Abstract
Podoplanin (RTI40, aggrus, T1α, hT1α-2, E11, PA2.26, RANDAM-2, gp36, gp38, gp40, OTS8) is a type I cell marker in rat lung. We show that a bacterial artificial chromosome vector containing the rat podoplanin gene (RTIbac) delivers a pattern of transgene expression in lung that is more restricted to mouse type I cells than that of the endogenous mouse podoplanin gene. RTIbac-transgenic mice expressed rat podoplanin in type I cells; type II cells, airways, and vascular endothelium were negative. A modified bacterial artificial chromosome containing internal ribosome entry site (IRES)-green fluorescent protein (GFP) sequences in the podoplanin 3′UTR expressed rat podoplanin and transgenic GFP in type I cells. RTIbac transgene expression was absent or reduced in pulmonary pleura, lymphatic endothelium, and putative lymphoid-associated stromal tissue, all of which contained abundant mouse podoplanin. Rat podoplanin mRNA levels in normal rat lung and RTIbac transgenic lung were 25-fold higher than in corresponding kidney and brain samples. On Western blots, transgenic rat and endogenous mouse podoplanin displayed very similar patterns of protein expression in various organs. Highest protein levels were observed in lung with 10- to 20-fold less in brain; there were low levels in thymus and kidney. Both GFP and rat podoplanin transgenes were expressed at extrapulmonary sites of endogenous mouse podoplanin gene expression, including choroid plexus, eye ciliary epithelium, and renal glomerulus. Because their pulmonary expression is more restricted than endogenous mouse podoplanin, RTIbac derivatives should be useful for mouse type I cell–specific transgene delivery.
Keywords: alveolar type I cells, transgenes, podoplanin, bacterial artificial chromosome, alveolar epithelium
Transgene expression strategies targeting alveolar type II cells and Clara cells have significantly enhanced our understanding of pulmonary function and development (reviewed in Ref. 1). In contrast, alveolar type I cells have proven recalcitrant to these approaches. These cells cover more than 95% of the internal lung surface area (2) and exhibit a morphology highly distinct from other lung epithelial cells, yet few type I cell–specific promoters suitable for transgene expression vectors have been identified. For a number of years, we have studied podoplanin, an apical membrane glycoprotein previously known as RTI40 that is preferentially expressed in type I cells in rat lung (3–5). This expression pattern suggested the use of the podoplanin gene in type I cell–specific transgene vectors.
Podoplanin, the name originally used by Breiteneder-Geleff and colleagues, is now the accepted descriptor of this gene (6); however, many groups have described orthologous or identical genes (TIα [7], hTIα-2 [8], PA2.26 [9], RANDAM-2 [10], E11 [11, 12], OTS8 [13], Aggrus [14], gp38 [15], gp40 [16], and gp36 [17]). A clear in vivo function for podoplanin has remained elusive despite many genetic and biochemical investigations into properties of the protein (18–23). Several of the above reports described podoplanin expression in extrapulmonary sites such as renal podocytes, thymic stromal cells, and lymphatic vessel endothelium. In an effort to reconcile these varied reports with our own experience of restricted podoplanin expression in rat lung, we hypothesized that transgenes employing the rat gene would have a more restricted expression range in lung than the endogenous mouse gene.
Short podoplanin promoter fragments have not proven effective at driving accurate transgene expression. Although transfection results defined a 1.1-kbp basal promoter for the rat gene, both 1.3-kbp and longer DNA fragments did not confer high-level type I cell–specific expression in adult transgenic mice (24–26; J. N. Vanderbilt and L. G. Dobbs, unpublished observations). Lacking detailed knowledge on the fully functional podoplanin promoter, we turned to bacterial artificial chromosomes (BACs), extremely large (100–300 kbp), modifiable, circular vectors available as isolates from arrayed libraries spanning the genome of a given species (27). We used a BAC vector (RTIbac) containing the rat podoplanin gene flanked by 106 kbp of upstream and 27 kbp of downstream DNA sequence to deliver transgenes to type I cells in the mouse alveolar epithelium. We also modified RTIbac to express green fluorescent protein (GFP) from an internal ribosome entry site (IRES) inserted into the podoplanin 3′UTR. Both BACs conferred selective transgene expression in type I cells in a more restricted pattern than the endogenous mouse podoplanin gene. Transgene expression was absent or reduced in three pulmonary locations that contained abundant mouse podoplanin: pleura, lymphatic endothelium, and lymphoid-associated stromal tissue. Because their pulmonary expression is more restricted than endogenous mouse podoplanin, RTIbac derivatives should be useful for mouse type I cell–specific transgene delivery. Portions of these results were published previously in abstract form (28).
MATERIALS AND METHODS
Transgenic Animals
BAC DNA microinjection into fertilized mouse eggs followed by transplantation into pseudopregnant females was performed by the Stanford University Transgenic Research Facility (RTIbac into FVB/N mice) or by us (RTIbacGFP into C57bl6 mice) using standard protocols. Before weaning of pups, tail clip DNA was screened by PCR for the presence of RTIbac or RTIbacGFP using a primer pair positioned upstream of the podoplanin gene (JV984: 5′-GGATTATGACTGACAGG-3′; JV985: 5′-TGCTTATCGTGAGAAGG-3′). The copy number of RTIbac transgenic lines was estimated by comparison to normal rat DNA in PCR reactions over a variable number of amplification cycles.
BAC Isolation and Purification
Two DNA fragments corresponding to rat podoplanin genomic sequence −2487 to −1765 and 33273–33503 (26) were used to screen a rat BAC library (screening performed by Genome Systems, St. Louis, MO). RTIbac (clone address 240G15) hybridized to both probes and contains an insert in the pBeloBAC11 vector (29) of approximately 168 kbp corresponding to nucleotides 162,114,322 to 162,282,008 on the rat chromosome 5 genomic map (www.ncbi.nlm.nih.gov/genome/guide/rat/). RTIbac was maintained in the Escherichia coli strain DH10B. BAC DNA was isolated from overnight cultures in Luria Bertrani media containing 12.5 μg/ml chloramphenicol. Bacterial pellets were disrupted by alkaline lysis, and the crude BAC DNA was subjected to two rounds of cesium chloride equilibrium density gradient centrifugation. For microinjection into fertilized mouse eggs, DNA was concentrated by centrifugation through a Centriprep-30 membrane (Amicon, Beverly, MA) including two sequential washes with 10 mM Tris-HCl pH 7.4, 0.1 mM EDTA (pH 8.0) in the external chamber until the final volume in the centriprep unit was 500 μl. The DNA was filtered through a 0.45-μm filter (SLHV004NL; Millipore, Billerica, MA) and analyzed by ultraviolet spectrophotometry and gel electrophoresis.
BAC Modification
RTIbac was modified in DH10B as described by Gong and colleagues (30). The process involves two key recA-mediated recombination events between the BAC resident in E. coli and an introduced shuttle vector containing both the desired modification (IRES-GFP) and specialized functionalities that effect and select for recombination (recA, sacB, and ampr genes and an R6Kγ origin). Two DNA segments of about 300 bp each (A and B homology boxes), corresponding to BAC DNA sequences 5′ and 3′ of the desired modification site in exon 6 of the podoplanin gene, were cloned 5′ and 3′ of the IRES-GFP sequences in the shuttle vector to create targets for homologous recombination. Shuttle vector cloning details are provided below.
The first recombination event occurs after electroporation of the shuttle vector into RTIbac/DH10B cells and subsequent production of recA. Recombination events between either A or B boxes on the shuttle vector and the homologous sequences in the BAC generate cointegrate molecules containing the BAC with the complete shuttle vector positioned between duplications of the homology boxes. Shuttle vectors that have not recombined do not replicate because their R6Kγ origin is nonfunctional in DH10B cells (31). Two classes of cointegrate result, one from recombination through the A box, the other through the B box. Both classes are selected by growth on chloramphenicol plus ampicillin, then screened by PCR and Southern blotting for either of the two equally acceptable cointegrate possibilities.
A second recombination, this time intramolecular within a cointegrate molecule, has two possible outcomes depending on which pair of duplicated homology boxes recombine. One reaction deletes the entire shuttle vector to regenerate the starting BAC. The other reaction, preferable to the investigator but indifferent to the cell, retains the desired IRES-GFP modification while deleting the remaining shuttle vector sequences (recA, sacB, and ampr and R6Kγ origin). Both recombination events were selected by growth on chloramphenicol plus 4.5% sucrose (the sacB gene product metabolizes sucrose to a toxic product). The two possible outcomes were then distinguished by PCR. Candidate modified BACs were further analyzed by Southern blotting, restriction enzyme digestion and partial sequencing, always by reference to the parental RTIbac.
Shuttle Vector Construction
The general configuration of the shuttle vector was described above in the BAC modification section. The A and B homology boxes were PCR amplified from RTIbac DNA and cloned into pCR-Script (Stratagene, Cedar Creek, TX) using primers JV0303, 5′-GACATCAGGCGCGCCCTTTTTACAAGCCTCCAAACCCTC-3′ with added 5′ Asc I site (italics) and JV0304, 5′-CATCAAACCTTTCCATCCACCAC-3′ for the A box and JV0305, 5′-GACATCATTAATTAAGTCTTCGTGACATACCACATCTTCC-3′ with added Pac I site (italics) and JV0306 5′-GACACACACCTTTAGTCCCACTGC-3′ for the B box. The A box fragment was excised with Sma I and Asc I and ligated into identically cut PLD53SCAEB (30) to create p120.18. The B box fragment was excised with Sma I and Pac I then inserted in p120.18 between Pac I and T4 DNA Polymerase blunted Fse I sites to create p122.1. The IRES sequence was excised from pIRES2EGFP (BD Biosciences Clontech, San Jose, CA) and inserted as a 0.65-kbp Bgl II-Nco I fragment into pLitmus28i (New England Biolabs, Ipswich, MA) to make pLIRES. The Nco I to Sac I fragment (0.65 kb) from pLIRES containing the IRES element was linked to a 1.4-kbp Nco I–Bam HI EGFP-B-box fragment from p122.1 between Bam HI and Sac I sites in pBluescript SK- (Stratagene) to create the cloning intermediate p124B. Finally, a 1.6-kbp Bam HI–Pac I fragment from p124B was cloned in a triple ligation with 6.5 kbp Pac I–Asc I and 0.3 kbp Asc I–Bam HI fragments from p122.1 to generate p126B3, the shuttle vector for recombination with RTIbac. All R6Kγ-based plasmids were grown in pir2 cells (Invitrogen, Carlsbad, CA) and purified from 1-liter cultures using alkaline lysis and two rounds of CsCl equilibrium density gradient centrifugation. All other plasmids were maintained in DH5α cells and isolated with Qiagen Midi kits (Qiagen, Valencia, CA)
Real-Time RT-PCR
Total RNA, extracted from tissues with RNeasy kits (Qiagen) including on-column digestion with DNase I, was reverse transcribed with AMV reverse transcriptase (Invitrogen) and analyzed in triplicate on an ABI PRISM 7700HT Sequence Detector System as described using primers (forward, 5′-CAGTGTTGCTCTGGGTTTTGG-3′; reverse, 5′-AGACCTGGGTTCACCATGTCA-3′) and detection probe (6FAM-ATGGCCCCTCCCTGCGCTGA-TAMRA) specific for rat podoplanin. The data were normalized to the amount of 18S ribosomal RNA in each sample (TaqMan Ribosomal RNA control reagents, Applied Biosystems, Foster City, CA) then compared with a standard curve of rat podoplanin plasmid to determine the amount of each mRNA in the starting sample. RNAs isolated from the organs of three animals were each assayed in triplicate and the data were averaged.
Western Blots
After collection, tissues were minced with a surgical scalpel, then dispersed by pipetting 20 times through a wide-bore 1,000-μl tip in 400 μl extraction buffer (50 mM Tris Base, 4M urea, 20% glycerol, 2 mM EDTA, 2 μg/ml DNase I, 0.5 mM PMSF) and homogenizing (setting 3 for 10 s) with a PowerGen 700 tissue homogenizer (Thermo Fisher Scientific, Waltham, MA). Each sample received an equal volume of 2× Laemmli sample buffer followed by 10 passes through a 25-gauge needle. Samples were spun (14,000 × g for 2 min), the supernatant heated (95°C for 4 min) and spun again. Extracted proteins (160 μg per lane for liver, kidney, thymus, and brain, 16 μg for lung as determined by BCA assay [Pierce, Rockford, IL]) were run on SDS-polyacrylamide gels (12%) and electroblotted to nitrocellulose. Membranes were incubated in 15% hydrogen peroxide for 10 minutes and blocking buffer (Tris buffered saline with 1.0% nonfat dried milk, 0.4% fish tail gelatin, and 0.1% bovine serum albumin) for 2 hours. All subsequent incubations and washes were in TBS/T (Tris buffered saline with 0.1% Triton X-100). Primary and secondary antibody incubations (1:3,000 dilution for each) were for 40 minutes each, followed by 20 washes of 50 ml TBS/T for 2 minutes each. Labeled secondary antibodies were detected on film with ECLplus (GEhealthcare, Piscataway, NJ) per the manufacturer's recommendations. Chemiluminesence signals were collected on a Molecular Dynamics Storm 840 phosphoimager and quantified with ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Microscopy
Tissues were fixed, frozen, and sectioned as described previously (32). Two-micrometer cryosections were stained with antibodies to rat podoplanin (1:1,000 dilution of antibody referred to as anti-RTI40 [3]) or mouse podoplanin (1:200; #8.1.1 from Developmental Studies Hybridoma Bank, University of Iowa. This is the original anti-gp38 monoclonal antibody described by Farr and colleagues [15]). The antibody against rat podoplanin does not recognize the mouse protein and vice versa (Figures 2B and 2E). Other antibodies included anti-GFP (1:200; A-21311 from Invitrogen) and anti-LYVE-1 (1:1,000; #07-538 from Upstate Biochemicals, Lake Placid, NY [33]). Antigen retrieval was necessary to visualize LYVE-1 staining, consistent with previous studies (33). For these sections, slides were incubated for 10 minutes in Dako Target Retrieval Solution (Dako, Carpenteria, CA) while immersed in a 95°C water bath, then cooled and rinsed with distilled water. GFP antibody was directly conjugated to Alexa 488; all other antibodies were followed by the appropriate goat anti-mouse, goat anti-rabbit, or goat anti-hamster IgG Alexa 594– or Alexa 488–conjugated secondary antibodies at 1:3,000 (Invitrogen). Nuclei were visualized with DAPI stain. Slides were mounted in Prolong (Invitrogen) and examined on a Leica Orthoplan fluorescent microscope. Color images (2,600 × 2,060 dpi) were captured in separate channels by a Leica DC500 digital camera. For the composite images shown in Figures 2, 3, and 4, individual fluorescent signals were captured, then merged with the use of Photoshop CS software (Adobe, San Jose, CA).
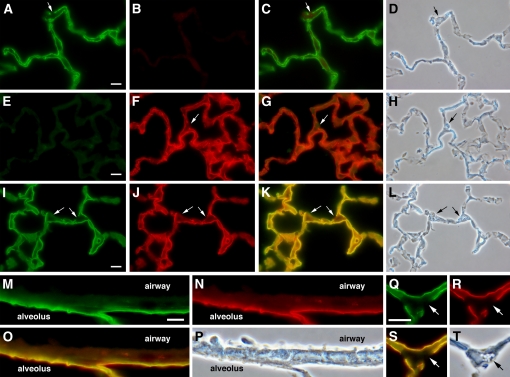
Figure 2.
Expression of transgenic rat podoplanin in type I alveolar epithelial cells of line 9 mice. Lung sections from normal rat (A–D), wild-type mouse (E–H), and line 9 transgenic mouse (I–T) were double-antibody stained with anti-rat (green) and anti-mouse (red) podoplanin antibodies. Separate channels are shown for green (A, E, I, M, and Q) and red (B, F, J, N, and R) staining along with the corresponding merged (C, G, K, O, and S) and phase contrast images (D, H, L, P, and T). Note higher magnification for M–T. Scale bar indicates 10 μm. Merged images were created from separately collected channels as described in Materials and Methods. Control reactions on normal rat (B) and wild-type mouse (E) lung demonstrate species specificity of the antibodies. Transgenic rat and endogenous mouse podoplanin overlap in line 9 type I cells with no expression in type II cells (arrows) or airway epithelium.
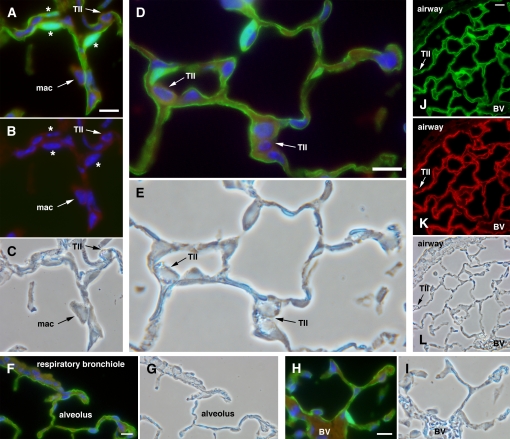
Figure 3.
RTIbacGFP containing an internal ribosome entry site (IRES)-green fluorescent protein (GFP)–modified rat podoplanin transgene expresses GFP and rat podoplanin in the transgenic alveolar epithelium. Shown is staining for GFP (green), rat podoplanin (red), and nuclei (DAPI, blue). A–I show restriction of GFP expression to type I cells with no staining in type II cells (“TII”) or alveolar macrophages (“mac”). A and D demonstrate that GFP labels the entire type I cell, including thin cytoplasmic extensions and perinuclear cytoplasm. B shows the isolated blue (DAPI) channel from A. In A and B, concomitant staining with GFP and DAPI identifies type I cell nuclei (asterisks). (C) Corresponding phase contrast image for A and B. (E) Phase contrast image corresponding to D. Neither respiratory bronchioles (F) nor blood vessels (“BV” in H and L) express detectable GFP. G and I are phase contrast images corresponding to F and H, respectively. (J–L) Low-magnification view demonstrating co-localization of GFP (J) and rat podoplanin (K) in RTIbacGFP-transgenic mice; phase contrast image is in L. Both proteins are expressed in type I cells but not in type II cells (“TII”) or airway cells. Scale bar indicates 10 μm.
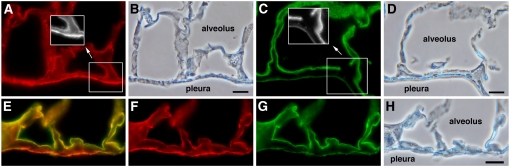
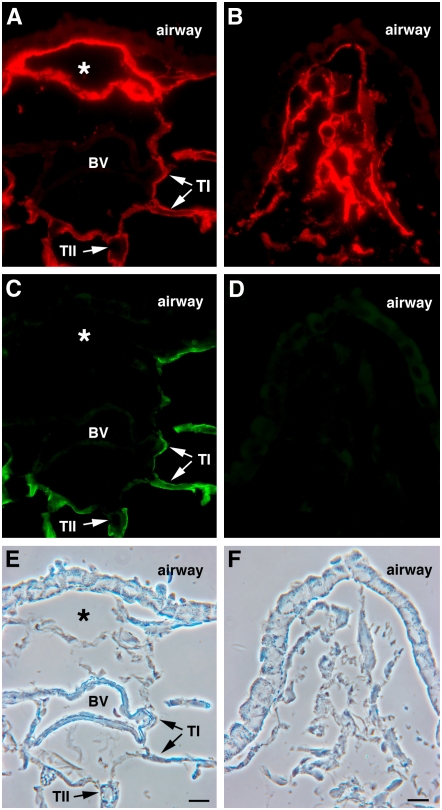
Figure 4.
Distinct patterns of rat and mouse podoplanin expression in pleura. (A and B) Paired immunofluorescence and phase contrast images of wild-type mouse lung stained for mouse podoplanin (red). The grayscale inset in A shows higher expression in pleura than in alveoli. (C and D) Paired immunofluorescence and phase contrast images of normal rat lung stained for rat podoplanin (green). The grayscale inset in C shows lower expression in pleura than in alveoli. (E–H) Line 9 mouse lung co-stained for transgenic rat (green) and endogenous mouse (red) podoplanin. (E) Merged image created from separately collected red and green channel images in F and G, respectively. Note low rat podoplanin and high mouse podoplanin expression in pleura relative to type I cell staining in alveoli. H shows the paired phase contrast image. Scale bar indicates 10 μm.
RESULTS
Transgenic Mouse Lines Expressing the Rat Podoplanin Gene
For a number of years, we have used rat podoplanin protein and mRNA expression, under the name RTI40, as markers of the alveolar type I cell phenotype (3, 4, 34). In contrast to mouse podoplanin expression in lymphatics (22), we observed that the rat podoplanin protein in lung appeared to be type I cell–restricted (see Figure 3 of Ref. 3). To test the idea that a transgene vector containing the rat podoplanin gene and promoter might confer a more selective type I cell expression profile than the endogenous mouse podoplanin gene, we isolated a BAC, designated here as RTIbac, from a rat genomic library using sequential screens with two widely separated podoplanin gene probes. Mapping and partial sequencing positioned the 33-kbp podoplanin gene (26) midway on the RTIbac insert with 106 kbp and 27 kbp of upstream and downstream genomic DNA, respectively. With the release of the rat genome sequence, we were able to identify the RTIbac insert on chromosome 5 between nucleotides 162,114,322 and 162,282,008.
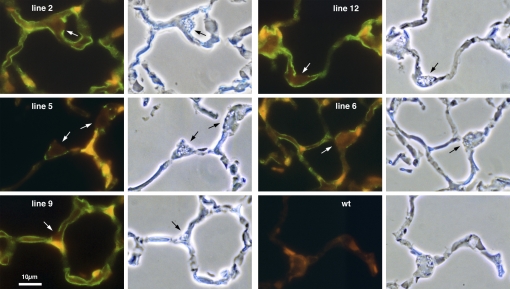
Among 18 pups produced after pronuclear injection of circular BAC DNA, we identified 6 transgenic founders whose DNA contained RTIbac sequences. All six transmitted RTIbac DNA to their offspring, thereby establishing six independent RTIbac transgenic lines (lines 2, 5, 6, 9, 12, and 14, named after the pup number in the original litter). As shown in Figure 1, five of these (lines 2, 5, 6, 9, and 12) expressed rat podoplanin in alveolar epithelial type I cells but not type II cells when screened by immunofluorescence with an anti-rat podoplanin monoclonal antibody (3) that did not cross react with mouse podoplanin (Figure 1, wt). Line 9 generated the highest level of type I cell staining; lines 2, 6, and 12 were slightly lower; and line 5 staining was weak. Line 14 was the only line that did not express detectable levels of the rat protein (not shown).
Figure 1.
Alveolar epithelial expression of rat podoplanin in five different rat podoplanin gene (RTIbac) transgenic mouse lines. Lungs from the indicated transgenic lines, numbered after the founding pup in the original litter, or wild-type control (wt) were stained with a monoclonal antibody against rat podoplanin (green). The antibody labels type I cells in the alveolar epithelium. Type II cells (arrows) do not stain. There was no staining in wild-type mouse lung.
Type I Cell–Specific Podoplanin Transgene Expression in the Alveolar Epithelium
Line 9 was chosen for further analysis based on the strong and uniform immunofluorescent signal in lung sections stained with the anti-rat podoplanin antibody (Figure 1). Using comparative genomic PCR reactions, we estimated that line 9 has between 4- and 8-fold higher RTIbac insert copy number than normal rat DNA (described in Materials and Methods). In Figure 2, we used double antibody staining to compare expression of the rat transgene with endogenous mouse podoplanin, also expressed in type I cells (35, 36). Each antibody identified its target protein with no cross-reaction on the appropriate normal control tissue (Figures 2A–2H). Because two different antibodies were used to label the two proteins, it is not possible to compare relative levels of rat and mouse podoplanin expression directly. However, qualitative comparison of tissues labeled with the same antibody does seem justified.
Examination of the separate channel images for each immunofluorescent probe in Figure 2 (rat podoplanin, green in Figures 2I, 2M, and 2Q; mouse podoplanin, red in Figures 2J, 2N, and 2R) as well as the merged images of the two channels (Figures 2K, 2O, and 2S) revealed that both proteins were present on all line 9 type I cells in these images. All type II cells, identified by the presence of phase-lucent lamellar bodies, were negative for both proteins. The relative strengths of rat podoplanin staining in line 9 (Figure 2I) versus normal rat (Figure 2A) lung were very similar, as was mouse podoplanin staining in line 9 (Figure 2J) and wild-type (Figure 2F) mouse lung. Higher magnification, dual stained images of type I cells, airway and a type II cell from line 9 are shown in Figures 2M through 2T. Expression of both proteins appeared to co-localize to a region consistent with the previously described type I cell apical plasma membrane expression of native rat podoplanin (3). In striking contrast, the airway epithelium and the type II cell were negative for expression of either protein. From these images and others covering broader areas of the lung (not shown), we conclude that most, if not all, type I cells in line 9 lung express rat podoplanin.
Modification of RTIbac to Express GFP in Type I Cells
The results with line 9 highlighted the potential of RTIbac derivatives to deliver functional gene products to type I cells. With this goal in mind, we modified RTIbac to express green fluorescent protein (GFP) under control of an internal ribosome entry site (IRES) (30). We positioned GFP in the 3′UTR under IRES control to minimize disruption of potential podoplanin regulatory sequences while maintaining integrity of the podoplanin marker gene. In addition, this approach provides an independent marker of podoplanin gene expression on histologic sections. The modified BAC should direct two proteins, each translated independently from a common transcript, to distinct locations in type I cells: GFP to the cytoplasm and podoplanin to the apical plasma membrane. From 21 pups, we identified 7 transgenic founders, two of which were established as stable RTIbacGFP lines. One of the lines was characterized in detail although both produced similar levels of GFP expression in lung.
In Figure 3, we show immunofluorescence staining of RTIbacGFP transgenic mouse lung sections. We used an anti-GFP antibody to better visualize the protein because native GFP fluorescence was weak, although detectable. Both transgenic proteins, GFP and rat podoplanin, were expressed on all type I cells, with no expression in type II cells, airway epithelium, or blood vessel endothelium. In sharp contrast to the apparent apical membrane localization of transgenic rat podoplanin (Figure 3K), GFP defined the entirety of the type I cell, including thin cytoplasmic extensions, nucleus, and, particularly striking, the thickened perinuclear region (Figures 3A, 3D, 3F, 3H, and 3J). Interaction of GFP and DAPI fluorescence imparted a bluish-green color to many type I cell nuclei (Figures 3A and 3B, asterisks). We conclude that RTIbac can be modified within the 3′UTR of the rat podoplanin gene with exogenous gene sequences under IRES control to deliver transgenes to type I cells while maintaining expression of the rat podoplanin marker.
Increased Specificity of Transgenic Versus Endogenous Podoplanin Expression in Lung
Throughout the alveolar epithelium that we examined, transgenic rat podoplanin was co-expressed with endogenous mouse podoplanin. We did observe, however, three regions in the lung where staining for the two proteins was different, notably in pleura, lymphatic vessels, and lymphoid-associated stromal tissue.
Pleura.
Two recent reports described podoplanin expression in human and mouse pleura (35, 37). Our experience with adult rats is that podoplanin staining, when observed in the pleura, is very weak relative to surrounding type I cells. To explore this issue, we directly compared pleural and alveolar podoplanin staining in rat, line 9, and wild-type mouse lung. In rat lung (Figures 4C and 4D), pleural staining was much weaker than in adjacent alveolar regions. In wild-type mouse lung (Figures 4A and 4B) the reverse was apparent, with stronger pleural than alveolar staining, a contrast more apparent in the grayscale insets within the images. Divergent pleural podoplanin expression patterns of both species were evident in line 9 lung, with the mouse protein stronger (Figure 4F) and the rat protein weaker (Figure 4G) in pleura compared with type I cell staining. The difference is more evident in the merged image (Figure 4E), where the red staining of mouse podoplanin largely dominates in pleura, whereas the alveolar region displays either a yellow or greenish-yellow color shift, indicating co-expression of both proteins.
Lymphatic endothelium.
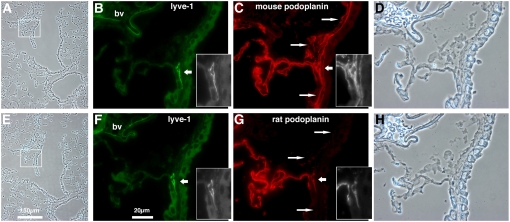
Podoplanin is a widely used marker of lymphatic vascular endothelial cells (11, 22, 38). Although relatively rare in rodent distal lung, lymphatic capillaries and vessels form a meshwork within the connective tissue surrounding respiratory bronchioles and larger airways (39). Podoplanin identification in pulmonary lymphatics is difficult given its abundant and pervasive expression in the enveloping alveolar epithelium. In addition, a discontinuous basement membrane renders lymphatic capillaries and vessels particularly susceptible to collapse unless special techniques are employed to maintain patency during fixation (39). We used an antibody to LYVE-1 (33) to identify lymphatic endothelium with the caveat that in mouse lung, LYVE-1 also stains some small blood vessels (40).
In Figure 5, we double antibody stained two serial sections of RTIbacGFP transgenic lung, the first for LYVE-1 and mouse podoplanin (Figures 5B and 5C) and the second for LYVE-1 and rat podoplanin (Figures 5F and 5G). In both serial sections, LYVE-1 stained the same two structures, one a blood vessel containing numerous erythrocytes (Figure 5, “bv”), the other consistent with a lymphatic capillary, based both on its location and its appearance (Figure 5, grayscale insets). Mouse podoplanin staining in the LYVE-1-negative, lymphatic-like structures (long arrows in Figure 5) is discussed in the following section. The blood vessel did not stain for either rat or mouse podoplanin. The presumptive lymphatic stained for both but, as in pleura (Figure 4), there were qualitative differences; these are evident in the grayscale insets within Figure 5. Rat podoplanin staining was lower in the LYVE-1–positive lymphatic than in nearby type I cells (Figure 5G). In contrast, mouse podoplanin staining appeared equivalent to type I cell staining (Figure 5C). Similar patterns were observed for several serial section pairs stained in the same manner (not shown). This observation was not an artifact of subtle differences in serial section position, because not only was LYVE-1 staining consistent from section to section, but also rat podoplanin staining was always less prevalent than that for mouse. The reverse, as would occur if the staining bias were due to subtle differences in vessel structure between consecutive sections, was never observed.
Figure 5.
Differences between endogenous mouse and transgenic rat podoplanin expression in lymphatic vessels and lymphoid associated stroma of RTIbacGFP-transgenic mouse lung. Two serial sections of RTIbacGFP-transgenic lung were co-stained for LYVE-1 (green in B and F) and either mouse (red in C) or rat podoplanin (red in G). Boxed areas in low-magnification phase contrast images (A and E) are enlarged in the three images to the right of each, including enlarged phase contrast images in D and H. Insets show grayscale images of an LYVE-1 staining lymphatic (B and F) and the same structure stained for mouse (C) and rat (G) podoplanin. Note the reduced rat podoplanin staining in the LYVE-1–positive lymphatic relative to alveolar staining. Long arrows identify LYVE-1–negative airway apposed structures staining for mouse but not rat podoplanin that are consistent in location with lymphoid-associated stroma (center long arrow) and peribronchial lymphatics (upper and lower long arrows). Scale bars are as indicated; bv, blood vessel.
In Figures 6A, 6C, and 6E, a distinct pulmonary section stained for endogenous mouse podoplanin (Figure 6A) and transgenic GFP (Figure 6C) showed very clear differences between the two labels. A large, open, lymphatic-like structure closely apposed to an airway, was strongly positive for mouse podoplanin but negative for GFP (asterisks in Figure 6). Neighboring alveoli contained both endogenous mouse podoplanin and transgenic GFP in type I cells. This structure appears to be a lymphatic vessel, although we could not use LYVE-1 staining to confirm this because the antigen retrieval process used for LYVE-1 detection (33) destroyed native GFP fluorescence and antigenicity.
Figure 6.
Pulmonary expression of GFP transgene is more restricted than the endogenous mouse podoplanin gene in RTIbacGFP-transgenic lung. RTIbacGFP-transgenic lung sections were co-stained for mouse podoplanin (red in A and B) and GFP (green in C and D). The corresponding phase contrast images are in E and F. Note extensive mouse podoplanin staining (red) in lymphatic-like structure (asterisk in A) and lymphoid-associated stroma in B. Both are negative for GFP expression. GFP and mouse podoplanin are co-expressed in type I cells within alveoli (A and C); TI, type I cells; TII, type II cells; BV, blood vessel. Scale bar indicates 10 μm.
Lymphoid-associated stroma.
Among the early descriptions of mouse podoplanin was gp38, a glycoprotein expressed in T cell zone stroma of lymphoid organs (15). Development and function of this important lymphoid tissue component are routinely monitored with the same anti-mouse podoplanin (gp38) monoclonal antibody that we have used here (41, 42). Although pulmonary foci of lymphocytic infiltration are well described (for example, see Ref. 43), the associated stromal regions that support these infiltrates have not, to our knowledge, been characterized but would likely express mouse podoplanin. We emphasize the putative nature of this pulmonary T cell zone stroma because, until it is characterized in more detail, it is only postulated to exist.
We noted mouse podoplanin staining over several areas consistent with lymphoid-associated stroma (43–45). One (Figures 6B, 6D, and 6F) was airway-apposed and contained a loose reticular network of mouse podoplanin-positive cells (Figure 6B) that were negative for GFP (Figure 6D). There were similar areas in the two serial sections shown in Figure 5. As described previously, LYVE-1 labeled the lymphatic endothelium in both sections (Figures 5B and 5F). One section was labeled for endogenous mouse podoplanin (Figure 5C) and the other for transgenic rat podoplanin (Figure 5G). An extensive mouse podoplanin-positive area immediately adjacent to the airway epithelium (Figure 5C, long arrows) was negative for both rat podoplanin (Figure 5G, long arrows) and LYVE-1 (Figure 5F). The center of this region (Figure 5B, middle long arrow), closely associated with the LYVE-1–positive lymphatic, had a reticular pattern similar to that shown in Figure 6B. In contrast, on either side of the reticular area, structures resembling collapsed lymphatic vessels extended parallel to the adjacent airway epithelium but, again, did not stain for LYVE-1 (Figure 5B, top and bottom long arrows; see also Ref. 39). Thus, in this transgenic line, endogenous mouse podoplanin but not transgenic rat podoplanin or its GFP surrogate identified a cellular compartment spatially distinct from alveoli that may correlate with T cell zone stroma described in lymphoid organs (15, 41, 42). Coupled with the reduced staining for transgene expression in pleura and lymphatic endothelium, the complete lack of rat podoplanin staining in this putative lymphoid associated stromal compartment further supports our conclusion that RTIbac confers a more restrictive pattern of podoplanin expression in pulmonary tissue than does the endogenous mouse gene. Table 1 summarizes these observations.
TABLE 1.
DIFFERENCES BETWEEN ANTIBODY STAINING FOR RAT AND MOUSE PODOPLANIN IN TRANSGENIC LUNG
| Site | Mouse Podoplanin | Rat Podoplanin |
|---|---|---|
| Alveolar epithelium | Strong | Strong |
| Lymphoid-associated stroma | Strong | Absent |
| Pleura | Very strong | Weak |
| Lymphatic endothelium | Strong | Moderate |
Transgene Expression in Extrapulmonary Organs
We compared rat podoplanin mRNA levels in lung, brain, and kidney from normal rat, line 9, and wild-type mice using rat-specific primers in real time RT-PCR reactions (Table 2). We expressed our results as the percentage of podoplanin RNA in lung (either rat or line 9) following standardization to the amount of 18S ribosomal RNA in each sample. The data showed that podoplanin mRNA levels in normal rat brain and kidney were 4.2% and 3.6% of those of rat lung. Similarly, rat podoplanin mRNA levels in line 9 brain and kidney were 4.8% and 3.0% of those in line 9 lung. On the other hand, absolute levels of rat podoplanin mRNA expression in line 9 lung, brain, and kidney were 5.9-, 4.9-, and 6.6-fold higher than in the corresponding rat organs. This generally uniform increase in transgene transcript level may reflect the corresponding increase in RTIbac insert copy number in line 9. We detected no rat podoplanin mRNA in wild-type mouse organs, thereby demonstrating the specificity of the PCR primers and probe (not shown).
TABLE 2.
REAL-TIME RT-PCR MEASUREMENT OF RAT PODOPLANIN mRNA LEVELS IN NORMAL RAT AND LINE 9 MOUSE TISSUES
| Normal Rat
|
Line 9 Mouse
|
Ratio
|
|||
|---|---|---|---|---|---|
| Tissue | pg* (± SD) | % of Lung | pg* (± SD) | % of Lung | Line 9:Normal Rat† |
| Lung | 17.1 ± 3.7 | 100 | 100.7 ± 25.6 | 100 | 5.9 |
| Brain | 0.7 ± 0.02 | 4.2 | 4.8 ± 2.2 | 4.8 | 4.9 |
| Kidney | 0.6 ± 0.06 | 3.6 | 3.0 ± 0.5 | 3.0 | 6.6 |
Picograms of rat podoplanin cDNA per 50 ng total cDNA transcribed from total RNA of the indicated normal rat or line 9 mouse tissue. Data represent the average ± SD on triplicate assays of the RNA isolated from each of three organ samples, each isolated from a different animal.
Ratio of rat podoplanin cDNA measured in line 9 tissue to that in normal rat tissue.
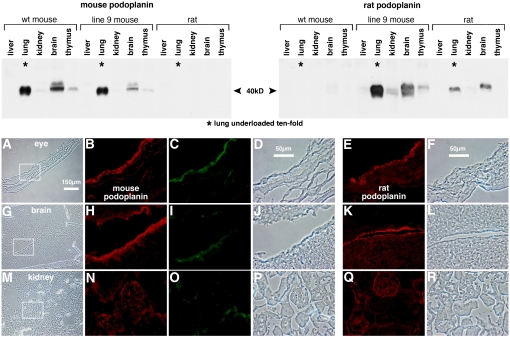
Rat and mouse podoplanin protein expression in various organs of line 9 and wild-type mice as well as normal rats was examined on the Western blots shown at the top of Figure 7. Both proteins migrated on SDS gels with apparent molecular weights of 40 kD (5, 15) and were detected separately by the two antibodies with no evidence of cross reactivity on podoplanin of the inappropriate species. Extremely strong podoplanin expression in lung compared with other organs led us to load 10-fold less protein on lung lanes to quantitate the relative band intensities lung and brain. The signals in the other lanes (liver, kidney, and thymus) were too low for reliable sample quantitation. The pattern of rat podoplanin expression was very similar between rat and line 9 mouse organs with very high expression in lung, 10- to 20-fold less in brain, and extremely low levels in kidney and thymus. The expression pattern of the endogenous mouse protein was similar with highest expression in lung, 10- to 20-fold lower in brain, and extremely low levels in kidney and thymus (Figure 7). Expression of either protein in liver was consistently at background levels. Absolute levels of transgenic rat podoplanin in line 9 lung and brain were 2- to 4-fold higher than in the corresponding normal rat organs and correlated with similar increases in mRNA levels (Table 2). Signals in kidney, thymus, and liver were too low to make similar comparisons between line 9 and normal rat. Transgene overexpression did not appear to affect endogenous podoplanin gene expression, since levels of the mouse protein in the various organs were indistinguishable between line 9 and wild-type mice. We conclude that both genes are expressed in similar relative amounts in the organs examined and, furthermore, that the same relative pattern is expressed by the RTIbac rat podoplanin transgene. On a separate Western blot (not shown), we determined that the RTIbacGFP line expressed less rat podoplanin in lung than line 9 and approximately the same amount as in normal rat lung.
Figure 7.
Podoplanin expression in extrapulmonary organs. (Top) Western blot analysis of rat and mouse podoplanin expression in wild-type mouse, line 9 transgenic mouse, and normal rat organs. Duplicate blots of total protein extracted from the indicated organs were probed with antibodies against mouse (left) and rat (right) podoplanin. One hundred sixty micrograms of protein per lane were loaded except for lung lanes, which were 10-fold underloaded (16 μg) due to high podoplanin levels in this tissue, a difference indicated with asterisks in lung lanes. In lung and brain lanes, chemiluminesence in the 40-kD bands was quantified, corrected for the 10-fold loading factor and normalized to expression in wild-type mouse lung (for lanes in the left panel) or normal rat lung (for lanes in the right panel). The values for mouse podoplanin in wild-type mouse lung and brain are 100% and 7.5%, and for mouse podoplanin in line 9 lung and brain, 88% and 3.6%, respectively. The corresponding values for rat podoplanin in line 9 lung and brain are 416% and 20% and for rat podoplanin in normal rat lung and brain, 100% and 8.8%, respectively. The signal to noise ratios in the remaining lanes were too high to reliably quantitate protein levels in these organs. Rat and mouse podoplanin have similar patterns of expression in the organs examined. The RTIbac rat podoplanin transgene is expressed in a similar pattern. (Bottom) Cell specificity of mouse and rat podoplanin and GFP expression in RTIbacGFP transgenic organs. Frozen thin sections of eye (top row), brain (middle row), and kidney (bottom row) were co-stained for mouse podoplanin (red in B, H, and N) and GFP (green in C, I, and O); corresponding phase contrast images are in D, J, and P. These images are enlarged from the areas outlined in the low magnification phase contrast views in A, G, and M. Separate tissue sections stained for rat podoplanin are shown in E, K, and Q (red) along with corresponding phase contrast images in F, L, and R. The rat podoplanin transgene shares the same cell specificity of expression in these organs as the endogenous mouse podoplanin gene. GFP transgene expression in kidney was barely above background but overlapped with mouse podoplanin expression in eye and brain.
The transgenic mRNA and protein expression profile in extrapulmonary organs suggested that RTIbac transgene expression was accurately regulated in line 9. We were curious to determine if the BAC conveyed cell-specific expression within some of these organs. For this purpose, we examined RTIbacGFP mice at three well-characterized sites of podoplanin expression: choroid plexus, eye ciliary epithelium, and renal podocytes (Figure 7, bottom). Endogenous mouse (Figures 7B, 7H, and 7N) and transgenic rat (Figures 7E, 7K, and 7Q) podoplanin were detected in all three locations with prominent staining for both proteins in choroid plexus and ciliary epithelium and rather diffuse staining in kidney. These patterns were very similar to those reported previously for rat and mouse proteins (6, 11, 46). Background staining of the rat anti-podoplanin antibody on wild-type or transgenic mouse organs was higher than with the mouse anti-podoplanin antibody, perhaps due to Fc-receptor binding of the mouse anti-rat podoplanin monoclonal antibody (Figures 7E, 7K, and 7Q, wild-type staining not shown). GFP expression paralleled that of mouse and transgenic rat podoplanin in ciliary epithelium and choroid plexus, but was extremely weak in kidney. The combination of real-time PCR, Western blot, and immunofluorescence microscopy data (Table 2 and Figure 7) indicates that RTIbac and its IRES-GFP derivative convey cell-specificity of transgene expression in extrapulmonary organs that closely correlates with expression of the mouse and rat podoplanin genes.
DISCUSSION
RTIbac is an effective vector for directing transgene expression to type I cells in the mouse alveolar epithelium. Using the native rat podoplanin gene and IRES-GFP derivatives, we demonstrated prominent transgene expression in type I cells, whereas type II cells and airways were negative. Transgenes showed reduced or no expression in pleura, lymphatic endothelium, and lymphoid-associated stroma, all three sites of strong endogenous mouse podoplanin gene expression in the lung. In nonpulmonary tissues, expression was consistent with previous reports of podoplanin in brain choroid plexus, optic cup ciliary epithelium, and renal glomerulus. Levels of transgenic rat podoplanin mRNA and protein paralleled those of the native rat gene in all organs examined. The highly specific transgene expression in adult type I cells indicates that additional RTIbac derivatives will be useful for type I cell biology experiments requiring ectopic gene delivery to this important cell type that covers more than 95% of the alveolar epithelium.
Immunofluorescence microscopy identified three regional differences in lung between transgenic rat and endogenous mouse podoplanin expression that support our conclusion of heightened specificity for the rat transgene. First, the transgene was not expressed in a class of cells closely apposed to airway epithelium that was positive for mouse podoplanin. These cells did not express a lymphatic marker, LYVE-1, although LYVE-1–positive lymphatics were often in close proximity. A lymphoid-associated stromal identity for these cells, though speculative, is consistent with both expression of mouse podoplanin on T cell zone stroma in peripheral lymphoid tissues (15, 41, 42) and observations of lymphoid accumulations in similar subepithelial airway regions during infection and lung injury (43–45). A second difference was noted in lymphatic endothelium, where mouse podoplanin staining was equivalent to that of type I cells, whereas by the same standard, staining for the rat transgene was reduced. Finally, RTIbac conferred a rat pattern of low podoplanin expression in pleura relative to the adjacent alveolar epithelium. In contrast, expression of mouse podoplanin was consistently stronger in pleura than in type I cells. Therefore, of the four distinct pulmonary cell types that expressed mouse podoplanin, all at roughly comparable levels (type I, pleura, lymphatic endothelium, and presumptive lymphoid stroma), only type I cells expressed the rat transgene at high levels. The remaining four transgene-expressing lines were not extensively studied for pleura, lymphatic, and lymphoid stroma expression, although the preliminary screen of these lines (Figure 1) appeared similar to line 9 with the exception of type I cell expression levels.
Because our immunofluorescence microscopy used two different podoplanin antibodies, one for mouse, the other for rat, we could not directly compare staining for the two proteins on the same cell. Instead, we relied on reference to type I cell staining in nearby alveoli of the same section. In Figure 4, for example, we could not conclude that there was more mouse than rat podoplanin in line 9 pleura, only that there was more mouse podoplanin staining in pleura than in type I cells and, similarly, that there was less rat podoplanin staining in pleura than in type I cells. By using GFP as a second measure of transgene expression, we sought to minimize potential epitope access problems resulting, for example, from protein modification or alternative splicing. This approach succeeded to an extent; however, native GFP fluorescence was weak, especially in extrapulmonary organs, and the protein was best visualized with an anti-GFP antibody. Inefficient use of the IRES element in the transgenic mRNA is a possible explanation for this result (47). In principle, an RTIbac derivative containing a podoplanin–GFP fusion protein construct might produce a brighter GFP signal.
In contrast to greater type I cell specificity in lung, transgene expression in extrapulmonary organs appeared rather similar to the endogenous gene. By immunofluorescence microscopy, the rat podoplanin transgene and the endogenous mouse podoplanin gene were expressed in similar cell populations in choroid plexus, eye ciliary epithelium, and renal glomerulus. An exhaustive analysis of transgene expression in other mouse organs was beyond the scope of the present study. In particular, we did not examine expression in osteocytes (12, 13), keratinocytes (9), extrapulmonary lymphatics (11, 22, 38), glutamatergic neurons (10), or the stroma in spleen and thymus (15, 41, 42), all documented sites of either rat or mouse podoplanin expression. At the whole organ level, the ratio of lung to extrapulmonary organ expression on Western blots was generally similar for both proteins. Finally, the 25:1 ratio of lung to brain or kidney for rat podoplanin mRNA content was very similar between line 9 and normal rat organs, indicating that native and transgenic rat podoplanin genes are regulated similarly. Absolute mRNA levels of rat podoplanin were 4.9- to 6.6-fold higher in line 9 than in normal rat for all three organs examined, presumably due to increased podoplanin gene copy number in line 9. Our transgenic results are consistent with previously published descriptions of podoplanin levels in various organs of wild-type mice and rats (6, 10, 11, 13, 48). In contrast, the human and dog podoplanin genes have a broader range of tissue expression and lack the strong pulmonary bias of the mouse and rat genes (8, 17, 49). The mechanism of increased type I cell specificity of transgenic rat podoplanin expression is unknown. Isolated rat type I cells contain significantly greater amounts of podoplanin mRNA than isolated type II cells, indicating that regulation at the level of transcript initiation, processing, or stability contributes to differences in podoplanin levels between the two cell types (4). In addition, translational and/or post-translational mechanisms for podoplanin appear to exist since kidney and brain have nearly identical mRNA levels, yet express very different levels of podoplanin protein (Figure 7 and Table 2). Additional experiments with isolated type I cells as well as lymphatic endothelium, lymphoid stroma, and pleural cell populations from transgenic mice may be able to address some of these questions.
The function of podoplanin is unknown. Mice carrying homozygous deletions of the gene die shortly after birth from respiratory distress, but also have defects in lymphatic vessel function, the latter leading to severe edema (12, 18, 19). Epithelial mucin properties of podoplanin suggest roles in maintaining surface hydration or establishing protective zones above epithelial or endothelial cell layers (22). Overexpression of podoplanin in cultured cells generates changes in cell shape and motility, an activity mediated by interaction of ERM family proteins (Ezrin, Radixin, Moesin) with a defined motif in the cytoplasmic domain of podoplanin (21, 22). Podoplanin expression by tumor cells causes platelets to aggregate on the tumor cell surface, thereby facilitating metastases (20). Recent studies in a RIP-TAG transgenic tumor progression model implicate podoplanin in an unusual form of tissue invasion that does not require a canonical epithelial–mesenchymal transition (23). Whether these highly disparate properties reflect single or multiple cellular functions for podoplanin is unclear.
The transgenic lines described here overexpress podoplanin, albeit with differences in species of origin and regulation of expression. We have observed no obvious abnormalities in line 9 mice of up to a year in age and beyond, although more precise functional testing may be necessary to detect differences. This normal phenotype indicates that podoplanin overexpression is tolerated and, perhaps more importantly, that the remaining 140 kbp of nonpodoplanin genomic DNA in RTIbac are not deleterious to transgenic animals. In addition to podoplanin, RTIbac contains a downstream α-tubulin pseudogene, a truncated Pramel7 gene at one end of the insert and a complete but uncharacterized gene, LOC313681, positioned 50 kbp upstream from the podoplanin transcription start site. We have yet to investigate expression of these sequences in our transgenic lines.
A clear advantage of BAC vectors is their ability to confer accurate transgene expression without the complications of position effect and copy number variation inherent in small DNA fragment–mediated transgenesis (27). Transgenic RTIbac podoplanin expression was highly efficient, with five of six founder lines expressing podoplanin accurately in type I cells. The lone expression-negative founder line may reflect a disrupted podoplanin gene or regulatory element in the integrated RTIbac DNA. Line 9 has four to eight times more transgene copies than in the rat genome and expressed from 4.9- to 6.6-fold more podoplanin RNA in the three tissues examined. The other transgenic lines described in Figure 1 contained between one and eight times the copy number of the rat genome, but we have not physically characterized the integrated transgene locus in any of these lines.
The general utility of BAC vectors is somewhat hindered by the need for homologous recombination based strategies instead of traditional DNA ligase approaches to make even simple modifications (27). However, recent advances in recombination methodologies, particularly in the lambda RED/galK system, have dramatically simplified BAC modification (50). These techniques should ease replacement of podoplanin coding sequences in RTIbac with other cDNAs of interest, permit targeted mutation of selected regulatory elements to further improve the specificity of transgene expression in type I cells, and facilitate construction of RTIbac derivatives containing multiple functionalities on a single BAC. Our characterization of the ability of RTIbac vectors to deliver transgenes to type I cells represents an important step toward these goals.
Acknowledgments
The authors thank Nathaniel Heintz for providing pLD53SCAEB and advice on BAC modification.
This work was supported in part by National Institutes of Health grants HL-24075 and HL-57426 (to L.G.D.).
Originally Published in Press as DOI: 10.1165/rcmb.2008-0049OC on March 26, 2008
References
- 1.Whitsett JA, Glasser SW, Tichelaar JW, Perl AK, Clark JC, Wert SE. Transgenic models for study of lung morphogenesis and repair: Parker B. Francis lecture. Chest 2001;120:27S–30S. [DOI] [PubMed] [Google Scholar]
- 2.Stone KC, Mercer RR, Freeman BA, Chang LY, Crapo JD. Distribution of lung cell numbers and volumes between alveolar and nonalveolar tissue. Am Rev Respir Dis 1992;146:454–456. [DOI] [PubMed] [Google Scholar]
- 3.Dobbs LG, Williams MC, Gonzalez R. Monoclonal antibodies specific to apical surfaces of rat alveolar type I cells bind to surfaces of cultured, but not freshly isolated, type II cells. Biochim Biophys Acta 1988;970:146–156. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez R, Yang YH, Griffin C, Allen L, Tigue Z, Dobbs L. Freshly isolated rat alveolar type I cells, type II cells, and cultured type II cells have distinct molecular phenotypes. Am J Physiol Lung Cell Mol Physiol 2005;288:L179–L189. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez RF, Dobbs LG. Purification and analysis of RTI40, a type I alveolar epithelial cell apical membrane protein. Biochim Biophys Acta 1998;1429:208–216. [DOI] [PubMed] [Google Scholar]
- 6.Breiteneder-Geleff S, Matsui K, Soleiman A, Meraner P, Poczewski H, Kalt R, Schaffner G, Kerjaschki D. Podoplanin, novel 43-kd membrane protein of glomerular epithelial cells, is down-regulated in puromycin nephrosis. Am J Pathol 1997;151:1141–1152. [PMC free article] [PubMed] [Google Scholar]
- 7.Williams MC, Dobbs LG. Expression of cell-specific markers for alveolar epithelium in fetal rat lung. Am J Respir Cell Mol Biol 1990;2:533–542. [DOI] [PubMed] [Google Scholar]
- 8.Ma T, Yang B, Matthay MA, Verkman AS. Evidence against a role of mouse, rat, and two cloned human t1alpha isoforms as a water channel or a regulator of aquaporin-type water channels. Am J Respir Cell Mol Biol 1998;19:143–149. [DOI] [PubMed] [Google Scholar]
- 9.Gandarillas A, Scholl FG, Benito N, Gamallo C, Quintanilla M. Induction of PA2.26, a cell-surface antigen expressed by active fibroblasts, in mouse epidermal keratinocytes during carcinogenesis. Mol Carcinog 1997;20:10–18. [DOI] [PubMed] [Google Scholar]
- 10.Kotani M, Tajima Y, Osanai T, Irie A, Iwatsuki K, Kanai-Azuma M, Imada M, Kato H, Shitara H, Kubo H, et al. Complementary DNA cloning and characterization of RANDAM-2, a type I membrane molecule specifically expressed on glutamatergic neuronal cells in the mouse cerebrum. J Neurosci Res 2003;73:603–613. [DOI] [PubMed] [Google Scholar]
- 11.Wetterwald A, Hoffstetter W, Cecchini MG, Lanske B, Wagner C, Fleisch H, Atkinson M. Characterization and cloning of the E11 antigen, a marker expressed by rat osteoblasts and osteocytes. Bone 1996;18:125–132. [DOI] [PubMed] [Google Scholar]
- 12.Zhang K, Barragan-Adjemian C, Ye L, Kotha S, Dallas M, Lu Y, Zhao S, Harris M, Harris SE, Feng JQ, et al. E11/gp38 selective expression in osteocytes: regulation by mechanical strain and role in dendrite elongation. Mol Cell Biol 2006;26:4539–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nose K, Saito H, Kuroki T. Isolation of a gene sequence induced later by tumor-promoting 12-O-tetradecanoylphorbol-13-acetate in mouse osteoblastic cells (MC3T3–E1) and expressed constitutively in ras-transformed cells. Cell Growth Differ 1990;1:511–518. [PubMed] [Google Scholar]
- 14.Watanabe M, Okochi E, Sugimoto Y, Tsuruo T. Identification of a platelet-aggregating factor of murine colon adenocarcinoma 26: Mr 44,000 membrane protein as determined by monoclonal antibodies. Cancer Res 1988;48:6411–6416. [PubMed] [Google Scholar]
- 15.Farr AG, Berry ML, Kim A, Nelson AJ, Welch MP, Aruffo A. Characterization and cloning of a novel glycoprotein expressed by stromal cells in T-dependent areas of peripheral lymphoid tissues. J Exp Med 1992;176:1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmer G, Lottspeich F, Maisner A, Klenk HD, Herrler G. Molecular characterization of gp40, a mucin-type glycoprotein from the apical plasma membrane of Madin-Darby canine kidney cells (type I). Biochem J 1997;326:99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmer G, Oeffner F, Von Messling V, Tschernig T, Groness HJ, Klenk HD, Herrler G. Cloning and characterization of gp36, a human mucin-type glycoprotein preferentially expressed in vascular endothelium. Biochem J 1999;341:277–284. [PMC free article] [PubMed] [Google Scholar]
- 18.Ramirez MI, Millien G, Hinds A, Cao Y, Seldin DC, Williams MC. T1alpha, a lung type I cell differentiation gene, is required for normal lung cell proliferation and alveolus formation at birth. Dev Biol 2003;256:61–72. [DOI] [PubMed] [Google Scholar]
- 19.Schacht V, Ramirez MI, Hong YK, Hirakawa S, Feng D, Harvey N, Williams M, Dvorak AM, Dvorak HF, Oliver G, et al. T1alpha/podoplanin deficiency disrupts normal lymphatic vasculature formation and causes lymphedema. EMBO J 2003;22:3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunita A, Kashima TG, Morishita Y, Fukayama M, Kato Y, Tsuruo T, Fujita N. The platelet aggregation-inducing factor aggrus/podoplanin promotes pulmonary metastasis. Am J Pathol 2007;170:1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin-Villar E, Megias D, Castel S, Yurrita MM, Vilaro S, Quintanilla M. Podoplanin binds ERM proteins to activate RhoA and promote epithelial-mesenchymal transition. J Cell Sci 2006;119:4541–4553. [DOI] [PubMed] [Google Scholar]
- 22.Scholl FG, Gamallo C, Vilaro S, Quintanilla M. Identification of PA2.26 antigen as a novel cell-surface mucin-type glycoprotein that induces plasma membrane extensions and increased motility in keratinocytes. J Cell Sci 1999;112:4601–4613. [DOI] [PubMed] [Google Scholar]
- 23.Wicki A, Lehembre F, Wick N, Hantusch B, Kerjaschki D, Christofori G. Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell 2006;9:261–272. [DOI] [PubMed] [Google Scholar]
- 24.Ramirez MI, Cao YX, Williams MC. 1.3 kilobases of the lung type I cell T1alpha gene promoter mimics endogenous gene expression patterns during development but lacks sequences to enhance expression in perinatal and adult lung. Dev Dyn 1999;215:319–331. [DOI] [PubMed] [Google Scholar]
- 25.Ramirez MI, Rishi AK, Cao YX, Williams MC. TGT3, thyroid transcription factor I, and Sp1 elements regulate transcriptional activity of the 1.3-kilobase pair promoter of T1alpha, a lung alveolar type I cell gene. J Biol Chem 1997;272:26285–26294. [DOI] [PubMed] [Google Scholar]
- 26.Vanderbilt JN, Dobbs LG. Characterization of the gene and promoter for RTI40, a differentiation marker of type I alveolar epithelial cells. Am J Respir Cell Mol Biol 1998;19:662–671. [DOI] [PubMed] [Google Scholar]
- 27.Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat Rev Neurosci 2001;2:861–870. [DOI] [PubMed] [Google Scholar]
- 28.Vanderbilt JN, Allen L, Tigue Z, Gonzalez R, Edmondson J, Dobbs L. Directed expression of a transgene to type I cells in the mouse. Am J Respir Crit Care Med 2007;175:A743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA 1992;89:8794–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res 2002;12:1992–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filutowicz M, Rakowski SA. Regulatory implications of protein assemblies at the gamma origin of plasmid R6K: a review. Gene 1998;223:195–204. [DOI] [PubMed] [Google Scholar]
- 32.Wong CJ, Akiyama J, Allen L, Hawgood S. Localization and developmental expression of surfactant proteins D and A in the respiratory tract of the mouse. Pediatr Res 1996;39:930–937. [DOI] [PubMed] [Google Scholar]
- 33.Prevo R, Banerji S, Ferguson DJ, Clasper S, Jackson DG. Mouse LYVE-1 is an endocytic receptor for hyaluronan in lymphatic endothelium. J Biol Chem 2001;276:19420–19430. [DOI] [PubMed] [Google Scholar]
- 34.Johnson MD, Bao HF, Helms MN, Chen XJ, Tigue Z, Jain L, Dobbs LG, Eaton DC. Functional ion channels in pulmonary alveolar type I cells support a role for type I cells in lung ion transport. Proc Natl Acad Sci USA 2006;103:4964–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schacht V, Dadras SS, Johnson LA, Jackson DG, Hong YK, Detmar M. Up-regulation of the lymphatic marker podoplanin, a mucin-type transmembrane glycoprotein, in human squamous cell carcinomas and germ cell tumors. Am J Pathol 2005;166:913–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millien G, Spira A, Hinds A, Wang J, Williams MC, Ramirez MI. Alterations in gene expression in T1 alpha null lung: a model of deficient alveolar sac development. BMC Dev Biol 2006;6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ordonez NG. D2–40 and podoplanin are highly specific and sensitive immunohistochemical markers of epithelioid malignant mesothelioma. Hum Pathol 2005;36:372–380. [DOI] [PubMed] [Google Scholar]
- 38.Breiteneder-Geleff S, Soleiman A, Kowalski H, Horvat R, Amann G, Kriehuber E, Diem K, Weninger W, Tschachler E, Alitalo K, et al. Angiosarcomas express mixed endothelial phenotypes of blood and lymphatic capillaries: podoplanin as a specific marker for lymphatic endothelium. Am J Pathol 1999;154:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leak LV, Jamuar MP. Ultrastructure of pulmonary lymphatic vessels. Am Rev Respir Dis 1983;128:S59–S65. [DOI] [PubMed] [Google Scholar]
- 40.Favre CJ, Mancuso M, Maas K, McLean JW, Baluk P, McDonald DM. Expression of genes involved in vascular development and angiogenesis in endothelial cells of adult lung. Am J Physiol Heart Circ Physiol 2003;285:H1917–H1938. [DOI] [PubMed] [Google Scholar]
- 41.Ngo VN, Cornall RJ, Cyster JG. Splenic T zone development is B cell dependent. J Exp Med 2001;194:1649–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Withers DR, Kim MY, Bekiaris V, Rossi SW, Jenkinson WE, Gaspal F, McConnell F, Caamano JH, Anderson G, Lane PJ. The role of lymphoid tissue inducer cells in splenic white pulp development. Eur J Immunol 2007;37:3240–3245. [DOI] [PubMed] [Google Scholar]
- 43.Hawgood S, Ochs M, Jung A, Akiyama J, Allen L, Brown C, Edmondson J, Levitt S, Carlson E, Gillespie AM, et al. Sequential targeted deficiency of SP-A and -D leads to progressive alveolar lipoproteinosis and emphysema. Am J Physiol Lung Cell Mol Physiol 2002;283:L1002–L1010. [DOI] [PubMed] [Google Scholar]
- 44.Botas C, Poulain F, Akiyama J, Brown C, Allen L, Goerke J, Clements J, Carlson E, Gillespie AM, Epstein C, et al. Altered surfactant homeostasis and alveolar type II cell morphology in mice lacking surfactant protein D. Proc Natl Acad Sci USA 1998;95:11869–11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giannoni E, Sawa T, Allen L, Wiener-Kronish J, Hawgood S. Surfactant proteins A and D enhance pulmonary clearance of Pseudomonas aeruginosa. Am J Respir Cell Mol Biol 2006;34:704–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams MC, Cao Y, Hinds A, Rishi AK, Wetterwald A. T1 alpha protein is developmentally regulated and expressed by alveolar type I cells, choroid plexus, and ciliary epithelia of adult rats. Am J Respir Cell Mol Biol 1996;14:577–585. [DOI] [PubMed] [Google Scholar]
- 47.Jackson RJ, Howell MT, Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci 1990;15:477–483. [DOI] [PubMed] [Google Scholar]
- 48.Rishi AK, Joyce-Brady M, Fisher J, Dobbs LG, Floros J, VanderSpek J, Brody JS, Williams MC. Cloning, characterization, and development expression of a rat lung alveolar type I cell gene in embryonic endodermal and neural derivatives. Dev Biol 1995;167:294–306. [DOI] [PubMed] [Google Scholar]
- 49.Martin-Villar E, Scholl FG, Gamallo C, Yurrita MM, Munoz-Guerra M, Cruces J, Quintanilla M. Characterization of human PA2.26 antigen (T1alpha-2, podoplanin), a small membrane mucin induced in oral squamous cell carcinomas. Int J Cancer 2005;113:899–910. [DOI] [PubMed] [Google Scholar]
- 50.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 2005;33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]