Abstract
Rationale: We previously demonstrated pre–B-cell colony enhancing factor (PBEF) as a biomarker in sepsis and sepsis-induced acute lung injury (ALI) with genetic variants conferring ALI susceptibility.
Objectives: To explore mechanistic participation of PBEF in ALI and ventilator-induced lung injury (VILI).
Methods: Two models of VILI were utilized to explore the role of PBEF using either recombinant PBEF or PBEF+/− mice.
Measurements and Main Results: Initial in vitro studies demonstrated recombinant human PBEF (rhPBEF) as a direct rat neutrophil chemotactic factor with in vivo studies demonstrating marked increases in bronchoalveolar lavage (BAL) leukocytes (PMNs) after intratracheal injection in C57BL/6J mice. These changes were accompanied by increased BAL levels of PMN chemoattractants (KC and MIP-2) and modest increases in lung vascular and alveolar permeability. We next explored the potential synergism between rhPBEF challenge (intratracheal) and a model of limited VILI (4 h, 30 ml/kg tidal volume) and observed dramatic increases in BAL PMNs, BAL protein, and cytokine levels (IL-6, TNF-α, KC) compared with either challenge alone. Gene expression profiling identified induction of ALI- and VILI-associated gene modules (nuclear factor-κB, leukocyte extravasation, apoptosis, Toll receptor pathways). Heterozygous PBEF+/− mice were significantly protected (reduced BAL protein, BAL IL-6 levels, peak inspiratory pressures) when exposed to a model of severe VILI (4 h, 40 ml/kg tidal volume) and exhibited significantly reduced expression of VILI-associated gene expression modules. Finally, strategies to reduce PBEF availability (neutralizing antibody) resulted in significant protection from VILI.
Conclusions: These studies implicate PBEF as a key inflammatory mediator intimately involved in both the development and severity of ventilator-induced ALI.
Keywords: visfatin, acute lung injury, chemotaxis, apoptosis, mechanical ventilation
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Pre–B-cell colony enhancing factor (PBEF), whose gene variants are associated with ventilator-induced lung injury (VILI) susceptibility, has been identified as a potential VILI candidate gene and biomarker and has been localized to animal and human lung epithelium in acute lung injury. The mechanism of this association remains unknown.
What This Study Adds to the Field
This study implicates PBEF as a key inflammatory mediator in the development and severity of VILI in murine models of VILI. PBEF may be a molecular target for amelioration of VILI in critically ill patients.
Mechanical ventilation is a life-saving intervention in critically ill patients with respiratory failure due to acute lung injury (ALI), a devastating syndrome characterized by profound lung inflammation, vascular permeability, and protein-rich alveolar edema (2, 3). Unfortunately, mechanical ventilation also potentially contributes directly to lung injury, a process known as ventilator-induced lung injury (VILI), with augmented capillary leakage, acute inflammation, and increases in inflammatory cytokine expression (4–8). The clinical relevance of VILI was highlighted by the landmark Acute Respiratory Distress Syndrome Network trial, which reported decreased mortality in patients with acute respiratory distress syndrome (ARDS) placed on low tidal volume ventilation, accompanied by decreases in bronchoalveolar lavage (BAL) leukocytes and inflammatory cytokines (9, 10).
Despite improved understanding of ALI pathophysiology, the underlying mechanisms of the injurious effects of mechanical ventilation in the setting of ALI remain unclear and effective pharmacotherapy has not yet emerged. We previously used genomics-intensive approaches to identify potential ALI and VILI susceptibility candidate genes (11, 12). We determined that the gene encoding pre–B-cell colony enhancing factor (PBEF), a proinflammatory cytokine expressed in amniotic membranes during gestation (13), represented a potential VILI candidate gene and novel biomarker in sepsis and ALI (1). Both canine and murine ALI models revealed highly up-regulated PBEF lung gene expression, a finding confirmed by increased PBEF levels in BAL fluid obtained from patients with ARDS, with PBEF expression spatially localized to lung epithelium, endothelium, and leukocytes (1). Our genetic analyses revealed single nucleotide polymorphisms (SNPs) in the PBEF promoter, which were significantly associated with susceptibility to sepsis and ALI (1). Recently, these PBEF promoter variant associations were confirmed in a replicate ALI population and additionally associated with the number of ventilator-free days and overall ALI mortality (14).
Although information regarding PBEF involvement in ALI/VILI is increasing, there is limited information as to the mechanism of PBEF involvement in ALI pathophysiology (1) and in responses to mechanical stress leading to VILI. In the current study, we demonstrate that recombinant human PBEF (rhPBEF) is a direct neutrophil chemotaxin inducing both increases in BAL neutrophils (PMNs) and expression of murine PMN chemoattractants, keratinocyte (KC) and macrophage inflammatory protein (MIP)-2, after intratracheal administration of rhPBEF. PBEF was synergistic with mechanical ventilation in producing VILI-like lung injury with dramatic increases in BAL PMNs, BAL protein, and inflammatory cytokine levels, such as IL-6. Heterozygous PBEF+/− mice, with targeted deletion of a single PBEF allele, were significantly protected in a model of severe VILI (4 h, 40 ml/kg tidal volume). Bioinformatic analyses revealed strong PBEF-driven induction of ALI/VILI gene ontologies (apoptosis, leukocyte extravasation, Toll receptor signaling). Finally, strategies to reduce PBEF availability (neutralizing antibody) resulted in significant protection from VILI-induced inflammation. Together, these studies are consistent with PBEF as a key inflammatory mediator in the development and severity of VILI.
METHODS
Transmigration and Chemotaxis Assay
Chemotaxis of rat peritoneal neutrophils (calcein-acetoxymethyl ester [AM]) across 3-μm polycarbonate filters was assessed in response to rh PBEF/visfatin (rhPBEF; PeproTech, Rocky Hill, NJ) or casein (positive control) (see the online supplement Methods for additional details).
Experimental Protocols and Generation of PBEF+/− Transgenic Mice
C57BL/6J (B6) mice were housed under standard conditions with all procedures approved by the Animal Care and Use Committee (University of Chicago). To generate PBEF+/− mice, the 129/Sv/Ola-derived embryonic stem cell line (15) RR084, harboring the exon-trap vector pGT0lxf (16) in the seventh intron of the murine PBEF gene, was obtained (BayGenomics Consortium, San Francisco, CA). Transgenic mice were produced by microinjection of the embryonic stem cells into blastocysts derived from B6 mice, and screened by insertion junction-specific polymerase chain reaction (PCR) of tail DNA. Founder mice were out-crossed to B6 mice for four generations to reach 85% congenic status. All outcrosses to B6 mice were viable; however, in-crosses failed to produce homozygous knockout progeny (see the online supplement Methods).
Models of Ventilator-induced Murine Lung Injury
The first VILI model was designed to produce limited lung injury (VILIa; tidal volume, 30 ml/kg), thus allowing assessment of potential synergy with rhPBEF challenge. Male C57BL/6J B6 mice were anesthetized with ketamine/acepromazine, intubated, and administered rhPBEF (20 μg/mouse) via an intratracheal route approximately 30 minutes before ventilator placement (room air; tidal volume, 30 ml/kg; 65 breaths/min; 0 cm H2O positive end-expiratory pressure [PEEP]) for 4 hours as previously described (12, 17). Groups included a spontaneously breathing (SB) group, an SB group challenged with rhPBEF (SB-rhPBEF), a high tidal ventilation group (VILIa), and a high tidal ventilation group with rhPBEF challenge (VILIa-rhPBEF) (n = 4–6 for all groups).
Our second ventilation approach was designed to produce more severe lung injury (VILIb; tidal volume, 40 ml/kg) to assess potential protective effects of the single PBEF allele deletion in PBEF+/− mice. Transgenic PBEF+/− and wild-type (WT: PBEF+/+) mice were anesthetized and ventilated (room air; tidal volume, 40 ml/kg; 65 breaths/min; 0 cm H2O PEEP) for 4 hours and randomly allocated into four groups, which were either spontaneously breathing (SB-WT and SB-PBEF+/− groups) or exposed to high tidal ventilation (VILIb-WT and VILIb-PBEF+/− groups) (see the online supplement Methods).
BAL: Tissue Albumin and Cytokine Content
BAL fluid recovered as we previously described was used for multiple assays, including total BAL protein and BAL cell differentials (18). Tissue albumin content was assessed as previously described (12). Cytokine levels in BAL fluid (IL-1β, IL-6, KC, MIP-2, tumor necrosis factor [TNF]-α) were analyzed by multiplex assays (Bio-Rad, Hercules, CA). BAL PBEF levels were assayed by a C-terminal ELISA (Phoenix Pharmaceuticals, Inc., Belmont, CA) and analyzed using the SoftmaxPRO software (Molecular Devices Corp., Sunnyvale, CA) (see the online supplement Methods).
Statistical Analysis for Biomarker Data
Statistical analysis (mean ± SEM) was performed using SPSS 12.0 (SPSS, Inc., Chicago, IL) with one-way analysis of variance tests and post hoc multiple comparisons using Tukey's method. A P value of less than 0.05 was considered significant.
RNA Isolation and Microarray Analysis
Total lung RNA was isolated as described previously (12). Affymetrix Mouse430_2 arrays (Affymetrix, Inc., Santa Clara, CA) were used. Chip quality (19) and “present” or “absent” expression calls were determined by GeneChip Operating Software (GCOS) (GSE9368-rhPBEF, GSE9314-PBEF+/−). Intensities and normalization of probe sets were calculated by Bioconductor software (GCRMA package) (http://www.Bioconductor.org) (20). To identify differentially expressed genes, pairwise comparisons were conducted using Significance Analysis of Microarrays (SAM) as previously described (12). Gene filtering parameters and results are summarized in Table E2 of the online supplement. Differentially expressed genes displaying greater than twofold changes are referred to as “dysregulated genes.”
Gene Ontology and Ingenuity Pathways Analysis of Dysregulated Genes
Dysregulated gene functional profiles were analyzed using Onto-Express (http://vortex.cs.wayne.edu/projects.htm) with the number of genes in each Gene Ontology (GO) biological process category (21) compared with all genes on the Mouse432_2 chip to determine the significance of the GO category (22). Ingenuity Pathway Analysis (IPA) software (containing individually modeled relationships between gene objects; e.g., genes, mRNAs, proteins) was used to dynamically generate significant regulatory and signaling networks or pathways. The significance of a canonical pathway is controlled by the P value calculated using the right-tailed Fisher exact test for 2 × 2 contingency tables (see the online supplement Methods).
Experiments with PBEF Neutralizing Antibody
Polyclonal anti-PBEF antibodies were custom produced by Lampire Biological Laboratories, Inc. (Pipersville, PA), by immunization of a goat with full-length rhPBEF protein. PBEF antibodies (PBEF-Abs) were purified over a protein G column to a final concentration of 1.1 mg/ml and used for the in vivo neutralization studies to determine the optimal dose for subsequent experiments in our severe lung injury (VILIb) model. In specific experiments, PBEF-Ab or saline were injected intratracheally (70 μl), and after a 30-minute period, mice were ventilated with room air (SB) or VILIb (40 ml/kg, 4 h).
RESULTS
Effect of rhPBEF on Neutrophil Transmigration and Chemotaxis In Vitro
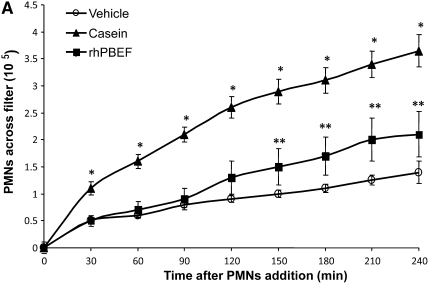
Prior reports have suggested an effect of PBEF gene and protein expression on neutrophil function (23). Given the implied pathologic role for PBEF in sepsis, ALI, and in preclinical models of mechanical ventilation–induced ALI (1), our initial studies were designed to investigate the possibility that PBEF directly serves as a neutrophil chemoattractant (Figure 1A). Migration of rat peritoneal neutrophils (∼90% purity) across transwell filters in response to lower chamber rhPBEF or casein was assessed for up to 4 hours. PBEF induced significant PMN migration beginning after 2 hours, whereas casein-mediated PMN migration was significant at all time points beginning at 30 minutes (Figure 1A), results consistent with a physiologic role for PBEF as a direct PMN chemoattractant.
Figure 1.
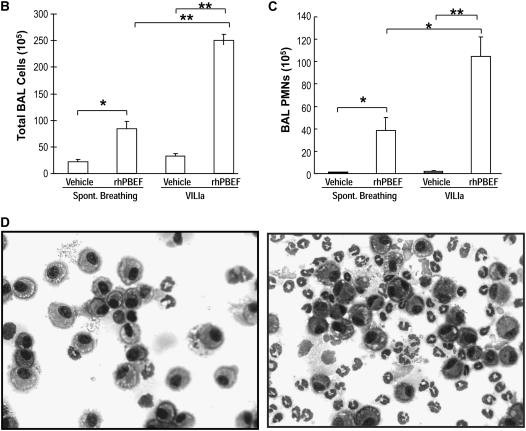
In vitro and in vivo effects of recombinant human pre–B-cell colony enhancing factor (rhPBEF) on neutrophil chemotaxis and lung leukocyte recruitment. (A) The migration of rat peritoneal neutrophils (90% purity of PMNs) across transwell filters in response to rhPBEF or casein (lower chamber) for up to 4 hours. rhPBEF induced significant PMN migration beginning at 2 hours (**P < 0.05), whereas casein-mediated PMN migration was significant at all time points beginning at 30 minutes (*P < 0.05) and diminished after 210 minutes. (B) rhPBEF instillation produces significant increases in number of bronchoalveolar lavage (BAL) cells in C57BL/6 mice. (vehicle, 22 × 105 ± 4, vs. rhPBEF, 83 × 105 ± 15, x̄ ± SEM; *P < 0.01) assessed 4 hours after instillation and compared with vehicle controls. BAL cell counts are further increased in mice receiving rhPBEF 30 minutes before mechanical ventilation (VILIa, tidal volume 30 ml/kg, 4 h) when compared with VILIa alone (VILIa, 33 × 105 ± 5, vs. VILIa-rhPBEF, 250 × 105 ± 21; **P < 0.001). (C) The significant increase in number of BAL neutrophils (PMNs) in rhPBEF-challenged mice (vehicle, 0.4 × 105 ± 0.2, vs. rhPBEF, 38. 8 × 105 ± 11.1; *P < 0.01) at 4 hours. BAL neutrophil counts were further increased in the VILIa-rhPBEF group compared with each group including VILIa alone (VILIa, 1.4 × 105 ± 0.7, vs. VILIa-rhPBEF, 104.4 × 105 ± 17.8; **P < 0.001). (D) A representative cytospin result (original magnification, ×400) with the typical predominance of alveolar macrophages in BAL from vehicle-treated mice (left panel). In contrast, there is a dramatic influx of neutrophils into the alveolar space (right panel) after exposure to 20 μg rhPBEF in the VILIa-rhPBEF group. n = 4 to ∼6 animals in all experimental groups.
In Vivo Effects of rhPBEF in SB Mice
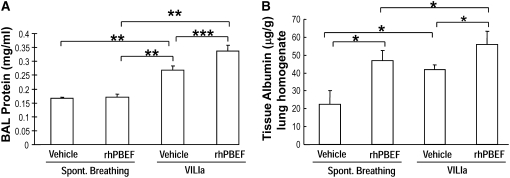
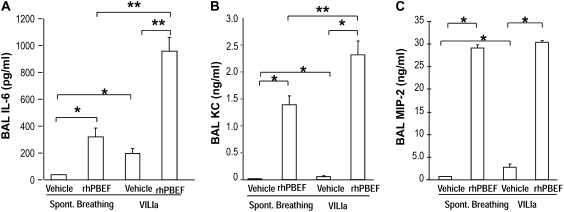
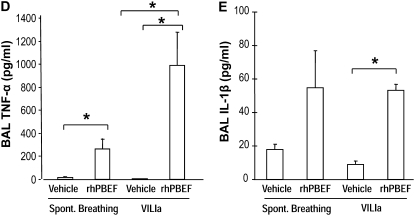
We next extended these in vitro findings to assess PBEF-induced PMN lung recruitment in SB B6 mice receiving intratracheal instillation of rhPBEF. We observed that PBEF produced significant increases in total BAL cells (P < 0.01; Figure 1B) and BAL neutrophils (P < 0.05; Figure 1C), findings confirmed by cytologic observations (Figure 1D). rhPBEF does not increase the level of BAL protein content in spontaneously breathing animals (Figure 2A) but does modestly increase levels of lung tissue albumin (P < 0.05; Figure 2B). Supporting the proinflammatory effects of PBEF, we noted PBEF-mediated increases in the BAL level of inflammatory cytokines such as IL-6 (P < 0.01), TNF-α (P < 0.05), and IL-1β (P = 0.01), as well as PMN chemokines KC (P < 0.001), and MIP-2 (P < 0.001) compared with controls (Figures 3A–3E).
Figure 2.
Effect of recombinant human pre–B-cell colony enhancing factor (rhPBEF) on bronchoalveolar lavage (BAL) protein levels and lung tissue albumin content in C57BL/6 mice. Instillation of rhPBEF did not produce increased permeability as reflected by BAL protein levels (vehicle, 0.17 ± 0.01, vs. rhPBEF, 0.17 ± 0.01 mg/ml; P = 0.77; A). However, rhPBEF significantly increased lung albumin content (vehicle, 22.5 ± 7.5, vs. rhPBEF, 47.0 ± 5.6 μg/g tissue; *P = 0.03; B), potentially reflecting the temporal course and/or anatomic location of edema formation. In contrast, VILIa (30 ml/kg, 4 h) induced significant increases in BAL protein compared with spontaneous breathing (**P < 0.001). In addition, the combined VILIa-rhPBEF challenge induced increases in BAL protein compared with VILIa and spontaneously breathing (SB)-rhPBEF mice (VILIa, 0.27 ± 0.02, vs. VILIa-rhPBEF, 0.34 ± 0.02 mg/ml; ***P < 0.05; A) (n = 4 to ∼6 animals in each group, all experiments).
Figure 3.
Effect of recombinant human pre–B-cell colony enhancing factor (rhPBEF) on bronchoalveolar lavage (BAL) cytokine levels in C57BL/6 mice. The levels of BAL IL-6 (A) were significantly elevated in the rhPBEF group when compared with vehicle group (vehicle, 36 ± 4, vs. rhPBEF, 319 ± 67 pg/ml; *P < 0.01). In addition, when mice were exposed to rhPBEF followed by mechanical ventilation, the VILIa-rhPBEF group had significantly elevated concentrations of IL-6 (VILIa, 194 ± 38, vs. VILIa-rhPBEF, 957 ± 103 pg/ml; **P < 0.001). Similarly, rhPBEF challenge significantly elevated the concentrations of the neutrophil (PMN) chemokines keratinocyte (KC) (vehicle, 12 ± 3, vs. rhPBEF, 1,388 ± 169 pg/ml; *P < 0.001; B) and macrophage inflammatory protein (MIP)-2 (vehicle, 676 ± 87, vs. rhPBEF, 29,026 ± 762 pg/ml; *P < 0.001; C) compared with the vehicle control group. The levels of KC were substantially elevated in VILIa-rhPBEF (VILIa, 58 ± 15, vs. VILIa-rhPBEF, 2,310 ± 263 pg/ml; **P < 0.001). MIP-2 levels were elevated in VILIa-rhPBEF group compared with VILIa (VILIa, 2,751 ± 821, vs. VILIa-rhPBEF, 30,480 ± 219 pg/ml; *P < 0.001). Finally, significantly elevated concentrations of tumor necrosis factor (TNF)-α (D) in the rhPBEF group were found (spontaneously breathing [SB], 17 ± 8, vs. SB-rhPBEF, 262 ± 94 pg/ml; *P < 0.05), as well as the VILIa-rhPBEF group of TNF-α (VILIa, 6 ± 2, vs. VILIa-rhPBEF, 985 ± 293 pg/ml; *P < 0.05). Significantly elevated concentrations of IL-1β (E) were also noted in the SB-rhPBEF (SB, 18 ± 3, vs. SB-rhPBEF, 55 ± 22 pg/ml) and VILIa-rhPBEF groups (VILIa, 9 ± 2, vs. VILIa-rhPBEF 53 ± 4 pg/ml; *P = 0.01). n = 4 to ∼6 animals per group.
Inflammatory Effects of rhPBEF in a Model of Ventilator-induced Murine Lung Injury
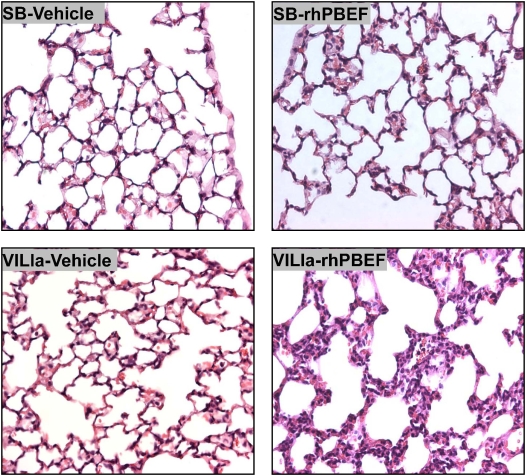
We next examined potential synergism between rhPBEF challenge and the mechanical stress produced in B6 mice by exposure to a limited lung injury model elicited by high tidal volume ventilation (VILIa, 4 h, 30 ml/kg tidal volume). There were no statistically significant differences in pH, PaO2, PaCO2, HCO3, and peak inspiratory pressures at the end of the mechanical ventilation period between the VILIa alone and VILIa-rhPBEF groups (Table E1). Compared with SB mice, mice exposed to VILIa alone (without intratracheal rhPBEF) exhibited increased BAL protein levels (P < 0.001), increased lung tissue albumin (P = 0.03), and increased levels of MIP-2 (P < 0.001), IL-6, KC, and IL-1β (Figures 1–3). However, the VILIa-rhPBEF group (intratracheal rhPBEF followed by mechanical ventilation) demonstrated dramatic increases in BAL total cells (P < 0.001), BAL PMNs (P < 0.001), BAL protein (P < 0.05), and levels of several cytokines inducing IL-6, TNF-α, MIP-2, IL-1β, and KC (all P < 0.05) compared with VILIa-challenged mice (Figures 1–3). Histologic assessment demonstrated that both SB-rhPBEF– and VILIa-exposed mice exhibited mild inflammation compared with the SB group. However, VILIa-rhPBEF–challenged mice produced greater alveolar wall thickening, and neutrophil infiltration into the lung interstitium and alveolar space (Figure 4), but with relative preservation of alveolar architecture and only modest alveolar and tissue edema.
Figure 4.
Histologic assessment of recombinant human pre–B-cell colony enhancing factor (rhPBEF) effects on ventilator-induced lung inflammation and injury. Lungs from each experimental group (n = 3) were inflated to 25 cm H2O with 0.2% of low-melting agarose and fixed in 4% paraformaldehyde at 4°C for histologic evaluation by hematoxylin-and-eosin staining. Histologic analysis of lung tissue (original magnification, ×40) obtained from control mice (spontaneously breathing [SB]-Vehicle, 30 μl vehicle intratracheal) demonstrated preserved lung parenchymal architecture. In contrast, mice exposed to either VILIa for 4.5 hours (VILIa-Vehicle) or, to a lesser degree, rhPBEF (20 μg/mouse dissolved in 30 μl of saline) (SB-rhPBEF) produced macrophage and neutrophil infiltration and edematous alveolar areas. Each of these features was dramatically augmented in mice receiving intratracheal rhPBEF 30 minutes before placement on mechanical ventilation for 4 hours (VILIa-rhPBEF).
Responses of PBEF+/− Heterozygous Mice to Severe Ventilator-induced Murine Lung Injury
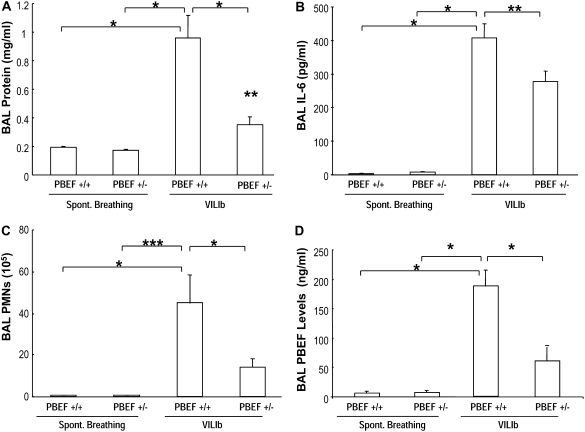
To further explore the in vivo contribution of PBEF generation to ventilator-induced lung injury, we next generated a heterozygous PBEF+/− mouse line with targeted deletion of a single PBEF allele, and subsequently exposed these mice to a model of severe VILI (VILIb, 4 h, 40 ml/kg tidal volume). WT and heterozygous PBEF+/− mice were statistically similar in acid-base parameters (pH, PaO2, PaCO2, HCO3) before and after exposure to 4 hours of mechanical ventilation and in the peak inspiratory pressures at the initiation of mechanical ventilation (Table E1). However, VILIb-exposed PBEF+/− mice were significantly protected from VILI, with significantly decreased inflammatory lung injury and lower peak inspiratory pressures at the end of the mechanical ventilation period compared with WT mice (22.0 ± 1.6 vs. 26.7 ± 2.9 mm Hg, P < 0.05) (Table E1). For example, the increase in BAL protein induced by VILIb was largely abrogated in PBEF+/− mice (Figure 5A; *P < 0.05) compared with WT mice and the increases in BAL IL-6 levels were also significantly reduced in VILIb-challenged PBEF+/− mice (Figure 5B; **P < 0.05). Additional indices of lung injury, such as total BAL PMNs, were significantly lower in PBEF+/− mice (1.37 × 105 ± 0.7 vs. 44.8 × 105 ± 26.9, ***P < 0.05) compared with control animals (Figure 5C). The levels of inflammatory cytokines (KC, MIP-2, IL-1β, TNF-α; data not shown) were all reduced in VILIb-challenged PBEF+/− mice but failed to reach statistical significance. As expected, compared with WT mice, PBEF+/− mice exposed to a model of severe ventilation-mediated lung injury (VILIb) demonstrated significant reductions in lung PBEF levels (187 ± 28 vs. 60 ± 30 ng/ml, P < 0.05) (Figure 5D). Basal PBEF levels in spontaneously breathing WT and PBEF+/− mice were negligible.
Figure 5.
Heterozygous pre–B-cell colony enhancing factor (PBEF+/−) mice are protected in a model of severe ventilation-induced lung injury (VILIb). (A) In contrast to the increase in bronchoalveolar lavage (BAL) protein induced by VILIb (40 ml/kg, 4 h) in wild-type (WT) PBEF+/+ mice (*P < 0.05), this effect was largely abrogated in the PBEF+/− mice (VILIb-PBEF+/+, 0.96 ± 0.16, vs. VILIb-PBEF+/−, 0.35 ± 0.06 mg/ml; **P < 0.05). (B) Evaluation of BAL cytokines in VILIb-challenged WT mice and VILIb-exposed PBEF+/− mice showed that PBEF+/− mice had negligible levels of IL-6 at baseline (spontaneously breathing [SB]-PBEF+/−, 8 ± 1) and significantly lower elevated IL-6 concentrations when compared with WT mice exposed to VILIb mechanical ventilation (VILIb-WT, 407 ± 43, vs. VILIb-PBEF+/−, 278 ± 30 pg/ml; *P < 0.05). Data represent mean values ± SEM, pg/ml, and n = 6 animals in all experiments. (C) Shown are comparisons of BAL neutrophil (PMN) counts in VILIb-WT (44.8 × 105 ± 26.9) versus VILIb-PBEF+/− mice (13.9 ± 4.4, *P < 0.05). PMN counts were dramatically reduced in PBEF+/− mice when compared with WT mice (***P < 0.0001). (D) Shown are the levels of PBEF in BAL fluid as determined by ELISA (see the online supplement Methods for details). As expected, levels of PBEF in mice exposed to a model of severe VILI (VILIb) were significantly higher in VILIb-challenged WT mice compared with VILIb-exposed PBEF+/− mice (187 ± 28 vs. 60 ± 30 ng/ml, *P < 0.05).
Genomic Analysis of WT and PBEF+/− Mice Exposed to rhPBEF and Mechanical Ventilation
We next attempted to determine molecular signatures that describe the direct effects of PBEF as well as the synergistic effects of rhPBEF in VILI-mediated inflammatory injury. We performed microarray analyses of SB, rhPBEF-challenged WT and heterozygous PBEF+/− mice as well as mice exposed to mechanical ventilation–induced lung injury. Dysregulated genes identified by pairwise comparison of two groups using SAM software are summarized in Table E2. The functional profiles of the dysregulated genes were also explored by IPA for canonical pathways and analyzed by Onto-Express for gene ontology assessment to identify the overrepresented biological processes (Tables E3 and E4).
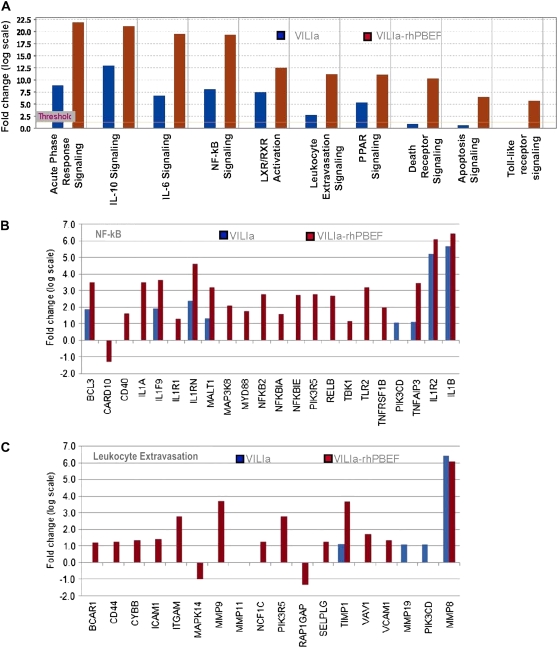
Interestingly, the majority of the signaling pathways deregulated by rhPBEF challenge (acute phase response signaling, IL-10, nuclear factor [NF]- κB, IL-6, leukocyte extravasation) (Figure E1) were identical to those pathways affected by VILIa treatment (Figure 6A), thus highlighting the potential involvement of PBEF in VILI pathogenesis. The potential for additive effects of rhPBEF on VILI-induced inflammation was confirmed by the greater number of dysregulated genes induced by the combined VILIa-rhPBEF exposure (690 genes) compared with VILIa alone (220 genes) (Table E2). The common canonical VILIa-induced signaling pathways, which were augmented by combined rhPBEF challenge, include the acute phase response signaling, IL-10, IL-6, NF-κB, peroxisome proliferator-activated receptors (PPARs), and the liver X receptor/retinoid X receptor ligand (LXR/RXR) signaling pathways (Figure 6A). Deregulated pathways, such as Toll-like signaling and apoptosis signaling pathways, were only induced by the combined challenge of VILIa and rhPBEF and not altered by VILIa alone (Figure 6A). The potent effects of rhPBEF on these processes are highlighted for the NF-κB signaling pathway in Figures 6A and 6B, for the leukocyte extravasation signaling pathway in Figures 6A and 6C, and for the apoptosis pathway in Figure 6A and Figure E2. These findings are consistent with the capacity of rhPBEF to induce lung inflammation, with the majority of genes in these pathways driven by rhPBEF administration and not by exposure to VILIa. For example, a total of 21 dysregulated genes in the NF-κB pathway were altered by the VILIa-rhPBEF combined exposure, whereas only 8 dysregulated genes were found in the VILIa group alone (each exhibiting much lower fold changes than VILIa-rhPBEF) (Figure 6B). Additional details of deregulation of the murine NF-κB and leukocyte extravasation pathways after exposure to VILIa-rhPBEF are depicted in Figures E3 and E4, respectively. Similar to the findings in canonical pathway analysis, the GO analysis revealed that combined VILIa-rhPBEF challenge induced the deregulation of several novel biological processes, such as chemotaxis, small GTPase-mediated signal transduction, and cytokine- and chemokine-mediated signaling pathways (Table E3).
Figure 6.
Ingenuity Pathway Analysis of recombinant human pre–B-cell colony enhancing factor (rhPBEF)–mediated dysregulated genes. The addition of rhPBEF to the limited ventilator-induced lung injury model (VILIa) produces a strong signature of dysregulated genes (see Table E2 for description of gene selections with Significance Analysis of Microarrays software; gene list 2 and 3 for VILIa and VILIa-rhPBEF treatment, respectively). (A) Significant canonical pathways enriched with dysregulated genes induced by VILIa or VILIa-rhPBEF treatment. The threshold line represents the Fisher exact P value of 0.05 (see the online supplement Methods). (B, C) Fold changes of the dysregulated genes induced by VILIa or VILIa-rhPBEF treatment in the nuclear factor (NF)-κB (B) and leukocyte extravasation pathway (C).
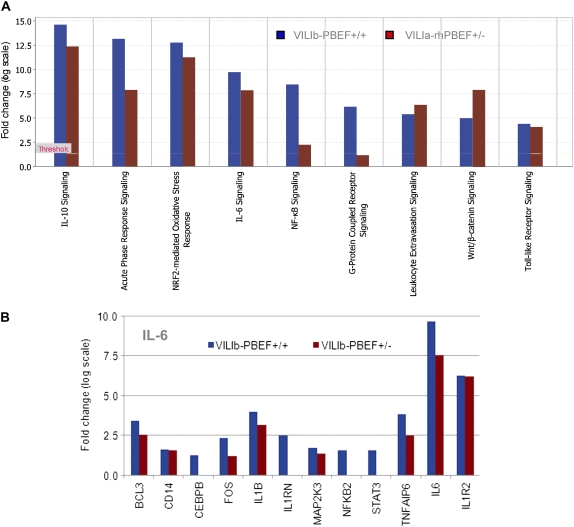
As noted above, we used two models of VILI: a model of limited VILI with a tidal volume of 30 ml/kg (i.e., VILIa) to interrogate the synergy between rhPBEF and VILI, and a second model of severe VILI with a tidal volume of 40 ml/kg (i.e., VILIb) to evaluate the potential protection afforded by deletion of a PBEF allele in PBEF+/− mice. Despite use of a stringent gene filtering criterion (see Table E2), WT mice exposed to the more severe VILIb protocol induced a greater number of dysregulated genes (748 genes) than the less injurious VILIa protocol (220 genes), indicating a clear dose-dependent effect of mechanical stress on gene dysregulation induced by VILI exposure. Murine expression profiling displayed minimal impact of the PBEF+/− genotype on basal global gene expression (Table E2) with only eight dysregulated genes (including PBEF, CYP1A1, and an uncharacterized cDNA, BC018473). However, consistent with the critical role of PBEF in VILI, specific deregulated pathways induced by VILIb in WT mice were absent in PBEF+/− mice, including the previously noted NF-κB and G-protein–coupled receptor signaling pathways (Figure 7A), indicating a clear protective effect of PBEF+/− genotype on VILIb-induced genomic alterations. For example, compared with VILIb-challenged WT mice, the expression of all VILIb-induced deregulated genes in the IL-6 pathway (e.g., IL1RN, NFKB2, STAT3, IL-6) was either reduced or failed to be dysregulated in PBEF+/− mice exposed to VILIb (Figure 7B). Results for selected genes (Cxcl1, Cxcl2, BCL3, Map3k8, MMP9, Il-6, Il-1β, TNFα, and BC018473) were further validated by ELISA and reverse transcriptase (RT)–PCR approaches (Table E5). VILIb-induced dysregulated genes in WT or PBEF+/− mice submitted to OntoExpress (to identify the overrepresented biological processes) demonstrated several deregulated biological processes induced by VILIb in WT mice, which were absent in the PBEF+/− group, including cytoskeleton organization and biogenesis, angiogenesis, and defense responses (Table E5).
Figure 7.
Ingenuity Pathway Analysis of dysregulated genes in heterozygous pre–B-cell colony enhancing factor (PBEF+/−) mice. (A) Listed are significant canonical pathways enriched with dysregulated genes induced by VILIb-PBEF+/+ and VILIb-PBEF+/−. (B) Depicted are fold changes of the dysregulated genes induced by VILIb-PBEF+/+ or VILIb-PBEF+/− in the IL-6 signaling pathway. See Table E2 for the description of gene selections with Significance Analysis of Microarrays software (gene list 6 and 7 for VILIb-PBEF+/+ and VILIb-PBEF+/− challenge, respectively).
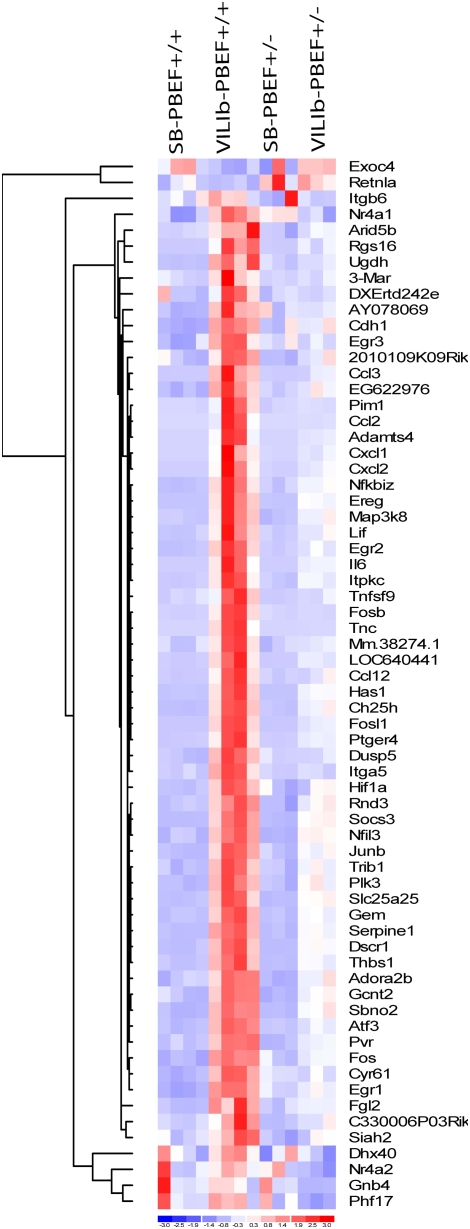
We next explored the impact of rhPBEF or PBEF+/− on the global gene expression pattern in VILI-treated animals. Sixty-five differentially expressed genes were identified by pairwise comparison between VILIa-rhPBEF and VILIa groups, with 43 genes demonstrating ranked expression levels—that is, an order of control < VILIa < < VILIa-rhPBEF (data not shown). Similarly, 66 genes were identified between VILIb-challenged PBEF+/− mice and VILIb-exposed WT mice (Table E2). Hierarchical clustering of the gene expression levels across all experimental groups are displayed in Figure 8, with 64 out of the 66 genes following the order of control < VILIb-PBEF+/− < < VILIb-PBEF+/+. Together, these findings support the notion that rhPBEF exaggerates VILI-induced gene dysregulation, whereas the PBEF+/− genotype attenuates this dysregulation.
Figure 8.
Gene expression pattern of differentially expressed genes in VILIb-challenged wild-type (WT) mice and VILIb-challenged heterozygous pre–B-cell colony enhancing factor (PBEF+/−) mice Differentially expressed genes between VILIb-WT and VILIb-PBEF+/− challenged mice were generated by Significance Analysis of Microarrays (Table E2). The expression levels of the differentially expressed genes between VILIb-WT and VILIb-challenged PBEF+/− mice are displayed by dChip hierarchical clustering (DNA and enrichment analysis) (http://biosun1.harvard.edu/complab/dchip/clustering.htm). Blue, white, and red represent expression levels below, at, and above mean levels, respectively.
Evaluation of Molecular Markers in VILI- and rhPBEF-challenged WT and PBEF+/− Mice
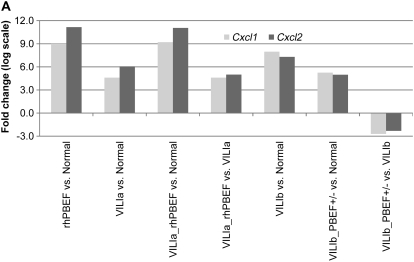
We assessed the intersection of the combined VILI and PBEF challenge by comparing the overlapping genes across all gene lists (Table E2). This subsequently identified two genes, Cxcl1 (or KC) and Cxcl2 (or MIP-2), which displayed significant alterations across each list (except gene list 5) (Figure 9A). Both Cxcl1 and Cxcl2 displayed tidal volume– and rhPBEF-dependent responses with fold changes in gene expression much greater in severe VILIb-challenged mice (Table E5; 251-fold increase, Cxcl1; 157-fold increase, Cxcl2) rather than limited VILIa (24-fold, Cxcl1; 67-fold, Cxcl2) and was much higher when mice were exposed to VILIa-rhPBEF (589-fold, Cxcl1; 2,128-fold, Cxcl2) compared with VILIa alone.
Figure 9.
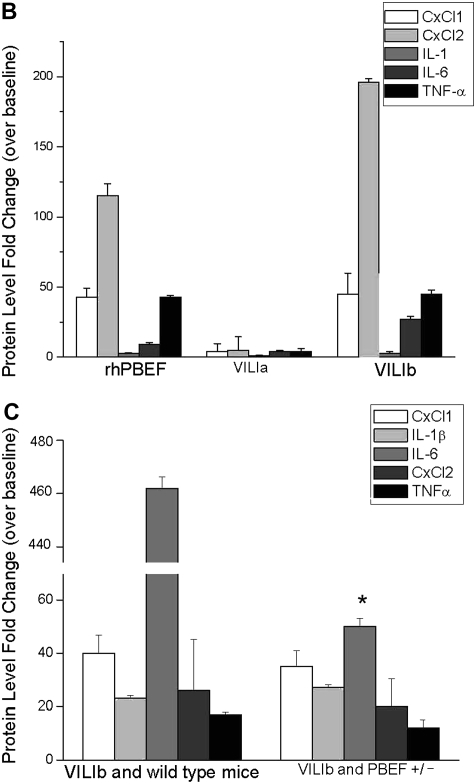
Validation of potential ventilator-induced lung injury (VILI) biomarkers in lung tissue and in bronchoalveolar lavage (BAL) fluid. (A) Two markers, Cxcl1 and Cxcl2, were selected by their presence across all gene lists (Table E2) (except list 5 for dysregulated genes in heterozygous pre–B-cell colony enhancing factor [PBEF+/−] mice). The levels of each gene mirrored the severity of VILI and/or the injury produced by VILI and recombinant human PBEF (rhPBEF) challenges, suggesting that Cxcl1 and Cxcl2 represent potential biomarkers in VILI. (B) VILIa- and rhPBEF-mediated challenge of wild-type B6 mice each induced significantly increased fold changes in CxCl1, CxCl2, IL-1β, IL-6, and TNF-α compared with spontaneously breathing controls (*P < 0.05) in BAL. Moreover, the combined challenge of VILIa-rhPBEF indicated an additive effect on the induction of these cytokines (*P < 0.05). (C) Exposure of wild-type B6 mice to VILIb induced significantly elevated fold changes in CxCl1, CxCl2, IL-1β, IL-6, and TNF-α relative to VILIb-PBEF+/− mice. Reductions in BAL cytokine production in VILIb-PBEF+/− mice are significant with IL-6 (*P < 0.05).
Complementing these changes in gene expression, BAL fluid was also assessed for levels of CxCl1, CxCl2, IL-1β, IL-6, and TNF-α (Figures 9B and 9C). Isolated challenge with rhPBEF or VILIa produced significantly increases in each cytokine compared with SB controls, whereas the combined challenge revealed an additive effect. In the VILIb model, the protective effect of PBEF+/− heterozygosity was again evident, with reductions in all cytokines in VILIb-PBEF+/− mice compared with the VILIb-WT group. Only IL-6 levels in the VILIb-PBEF+/− group remained significantly reduced compared with controls. Collectively, the effects of rhPBEF and PBEF+/− heterozygosity on the production of these inflammatory cytokines further underscores the important role of PBEF in VILI.
Effect of Reduced PBEF Availability on VILI-induced PMN Recruitment and Vascular Leakage
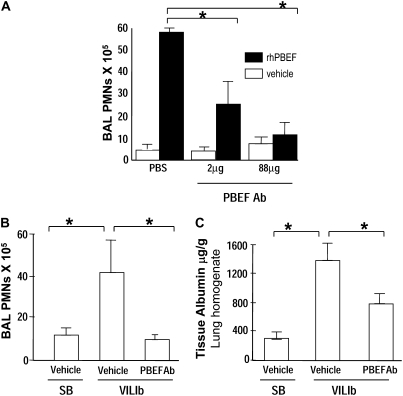
To address the potential for PBEF to serve as a therapeutic target in ameliorating VILI, we generated PBEF antisera and assessed the effect of PBEF neutralizing antibody on rhPBEF-stimulated PMN recruitment into the alveolar space (Figure 10A). Simultaneous instillation of rhPBEF (20 μg/mouse) and PBEF neutralizing antibody produced dramatic reductions in rhPBEF-induced PMN recruitment. We next assessed the ability of PBEF neutralizing antibody to reduce VILIb-mediated PMN recruitment and lung injury (40 ml/kg, 4 h). The intratracheal delivery of PBEF neutralizing antibody (30 min before mechanical ventilation) abolished VILIb-induced increases in total BAL cell counts and significantly decreased PMN influx into the alveolar space (Figure 10B) as well as VILI-mediated increases in lung tissue albumin (Figure 10C).
Figure 10.
Reductions in pre–B-cell colony enhancing factor (PBEF) availability is protective in ventilation-induced lung injury (VILI). (A) We initially assessed the effect of PBEF neutralizing antibody (PBEF Ab) on recombinant human PBEF (rhPBEF)–mediated neutrophil (PMN) influx into lung tissues and alveolar space (4.5 h, 20 μg/mouse). Simultaneous administration of rhPBEF and PBEF antibody (2 μg and 88 μg/mouse, ratio PBEF Ab/rhPBEF ratio of 0.1 and 4.4) resulted in marked reductions in PBEF-mediated accumulation of bronchoalveolar lavage (BAL) PMNs (*P < 0.05). (B, C) We next used PBEF neutralizing antibody (88 μg) via intratracheal delivery in VILIb-challenged B6 mice and determined that PBEF Ab reduces (B) VILI-induced PMN accumulation and (C) tissue albumin leakage (40 ml/kg) (n = 3–4), *P < 0.05. SB = spontaneously breathing mice.
DISCUSSION
Despite the recognized morbidity and mortality associated with the mechanical ventilation of critically ill patients, the fundamental basis for ventilator-evoked pathophysiology remains unclear. In vitro and in vivo studies have highlighted the potential for direct physical injury by excessive mechanical stress (24) as well as VILI-mediated transcriptional and translational induction of inflammatory cytokines such as TNF-α, IL-1β, and IL-6 (1, 4). Using a candidate gene approach, coupled to intense gene expression profiling and bioinformatic analysis, we previously demonstrated that PBEF is a novel biomarker in sepsis and sepsis-induced ALI with increased PBEF gene and protein expression spatially localized to epithelium, endothelium, and inflammatory leukocytes (1). Furthermore, genetic variants in the PBEF promoter region were determined to confer susceptibility to sepsis and sepsis-induced ALI (1) and to be significantly associated with the number of ventilator-free days and overall ALI mortality (14).
This report further characterizes the mechanistic participation of PBEF in ALI and VILI and extends understanding of the role of PBEF in lung inflammation beyond involvement in regulation of neutrophil apoptosis in the setting of sepsis (23). Tracheal injection of rhPBEF into spontaneously breathing B6 mice produces marked increases in BAL leukocytes (PMNs). Although the murine response to rhPBEF was accompanied by increased BAL levels of known PMN chemoattractants KC and MIP-2 (Figure 3), we also determined that rhPBEF is a direct neutrophil chemotactic factor (Figure 1A). Together, these findings are consistent with a potentially direct role for PBEF in ALI- and VILI-associated inflammatory responses. Despite the marked influx of inflammatory leukocytes, rhPBEF produces only modest lung injury defined by levels of protein-rich tissue and alveolar edema. The reduced level of lung edema, in face of significant neutrophil recruitment to the alveolar space, has been previously observed with other neutrophil chemoattractants, including leukotriene B4, in which intrapulmonary administration produced robust recruitment of neutrophils into human airspaces with minimal changes in lung protein permeability (25). Thus, PBEF does not to appear to serve a direct role in inflammation-induced lung endothelial and epithelial barrier dysfunction. In contrast, using a murine model of modest mechanical VILI (4 h, 30 ml/kg tidal volume), we observed significant additive/synergistic relationship between rhPBEF challenge and VILI with dramatic increases in BAL PMNs, BAL protein, and cytokines (IL-6, TNF-α, KC).
The key role of PBEF in VILI is further substantiated by the response of heterozygous PBEF+/− mice to VILI challenge where mice with targeted deletion of a single PBEF allele were protected from severe ventilator-mediated lung injury (4 h, 40 ml/kg tidal volume) with significant reductions in VILI-associated increases in BAL protein and levels of BAL IL-6 and a significant reduction in peak inspiratory pressure compared with WT littermate control animals. Detailed assessment of potential biomarkers in BAL across the two models, VILIa and VILIb, revealed rhPBEF to induce significantly increased levels of CxCl1, CxCl2, IL-1β, IL-6, and TNF-α (Figure 9B, *P < 0.01). Likewise, VILIb-exposed mice demonstrated reductions in all chemokines and cytokines when compared with WT mice, of which IL-6 (*P < 0.05) is the most representative (Figure 9C).
We used genomic approaches with microarray analyses in mice exposed to rhPBEF and VILI to mechanistically interrogate PBEF involvement in ALI and VILI inflammatory lung responses. We observed a potential molecular signature in rhPBEF-challenged mice that reflects PBEF-driven induction of several signaling pathways referable to ALI and VILI, including Toll-like receptor signaling, apoptosis, and leukocyte extravasation signaling. Common canonical pathways were deregulated in both the VILIa-alone and the VILIa-rhPBEF groups, with novel deregulated pathways only induced by the combined treatment of VILI and rhPBEF, including acute phase response, NF-κB, and leukocyte extravasation pathways (Figure 6). A striking finding was that, although the PBEF+/− genotype did not impact global gene expression in comparison to WT mice, this genotype provided dramatic protection for the dysregulation of gene expressions and the functional pathways induced by VILI. Specific deregulated pathways induced by exposure to VILIb in WT mice were absent in PBEF+/− mice (e.g., NF-κB and G-protein–coupled receptor signaling pathways) (Figure 7A), again indicating a protective effect of the PBEF+/− genotype against ventilator-induced functional alterations.
A number of well-known ALI candidate genes (26) were identified by the current bioinformatic approaches, including IL-6, a pleiotropic cytokine critically involved in a variety of immunologic processes (26–28). IL-6 gene expression was increased by approximately 900-fold by the combined challenge of VILIa and rhPBEF, a finding verified by IL-6 levels in BAL fluid from challenged mice (∼30-fold increase). These results are consistent with a recent report that showed that rhPBEF increases IL-6 expression and secretion in vitro and in vivo (29). The dysregulated genes in the IL-6 pathway displayed in WT mice were either expressed at lower fold changes or were not dysregulated in PBEF+/− mice, consistent with a strongly protective effect of the PBEF+/− functional genotype in mice exposed to VILI (Figure 7). Although controversy exists regarding the exact role of lung cytokines in VILI (30) (biotrauma vs. volutrauma), cytokine responses in our model were apparent not only in VILI alone but were significantly augmented in rhPBEF-challenged mice exposed to excessive mechanical ventilation.
The combination of VILI and rhPBEF exposure also induced gene dysregulation in the NF-κB pathway displaying either higher fold changes or unique up-regulation compared with VILI alone, indicating a synergistic effect of VILI and rhPBEF treatment in NF-κB pathway deregulation. The functional consequences of this synergy is that in the setting of mechanical stress, PBEF expressed and localized to lung tissue may induce NF-κB activation and subsequent induction of genes well known to be involved in ALI (31) via augmentation of the inflammatory process (32), including cell survival (33), and neutrophil chemotaxis (34). It is possible that the neutrophil chemoattractant effect of PBEF in this study may be related, at least in part, to its effect on the NF-κB signaling pathway. Intratracheal injection of rhPBEF into SB and VILI-exposed mice resulted in significantly elevated BAL IL-6 levels, with a synergistic effect of VILI and rhPBEF compared with VILI alone. Because NF-κB is known to induce IL-6 production in response to inflammatory stimuli (32), it is likely that the deregulated NF-κB signaling pathway detected by our microarray analysis plays a direct role in IL-6 production. The lower levels of BAL IL-6 in PBEF+/− mice compared with WT mice in the VILI setting may be related to a protective effect on the deregulation of the NF-κB pathway. Our study shows that the deregulation of the NF-κB signaling pathway known to be involved in ALI processes is related to the presence of PBEF in the setting of normal and acutely inflamed lung tissue.
Our utilization of the IPA allows for identification of pathways relevant to our genes of interest and experimental models (see Methods). The dysregulation of specific genes may be further visualized through the IPA software via biological pathway maps (Figures E3 and E4). The use of IPA validates our previous findings (i.e., the initial identification of PBEF involvement in animal models of ALI [1]) as well as the role of PBEF in endothelial barrier dysfunction and PBEF-mediated deregulation of the expression of inflammatory cytokines in the setting of mechanical stress (1). IPA additionally determined PBEF-dependent pathways to include cell survival, cell death, and cell proliferation, further validating its role in ALI. Besides contributing to the identification of the PBEF-specific pathways in ALI, the use of SAM software identified a novel gene, BC018473, as highly up-regulated in SB PBEF+/− mice, with up-regulation further confirmed by real-time RT-PCR (Table E5). We are currently exploring the nature and functional consequences of this novel gene up-regulation.
An unexplored function of PBEF in ALI and VILI may be related to the effect of PBEF in insulin receptor signaling (35, 36), a process we have not examined. The adipocyte cytokine visfatin, now known to be identical to PBEF, serves to regulate cytokine production in adipocytes (36). Fukahara and colleagues recently demonstrated that PBEF binds to and activates the insulin receptor and serves a regulatory role in blood glucose homeostasis (35). Furthermore, PBEF raises (nicotinamide adenine dinucleotide) (NAD+) levels in smooth muscle cells (37), suggesting yet another mechanism of action for this novel lung cytokine. Patrone and colleagues included PBEF among genes expressed after IFN-γ exposure of pre–B cells (38). In addition, PBEF gene expression is up-regulated in patients with psoriasis, again supporting a role for PBEF as an inflammatory cytokine (39). PBEF expression is enhanced by exposure of monocytic cells to nitric oxide (40), which is consistent with the notion that septic lung injury is associated with increases in intraalveolar nitric oxide. Although we failed to identify bioinformatic evidence of dysregulated genes in the insulin signaling pathway and the NAD biosynthesis pathway, further studies are needed to fully explore the potential influence of PBEF in the context of insulin receptor signaling and ventilator-mediated inflammatory lung injury.
Jia and colleagues demonstrated that PBEF inhibited neutrophil apoptosis in an LPS- inflammation model with increased levels of IL-1, granulocyte macrophage colony–stimulating factor (GM-CSF), IL-8, and TNF-α (23). Reductions in PBEF levels (antisense oligonucleotide) induced normal apoptosis patterns in neutrophils from severely septic patients. Our bioinformatic analyses have confirmed and extended these findings because death receptor signaling and apoptosis signaling were prominent deregulated pathways in both PBEF/VILI-challenged mice (Figure E1) and were attenuated in the PBEF+/− heterozygotes. This is directly evident in the overexpression of a member of the CxC chemokine family, CxCl1 (KC), in the PBEF/VILI-challenged mice and attenuation in the PBEF+/− mice, as detected by our microarray and RT-PCR studies (Table E5). The CxC chemokine family is known to be involved in inflammation, cell growth, and tumorigenesis (41–43), and has been implicated in ALI (44, 45). CxCl1 (KC, mouse homolog of human growth-regulated oncogene [GRO]-α) suppresses neutrophil apoptosis and may lead to further CxCl1 production through autocrine mechanisms, further prolonging neutrophil survival (46). Our results suggest that PBEF stimulates CxCl1 expression, leading to its known antiapoptotic and chemoattractant effects on neutrophils. The expression pattern of CxCl2 (MIP-2, mouse homolog of human GRO-β) was similar to that of CxCl1 in our analyses; however, its role is not as clear in our animal model. The capacity of CxCl2 to mobilize neutrophils and other hematopoietic cells is well recognized and is currently being evaluated for therapeutic peripheral blood stem cell mobilization. However, the stimulation of CxCl2 expression by PBEF, in concert with elevated levels of KC, likely results in the accumulation of neutrophils at sites of inflammation or in areas of high PBEF concentration. The PBEF-mediated induction of these chemokines may contribute to the pathogenesis of VILI via the antiapoptotic and chemoattractant effects on neutrophils. Future directed investigative pursuits will likely contribute to our understanding of these components in the development of ALI.
In summary, our studies exploring rhPBEF synergies with VILI support our prior implication of PBEF as a novel ALI biomarker and clearly define a role for PBEF as a direct neutrophil chemoattractant intimately involved in the pathogenesis of VILI. The present study does present several limitations, such as the lack of PEEP application in our model as typically used in the ventilator management of humans with ALI/ARDS. However, because the primary purpose of the present study was to evaluate the role of PBEF in ventilator-mediated lung injury, PEEP and low tidal volumes were intentionally not used. Moreover, regardless of the exact ventilator settings used in our model, we believe the mechanism of injury (i.e., excessive biomechanical forces) was entirely consistent with and relevant to that seen in human VILI, although murine lungs are more compliant to aggressive mechanical ventilation strategies. A second limitation is that the effects of PBEF were only assessed as a pretreatment in our VILI model. Future studies of PBEF will rely on the administration of PBEF after the initiation of mechanical ventilation in an effort to more fully characterize the deleterious effects of PBEF in this setting. Another potential limitation is that we failed to use hemodynamic monitoring in this model but instead relied on results of arterial blood gas analysis, which showed no statistically significant differences in pH, PaO2, PaCO2, or HCO3 between the experimental VILI groups. Finally, another potential limitation is that we failed to fully explore the function of PBEF in PBEF−/− mice. Unfortunately, PBEF−/− mice are not available because the double PBEF mutation is lethal. However, as depicted in Figure 10, reductions in PBEF availability via use of a PBEF neutralizing antibody serve to reduce PMN recruitment and vascular leakage in our model of VILI (VILIb). Together, these highly translational biochemical, molecular, and genomic studies confirm the prominent role of PBEF as a critical effector in the development of ventilator-induced lung pathobiology. Future studies should consider PBEF as a key inflammatory mediator and potential molecular target for the prevention and amelioration of severe ventilator-induced ALI.
Supplementary Material
Supported by the National Institutes of Health grants HL 73994 (J.G.N.G.), HL 80042 (S.Q.Y.), HL 88144 (S.M.D.), and HL 58064 (J.G.N.G.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200712-1822OC on July 24, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Ye SQ, Simon BA, Maloney JP, Zambelli-Weiner A, Gao L, Grant A, Easley RB, McVerry BJ, Tuder RM, Standiford T, et al. Pre–B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med 2005;171:361–370. [DOI] [PubMed] [Google Scholar]
- 2.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, Stern EJ, Hudson LD. Incidence and outcomes of acute lung injury. N Engl J Med 2005;353:1685–1693. [DOI] [PubMed] [Google Scholar]
- 3.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med 2000;342:1334–1349. [DOI] [PubMed] [Google Scholar]
- 4.Ranieri VM, Suter PM, Tortorella C, De Tullio R, Dayer JM, Brienza A, Bruno F, Slutsky AS. Effect of mechanical ventilation on inflammatory mediators in patients with acute respiratory distress syndrome: a randomized controlled trial. JAMA 1999;282:54–61. [DOI] [PubMed] [Google Scholar]
- 5.Dreyfuss D, Soler P, Basset G, Saumon G. High inflation pressure pulmonary edema: respective effects of high airway pressure, high tidal volume, and positive end-expiratory pressure. Am Rev Respir Dis 1988;137:1159–1164. [DOI] [PubMed] [Google Scholar]
- 6.Tremblay LN, Miatto D, Hamid Q, Govindarajan A, Slutsky AS. Injurious ventilation induces widespread pulmonary epithelial expression of tumor necrosis factor-alpha and interleukin-6 messenger RNA. Crit Care Med 2002;30:1693–1700. [DOI] [PubMed] [Google Scholar]
- 7.Slutsky AS, Tremblay LN. Multiple system organ failure: is mechanical ventilation a contributing factor? Am J Respir Crit Care Med 1998;157:1721–1725. [DOI] [PubMed] [Google Scholar]
- 8.Tremblay L, Valenza F, Ribeiro SP, Li J, Slutsky AS. Injurious ventilatory strategies increase cytokines and c-fos m-RNA expression in an isolated rat lung model. J Clin Invest 1997;99:944–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Acute Respiratory Distress Syndrome Network. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N Engl J Med 2000;342:1301–1308. [DOI] [PubMed] [Google Scholar]
- 10.Parsons PE, Eisner MD, Thompson BT, Matthay MA, Ancukiewicz M, Bernard GR, Wheeler AP. Lower tidal volume ventilation and plasma cytokine markers of inflammation in patients with acute lung injury. Crit Care Med 2005;33:1–6. [Discussion, pp. 230–232.] [DOI] [PubMed] [Google Scholar]
- 11.Garcia JG, Moreno Vinasco L. Genomic insights into acute inflammatory lung injury. Am J Physiol Lung Cell Mol Physiol 2006;291:L1113–L1117. [DOI] [PubMed] [Google Scholar]
- 12.Nonas SA, Moreno-Vinasco L, Ma SF, Jacobson JR, Desai AA, Dudek SM, Flores C, Hassoun PM, Sam L, Ye SQ, et al. Use of consomic rats for genomic insights into ventilator-associated lung injury. Am J Physiol Lung Cell Mol Physiol 2007;293:L292–L302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nemeth E, Tashima LS, Yu Z, Bryant-Greenwood GD. Fetal membrane distention: I. Differentially expressed genes regulated by acute distention in amniotic epithelial (wish) cells. Am J Obstet Gynecol 2000;182:50–59. [DOI] [PubMed] [Google Scholar]
- 14.Bajwa EK, Yu CL, Gong MN, Thompson BT, Christiani DC. Pre-B-cell colony-enhancing factor gene polymorphisms and risk of acute respiratory distress syndrome. Crit Care Med 2007;35:1290–1295. [DOI] [PubMed] [Google Scholar]
- 15.Nichols J, Evans EP, Smith AG. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development 1990;110:1341–1348. [DOI] [PubMed] [Google Scholar]
- 16.Ma SF, Grigoryev DN, Taylor AD, Nonas S, Sammani S, Ye SQ, Garcia JG. Bioinformatic identification of novel early stress response genes in rodent models of lung injury. Am J Physiol Lung Cell Mol Physiol 2005;289:L468–L477. [DOI] [PubMed] [Google Scholar]
- 17.Moitra J, Sammani S, Garcia JG. Re-evaluation of evans blue dye as a marker of albumin clearance in murine models of acute lung injury. Transl Res 2007;150:253–265. [DOI] [PubMed] [Google Scholar]
- 18.Peng X, Hassoun PM, Sammani S, McVerry BJ, Burne MJ, Rabb H, Pearse D, Tuder RM, Garcia JG. Protective effects of sphingosine 1-phosphate in murine endotoxin-induced inflammatory lung injury. Am J Respir Crit Care Med 2004;169:1245–1251. [DOI] [PubMed] [Google Scholar]
- 19.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 2001;98:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Team RDC (R Development Core). R. A language and environmental for statistical computing. 2005.
- 21.Wu Z, Irizarry R, Gentleman R, Martinez F, Spencer F. A model based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 2004;99:909–917. [Google Scholar]
- 22.Gene Ontology Consortium. The gene ontology (GO) project in 2006. Nucleic Acids Res 2006;34:D322–D326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD, Marshall JC. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest 2004;113:1318–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West JB. Cellular responses to mechanical stress: invited review: pulmonary capillary stress failure. J Appl Physiol 2000;89:2483–2489. [DOI] [PubMed] [Google Scholar]
- 25.Martin TR, Pistorese BP, Chi EY, Goodman RB, Matthay MA. Effects of leukotriene B4 in the human lung: recruitment of neutrophils into the alveolar spaces without a change in protein permeability. J Clin Invest 1989;84:1609–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.L'Her E, Deye N, Lellouche F, Taille S, Demoule A, Fraticelli A, Mancebo J, Brochard L. Physiologic effects of noninvasive ventilation during acute lung injury. Am J Respir Crit Care Med 2005;172:1112–1118. [DOI] [PubMed] [Google Scholar]
- 27.Kishimoto T, Akira S, Narazaki M, Taga T. Interleukin-6 family of cytokines and gp130. Blood 1995;86:1243–1254. [PubMed] [Google Scholar]
- 28.Peters M, Muller AM, Rose-John S. Interleukin-6 and soluble interleukin-6 receptor: direct stimulation of gp130 and hematopoiesis. Blood 1998;92:3495–3504. [PubMed] [Google Scholar]
- 29.Moschen AR, Kaser A, Enrich B, Mosheimer B, Theurl M, Niederegger H, Tilg H. Visfatin, an adipocytokine with proinflammatory and immunomodulating properties. J Immunol 2007;178:1748–1758. [DOI] [PubMed] [Google Scholar]
- 30.Dreyfuss D, Ricard JD, Saumon G. On the physiologic and clinical relevance of lung-borne cytokines during ventilator-induced lung injury. Am J Respir Crit Care Med 2003;167:1467–1471. [DOI] [PubMed] [Google Scholar]
- 31.Cho HY, Morgan DL, Bauer AK, Kleeberger SR. Signal transduction pathways of tumor necrosis factor–mediated lung injury induced by ozone in mice. Am J Respir Crit Care Med 2007;175:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanden Berghe W, Vermeulen L, De Wilde G, De Bosscher K, Boone E, Haegeman G. Signal transduction by tumor necrosis factor and gene regulation of the inflammatory cytokine interleukin-6. Biochem Pharmacol 2000;60:1185–1195. [DOI] [PubMed] [Google Scholar]
- 33.Kucharczak J, Simmons MJ, Fan Y, Gelinas C. To be, or not to be: NF-kappaB is the answer—role of rel/NF-kappaB in the regulation of apoptosis. Oncogene 2003;22:8961–8982. [DOI] [PubMed] [Google Scholar]
- 34.Blackwell TS, Holden EP, Blackwell TR, DeLarco JE, Christman JW. Cytokine-induced neutrophil chemoattractant mediates neutrophilic alveolitis in rats: association with nuclear factor kappa B activation. Am J Respir Cell Mol Biol 1994;11:464–472. [DOI] [PubMed] [Google Scholar]
- 35.Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, Kishimoto K, Matsuki Y, Murakami M, Ichisaka T, Murakami H, et al. Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science 2005;307:426–430. [DOI] [PubMed] [Google Scholar]
- 36.Tilg H, Moschen AR. Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat Rev Immunol 2006;6:772–783. [DOI] [PubMed] [Google Scholar]
- 37.van der Veer E, Nong Z, O'Neil C, Urquhart B, Freeman D, Pickering JG. Pre-B-cell colony-enhancing factor regulates NAD+-dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ Res 2005;97:25–34. [DOI] [PubMed] [Google Scholar]
- 38.Patrone L, Damore MA, Lee MB, Malone CS, Wall R. Genes expressed during the IFN gamma-induced maturation of pre-B cells. Mol Immunol 2002;38:597–606. [DOI] [PubMed] [Google Scholar]
- 39.Koczan D, Guthke R, Thiesen HJ, Ibrahim SM, Kundt G, Krentz H, Gross G, Kunz M. Gene expression profiling of peripheral blood mononuclear leukocytes from psoriasis patients identifies new immune regulatory molecules. Eur J Dermatol 2005;15:251–257. [PubMed] [Google Scholar]
- 40.Turpaev K, Bouton C, Diet A, Glatigny A, Drapier JC. Analysis of differentially expressed genes in nitric oxide-exposed human monocytic cells. Free Radic Biol Med 2005;38:1392–1400. [DOI] [PubMed] [Google Scholar]
- 41.Luster AD. Chemokines: chemotactic cytokines that mediate inflammation. N Engl J Med 1998;338:436–445. [DOI] [PubMed] [Google Scholar]
- 42.Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu Rev Immunol 2000;18:217–242. [DOI] [PubMed] [Google Scholar]
- 43.Wang JM, Deng X, Gong W, Su S. Chemokines and their role in tumor growth and metastasis. J Immunol Methods 1998;220:1–17. [DOI] [PubMed] [Google Scholar]
- 44.Reutershan J, Morris MA, Burcin TL, Smith DF, Chang D, Saprito MS, Ley K. Critical role of endothelial Cxcr2 in LPS-induced neutrophil migration into the lung. J Clin Invest 2006;116:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller EJ, Cohen AB, Nagao S, Griffith D, Maunder RJ, Martin TR, Weiner-Kronish JP, Sticherling M, Christophers E, Matthay MA. Elevated levels of NAP-1/interleukin-8 are present in the airspaces of patients with the adult respiratory distress syndrome and are associated with increased mortality. Am Rev Respir Dis 1992;146:427–432. [DOI] [PubMed] [Google Scholar]
- 46.Dunican A, Grutkoski P, Leuenroth S, Ayala A, Simms HH. Neutrophils regulate their own apoptosis via preservation of cxc receptors. J Surg Res 2000;90:32–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.