mutations proposes that the wild-type gene encodes a multimeric protein and that such mutations result from mixed oligomers in which the mutant subunit poisons the activity of the wild-type subunit (Herskowitz, 1987). We find that both Hsc3p and Hsc4p purify as monomers, similar to DnaK (Zylicz and Georgopoulos, 1984), and BiP purifies as a mixture of monomers, dimers, and oligomers (Blond-Elguindi et al., 1993). Most studies suggest that functional Hsc70 is monomeric. Dominant negative mutations can be found in genes encoding monomeric proteins if the mutant protein competes with the wild-type protein for substrates that may be limiting, thus preventing the wild-type protein from performing its proper function (Herskowitz, 1987). In the present case, dominant mutant Hsc70 could compete with the wild-type protein for either protein substrates or for the cochaperone proteins that regulate its ATPase cycle, which may be present in limiting concentrations. We show that mutant Hsc3p, produced at 25% the level of wild-type Hsc3p, was able to block ATP-induced substrate release by 40–60% (Figure 7B). These data are consistent with either a “mixed oligomer” model or a competition model if the mutant and wild-type proteins had different affinities for limiting amounts of substrates. Where it has been measured, different mutant proteins seem to have similar affinities for peptides. However, if mutant Hsc70s bind irreversibly to their protein substrates, they may sequester the substrates from the wild-type proteins at a level that is disproportionate to their abundance in the cell. This is likely the situation of mutant Hsc3p that cannot undergo ATP-induced substrate release and therefore bind substrates more stably than wild type. Of note, DnaK (D201N) (D206 in Hsc4p and D231 in Hsc3p) was shown to bind one of its cochaperones, GrpE, irreversibly (Kamath-Loeb et al., 1995), and therefore the same argument could be applied to limiting amounts of the cochaperones as well. And finally, we suggest that although Hsc70s may primarily exist as monomers, they may function in a sense as “oligomers.” For example, it is believed that Hsc70 must repeatedly bind and release unfolded proteins, until the final folded state is reached. This may require either simultaneous or sequential binding of multiple monomers to the same protein substrate. If any of these multiple interactions is with a mutant monomer that binds irreversibly to the substrate, that protein may never be able to fold correctly. Both models invoking an irreversibly binding mutant Hsc70 would explain how relatively low levels of mutant Hsc70 could severely interfere with wild-type functions and cause dominant lethality.
ACKNOWLEDGMENTS
We thank Drs. Susan Lindquist and Belinda Bullard for generously providing antibodies and Eric Fyrberg for providing MHC-β gal fly strains. We also thank Drs. David Rubin, Laurie Tompkins, and Robert Finkelstein for invaluable technical advice. Finally, we thank Drs. Harry Rappaport, Robert Finkelstein, and Jose Ramirez-Latorre for critical reading of the manuscript. This work was supported by a National Institute of Health grant GM-41000 to K.B.P.
REFERENCES
- Blond-Elguindi S, Fourie AM, Sambrook JF, Gething MJ. Peptide- dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem. 1993;268:12730–12735. [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;18:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Braun A, Lemaitre B, Lanot R, Zachary D, Meister M. Drosophila immunity: analysis of larval hemocytes by P-element-mediated enhancer trap. Genetics. 1997;147:623–634. doi: 10.1093/genetics/147.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Theyssen H, Schroder H, McCarty JS, Virgallita G, Milkereit P, Reinstein J, Bukau B. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J Biol Chem. 1995;270:16903–16910. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Valencia A, Mcmacken R, Sander C, Bukau B. The chaperone function of DnaK requires the coupling of ATPase activity with substrate binding through residue E171. EMBO J. 1994;13:1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Chappell TG, Welch WJ, Schlossman DM, Palter KB, Schlesinger MJ, Rothman JE. Uncoating ATPase is a member of the 70 kilodalton family of stress proteins. Cell. 1986;45:3–13. doi: 10.1016/0092-8674(86)90532-5. [DOI] [PubMed] [Google Scholar]
- Chirico WJ, Water MG, Blobel G. 70 K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Craig EA, Gross CA. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;10:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- Craig EA, Kramer J, Kosic-Smithers J. SSC1, a member of the 70-kDa heat shock protein multigene family of Saccharomyces cerevisiae, is essential for growth. Proc Natl Acad Sci USA. 1987;84:4156–4160. doi: 10.1073/pnas.84.12.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Kock BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–810. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Drysdale R, Rushton E, Bate M. Genes required for embryonic muscle development in Drosophila melanogaster. Roux’s Arch Dev Biol. 1993;202:282–295. doi: 10.1007/BF00363217. [DOI] [PubMed] [Google Scholar]
- Elguindie S, Fourie AM, Sambrook JF, Gething MJ. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem. 1993;268:12730–12735. [PubMed] [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat shock cognate protein. Nature. 1990;34:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Flaherty KM, Wilbanks SM, DeLuca-Flaherty C, McKay DB. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. J Biol Chem. 1994;269:12899–12907. [PubMed] [Google Scholar]
- Flynn GC, Chappell TG, Rothman SE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- Fyrberg EA, Bernstein SI, VijayRaghavan K. Basic methods for Drosophila muscle biology. Methods Cell Biol. 1994;44:237–256. doi: 10.1016/s0091-679x(08)60917-6. [DOI] [PubMed] [Google Scholar]
- Gao B, Greene L, Eisenberg E. Characterization of nucleotide-free uncoating ATPase and its binding to ATP, ADP, and ATP analogues. Biochemistry. 1994;33:2048–2053. doi: 10.1021/bi00174a010. [DOI] [PubMed] [Google Scholar]
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–34. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Ha J, McKay DB. Kinetics of nucleotide-induced changes in the tryptophan fluorescence of the molecular chaperone Hsc70 and its subfragments suggest the ATP-induced conformational change follows initial ATP binding. Biochemistry. 1995;34:11635–11644. doi: 10.1021/bi00036a040. [DOI] [PubMed] [Google Scholar]
- Ha JH, Hellman U, Johnson ER, Li L, McKay DB, Sousa MC, Takeda S, Wernstedt C, Wilbanks SM. Destabilization of peptide binding and interdomain communication by an E543K mutation in the bovine 70-kDa heat shock cognate protein, a molecular chaperone. J Biol Chem. 1997;272:27796–277803. doi: 10.1074/jbc.272.44.27796. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hendershot LM, Wei J, Gaut JR, Lawson B, Freiden PJ, Murti KG. In vivo expression of mammalian BiP ATPase mutants causes disruption of the endoplasmic reticulum. Mol Biol Cell. 1995;6:283–296. doi: 10.1091/mbc.6.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;6:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutation. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Mobley WC. Molecular studies in Alzheimer’s disease. Trends Biochem Sci. 1991;259:140–144. doi: 10.1016/0968-0004(91)90056-2. [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc Natl Acad Sci USA. 1982;7:2700–27004. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Zen lu, C, Suh WC, Lonetto MA, Gross CA. Analysis of three DnaK mutant proteins suggests that progression through the ATPase cycle requires conformational changes. J Biol Chem. 1995;270:30051–30059. doi: 10.1074/jbc.270.50.30051. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of the structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liberek K, Skowyra S, Zylicz M, Johnson C, Georgopolous C. The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J Biol Chem. 1991;22:14491–14496. [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Lis J, Wu C. Proteins traffic on the heat shock promoter, parking, stalling and trucking along. Cell. 1993;74:1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- Manseau L, Baradaran A, Brower S, Budhu A, Elefant F, Philip AP, Glover D, Kaiser K, Palter KB, Selleck S. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev Dyn. 1997;209:310–322. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Mehta A. Ph.D. Thesis. Philadelphia, PA: Temple University; 1993. Functional Properties and Protein Binding Studies of Drosophila 70 kDa Heat Shock Cognate Proteins. [Google Scholar]
- Morimoto RF. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259:1404–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- Morris JA, Dorner AJ, Edward CA, Hendershot LM, Kaufman RJ. Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J Biol Chem. 1997;272:4327–4334. doi: 10.1074/jbc.272.7.4327. [DOI] [PubMed] [Google Scholar]
- Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- O’Brien MC, Flaherty KM, McKay DB. Lysine 71 of the chaperone protein Hsc70 is essential for ATP hydrolysis. J Biol Chem. 1996;271:74–15878. doi: 10.1074/jbc.271.27.15874. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1974;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Palleros DR, Reid KL, Shi L, Welch WJ, Fink AL. ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- Palter KB, Watanabe M, Stinson L, Mahowald AP, Craig EA. Expression and localization of Drosophila melanogaster hsp70 cognate proteins. Mol Cell Biol. 1986;6:1187–1203. doi: 10.1128/mcb.6.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Doctor JS, Zhang K, Stinson L, Perrimon N, Craig EA. Molecular and developmental characterization of the heat shock cognate gene 4 of Drosophila melanogaster. Mol Cell Biol. 1990;10:3232–3238. doi: 10.1128/mcb.10.6.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Lanjuin A, Arnold C, Noll E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics. 1996;144:1681–1692. doi: 10.1093/genetics/144.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Philllis RW, Sclitz-Johnson DM. A stable source of P-element transposase in Drosophila. Genetics. 1988;118:120–128. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DM. Ph.D. Thesis. Philadelphia, PA: Temple University; 1993. Biochemical and Genetic Analysis of Drosophila Heat Shock Cognate 70 Proteins with Substitutions of Amino Acid Implicated in ATP Binding and Hydrolysis. [Google Scholar]
- Rubin DM, Mehta AD, Zhu J, Shoham S, Chen X, Wells QR, Palter KB. Genomic structure and sequence analysis of Drosophila melanogaster HSC70 genes. Gene. 1993;128:155–163. doi: 10.1016/0378-1119(93)90558-k. [DOI] [PubMed] [Google Scholar]
- Sadis S, Hightower LE. Unfolded proteins stimulate molecular chaperone Hsc70 ATPase by accelerating ADP/ATP exchange. Biochemistry. 1992;31:9406–9412. doi: 10.1021/bi00154a012. [DOI] [PubMed] [Google Scholar]
- Schmid D, Baici A, Gehring H, Christe P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Jackle H, Gaul U. Genetic analysis of the larval optic nerve projection in Drosophila. Development. 1997;124:937–948. doi: 10.1242/dev.124.5.937. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Cox JS, Walter P. The unfolded-protein-response pathway in yeast. Trends Cell Biol. 1994;4:56–60. doi: 10.1016/0962-8924(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Misra LM, Rose MR. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Biol Chem. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Gaut JR, Hendershot LM. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- Wei J, Hendershot LM. Characterization of the nucleotide binding properties and ATPase activity of recombinant hamster BiP purified from bacteria. J Biol Chem. 1995;270:26670–26676. doi: 10.1074/jbc.270.44.26670. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of HSP70 genes in Saccharomyces cerevisae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbanks SM, DeLuca-Flaherty C, McKay DB. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. J Biol Chem. 1994;269:12893–12898. [PubMed] [Google Scholar]
- Wild J, Kamath-Loeb A, Ziegelhoffer E, Lonetto M, Kawasaki Y, Gross CA. Partial loss of function mutations in DnaK, the Escherichia coli homologue of the 70-kDa heat shock proteins, affect highly conserved amino acids implicated in ATP binding and hydrolysis. Proc Natl Acad Sci USA. 1992;89:7139–7143. doi: 10.1073/pnas.89.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylicz M, Georgopoulos G. Purification and properties of the Escherichia coli DnaK replication protein. J Biol Chem. 1984;14:8820–8825. [PubMed] [Google Scholar]