Abstract
Background
The insect cuticle is composed of various proteins and formed during the molt under hormonal regulation, although its precise composition and formation mechanism are largely unknown. The exhaustive catalogue of genes expressed in epidermis at the molt constitutes a massive amount of information from which to draw a complete picture of the molt and cuticle formation in insects. Therefore, we have catalogued a library of full-length cDNAs (designated epM) from epidermal cells during the last larval molt of Bombyx mori.
Results
Of the 10,368 sequences in the library, we isolated 6,653 usable expressed sequence tags (ESTs), which were categorized into 1,451 nonredundant gene clusters. Seventy-one clusters were considered to be isoforms or premature forms of other clusters. Therefore, we have identified 1,380 putative genes. Of the 6,653 expressed sequences, 48% were derived from 92 cuticular protein genes (RR-1, 24; RR-2, 17; glycine-rich, 29; other classes, 22). A comparison of epM with another epidermal EST data set, epV3 (feeding stage: fifth instar, day 3), showed marked differences in cuticular protein gene. Various types of cuticular proteins are expressed in epM but virtually only RR-1 proteins were expressed in epV3. Cuticular protein genes expressed specifically in epidermis, with several types of expression patterns during the molt, suggest different types of responses to the ecdysteroid pulse. Compared with other Bombyx EST libraries, 13 genes were preferentially included in epM data set. We isolated 290 genes for proteins other than cuticular proteins, whose amino acid sequences retain putative signal peptides, suggesting that they play some role in cuticle formation or in other molting events. Several gene groups were also included in this data set: hormone metabolism, P450, modifier of cuticular protein structure, small-ligand-binding protein, transcription factor, and pigmentation genes.
Conclusion
We have identified 1,380 genes in epM data set and 13 preferentially expressed genes in epidermis at the molt. The comparison of the epM and other EST libraries clarified the totally different gene expression patterns in epidermis between the molting and feeding stages and many novel tissue- and stage-specifically expressed epidermal genes. These data should further our understanding of cuticle formation and the insect molt.
Background
Insect cuticles are formed during molting under hormonal regulation and are composed of complex and composite materials, made mainly of chitinous filaments embedded in proteinaceous layers (see [1] for review). The cuticle acts as both skin and exoskeleton, and has diversified in its mechanical properties for optimal biological functions in various insects. The differences in the mechanical properties of the exoskeleton are probably dependent on the respective features of various cuticular proteins and chitin itself, on the precise combination of different cuticular proteins, and on their secondary stabilization, called "sclerotization". However, the number of types of cuticular proteins included in the cuticle has not yet been resolved, nor how their coordinated expression in epidermal cells and their excretion into the cuticle are precisely controlled during molting. These issues are very important for any understanding of the mechanisms underlying the formation of the highly ordered and layered structure of the cuticle.
The amino acid sequences of cuticular proteins have been reported for a wide variety of insects, by directly sequencing the purified cuticular proteins or by their deduction from the corresponding cDNA sequences [1-3]. However, most excreted cuticle components are cross-linked, making them unextractable [2]. Therefore, we infer that many other cuticular proteins are yet to be identified. Previously reported information on cuticular protein sequences has revealed several conserved motifs, such as the R&R consensus [1], Tweedle [4], and a 44-amino-acid motif [5]. The R&R consensus sequence is the most prevalent motif, and was first reported by Rebers and Riddiford [6]. An extended version of this consensus sequence was subsequently described and is generally referred to as the R&R consensus, which is known to bind chitin [1,7,8]. Three distinct forms of this consensus are recognized: RR-1, RR-2, and RR-3 [1,9]. RR-1 is characteristic of proteins in soft and flexible cuticles, and RR-2 proteins are associated with stiff and hard cuticles in general, although this classification is tentative [1,10]. Many other cuticular proteins lacking these motifs have structures containing other repeated structures, such as GGX or AAP(A/V) [1,11,12].
The comprehensive identification of cuticular proteins with an R&R motif has been attempted in Drosophila melanogaster [13], Apis mellifera [14], and Anopheles gambiae [15], based on their genome sequences. Recently, we identified 220 putative cuticular protein genes (RR-1 56, RR-2 89, RR-3 3, Tweedle 4, CPF 1, CPFL 4, glycine-rich 29, and 34 other genes) in the silkworm Bombyx mori [12]. A key question is whether each cuticular protein gene is expressed in a tissue- or stage-specific manner. Togawa et al. [16] investigated the developmental expression patterns of all Anopheles cuticular protein genes with R&R consensus motifs, using real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR). They found that all the genes are expressed and can be grouped into 21 clusters with different developmental expression profiles, although their tissue specificities were not examined.
Bombyx has an advantage in the study of tissue specificity and the expression profiles of each tissue because its tissues are relatively large and it is easy to construct a tissue-specific cDNA library. More than 40 expressed sequence tag (EST) databases for various tissues at the different developmental stages of Bombyx are available [17-19]. Exhaustive sequencing of the full-length cDNAs expressed in epidermal cells is an effective method of gaining an overview of cuticular proteins, because it avoids directly sequencing the barely extractable cuticular proteins. Therefore, we constructed a library of full-length cDNAs, using a G-capping method [20], from mRNAs expressed in the larval epidermal cells of the silkworm, B. mori, during the fourth larval molt, when the subsequent cuticle of the fifth instar is formed.
We sequenced over 10,000 clones randomly selected from the library described above, and isolated 6,653 ESTs belonging to 1,451 nonredundant gene clusters. Seventy-one clusters were considered to be isoforms or premature forms of other clusters. Therefore, we identified 1,380 putative genes. About half of these ESTs encoded cuticular proteins, representing 92 genes. In addition to cuticular proteins, we also identified 290 genes encoding the amino acid sequences of their putative signal peptides, suggesting that they play some role in cuticle formation or other molting events. Here, we list those cuticular protein genes, secreted protein genes, and other epidermal protein genes, including those encoding transcription factors and many enzymes. These data should be useful in understanding cuticle formation and the insect molt.
Results & Discussion
Construction and characterization of the epM EST data set
To exhaustively identify the cuticular protein components and epidermal genes expressed during the larval molting stages of the silkworm, we constructed a library of full-length cDNAs by mixing the RNAs of eight consecutive stages (C1, C2, D1, D2, D3, E1, E2, and F, according to [20]). We designated this cDNA library epM (fourth instar epidermis in Molting). We randomly sequenced 10,368 clones, excluded any short insert sequences, and isolated 6,653 ESTs (GenBank accession numbers DC432783–DC439435; Table 1, Additional File 1). These ESTs were classified into 1,451 nonredundant EST clusters/singletons. Seventy-one clusters were considered to be isoforms or premature forms of other clusters (Additional File 2). Excluding these 71 clusters, we identified 1,380 putative gene clusters, 885 of which were singletons (only one clone included in the EST data set). Among the 1,380 gene clusters, 1,025 clusters had sequence similarities to Drosophila genes (P < 1e-05). We categorized the gene clusters of epM using the criteria for the gene ontology (GO) terms used for Drosophila [21] (Figure 1). Figure 1B shows the numbers of gene cluster types (number of genes; left column), some of which are categorized by multiple criteria, and the numbers of total EST clones included under those criteria, which represent their levels of expression (number of ESTs; right column). Figure 1A shows several major categories in the same ratios as shown in Figure 1B, but the total percentages, summing all categories, are shown as 100 in Figure 1A (total epM genes 131%, total epM ESTs 127%, in Figure 1B). It is remarkable that structural protein genes are expressed so abundantly (60% of the total ESTs) (Figure 1A, 1B, category 8), many of which encode cuticular proteins (Figure 1B, highlights; 48% of total ESTs), which suggests the active translation of cuticle components in the epidermal cells during the molt (see below).
Table 1.
Summary of epM EST data set
| Number of sequenced bacterial clones | 10,368 |
| Number of ESTs | 6,653 |
| Number of putative genes (nonredundant clusters)*1 | 1,380 (1,451) |
| Number of genes with similarity to Drosophila genes *2 | 1,025 |
*1: 71 clusters were considered to be isoforms/premature forms of other clusters.
*2: The threshold maximum E-value was set to 1e-05 by Blastx search.
Figure 1.
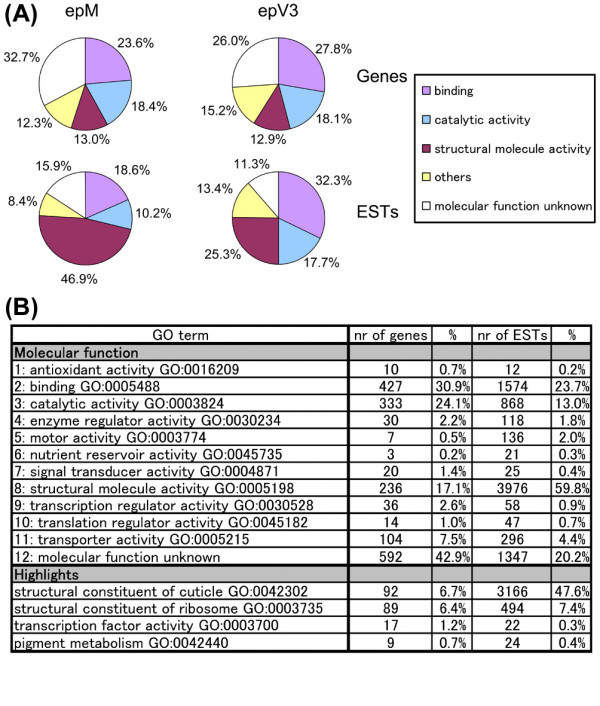
Assignment of Gene Ontology (GO) molecular function terms to the two epidermal EST data sets of Bombyx mori. (A) The ratio of gene numbers (upper) and total EST clones (lower) of the representative 4 GO terms are shown. (B) The ratio of genes classified into each GO term. The percentage of gene numbers (left) and total EST clones (right) were shown. Because one cluster can be associated with more than one GO term and total percentages summing all categories are shown as 100 in (A), percentage in (B) is different from that in (A). Highlights show the characteristic gene families such as cuticular proteins and transcription factors.
Following the structural protein genes, the genes in the categories "binding activity" (Figure 1B, category 2) and "catalytic activity" (Figure 1B, category 3) were the most abundant, in both the number of species (31% and 24% genes, respectively) and the level of expression (24% and 13%, respectively) (Figure 1B). Considerable numbers of molecules in these categories are thought to be involved in the formation of the cuticle structure during the molt. In the molting stages, new cuticle is synthesized and constructed to replace the old cuticle during its rapid apolysis. More than 30 species of proteolytic molecules (see below), ubiquitin cascades, and various degradation pathways are involved in the apolysis of the old cuticle structure. In contrast, many enzymes involved in glycolysis, ATP synthesis, and electron transport produce the energy for the amino acid metabolism and protein synthesis required for new cuticle formation. Chitin-binding proteins, phenoloxidase-activating enzymes, and laccase are also involved in the modification of cuticular proteins (sclerotization), and signal peptidase and several protein transporters are involved in the transport of cuticular components (Additional File 3). We also found putative genes related to insect hormone metabolism, small-ligand binding, and transcription (Additional File 3), which are summarized below.
A comparison of the categorized gene clusters of epM and epV3 shows that the percentage of structural protein genes expressed in epM is much larger than that in epV3 (Figure 1A), and that more different types of cuticular protein genes are represented in epM than in epV3 (Figure 2). This observation suggests that many types of cuticular proteins are involved in cuticle formation in the molting stages, whereas the intermolt cuticle is built of only a few kinds of cuticular proteins (most of them are class RR-1 proteins; Figure 2), which may contribute to the thickening of the endocuticle layer.
Figure 2.
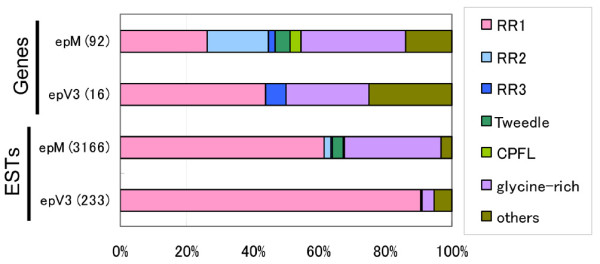
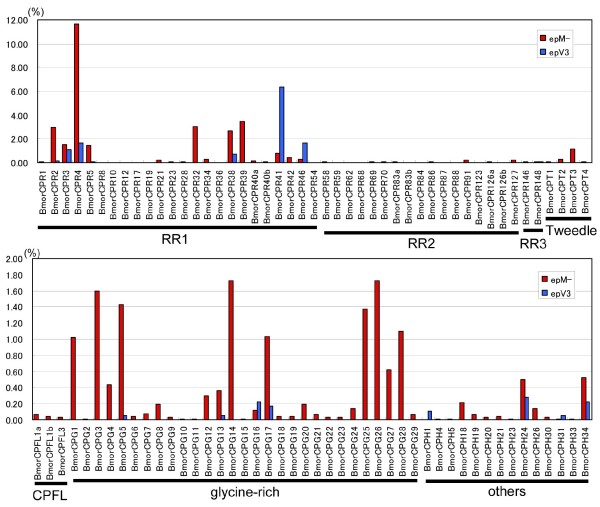
Composition of cuticular protein genes in two epidermal EST libraries (epM, epV3). The percentages of gene numbers (upper) and total clone numbers (lower) of each motif are shown. The numbers of gene numbers and total ESTs are shown in parentheses.
Species, structure, and expression of cuticular proteins encoded by genes in epM
We identified 92 cuticular protein genes in the epM library (Figures 2 and 3, Additional File 4). These genes correspond to 88 cuticular protein genes identified in the Bombyx p50 strain [12]. In four cases (BmorCPFL1, BmorCPR40, BmorCPR83, and BmorCPR126), two different genes in the epM data set corresponded to the same gene of the p50 strain. Because we have repeatedly mated the +p strain with its pS sibling (sibmating), the two strains have similar genetic backgrounds. Therefore, the sequence variations identified in this study may result from the differences between +p (or pS) and p50. It has been reported that the copy numbers of cuticular protein genes vary even among strains of D. melanogaster [22] and A. gambiae [15], suggesting that the copy numbers of cuticular protein genes also vary among Bombyx strains in the four cases cited above, although we cannot exclude the possibility that sequence differences are the result of strain differences between the +p and pS strains.
Figure 3.
The ratio (ESTs of each gene/total ESTs) of each cuticular protein gene in two Bombyx epidermal ESTs. Red columns indicate the ratio of epM library and blue columns indicate that of epV3 library. Cuticular motif of each gene was also shown below the lines.
Transcripts corresponding to 43 cuticular proteins in the epM cDNA library are predicted to have the R&R consensus motif in their amino acid sequences (Figure 3, Additional File 4). Twenty-four were RR-1, 17 were RR-2, and two were RR-3 proteins. A comparison of the R&R protein transcripts in epM and epV3 demonstrated totally different expression patterns. In the molting stages (epM), transcripts for the 24 RR-1 protein genes comprised 61% of the total ESTs for cuticular protein genes. However, in the intermolt of the fifth instar stages (epV3), only seven RR-1 protein transcripts comprised 91% of the total ESTs of the cuticular protein genes (Figure 2). In contrast, 17 RR-2 protein transcripts were found in epM (2.0% of the total cuticular protein ESTs), whereas no RR-2 protein transcripts were found in epV3 (Figure 2, Additional File 4). Cox and Willis [23] reported that the protein composition of the cuticle correlates with the flexibility of the mature cuticle. We found that RR-1 protein genes were abundantly expressed in epM compared with their expression in other EST libraries [12], which is consistent with the general claim that RR-1 proteins are involved in the flexible cuticle. The dominant expression in epV3 of RR-1 proteins, which affect the thickness of the endocuticle region during the intermolt, may contribute to the flexible cuticle structure in the feeding stages. The lack of RR-2 protein genes in epV3 suggests that these proteins are mainly involved in the formation of the outer cuticle region, the exocuticle or epicuticle layers, during the molting stages. It is noteworthy that transcripts corresponding not only to R&R proteins but also to other types of cuticular proteins (see below) are abundant in the epM library (Figure 2), and may therefore be essential for the formation of new cuticle.
Glycine-rich cuticular protein genes
As well as those with R&R consensus motifs, cuticular proteins with the glycine-rich motif (CPG) are often observed in Bombyx [12,24,25]. We found 29 types of CPG transcripts in the epM library (29% of the total cuticular protein genes; Figure 2). In the epV3 library, only four CPG protein transcripts were found, and at lower levels (3.9% in total; Figure 2). Andersen et al. [2] predicted that Gly-Gly (GG) repeats would form turn structures in proteins, and some proteins contain glycine-rich regions that include GG repeats [24-27]. We previously reported that the expression of the cuticular protein BMCPG1 (BmorCPG1 in this study), which has many Gly-Gly-Tyr (GGY) repeats and sequence similarity to Drosophila EDG91, is dependent on the ecdysteroid pulse during the fourth molt [26]. We also found that the tyrosine residues of GGY repeats were cross-linked to di-DOPA by tyrosinase treatment [28] (Fujiwara et al., in preparation), suggesting that the GGY motif is involved in protein cross-linking during sclerotization [29].
A comparison with other EST libraries showed that 12 CPG genes (BmorCPG2, BmorCPG3, BmorCPG7, BmorCPG8, BmorCPG15, BmorCPG19, BmorCPG21, BmorCPG23, BmorCPG25, BmorCPG26, BmorCPG27, and BmorCPG28) are specifically expressed in the epM library (Additional File 1). Transcripts of the CPG proteins in other libraries were found mainly in epithelial cells, such as the antenna, compound eye, and wing disc, suggesting that CPG proteins are components of body surfaces in general [12]. It is interesting that CPG proteins are usually positively charged, with a high isoelectric pH (> 8.0), and that these features are not observed in other cuticular proteins. This property of CPG proteins may contribute to their physical interaction with other types of cuticular proteins and between CPGs in the cuticle layer.
Other cuticular protein genes
Recently, other types of cuticular protein genes have been reported. Tweedle proteins, which are suggested to interact directly with chitin, are observed in the epidermis, tracheal tree, foregut, and wing discs in D. melanogaster [4]. There are four Tweedle protein genes in Bombyx (BmorCPT1-BmorCPT4) [12], and transcripts of all four Tweedle genes are found in the epM library (3.3% of the total cuticular protein genes), but are not present in the epV3 library (Figure 2). Togawa et al. [5] reported a cuticular protein with a 44-amino-acid motif (CPF) and CPF-like proteins (CPFL). There are one CPF and four CPFL genes in the Bombyx p50 strain [12], and we found three CPFL protein transcripts in the epM library (two of them are very similar and correspond to the gene BmorCPFL1 in the p50 strain). These CPFL protein transcripts occur in small amounts in epM (0.3%), whereas they are not present at all in the epV3 EST data set (Figures 2 and 3).
In this study, we also found 13 hypothetical cuticular proteins (CPH, Additional File 4) [12]. Some CPHs have the conserved motif. BmorCPH30 and BmorCPH31 have an 18-residue motif (PV)xDTPEVAAA(KR)AA(HF)xAA(HY), and seven CPH proteins (BmorCPH18, BmorCPH19, BmorCPH21, BmorCPH23, BmorCPH24, BmorCPH26, and BmorCPH34) have the AAP(A/V) motif, suggesting common roles in cuticle formation. However, CPH proteins have various structural features, amino acid compositions, repeated structures, and isoelectric points, so we cannot summarize their functional roles at present.
Highly expressed cuticular protein genes in the epM data set
Table 2 shows the top 50 genes most abundantly expressed in the epM library. Of these 50 genes, 29 are cuticular protein genes. BmorCPR4 (formally known as LCP18) is the most abundantly expressed (776 clones corresponding to 11.7% of the total epM ESTs and 24.5% of the epM cuticular protein transcripts), followed by BmorCPR39, BmorCPR32, BmorCPR2 (formally known as LCP17), and BmorCPR38 (formally known as LCP22). BmorCPR4 is also expressed abundantly in epV3 (30 clones; 12.9% of the epV3 cuticular protein transcripts), which is consistent with the former observation that this gene is also expressed in the intermolt stage [30]. BmorCPR4, the main component of the larval cuticle, is suggested to be orthologous to Hyalophora cecropia HCCP12 and Manduca sexta MSCP14.6 [30]. Similarly, BmorCPR38 and BmorCPR3 (5.7% and 3.1% of the epM cuticular protein transcripts, respectively) are abundantly expressed not only in epM but also in epV3 (5.6% and 8.6% of the epV3 cuticular protein transcripts, respectively; Figure 3). These cuticular proteins are constitutively expressed during both the molt and intermolt stages. In contrast, BmorCPR41 (formally known as LCP30) and BmorCPR46 (formally known as Bmwcp11) are more abundantly expressed during the intermolt stages (49.4% and 12.9% of the epV3 cuticular protein transcripts, respectively) than during the molt (1.7% and 0.6% of the epM cuticular protein transcripts, respectively). BmorCPR41 and BmorCPR46 comprise more than 62% of the cuticular protein transcripts in epV3, and these are therefore intermolt-specific genes. This observation is consistent with the fact that BmorCPR46 is mainly expressed at the intermolt stage in the wing disc [12].
Table 2.
Top 50 EST genes
| Cluster No. | Total ESTs | Gene Name | Drosophila CG Number | e-value* | score* |
| 1 | 776 | BmorCPR4 | CG32405 | 2E-24 | 108 |
| 2 | 230 | BmorCPR39 | CG30045 | 6E-25 | 111 |
| 3 | 202 | BmorCPR32 | CG30045 | 1E-39 | 159 |
| 4 | 195 | BmorCPR2 | CG30045 | 2E-26 | 115 |
| 5 | 180 | BmorCPR38 | CG30045 | 9E-25 | 110 |
| 6 | 116 | - | - | - | - |
| 7 | 115 | BmorCPG26 | CG14191 | 6E-22 | 100 |
| 8 | 115 | BmorCPG14 | CG2150 | 1E-21 | 100 |
| 9 | 106 | BmorCPG3 | CG15597 | 2E-22 | 102 |
| 10 | 100 | Actin 57B | CG10067 | 0 | 756 |
| 11 | 99 | BmorCPR3 | CG30045 | 2E-25 | 112 |
| 12 | 97 | BmorCPR5 | CG32405 | 3E-20 | 94.7 |
| 13 | 95 | BmorCPG5 | CG14191 | 7E-18 | 87.4 |
| 14 | 91 | BmorCPG25 | CG13050 | 6E-13 | 70.9 |
| 15 | 87 | Myosin light chain 2 | CG2184 | 7E-69 | 257 |
| 16 | 76 | BmorCPT3 | CG5812 | 5E-83 | 305 |
| 17 | 73 | BmorCPG28 | CG15597 | 9E-20 | 93.2 |
| 18 | 69 | BmorCPG17 | CG5225 | 7E-08 | 53.5 |
| 19 | 68 | BmorCPG1 | CG14191 | 4E-24 | 107 |
| 20 | 63 | Tropomyosin 1 | CG4898 | 9E-140 | 493 |
| 21 | 60 | CG32774 | CG32774 | 1E-06 | 50.8 |
| 22 | 53 | BmorCPR41 | CG30042 | 1E-30 | 130 |
| 23 | 47 | CG13731 | CG13731 | 2E-19 | 94.4 |
| 24 | 41 | BmorCPG27 | CG14191 | 5E-21 | 97.4 |
| 25 | 35 | Troponin C at 73F | CG7930 | 3E-65 | 244 |
| 26 | 35 | BmorCPH34 | CG34333 | 7E-33 | 137 |
| 27 | 35 | wings up A | CG7178 | 2E-98 | 355 |
| 28 | 34 | Myosin alkali light chain 1 | CG5596 | 2E-50 | 196 |
| 29 | 33 | BmorCPH24 | CG32564 | 4E-10 | 61.2 |
| 30 | 31 | - | - | - | - |
| 31 | 29 | lethal (2) essential for life | CG4533 | 3E-55 | 211 |
| 32 | 29 | BmorCPG4 | CG32564 | 1E-37 | 154 |
| 33 | 28 | BmorCPR42 | CG7658 | 2E-21 | 99 |
| 34 | 24 | BmorCPG13 | CG30101 | 2E-91 | 333 |
| 35 | 24 | mitochondrial ATPase subunit 6 | CG34073 | 4E-51 | 198 |
| 36 | 22 | Muscle LIM protein at 60A | CG33149 | 8E-41 | 163 |
| 37 | 22 | upheld | CG7107 | 3E-171 | 598 |
| 38 | 22 | - | - | - | - |
| 39 | 21 | BmorCPT2 | CG5812 | 8E-70 | 261 |
| 40 | 20 | Ejaculatory bulb protein III | CG11390 | 2E-24 | 109 |
| 41 | 20 | BmorCPR46 | CG2555 | 3E-16 | 81.6 |
| 42 | 20 | BmorCPG12 | CG16886 | 2E-66 | 250 |
| 43 | 19 | BmorCPR34 | CG8515 | 7E-42 | 168 |
| 44 | 18 | Kaz1-ORFB | CG1220 | 4E-12 | 67.8 |
| 45 | 18 | Gasp | CG10287 | 6E-137 | 484 |
| 46 | 18 | CG31997 | CG31997 | 2E-48 | 189 |
| 47 | 17 | Tropomyosin 2 | CG4843 | 2E-131 | 465 |
| 48 | 17 | Translationally controlled tumor protein | CG4800 | 3E-78 | 288 |
| 49 | 17 | Ribosomal protein S20 | CG15693 | 1E-51 | 198 |
| 50 | 15 | BmorCPR21 | CG12330 | 2E-23 | 106 |
| 51 | 15 | CG17127 | CG17127 | 4E-31 | 131 |
| 52 | 15 | Ribosomal protein LP2 | CG4918 | 4E-39 | 157 |
| 53 | 15 | BmorCPR127 | CG13935 | 4E-36 | 148 |
| 54 | 15 | Ribosomal protein L5 | CG17489 | 2E-137 | 485 |
| 55 | 15 | LDLa domain containing chitin binding protein 1 | CG8756 | 1E-112 | 404 |
* E-value and score are calculated in Drosophila homologs.
As shown in Figure 3, most cuticular protein genes are molt specific. The second most abundantly expressed, BmorCPR39 (7.3% of the epM cuticular protein transcripts), and the third most abundantly expressed, BmorCPR32 (6.4% of the epM cuticular protein transcripts), of the epM ESTs were not found in epV3 (Additional File 4). Of the 29 cuticular protein genes in the list of the 50 most abundantly expressed genes (Table 2), 17 are not found in epV3 (Figure 3, Additional File 4). The fact that many molt-specific cuticular protein genes are identified here supports the effectiveness of the cDNA-based screening of cuticle components.
Temporal and tissue-specific expression patterns of major cuticular protein genes during the molt
To determine the detailed expression profiles of several major cuticular protein genes during the last larval molt, we used semiquantitative RT-PCR analysis of epidermal mRNAs at eight developmental stages, A-E2 of the fourth instar, F (just after the fourth ecdysis), and A (day 3) of the fifth instar (Figure 4A, [20]). The expression of most cuticular protein genes is repressed at D1 with the peak of ecdysteroid during the molt, suggesting that a high dose of ecdysteriod suppresses their expression. The expression patterns of cuticular protein genes during the molt can be grouped into four divisions: first group, expressed after the decay of ecdysteroid, as well as in the intermolt phase (BmorCPR3, BmorCPR4, BmorCPR5, BmorCPR38, BmorCPR41, BmorCPR10, BmorCPR39, BmorCPG4, BmorCPG5 and BmorCPG26); second group, expressed after the decay of ecdysteroid with their disappearance at mid fifth instar (BmorCPR83a, BmorCPR91, BmorCPG1, BmorCPG8, and BmorCPG28); third group, not expressed during the molt (C1-E2) (BmorCPFL1a); and fourth group, ubiquitously expressed throughout the fourth and fifth instars (BmorCPR148). We speculate that the first and fourth classes are generally involved in thickening the cuticle layer, but that the second class of dominantly expressed genes during molt are mainly involved in newly synthesized cuticle formation. The second group seems to be induced by the decay of ecdysteroid. We have already reported that the expression of the second class gene BMCPG1 (BmorCPG1 in this study) is induced by an ecdysteroid pulse and may be controlled by the FtzF1 function [26], which supports the idea outlined above. It is noteworthy that the RR-2 and CPG proteins are mainly categorized in the second class, supporting the idea that these proteins form the outer layer of the cuticle, as proposed above.
Figure 4.
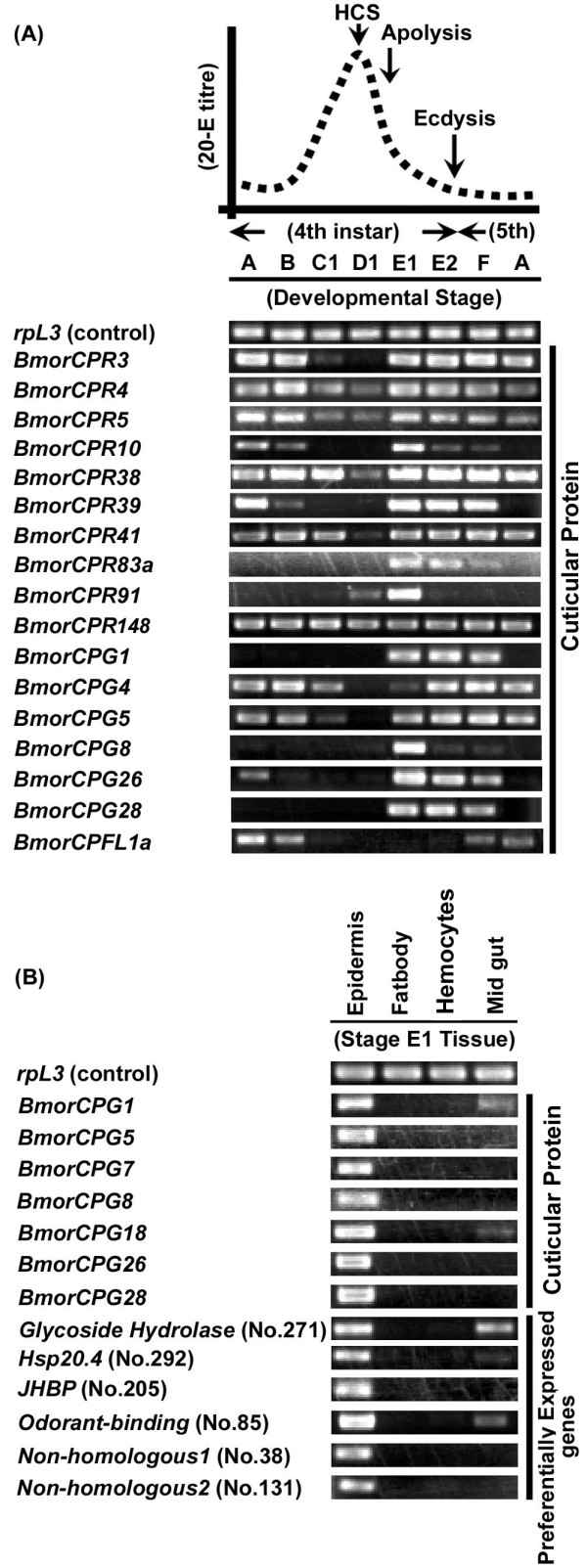
Stage and Tissue specific expression of epidermal genes isolated in epM data set by RT-PCR. (A) Expression profile of several cuticular protein genes by RT-PCR analysis. Relative hemolymph ecdysteroid titer is also shown according to Kiguchi and Agui, 1981 [20], and indicates the stage of epidermis (4A to 5A). The gene for ribosomal protein L3 (rpL3) was used as an internal control (see details in materials & methods). (B) Tissue specific expression of cuticular protein genes and preferentially expressed genes identified in epM data set. Four tissues (epidermis, fat body, hemolymph, and midgut) on fourth E1 stages were used.
A comparison of various EST libraries showed the tissue specificity of each cuticular protein gene. Of these 92 cuticular protein genes, transcripts for 28 genes were only found in the epM library (Additional Files 1 and 4). In particular, BmorCPR39, BmorCPG26, BmorCPG3, BmorCPG25, BmorCPG28, and BmorCPG27 were included in the 50 most abundantly expressed ESTs in epM (Table 2), but were not found in any other libraries, suggesting that they are mainly involved in the construction of the newly synthesized cuticle of the epidermis. As described above, transcripts for 12 CPG proteins were only found in epM, and transcripts for another five CPG genes (BmorCPG1, BmorCPG14, BmorCPG17, BmorCPG20, and BmorCPG29) were predominantly found in epM (over 75% of the total ESTs). To confirm the tissue-specific expression of several CPG genes, we used RT-PCR analyses of the mRNAs of the molting stage (E1) in four different tissues (epidermis, fat body, hemocytes, and midgut) and found that all of them were abundantly expressed in the epidermis in the molting stage (Figure 4B). In contrast, several cuticular proteins were predominantly expressed in tissues other than the epidermis, and in some cases, transcripts were also found in internal organs [12]. The cuticular protein genes may be expressed in different tissues in a different manner, and their composition and functional roles in tissue formation may be divergent.
EST clones preferentially expressed in epM
In addition to the genes for cuticular proteins, many other transcripts were preferentially found in the epM library. We identified 120 clusters (Additional File 1) using the criterion that the clone was more than threefold enriched in the epM data set compared with its occurrence in other EST libraries (over 75% of ESTs were found in the epM library). Thirteen of these clusters were represented by more than four clones in the epM library (Table 3). Nine of them were already known or homologous genes, although some of them were functionally undefined. The remaining four clusters had no sequence similarity to any known proteins. We also analyzed the epidermis-specific expression of four genes (Figure 4B) and confirmed their tissue specificity. These results suggest that comparing various EST libraries is useful in screening for tissue-specific genes.
Table 3.
Preferentially expressed genes in epM data set compared to other Bombyx EST libraries.
| Cluster No. | Total ESTs | Gene name | CG No. | E-value* | Score* | OtherLibrary** | |
| Homology with known proteins | |||||||
| 21 | 60 | CG32774 | CG32774 | 1E-06 | 50.8 | 5 (vg4M) | |
| 23 | 47 | CG13731 | CG13731 | 2E-19 | 94.4 | 4(vg4M, e100, ceN) | |
| 85 | 10 | Ejaculatory bulb protein III | CG11390 | 1E-20 | 95.9 | 2 (epV3, mxg) | |
| 91 | 10 | FG10924.1 (Gibberella zeae PH-1) | 0 | ||||
| 157 | 6 | Osiris 9 | CG15592 | 8E-22 | 100 | 1 (wdV3) | |
| 197 | 5 | Osiris 9 | CG15592 | 1E-09 | 60.5 | 0 | |
| 205 | 5 | CG10264 | CG10264 | 1E-13 | 73.6 | 0 | |
| 207 | 5 | TpnC25D | CG6514 | 9E-48 | 186 | 0 | |
| 216 | 4 | fungal protease-specific inhibitor-F (Bombyx mori) | |||||
| No homologous genes | |||||||
| 38 | 22 | no homology | 5 (phe, MFB, epV3) | ||||
| 131 | 7 | no homology | 0 | ||||
| 175 | 5 | no homology | 0 | ||||
| 199 | 5 | no homology | 0 | ||||
* E-value and score are calculated in Drosophila homologs.
**Abbreviation: ceN (compound eyes, 5th-pupa), e100 (embryo, 100 hr after fertilization), epV3 (epidermis, 5th day 3), MFB (fat body, 5th-instar larva microbe-infected), mxg (maxillary galea, 5th-instar day-3 larva), phe (pheromone gland, newly-eclosed adult), vg4M (Verson's gland, 4th molting stage), wdV3 (wing disc, spinning stage day-3)
Proteins with putative signal peptides at their N-termini
After the translation of their respective mRNAs, the cuticular proteins are transported outside the cell to the cuticle, through the function of the signal peptides at their N-termini. We found that all the cuticular protein genes identified here encoded the putative N-terminal signal peptide sequences, as characterized by the program SignalP3.0 [31] (Additional File 1) [12]. Interestingly, this program also revealed that 290 genes in epM, other than cuticular protein genes, encode N-terminal signal peptides (Additional File 1).
Willis et al. [1] summarized the nonstructural proteins found in the cuticle into four categories: (1) pigments, (2) enzymes, (3) defense proteins, and (4) arylphorins. In the first class, we found three yellow family proteins (pigmentation genes, discussed below). In the second class, we found several enzymes that are associated with sclerotization and cuticle digestion. Although we have no evidence of their transport outside (cuticle) or inside (hemolymph) the epidermal cell, at least some of these proteins seem to be transported into the cuticle: chitinase, chitinase precursor, and four chitin-binding proteins, which could be essential for cuticle formation. We also identified the Gasp gene (no. 1451), which is predicted to have four type-2 chitin-binding domains. Gasp is found in the tracheae that are present in the cast cuticles [3]. Molting-fluid carboxypeptidase [32] and a dozen molting-fluid carboxypeptidases (non-serine-type peptidases) may be involved in digesting the old cuticle in apolysis. Two EST clusters (nos 331 and 1230) are predicted to contain N-terminal signal peptides and to encode novel Bombyx peptidase proteins, which may participate in apolysis. The insect epithelium secretes innate immunity components, antimicrobial peptides, and clip-domain serine proteases, which react with hemolymph prophenoloxidase [33-36]. Among the third-class proteins encoded in the epM data set, there are six serine proteases, which are thought to be involved in immunity (Additional File 3). Their inhibitors, Serpin (a Kazal-type serine protease inhibitor) and the Kunitz family of serine protease inhibitors are expressed at the same time (Additional File 3). In the fourth class, we found three arylphorin genes (nos 109, 114, and 239). The role of arylphorin remains unknown, although it is generally assumed to participate in sclerotization because of its high tyrosine content [1]. Apart from these four classes, we also found several genes with signal peptides at their N-termini, many of which are small-ligand-binding proteins (see below).
Hormone metabolism and the P450 family
The epM data set contains already-known genes and putative homologous genes that encode proteins involved in ecdysteroid and steroid metabolism (Additional File 3). During the molting process, ecdysteroid is finally inactivated by enzymes [37-41]. These genes may encode enzymes that inactivate ecdysteroid to terminate molting. There are also genes putatively involved in juvenile hormone (JH) metabolism and P450 genes in the epM data set. In the list of epM clusters encoding proteins with signal peptides are many known sequences (immune proteins, enzymes involved in various metabolic pathways) and proteins with putative functional domains (JH-binding proteins and odorant-binding proteins, etc.), but their actual roles in the molting stage of the epidermis remain ambiguous.
Modifiers of cuticular protein structure
Enzymes involved in cuticular sclerotization may be excreted into the cuticular layer. Candidate genes are also included in the epM data set. Laccase 2 is involved in cuticular sclerotization in the red flour beetle, Tribolium castaneum [29]. Although the sequence of Bombyx laccase 2 in epM (cluster no. 549) is a truncated sequence, we obtained the complete sequence with 5', 3' rapid amplification of cDNA ends and analyzed its expression dynamics in the larval molting stages. Bombyx laccase 2 is expressed in the late molting period, when the cuticle is sclerotized (data not shown), suggesting that laccase 2 also functions in cuticular sclerotization in Bombyx.
Prolyl 4-hydroxylase catalyzes peptidyl-proline hydroxylation to 4-hydroxy-L-proline in collagen proteins in vertebrates [42]. Together with peptidyl-prolyl cis-trans isomerase and disulfide isomerase, it is considered to have at least three functions: (i) catalyzing the formation of intrachain and interchain disulfide bonds; (ii) as the β-subunit of collagen prolyl 4-hydroxylases; and (iii) as a chaperone that binds nascent collagen chains and prevents their aggregation [42]. Collagens are the most abundant proteins in the human body, constituting ~30% of its protein mass. In contrast, the Drosophila genome has only three collagen-like genes, and it is considered that these protein-modifying enzymes modify proteins other than collagen [42]. Perhaps these insect cuticular proteins are candidate targets of these protein-modifying proteins. The battery of genes included in epM supports this proposition and the general cuticle (R&R-type) proteins have 5%–18% proline in their primary sequences. Prolyl-isomerization may also play an important role in the modification of the tertiary structures of cuticular proteins.
Small-ligand-binding proteins
The epM data set includes various putative small-ligand-binding protein genes (Additional File 3). JH-binding proteins (JHBPs) are secreted into the extracellular region, bind to JH, and are regulated by other insect hormones and nutrients [43]. JH also acts on the epidermis to prevent the ecdysteroid-induced expression of the Broad-Complex, and probably maintains larval traits [43-47]. Recently, we found that JH also regulates the larval pattern switch in the swallowtail butterfly, Papilio xuthus [48]. Ten JHBPs (or takeout) family proteins have been reported in Bombyx [49]. These proteins are assumed to bind hydrophobic ligands, although the precise ligands of the gene family are not known except for one JHBP that binds JH. Intriguingly, 16 putative JHBP genes occur in the epM data set (Additional File 3). Twelve of them are novel (nos 117, 205, 268, 337, 448, 479, 535, 646, 665, 737, 951, and 967), and five of them (nos 205, 479, 665, 951, and 967) are not included in other Bombyx EST data sets (Additional File 1). The sum of their clone numbers is 43. These JHBPs may be expressed and function cooperatively to maintain fourth-instar larval traits. These proteins have signal peptides at their N-termini, and the carotenoid-binding protein found in the cuticle has sequence similarity to JHBPs [50], suggesting that some JHBPs may also be secreted into the cuticle.
Unexpectedly, there are 14 genes for odorant-binding proteins in the epM data set, all of which have signal peptides at their N-termini. These proteins are known to localize to the sense organs (antennae) or pheromone glands (genitalia) in insects [51-53]. It is possible that they are associated with the sensory hairs on the epidermal surface. Although the functions of these proteins in the epidermis remain unclear, hydrophobic molecules may play important roles in molting.
A vitamin E (an antioxidant molecule)-binding protein was also isolated in this study. There have been few reports of invertebrate vitamin E (tocopherol derivatives) or α-tocopherol, which is the form mainly synthesized and accumulated in D. melanogaster [54]. Some tocopherol-metabolizing enzymes also occur in epM, so tocopherol may be synthesized and function as an antioxidant in the epidermis of B. mori.
There are some putative small-ligand-binding proteins that retain their N-terminal signal peptides. Genes for putative cellular retinaldehyde-binding proteins (seven genes) and FK506-binding proteins (two genes) are also included in the epM data set. Cellular retinaldehyde-binding protein (CRABP) has been reported to interact with all-trans retinoic acid, which is a structural derivative of ecdysone [55]. The overexpression of CRABP leads to the inhibition of growth in the Bombyx cultured cell line, BmN [55]. The epM data set also includes genes encoding FK506-binding protein (FKBP) [56], which is a member of the Drosophila protein family, FKBP39, and binds the immunosuppressive drug FK506. Recently, a member of the FKBP39 family was included in the ecdysone receptor and putative JH receptor Methoprene-tolerant complex [57]. Although the target substance is as yet unknown, these proteins may modify the ecdysone/JH signal cascade.
Transcription factors
The epM data set includes 17 transcripts for genes encoding proteins homologous or similar to known transcription factors or nucleic-acid-binding proteins (Figure 1B, Additional File 3). They correspond to about 1.2% of the total nonredundant clusters. Three clones are modifier of mdg4, encoding the BTB/POZ domain protein of Bombyx, which is predicted to encode a zinc-finger protein. However, because this gene is also found in many other libraries, it may be expressed ubiquitously. Consistent with this observation, the Modifier of mdg4 protein functions with chromatin proteins in many types of tissue in Drosophila [58]. Two proteins known to act in the Notch pathway, E(spl)mbeta and Suppressor of Hairless (Su(H)), are also found in epM. The Notch pathway is implicated in the process of lateral inhibition and is required for the epidermal/neural fate decision in the stereotyped patterning of the sensory organs on the epidermis. Su(H) and proteins of the E(spl) complex are required for the epidermal fate decision in Drosophila [59]. It is noteworthy that we found a homologue of Drosophila drumstick (drm) in epM, which encodes a zinc-finger protein. The Drm protein allows the accumulation of Bowl, a zinc-finger protein whose mRNA is ubiquitously expressed, by sequestering Lines, which otherwise reduces the abundance of Bowl by binding directly to it. The patterning across the dorsal epidermis of the Drosophila larva is organized by the regulation of drm expression by Hedgehog and Wingless, which are secreted from adjacent sources flanking the parasegment boundary [60]. The inclusion of drm in the epM data set implies that the Drm/Lines/Bowl regulatory cassette may also function in patterning the larval epidermis of the silkworm. The gene for slow border cells encodes a protein that functions in choriogenesis in both Bombyx and Drosophila [61]. Intriguingly, there are features common to choriogenesis and cuticle formation. Both of them involve the formation of rigid exoskeletons with excreted proteins. This transcription factor may be a central regulator or maintain the identity of the integument in both choriogenesis and cuticle formation. These proteins and/or some hormonal regulators cooperatively specify the integument as the larvae periodically shed their egg cases.
Pigmentation genes
The epM data set includes transcripts for nine genes encoding proteins homologous or similar to known pigmentation genes (Figure 1B, Additional File 3). In the silkworm larval body, three types of coloration are typically observed: black (probably melanin), white (probably uric acid and pteridine), and brown (probably ommochrome) [62].
Melanin biosynthesis
Bombyx mori larvae have black markings on their dorsal integuments (segments 2, 5, and 8 of the +p strain and in all segments of the pS strain). Because we prepared RNA from the pS strain, which shows bold black stripes as the larval markings, the epM library was expected to contain several melanin synthesis genes. Six genes (encoding phenylalanine hydroxylase [PAH], tyrosine hydroxylase [TH], yellow, yellow-f, yellow-f2, and tan) are involved in the melanin synthesis pathway. In the larvae of the swallowtail butterfly, P. xuthus, both TH and yellow are associated with the stage-specific black larval markings [48,63,64], suggesting that these genes are associated with the Bombyx black markings. In contrast, PAH is a rather ubiquitous enzyme in insect tissues [65], and is broadly expressed in the larval epidermis of P. xuthus [66]. In D. melanogaster, Yellow-f and Yellow-f2 act as dopachrome-conversion enzymes downstream from dopa [67], and Tan acts as an N-β-alanyl-dopamine hydrolase, which converts N-β-alanyl-dopamine to dopamine [68]. These proteins may also function in melanin synthesis and/or black pattern regulation.
Ommochrome biosynthesis
Two genes, cinnabar and ruby, are associated with the ommochrome biosynthetic process. Cinnabar acts as a kynurenine 3-monooxygenase, which catalyzes the oxidation of kynurenine to 3-hydroxykynurenine [69]. Ruby is involved in vesicle trafficking [70]. Both proteins only affect eye coloration in Drosophila. Recently, ommochrome synthesis proteins have been associated with the adult wing coloration of the butterfly Heliconius [71,72]. cinnabar expression is associated with the forewing band, regardless of the pigment color [71]. Because ommochrome is abundant in the larval epidermis of Bombyx, these proteins may be involved in the larval brown coloration, although loss of function of cinnabar results in egg and eye coloration but not in larval body coloration [73].
Uric acid biosynthesis
The epM library contains one clone of the rosy gene. Rosy acts as a xanthine dehydrogenase, which is involved in uric acid synthesis. In Bombyx larvae, its mutation results in oily skin color [74,75]. Two xanthine dehydrogenase genes are highly expressed in the Malpighian tubules, fat body, and midgut [75]. Our results suggest that xanthine dehydrogenase functions in de novo uric acid synthesis in the epidermis.
Conclusion
By exhaustive sequencing and the analysis of full-length cDNA clones, we identified 1,380 putative genes expressed in the silkworm epidermis during the larval molt. Ninety-two of these encode putative cuticular proteins, and a comparison of the epM and epV3 ESTs revealed that the expression of the cuticular protein genes is markedly different in the molting and intermolt periods. Compared with other EST data sets, we identified 13 genes preferentially expressed in the epidermis during the molt. We identified 290 other genes with signal peptide sequences, in addition to the cuticular protein genes, suggesting that some of them play novel roles in cuticle formation and the molt. In this study, we catalogued the genes expressed in the epidermis during the larval molt, which should be helpful in gaining an overview of cuticle formation and the insect molt.
Methods
Experimental animals and developmental stages
The laboratory silkworm stocks +p (normal) and their sib-mating strain, pS (Striped) were reared on the artificial diet Silkmate 2 M (Kyodo Shiryo, Yokohama) under a 16 h: 8 h L:D photoperiod at 25°C. The staging of molting period was based on the spiracle index, which represented the characteristic sequence of new spiracle formation [76]. Based on the visible characteristics, 10 morphological larval stages (A, B, C1, C2, D1, D2, D3, E1, E2 [A-E, fourth instar larva] and F [fifth]) could be distinguished, which are referred as the spiracle index. C1 is a start stage of the fourth molting, C2 is a peak stage for the ecdysteroid titre and F is just after the molt [76]. The newly molted fourth and fifth instar larvae were segregated immediately after the onset of the photophase (this day was designated day 0).
Construction and sequencing of the full-length cDNA library
We dissected epidermal tissues from dorsal integument of segments 5, 6 of the silkworm stocks +p and the sib-mating strain, pS. The attached muscle, fat body and Verson's glands were removed by forceps from epidermal tissues. Total epidermal RNAs of each molting stages (C1, C2, D1, D2, D3, E1, E2 and F; 8 stages) were isolated from one larva of each silkworm strain, using TRI reagent (Sigma) according to the manufacturer's instructions. Total RNAs (3 μg each) of 8 molting stages (16 samples) were mixed and subjected to full-length cDNA library construction, which was performed by Hitachi Science Systems, Ltd. Japan. The construction of the full-length cDNA library was made by a G-capping method previously reported by Ohtake et al., 2004 [20], which enables relatively long insertions. We named this library as epM (fourth instar epidermis in Molting). After construction of the cDNA library, the library quality was evaluated by 96-clone test sequencing (Library size, 1.54 × 105; Ratio of insert-including bacterial clone, 79%; Ratio of full-length cDNA insert, 76%; Average of insert length, 0.9 kb). Judgment of the full-length cDNA was followed with Ohtake et al., 2004 [20]. More than 10,000 cDNA clones were picked up randomly from the epM library, and obtained a sequence of ~650 nucleotides from the 5' end of each cDNA in average.
Sequence Analysis and characterization of epM data set
The sequences more than 300 nucleotide length were taken as EST in the epM dataset. Nonredundant EST clusters were identified by clustering with EST clones of the other libraries with the criterion previously described [19]. By comparison of the deduced amino acid sequences with public protein databases InterProScan: [77], gene classification was assigned by using a criterion of homology of > 30% identity in a sequence > 100 amino acids as well as an E value lower than e-15 in a BLAST search (previously described [19]). Gene category was classified with GO term [78] or with FlyBase GO classification [21]. Identification and naming of cuticular protein genes were referred as Futahashi et al., 2008 [12], and cuticular protein genes were classified as structural molecule activity (GO:0005198) and structural constituent of cuticle (GO:0042302) in GO term. SignalP 3.0 [31] was used for signal peptide prediction.
Reverse transcription polymerase chain reaction (RT-PCR) analyses
To determine expression levels of cuticular protein genes, total epidermal RNA from fourth instar stages (A, B, C1, D1, E1, E2) and fifth (F and day3) were extracted. To detect tissue-specificity of EST clones, total RNA of fourth instar (stage E1) epidermis, fat body, hemocytes, and midgut were extracted. Total RNA was treated with RNase-free DNase (TaKaRa), and 1 μg of total RNA was reverse transcribed using First-Strand cDNA Synthesis Kit (Amersham). The primer sets were described in Additional File 5. The PCR conditions used were 96°C for 2 min followed by 30 (or 33) cycles of 96°C for 15 sec, 50°C (or 52°C) for 15 sec, and 72°C for 1 min. The reactions were kept at 72°C for 1 min after the last cycle. The gene for the ribosomal protein L3 (rpL3) [79] that is expressed constitutively in the cell was used as an internal control for normalization of equal sample loading.
Abbreviations
CPF: cuticular protein(s) with 44-amino acid motif; CPFL: CPF-like protein; CPG: cuticular protein glycine-rich; CPH: cuticular protein hypothetical; CPR: cuticular protein(s) with the R&R Consensus; CPT: cuticular protein(s) with a Tweedle motif; EST: expressed sequence tag; FKBP: FK506 binding protein; GO: gene onthology; HCS: head capsule slippage; JH: juvenile hormone; JHBP: JH binding protein; R&R Consensus: Rebers and Riddiford Consensus; RR-1, RR-2, and RR-3: three class of CPR proteins; RT-PCR: reverse transcription and polymerase chain reaction; 20-E: 20-hydroxyecdysone.
Authors' contributions
SO carried out the molecular genetic studies, participated in the construction of EST library and performed RT-PCR analysis, and wrote the manuscript; RF performed sequence data analysis and wrote the manuscript; TK wrote and assisted preparation of manuscript; KM directed the sequencing and data analysis; HF directed the experimental work on EST construction and data analysis and completed the manuscript. The project was conceived and coordinated by KM and HF. All authors have read and approved the final version of this manuscript.
Supplementary Material
List of nonredundant gene sequence and its number of epM data set. 1–1380 nonredundant sequences in epM were listed. The column of "Other libraries" indicates the numbers of the corresponding sequence found in the silkworm EST libraries constructed by Japanese groups, other than the epM library. "epM/total" represents the percentage of EST numbers of epM to total silkworm EST data set. DmGene represents an orthologous gene in Drosophila melanogaster
List of identified putative isoforms or premature transcript.
List of characteristic gene groups identified in epM dataset.
List of identified cuticular proteins and comparison with epV3 library. The number in epM and epV3 columns indicates the genes expressed in the respective data sets.
Oligonucleotide primers used for analysis of tissue- and stage-specific expression of selected genes.
Acknowledgments
Acknowledgements
This work was supported in part by grants in aid for scientific research from the Ministry of Education, Science, and Culture of Japan, National Bio-resource Project (NBRP), and Culture of Japan, PROBRAIN, the Basic Research Program to HF, and Research Fellowship of Japan Society for the Promotion of Science for Young Scientists to RF. EST sequencing was partly supported by the Insect Genome Project of the Ministry of Agriculture, Forestry and Fisheries of Japan. We are very grateful to Michihiko Shimomura and Kazutoshi Yoshitake for data analysis.
Contributor Information
Shun Okamoto, Email: kk57516@mail.ecc.u-tokyo.ac.jp.
Ryo Futahashi, Email: ryo284@k.u-tokyo.ac.jp.
Tetsuya Kojima, Email: tkojima@k.u-tokyo.ac.jp.
Kazuei Mita, Email: kmita@nias.affrc.go.jp.
Haruhiko Fujiwara, Email: haruh@k.u-tokyo.ac.jp.
References
- Willis JH, Iconomidou VA, Smith RF, Hamodrakas SJ. Cuticular proteins. In: Gilbert LI, Iatrou K, Gill S, editor. Comprehensive Insect Science. Vol. 4. Oxford, Elsevier; 2005. pp. 79–109. [Google Scholar]
- Andersen SO, Højrup P, Roepstorff P. Insect cuticular proteins. Insect Biochem Mol Biol. 1995;25:153–176. doi: 10.1016/0965-1748(94)00052-j. [DOI] [PubMed] [Google Scholar]
- He N, Botelho JM, McNall RJ, Belozerov V, Dunn WA, Mize T, Orlando R, Willis JH. Proteomic analysis of cast cuticles from Anopheles gambiae by tandem mass spectrometry. Insect Biochem Mol Biol. 2007;37:135–146. doi: 10.1016/j.ibmb.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Guan X, Middlebrooks BW, Alexander S, Wasserman SA. Mutation of TweedleD, a member of an unconventional cuticle protein family, alters body shape in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16794–16799. doi: 10.1073/pnas.0607616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa T, Augustine Dunn W, Emmons AC, Willis JH. CPF and CPFL, two related gene families encoding cuticular proteins of Anopheles gambiae and other insects. Insect Biochem Mol Biol. 2007;37:675–688. doi: 10.1016/j.ibmb.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Rebers JE, Riddiford LM. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. Journal of molecular biology. 1988;203:411–423. doi: 10.1016/0022-2836(88)90009-5. [DOI] [PubMed] [Google Scholar]
- Rebers JE, Willis JH. A conserved domain in arthropod cuticular proteins binds chitin. Insect Biochem Mol Biol. 2001;31:1083–1093. doi: 10.1016/s0965-1748(01)00056-x. [DOI] [PubMed] [Google Scholar]
- Togawa T, Nakato H, Izumi S. Analysis of the chitin recognition mechanism of cuticle proteins from the soft cuticle of the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2004;34:1059–1067. doi: 10.1016/j.ibmb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Andersen SO. Studies on proteins in post-ecdysial nymphal cuticle of locust, Locusta migratoria, and cockroach, Blaberus craniifer. Insect Biochem Mol Biol. 2000;30:569–577. doi: 10.1016/s0965-1748(00)00029-1. [DOI] [PubMed] [Google Scholar]
- Iconomidou VA, Willis JH, Hamodrakas SJ. Is beta-pleated sheet the molecular conformation which dictates formation of helicoidal cuticle? Insect Biochem Mol Biol. 1999;29:285–292. doi: 10.1016/s0965-1748(99)00005-3. [DOI] [PubMed] [Google Scholar]
- Magkrioti CK, Spyropoulos IC, Iconomidou VA, Willis JH, Hamodrakas SJ. cuticleDB: a relational database of Arthropod cuticular proteins. BMC Bioinformatics. 2004;5:138. doi: 10.1186/1471-2105-5-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futahashi R, Okamoto S, Kawasaki H, Zhong YS, Iwanaga M, Mita K, Fujiwara H. Genome-wide identification of cuticular protein genes in the silkworm, Bombyx mori. Insect Biochem Mol Biol. [DOI] [PubMed]
- Karouzou MV, Spyropoulos Y, Iconomidou VA, Cornman RS, Hamodrakas SJ, Willis JH. Drosophila cuticular proteins with the R&R Consensus: annotation and classification with a new tool for discriminating RR-1 and RR-2 sequences. Insect Biochem Mol Biol. 2007;37:754–760. doi: 10.1016/j.ibmb.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Consortium THGS. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Togawa T, Dunn WA, He N, Emmons AC, Willis JH. Annotation and analysis of a large cuticular protein family with the R&R Consensus in Anopheles gambiae. BMC Genomics. 2008;9:22. doi: 10.1186/1471-2164-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa T, Dunn WA, Emmons AC, Nagao J, Willis JH. Developmental expression of cuticular protein genes with the R&R consensus from Anopheles gambiae. Insect Biochem Mol Biol. 2008 doi: 10.1016/j.ibmb.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Ote M, Okano K, Shimada T, Guo-Xing Q, Mita K. Change in the expressed gene patterns of the wing disc during the metamorphosis of Bombyx mori. Gene. 2004;343:133–142. doi: 10.1016/j.gene.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Kinjoh T, Kaneko Y, Itoyama K, Mita K, Hiruma K, Shinoda T. Control of juvenile hormone biosynthesis in Bombyx mori: cloning of the enzymes in the mevalonate pathway and assessment of their developmental expression in the corpora allata. Insect Biochem Mol Biol. 2007;37:808–818. doi: 10.1016/j.ibmb.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Mita K, Morimyo M, Okano K, Koike Y, Nohata J, Kawasaki H, Kadono-Okuda K, Yamamoto K, Suzuki MG, Shimada T, Goldsmith MR, Maeda S. The construction of an EST database for Bombyx mori and its application. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14121–14126. doi: 10.1073/pnas.2234984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake H, Ohtoko K, Ishimaru Y, Kato S. Determination of the capped site sequence of mRNA based on the detection of cap-dependent nucleotide addition using an anchor ligation method. DNA Res. 2004;11:305–309. doi: 10.1093/dnares/11.4.305. [DOI] [PubMed] [Google Scholar]
- FlyBase http://flybase.bio.indiana.edu/
- Charles JP, Chihara C, Nejad S, Riddiford LM. A cluster of cuticle protein genes of Drosophila melanogaster at 65A: sequence, structure and evolution. Genetics. 1997;147:1213–1224. doi: 10.1093/genetics/147.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DC, Willis JH. The cuticular proteins of Hyalophora cecropia from different anatomical regions and metamorphic stages. Insect Biochem. 1985;15:349–362. [Google Scholar]
- Charles JP, Bouhin H, Quennedey B, Courrent A, Delachambre J. cDNA cloning and deduced amino acid sequence of a major, glycine-rich cuticular protein from the coleopteran Tenebrio molitor. Temporal and spatial distribution of the transcript during metamorphosis. European journal of biochemistry / FEBS. 1992;206:813–819. doi: 10.1111/j.1432-1033.1992.tb16989.x. [DOI] [PubMed] [Google Scholar]
- Krogh TN, Skou L, Roepstorff P, Andersen SO, Hojrup P. Primary structure of proteins from the wing cuticle of the migratory locust, Locusta migratoria. Insect Biochem Mol Biol. 1995;25:319–329. doi: 10.1016/0965-1748(94)00056-n. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Matsuoka T, Iimura Y, Fujiwara H. Ecdysteroid-dependent expression of a novel cuticle protein gene BMCPG1 in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2002;32:599–607. doi: 10.1016/s0965-1748(01)00136-9. [DOI] [PubMed] [Google Scholar]
- Zhong YS, Mita K, Shimada T, Kawasaki H. Glycine-rich protein genes, which encode a major component of the cuticle, have different developmental profiles from other cuticle protein genes in Bombyx mori. Insect Biochem Mol Biol. 2006;36:99–110. doi: 10.1016/j.ibmb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Held MA, Tan L, Kamyab A, Hare M, Shpak E, Kieliszewski MJ. Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. The Journal of biological chemistry. 2004;279:55474–55482. doi: 10.1074/jbc.M408396200. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11337–11342. doi: 10.1073/pnas.0504982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shofuda K, Togawa T, Nakato H, Tomino S, Izumi S. Molecular cloning and characterization of a cDNA encoding a larval cuticle protein of Bombyx mori. Comparative biochemistry and physiology. 1999;122:105–109. doi: 10.1016/s0305-0491(98)10151-7. [DOI] [PubMed] [Google Scholar]
- SignalP 3.0 http://www.cbs.dtu.dk/services/SignalP/
- Ote M, Mita K, Kawasaki H, Daimon T, Kobayashi M, Shimada T. Identification of molting fluid carboxypeptidase A (MF-CPA) in Bombyx mori. Comparative biochemistry and physiology. 2005;141:314–322. doi: 10.1016/j.cbpc.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Abraham EG, Pinto SB, Ghosh A, Vanlandingham DL, Budd A, Higgs S, Kafatos FC, Jacobs-Lorena M, Michel K. An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16327–16332. doi: 10.1073/pnas.0508335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Ashida M. Cuticular pro-phenoloxidase of the silkworm, Bombyx mori. Purification and demonstration of its transport from hemolymph. The Journal of biological chemistry. 2001;276:11100–11112. doi: 10.1074/jbc.M008426200. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunological reviews. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- Gilbert LI. Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Molecular and cellular endocrinology. 2004;215:1–10. doi: 10.1016/j.mce.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Rigden DJ, Ebrahimi B, Turner PC, Rees HH. Regulation of ecdysteroid signalling during Drosophila development: identification, characterization and modelling of ecdysone oxidase, an enzyme involved in control of ligand concentration. The Biochemical journal. 2005;389:637–645. doi: 10.1042/BJ20050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb TJ, Powls R, Rees HH. Enzymes of ecdysteroid transformation and inactivation in the midgut of the cotton leafworm, Spodoptera littoralis: properties and developmental profiles. The Biochemical journal. 1995;312 ( Pt 2):561–568. doi: 10.1042/bj3120561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Chen JH, Fisher MJ, Rees HH. Induction of enzymes involved in molting hormone (ecdysteroid) inactivation by ecdysteroids and an agonist, 1,2-dibenzoyl-1-tert-butylhydrazine (RH-5849) The Journal of biological chemistry. 1997;272:8427–8432. doi: 10.1074/jbc.272.13.8427. [DOI] [PubMed] [Google Scholar]
- Williams DR, Fisher MJ, Rees HH. Characterization of ecdysteroid 26-hydroxylase: an enzyme involved in molting hormone inactivation. Archives of biochemistry and biophysics. 2000;376:389–398. doi: 10.1006/abbi.2000.1731. [DOI] [PubMed] [Google Scholar]
- Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Du J, Hiruma K, Riddiford LM. A novel gene in the takeout gene family is regulated by hormones and nutrients in Manduca larval epidermis. Insect Biochem Mol Biol. 2003;33:803–814. doi: 10.1016/s0965-1748(03)00079-1. [DOI] [PubMed] [Google Scholar]
- Riddiford LM, Hiruma K, Zhou X, Nelson CA. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem Mol Biol. 2003;33:1327–1338. doi: 10.1016/j.ibmb.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Zhou B, Hiruma K, Shinoda T, Riddiford LM. Juvenile hormone prevents ecdysteroid-induced expression of broad complex RNAs in the epidermis of the tobacco hornworm, Manduca sexta. Developmental biology. 1998;203:233–244. doi: 10.1006/dbio.1998.9059. [DOI] [PubMed] [Google Scholar]
- Zhou B, Riddiford LM. Hormonal regulation and patterning of the broad-complex in the epidermis and wing discs of the tobacco hornworm, Manduca sexta. Developmental biology. 2001;231:125–137. doi: 10.1006/dbio.2000.0143. [DOI] [PubMed] [Google Scholar]
- Zhou X, Riddiford LM. Broad specifies pupal development and mediates the 'status quo' action of juvenile hormone on the pupal-adult transformation in Drosophila and Manduca. Development (Cambridge, England) 2002;129:2259–2269. doi: 10.1242/dev.129.9.2259. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Fujiwara H. Juvenile hormone regulates butterfly larval pattern switches. Science. 2008;319:1061. doi: 10.1126/science.1149786. [DOI] [PubMed] [Google Scholar]
- Saito K, Su ZH, Emi A, Mita K, Takeda M, Fujiwara Y. Cloning and expression analysis of takeout/JHBP family genes of silkworm, Bombyx mori. Insect molecular biology. 2006;15:245–251. doi: 10.1111/j.1365-2583.2006.00612.x. [DOI] [PubMed] [Google Scholar]
- Wybrandt GB, Andersen SO. Purification and sequence determination of a yellow protein from sexually mature males of the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol. 2001;31:1183–1189. doi: 10.1016/s0965-1748(01)00064-9. [DOI] [PubMed] [Google Scholar]
- Bohbot J, Vogt RG. Antennal expressed genes of the yellow fever mosquito (Aedes aegypti L.); characterization of odorant-binding protein 10 and takeout. Insect Biochem Mol Biol. 2005;35:961–979. doi: 10.1016/j.ibmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Jin X, Brandazza A, Navarrini A, Ban L, Zhang S, Steinbrecht RA, Zhang L, Pelosi P. Expression and immunolocalisation of odorant-binding and chemosensory proteins in locusts. Cell Mol Life Sci. 2005;62:1156–1166. doi: 10.1007/s00018-005-5014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung O, Wolfner MF. Identification and characterization of the major Drosophila melanogaster mating plug protein. Insect Biochem Mol Biol. 2001;31:543–551. doi: 10.1016/s0965-1748(00)00154-5. [DOI] [PubMed] [Google Scholar]
- Miquel J, Fleming J, Economos AC. Antioxidants, metabolic rate and aging in Drosophila. Archives of gerontology and geriatrics. 1982;1:159–165. doi: 10.1016/0167-4943(82)90016-4. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Chen J, Lv ZB, Nie ZM, Wang D, Shen HD, Wang XD, Wu XF, Zhang YZ. Expression and functional analysis of the cellular retinoic acid binding protein from silkworm pupae (Bombyx mori) Journal of cellular biochemistry. 2007;102:970–979. doi: 10.1002/jcb.21333. [DOI] [PubMed] [Google Scholar]
- Theopold U, Dal Zotto L, Hultmark D. FKBP39, a Drosophila member of a family of proteins that bind the immunosuppressive drug FK506. Gene. 1995;156:247–251. doi: 10.1016/0378-1119(95)00019-3. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Robinson GE, Palli SR. Identification and characterization of a juvenile hormone response element and its binding proteins. The Journal of biological chemistry. 2007;282:37605–37617. doi: 10.1074/jbc.M704595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner K, Roth P, Schotta G, Krauss V, Saumweber H, Reuter G, Dorn R. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics. 2000;155:141–157. doi: 10.1093/genetics/155.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development (Cambridge, England) 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Hatini V, Green RB, Lengyel JA, Bray SJ, Dinardo S. The Drumstick/Lines/Bowl regulatory pathway links antagonistic Hedgehog and Wingless signaling inputs to epidermal cell differentiation. Genes & development. 2005;19:709–718. doi: 10.1101/gad.1268005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourmeli S, Papantonis A, Lecanidou R. A novel role for the Bombyx Slbo homologue, BmC/EBP, in insect choriogenesis. Biochemical and biophysical research communications. 2005;337:713–719. doi: 10.1016/j.bbrc.2005.09.103. [DOI] [PubMed] [Google Scholar]
- Kato T, Sawada H, Yamamoto T, Mase K, Nakagoshi M. Pigment pattern formation in the quail mutant of the silkworm, Bombyx mori: parallel increase of pteridine biosynthesis and pigmentation of melanin and ommochromes. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2006;19:337–345. doi: 10.1111/j.1600-0749.2006.00316.x. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Fujiwara H. Melanin-synthesis enzymes coregulate stage-specific larval cuticular markings in the swallowtail butterfly, Papilio xuthus. Development genes and evolution. 2005;215:519–529. doi: 10.1007/s00427-005-0014-y. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Fujiwara H. Regulation of 20-hydroxyecdysone on the larval pigmentation and the expression of melanin synthesis enzymes and yellow gene of the swallowtail butterfly, Papilio xuthus. Insect Biochem Mol Biol. 2007;37:855–864. doi: 10.1016/j.ibmb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Kramer KJ., Hopkins, T.L. Tyrosine metabolism for insect cuticle tanning. Arch Insect Biochem Physiol. 1987;6:279–301. [Google Scholar]
- Futahashi R, Fujiwara H. Expression of one isoform of GTP cyclohydrolase I coincides with the larval black markings of the swallowtail butterfly, Papilio xuthus. Insect Biochem Mol Biol. 2006;36:63–70. doi: 10.1016/j.ibmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Han Q, Fang J, Ding H, Johnson JK, Christensen BM, Li J. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. The Biochemical journal. 2002;368:333–340. doi: 10.1042/BJ20020272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Yeh SD, Hovemann BT, Kemme T, Meinertzhagen IA, Edwards TN, Liou SR, Han Q, Li J. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS genetics. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DT, Kitos RJ, Sullivan MC. Developmental and genetic studies on kynurenine hydroxylase from Drosophila melanogaster. Genetics. 1973;75:651–661. doi: 10.1093/genetics/75.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar D, Poeck B, Roth H, Ernst R, Keller A, Porsch M, Strauss R, Pflugfelder GO. Defective pigment granule biogenesis and aberrant behavior caused by mutations in the Drosophila AP-3beta adaptin gene ruby. Genetics. 2000;155:213–223. doi: 10.1093/genetics/155.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RD, McMillan WO, Nagy LM. Gene expression underlying adaptive variation in Heliconius wing patterns: non-modular regulation of overlapping cinnabar and vermilion prepatterns. Proceedings. 2008;275:37–45. doi: 10.1098/rspb.2007.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RD, Nagy LM. Evolutionary redeployment of a biosynthetic module: expression of eye pigment genes vermilion, cinnabar, and white in butterfly wing development. Evolution & development. 2005;7:301–311. doi: 10.1111/j.1525-142X.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- Quan GX, Kim I, Komoto N, Sezutsu H, Ote M, Shimada T, Kanda T, Mita K, Kobayashi M, Tamura T. Characterization of the kynurenine 3-monooxygenase gene corresponding to the white egg 1 mutant in the silkworm Bombyx mori. Mol Genet Genomics. 2002;267:1–9. doi: 10.1007/s00438-001-0629-2. [DOI] [PubMed] [Google Scholar]
- Komoto N. A deleted portion of one of the two xanthine dehydrogenase genes causes translucent larval skin in the oq mutant of the silkworm (Bombyx mori) Insect Biochem Mol Biol. 2002;32:591–597. doi: 10.1016/s0965-1748(01)00134-5. [DOI] [PubMed] [Google Scholar]
- Yasukochi Y, Kanda T, Tamura T. Cloning of two Bombyx homologues of the Drosophila rosy gene and their relationship to larval translucent skin colour mutants. Genetical research. 1998;71:11–19. doi: 10.1017/s0016672397003078. [DOI] [PubMed] [Google Scholar]
- Kiguchi K., Agui, N Ecdysteroid levels and developmental events during larval moulting in the silkworm, Bombyx mori. J Insect Physiol. 1981;27:805–812. [Google Scholar]
- InterProScan http://www.ebi.ac.uk/InterProScan/
- Gene Ontology http://www.geneontology.org/
- Matsuoka T, Fujiwara H. Expression of ecdysteroid-regulated genes is reduced specifically in the wing discs of the wing-deficient mutant (fl) of Bombyx mori. Development genes and evolution. 2000;210:120–128. doi: 10.1007/s004270050018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of nonredundant gene sequence and its number of epM data set. 1–1380 nonredundant sequences in epM were listed. The column of "Other libraries" indicates the numbers of the corresponding sequence found in the silkworm EST libraries constructed by Japanese groups, other than the epM library. "epM/total" represents the percentage of EST numbers of epM to total silkworm EST data set. DmGene represents an orthologous gene in Drosophila melanogaster
List of identified putative isoforms or premature transcript.
List of characteristic gene groups identified in epM dataset.
List of identified cuticular proteins and comparison with epV3 library. The number in epM and epV3 columns indicates the genes expressed in the respective data sets.
Oligonucleotide primers used for analysis of tissue- and stage-specific expression of selected genes.