Abstract
The molecular mechanisms that govern the transcription of human extracellular superoxide dismutase (EC-SOD), the major extracellular antioxidant enzyme, are largely unknown. To elucidate the mechanisms involved in human EC-SOD gene regulation and expression, we localized multiple transcription start sites to a finite region located 3.9 kb upstream of the ATG initiation codon. Within this segment, we subcloned a 2.7-kb fragment upstream of a luciferase reporter gene; the resulting construct exhibited strong in vivo promoter activity in two lung-derived cell lines. Deletion analysis of the EC-SOD 5′-flanking sequences identified a minimal 0.3-kb region that had strong basal promoter activity. Computer sequence analysis revealed a putative Sp1-like binding site within the EC-SOD proximal promoter region that lacked a TATA-box and showed a high frequency of GC nucleotides. Binding of Sp1 and Sp3 transcription factors to the EC-SOD promoter was confirmed by DNase I footprint analysis, electophoretic mobility shift assay, and competition and supershift assays. Cotransfection of the EC-SOD promoter–luciferase reporter constructs with plasmids encoding Sp1 and Sp3 into Sp-deficient insect SL2 cells showed strong activation of luciferase gene expression. The occupancy of the EC-SOD promoter by Sp1/Sp3 and RNA polymerase II in vivo was determined by chromatin immunoprecipitation assay and correlated well with levels of EC-SOD expression in lung epithelial cells (A549) and pulmonary fibroblasts (MRC5). Collectively, our results demonstrate the important role Sp1 and Sp3 plays in regulating the expression of human EC-SOD in the lung.
Keywords: extracellular superoxide dismutase, promoter, transcription, Sp1 gene family, antioxidant
CLINICAL RELEVANCE
We have identified key regulatory elements for human extracellular superoxide dismutase (EC-SOD), the major antioxidant enzyme in the lung. Since oxidative stress contributes to the development of lung disease, understanding factors that regulate EC-SOD may provide novel therapeutic strategies.
Superoxide dismutases (CuZn-SOD, Mn-SOD, and EC-SOD) represent major defenses against oxidative stress in mammals, and a deficiency in these enzymes may cause or exacerbate a variety of diseases (1, 2). Extracellular superoxide dismutase (EC-SOD) is the least studied enzyme, but recent data support an important role for EC-SOD in maintaining oxidative homeostasis within extracellular matrix elements and in other extracellular spaces. Posttranslational modification of EC-SOD involves glycosylation in the Golgi apparatus, proteolytic processing by a furin protease, and secretion into the extracellular milieu were it exists in solution or bound to extracellular matrix components (such as collagen) via its C-terminal, negatively charged heparin binding domain (3). It has been postulated that a high local concentration of EC-SOD near extracellular matrix elements and cell surfaces helps protect against oxidative stress by lowering levels of superoxide anion radicals.
Expression of EC-SOD has been localized to specific cells and tissues, with the highest expression occurring in lung, heart, kidney, and vasculature. Disruption of the EC-SOD gene in mice does not produce obvious pathologies under normal conditions, but these mice are more prone to environmental stressors after exposure to hyperoxia (4, 5), radiation (6), or pulmonary fibrogenic chemicals (7, 8). Recently, we reported that basal and inducible transcription of the murine EC-SOD gene is regulated, at least in part, by interactions between a proximal promoter element involving Sp1/Sp3 transcription factors (9), with distal promoter elements involving Ets, Kruppel, and MZF-1 transcription factors (10). The gene for human EC-SOD is located on chromosome 4 (region 4p-q21) and spans approximately 5,900 bp. It consists of three exons separated by two introns (11, 12). The complete coding region is located within exon 3 and spans 1,336 bp. Exon 1 is 5 bp long, and it is separated from exon 2 by 572 bp of intron 1. Based on primitive DNA sequence analysis, it was predicted that the human EC-SOD promoter was TATA-less but that it contained two CAAT box elements located 20 bp and 335 bp upstream of exon 1. The short sequence of exon 1 raised questions about its physiologic relevance and initiated our search for alternative transcriptional start sites and our attempt to map more clearly the promoter that regulates human EC-SOD transcriptional expression.
In this study, we have identified new and dominant transcriptional start sites for the human EC-SOD gene and have identified and characterized its promoter, which is important in regulating basal transcription in lung cell lines and tissue. We found that Sp1/Sp3 transcription factors are the major transactivating factors that bind to a specific cis-element within the human EC-SOD promoter, activating basal gene transcription. Using chromatin immunoprecipitation assays, we show that Sp1/Sp3 interacts with the human EC-SOD promoter in vivo but only in lung fibroblasts that express EC-SOD at a high level.
MATERIALS AND METHODS
Reagents
Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). α-32P-dCTP (3,000 Ci/mmol) was purchased from Amersham (Arlington Heights, IL). Anti-Sp1 (sc-59X), anti-Sp2 (sc-643X), and anti-Sp3 (sc-644X) IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-RNA polymerase II IgG (clone CTD4H8) were purchased from Upstate (Lake Placid, NY). All other chemicals and enzymes were from Boehringer Mannheim (Indianapolis, IN), Sigma Chemical Co. (St. Louis, MO), or Gibco BRL (Gaithersburg, MD).
Cell Culture and Transfections
MRC-5 (human embryonic lung fibroblast cell line), A549 (human lung epithelial cells line), Hep3B (human hepatocellular carcinoma cell line), and 769-P (human kidney renal carcinoma cell line) were obtained from ATCC (Manassas, VA) via the Duke University Cell Culture Facility. Hep3B cells were cultured in Eagle's minimum essential medium supplemented with nonessential amino acids and 10% FBS. A549 cells were cultured in Ham's F12K Nutrient Mixture (Kaighn's modification) with 2 mM l-glutamine and 10% FBS. MRC-5 cells were cultured in Eagle's minimal essential medium with Earle's balanced salt solution and 2 mM l-glutamine supplemented with 10% FBS. 769-P cells were cultured in RPMI 1640 medium with 25 mM Hepes and l-glutamine containing 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, 1 mM sodium pyruvate, and 10% FBS. Cells were maintained at 37°C and 5% CO2 in a humidified incubator.
Transfection assays were performed using LipofectAmine reagent (Invitrogen, Carlsbad, CA). Briefly, A549 and MRC5 cells were seeded at 90% confluence on 24-well plates, incubated overnight before transfection, and treated with LipofectAmine for 5 hours according to the manufacturer's protocol. Twenty-four hours after transfection, cell extracts were assayed for luciferase activity using a dual-luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer's instructions. All experiments were performed at least in triplicate, and the data were normalized to Renilla luciferase to control for differences in transfection efficiency by co-transfecting pRL-CMV plasmid (Promega).
Drosophila SL2 cells were maintained in Schneider's medium supplemented with 10% heat-inactivated FBS at 23°C in air. One day before transfection, SL2 cells were seeded in 24-well plates at 7.8 × 105 cells per well. Cells were transfected with indicated plasmids using LipofectAmine reagent (Invitrogen). Cell extracts were prepared 24 hours after transfection, and promoter activities were determined with the Luciferase Assay System (Promega) and normalized to protein concentration in lysates.
RNA Ligase-Mediated Rapid Amplification of cDNA Ends to Define the Transcription Start Sites for the Human EC-SOD Gene
Ligase-mediated rapid amplification of cDNA ends (RLM-RACE) was performed using the First Choice RLM-RACE Kit (Ambion, Austin, TX) according to the manufacturer's instructions. In brief, total RNA was extracted from MRC5, Hep3B, or 769-P cells or from human heart and lung tissues using Trizol. RNA was treated with calf intestine phosphatase to remove 5′-phosphate from degraded mRNA, rRNA, and DNA and was treated with tobacco acid pyrophosphatase to remove the cap from full-length, intact mRNA. RNA adaptors were ligated to decapped mRNA with T4 RNA ligase and reverse transcribed with Moloney murine leukemia virus reverse transcriptase primed with random decamers. Nested PCR was conducted to amplify the 5′-end of a specific transcript using two nested primers. The primary PCR was conducted using antisense gene-specific primer (5′-GCG AAG AAG GCG TCG AGC T-3′, complementary to nucleotides −293 to −275, with the A nucleotide of ATG designated as position +1) and outer adapter primer 1. The secondary PCR was conducted using antisense gene-specific primer 2 (5′-CCG ACG GCT GCA CCT GGC A-3′, complementary to nucleotides −205 to −187) and inner adapter primer 2. The PCR conditions for primary and secondary PCR were as follows: 95°C for 5 minutes; 35 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute; and 72°C for 7 minutes. The PCR reactions were analyzed on 1.5% agarose gel, and the DNA bands were excised, extracted, and cloned into pCR4-BluntII-TOPO (Invitrogen). The initiation start sites were determined by DNA sequence analysis.
RT-PCR
Total RNA was prepared from the cultured cells using Trizol reagent (Invitrogen). The synthesis of single-stranded DNA from RNA was performed using SuperScript First-Strand Synthesis System for RT-PCR and random hexamers (Invitrogen), according to the protocol provided by manufacturer. The level of mRNA-derived cDNA for a specific gene was quantified using the following gene specific primers: EC-SOD (forward 5′-TGC CCC GCG TCT TCA G-3′ and reverse 5′-CCA AAC ATT CCC CCA AAG G-3′); GAPDH (forward 5′-CAT GGA CTG TGG TCA TGA GT-3′ and reverse 5′-CCA TGT TCG TCA TGG GTG TGA-3′). After amplification, PCR products were separated on a 1.2% agarose gel and visualized using ethidium bromide and UV-light.
Quantitative RT-PCR
To quantitate the abundance of mRNA specific for Sp1 and Sp3 in A549 and MRC5 cells, quantitative PCR was undertaken using the iCycler iQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA) and an iQ SYBR Green Master Mix. The PCR cycles were 95°C for 3 minutes followed by 40 cycles of 95°C for 15 seconds 60°C for 1 minute. The Sp1 primers were forward (5′-TTG AAA AAG GAG TTG GTG GC-3′) and reverse (5′-TGC TGG TTC TGT AAG TTG GG-3′); the Sp3 primers were forward (5′-CCA GGA TGT GGT AAA GTC TA-3′) and reverse (5′-CTC CAT TGT CTC ATT TCC AG-3′). PCR assays were run in triplicate, and Sp1/Sp3 mRNA levels were normalized to GAPDH mRNA levels as described previously.
Plasmids
The human EC-SOD gene's 5′-flanking sequences were generated by a standard PCR technique using Pfu Turbo polymerase (Stratagene, La Jolla, CA) and corresponding primer pairs containing Kpn I and Bgl II restriction sites for the 5′ and 3′ primers, respectively. The amplified DNA was cloned using a pCR-Blunt II-TOPO kit (Invitrogen). The cloned EC-SOD promoter regions were recloned into pGL3-Basic vector (Promega) at Mlu I and Bgl II restriction sites. DNA sequencing confirmed the identity of all constructs.
DNase I Footprinting Assay
A fragment of the EC-SOD gene promoter region (−263 to +45) was amplified from pGL3-hSOD3(−1106/+45) plasmid using corresponding primers with a Mlu I restriction site introduced at the 5′ or 3′ end. Amplified fragments were cloned into pCR2.1-TOPO vector using the TOPO TA cloning kit (Invitrogen). The resulting fragment was excised from plasmid using Mlu I and EcoR I restriction enzymes, separated on a 1.2% agarose gel, and purified using GelExtraction Kit (Qiagen). DNA probes were labeled using 32P-dCTP and Klenow enzyme. A DNase I footprinting assay was performed by incubating a 32P-labeled probe (50,000 cpm) with 10 μg of BSA or nuclear extract from MRC-5 cells at room temperature for 30 minutes in 20 mM Hepes (pH 7.9), 1 mM DTT, 5% glycerol, 0.2 mM EDTA, 0.05% NP-40, and 500 ng poly(dI·dC) in a total volume of 50 μl. After incubation, the samples were treated with three units of DNase I for 5 minutes at room temperature. All samples were phenol extracted, ethanol precipitated, and loaded on a 6% acrylamide, 8 M urea sequencing gel. A sequencing ladder was obtained by loading a sample of the probe that had been treated with formic acid and piperidine to cleave at G and A residues.
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay was performed in 10 μl of 20 mM Hepes (pH 7.9), 1 mM DTT, 5% glycerol, 0.2 mM EDTA, 0.05% NP-40, 32P–end-labeled oligonucleotide probe (40,000 cpm), and 100 ng poly(dI·dC). The reaction was started by adding 2 μg of BSA or nuclear extract. In competition experiments, a 50× and 200× molar excess of unlabeled oligonucleotide was added to the mixtures before the start of the binding reaction. In supershift experiments, the nuclear extracts were preincubated with corresponding IgG for 30 minutes on ice. The free and protein-bound probes were separated on 5% polyacrylamide gel in 0.25× Tris/Borate/EDTA buffer (TBE), then dried and exposed to X-ray film.
Chromatin Immunoprecipitation
The chromatin immunoprecipitation was performed as described previously (9). Briefly, A549 and MRC5 protein–DNA complexes were crosslinked using formaldehyde. Lysed cells were treated with micrococcal S7 nuclease for 6 minutes at 37°C to achieve an estimated DNA size range from 150 bp to 1 kb. The final lysate was incubated with normal RNA polymerase II, Sp1- or Sp3-specific antibodies and precipitated with Protein G-Plus Agarose (Santa Cruz Biotechnology). After extensive washing, DNA-protein complexes were eluted and reverse-crosslinked by incubation at 65°C overnight. DNA was purified using a QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and eluted in 50 μl of Tris/EDTA buffer. The abundance of EC-SOD and GAPDH promoter regions in ChIP precipitates was quantified using PCR and specific primers: EC-SOD primers were sense (5′-AAT CCT TGG CCC GAA AAC CC-3′) and antisense (5′-CAG CCA GCC CAG GAA CGC AG-3′) and amplified region from −237 to −10 bp relative to the transcription initiation start site; GAPDH primers were sense (5′-TAC TAG CGG TTT TAC GGG CG-3′) and antisense (5′-TCG AAC AGG AGG AGC AGA GAG CGA-3′). Products of PCR amplification were separated on 1.2% agarose gel and visualized using ethidium bromide and UV light.
Western Blot
Nuclear extracts from A549 and MRC5 cells were separated on an 8 to 16% SDS-PAGE gel (Invitrogen), transferred to polyvinylidene difluoride membrane, and stained using anti-Sp3 antibodies. The specific protein bands were visualized using a chemiluminescence development kit.
RESULTS
Analysis of EC-SOD mRNA Expression in Different Cells
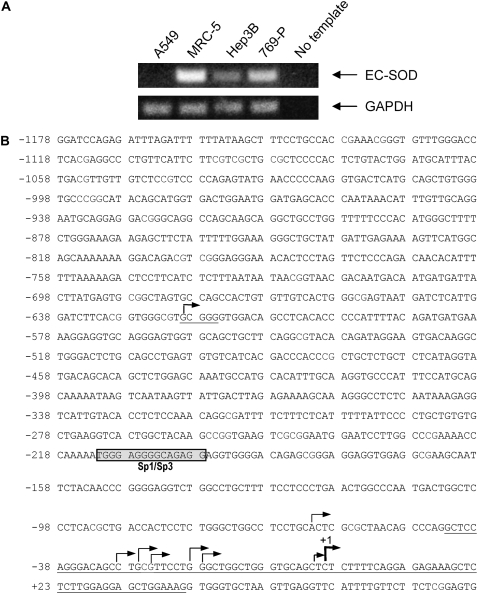
To evaluate baseline levels of EC-SOD expression, we used several distinct human cell lines: human lung airway epithelial carcinoma cells (A549), embryonic lung fibroblasts (MRC-5), hepatocarcinoma (Hep3B), and kidney renal carcinoma (769-P). The level of EC-SOD mRNA was highest in MRC-5 cells, followed by intermediate levels in 769-P and Hep3B and no detectable levels in A549 cells (Figure 1A). To identify and characterize molecular mechanisms involved in governing cell-specific expression of EC-SOD, we studied the two cell lines that express EC-SOD at very high (MRC-5) and very low (A549) levels.
Figure 1.
Extracellular superoxide dismutase (EC-SOD) expression, genomic sequence, and transcriptional start sites. (A) EC-SOD mRNA levels in different cells. Total RNA was purified, reverse transcribed, and amplified with primers specific for EC-SOD or GAPDH. Amplified products were separated on 1.2% agarose gel and visualized with ethidium bromide. (B) Nucleotide sequence of 5′-flanking region of EC-SOD gene. Transcriptional start sites are depicted as arrows, exon sequences are underlined, and the Sp1/Sp3 protected region is boxed. The previously identified transcriptional start site, depicted at position −620, was not confirmed in our experiments.
Identification of Multiple Transcription Start Sites for Human EC-SOD
To determine the transcription start site(s) for the human EC-SOD gene, we used the RLM-RACE method. We analyzed the transcriptional start sites of EC-SOD in MRC-5, Hep3B, and 769-P cells and in human heart and lung tissues. Direct DNA sequence analysis revealed that the transcription start sites for the EC-SOD gene begins at several distinct locations 102, 103, 126, 134, 136, and 137 nucleotides upstream of the ATG start site, with a similar frequency among all cells and tissues analyzed (Figure 1B). Increasing the extension time of the nested PCR did not produce larger PCR products (data not shown).
Functional Characterization of the 5′-Flanking Region of the Human EC-SOD Gene
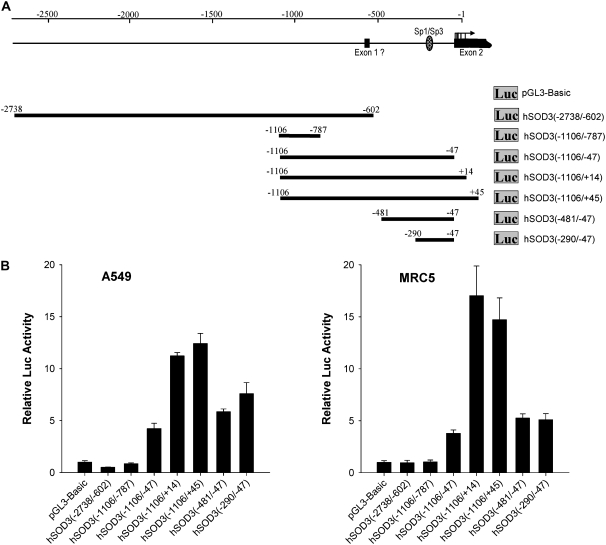
We analyzed if the sequence located upstream of the previously identified transcriptional start site (located at position −620) is able to direct transcription of the reporter gene. For this purpose, DNA containing the 5′-flanking region of the human EC-SOD gene was amplified with a PCR-based method and inserted in front of a luciferase reporter gene (pGL3-Basic). The construct spanning the region from −2738 to −602 of the EC-SOD promoter showed no increase in reporter activity, similar to that seen with the shorter sequence from −1106 to −787 after transfection into A549 and MRC-5 cell (Figure 2A). The extension of this fragment to −47 resulted in significant increases in luciferase reporter activity. Further extension of the 3′ end from −47 to +14 and then +45 gradually increases promoter/reporter activity in both cell lines. Truncations of the sequence at the 5′ end from −1106 to −481 and then to −290 slightly reduced the promoter/reporter activity (Figure 2B). It is not clear from this experiment whether increases in reporter activity observed with the constructs −1106/+14 and −1106/+45 was due to introduction of additional cis-elements, a physical positioning of the transcription initiation complex, or both. These experiments show that major cis-element(s) driving transcription of the promoter/reporter constructs, and probably basal transcription of the EC-SOD gene, is located between nucleotides −290 and −47.
Figure 2.
Functional properties of the EC-SOD promoter fragments in A549 and MRC5 cells. (A) 5′ regions of EC-SOD promoter were fused with the luciferase reporter gene in pGL3-Basic vector. Transcription start sites are depicted as arrows. A Dashed oval represents the putative Sp1/Sp3 binding site. (B) All constructs were transiently transfected into A549 or MRC5 cells, and luciferase activity was measured 24 hours later. Firefly luciferase activities are normalized to Renilla luciferase activity produced by co-transfecting the control plasmid pRL-CMV. Results shown are mean ± SD from at least two independent transfection experiments, each performed in quadruplicate.
Identification of cis-Elements and Trans-Factors that Control Expression of the EC-SOD Gene
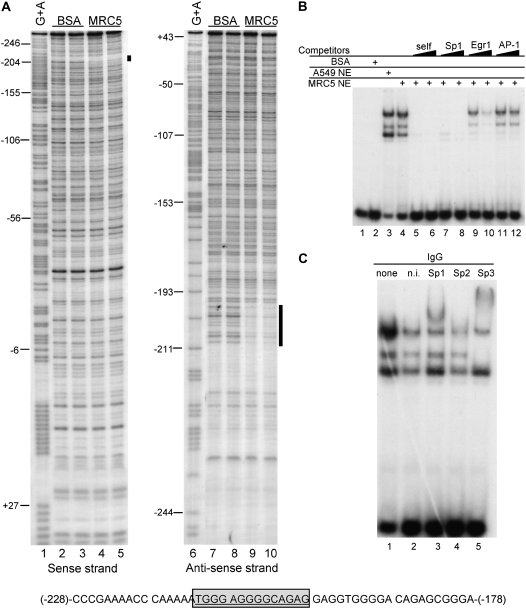
To identify the location of cis-elements in the EC-SOD promoter that interacts with nuclear transcription factors, we performed DNase I footprint analysis using a 32P-labeled region of the EC-SOD promoter containing the nucleotides −263 to +45. Nuclear extracts from MRC-5 cells protected a well defined region on both DNA strands, spanning nucleotides −195/−210 (Figure 3A, Lanes 4–5 and 9–10). The addition of BSA into this reaction did not produce any protected regions (Figure 3A, Lanes 2–3 and 7–8). No protected regions were observed between nucleotides −47 to +47, suggesting that the increase in promoter/reporter activity for these constructs was probably due to a physical positioning effect of transcription initiation complexes.
Figure 3.
Identification of trans-factors that bind to EC-SOD proximal promoter. (A) An end-labeled −263/+45 bp fragment of the human EC-SOD promoter was incubated with BSA (Lanes 2–3 and 7–8) or MRC5 nuclear extract (Lanes 4–5 and 9–10) and digested with increasing amount of DNase I. The single protected region (marked as black boxes on the right side of the gel) is identified at positions −198/−211. The nucleotide sequence of protected region shown at the bottom. Lanes 1 and 6, G+A, Maxam-Gilbert sequencing ladder of the probe DNA. (B) Binding of transcription factors from Sp-like family to the EC-SOD promoter. An end-labeled hSOD3 (−228/−178) oligonucleotide was incubated with A549 or MRC5 nuclear extract. Competition assays were performed with 50- and 200-fold excesses of unlabeled oligonucleotides to analyze the specificity of Sp1/Sp3 binding. The self and Sp1 consensus oligonucleotides eliminated shifted bands (Lanes 5–8). Oligonucleotides representing the consensus binding site for Egr-1 show some competition for DNA-protein complexes (Lanes 9 and 10), whereas the AP-1–specific probe shows no competition (Lanes 11 and 12). (C) For supershift experiments, antibodies specific for Sp1, Sp2, Sp3, or nonimmune (n.i.) IgG were added to the reaction mixture. DNA-protein complexes were separated on 5% nondenaturing polyacrylamide gel, dried, and exposed to X-ray film.
To identify the transcription factor that binds to the region −195/−210, we labeled a double-stranded oligonucleotide representing this protected region with 32P and incubated it with nuclear extracts from A549 or MRC-5 cells. Nuclear extracts from both cells produce slow migrating bands with approximately the same intensity, whereas the addition of BSA does not produce any shifted bands (Figure 3B, Lanes 2–4).
Competition experiments show that all bands were abolished by a 50 and 200 molar excess of cold self-nucleotide (Figure 3B, Lanes 5 and 6). To identify the specific transcription factors that produce these slowly migrating complexes, several oligonucleotides that encode consensus binding sequences for Sp1, Egr-1, and AP-1 transcription factors were used in competition experiments. Among them, only the Sp1 oligonucleotide eliminated specific bands at 50 and 200 molar excess (Figure 3B, Lanes 7 and 8). Oligonucleotides specific for AP-1 did not affect the intensity of the slowly migrating upper bands but reduced the intensity of the lower band (Figure 3B, Lanes 11 and 12). Egr-1–specific oligonucleotides showed partial competition for binding (Figure 3B, Lanes 9 and 10). Overall, these data indicate that members of the Sp1 family of transcription factors seem to be the best candidates for trans-acting factors that interact with the −195/−210 protected region. The partial competition seen when using Egr-1 oligonucleotides is not surprising because they belong to the same Kruppel-like family of transcription factors and share similar consensus binding sites with the Sp1 family.
Because the Sp1 family consists of at least three distinct proteins (Sp1, Sp2, and Sp3), each with different DNA-binding and transactivation properties, we performed supershift experiments with antibodies specific for each of these proteins. Only Sp1- and Sp3-specific antibodies supershifted these slowly migrating complexes (Figure 3, Lanes 3 and 5), whereas Sp2-specific antibodies did not produce supershifted bands (Figure 3, Lane 4). These data suggest that Sp1 and Sp3 transcription factors are responsible for protecting the EC-SOD promoter in DNase I foot-printing assays.
Stimulation of Transcription from EC-SOD Promoter by Sp1 and Sp3 in SL2 Cells
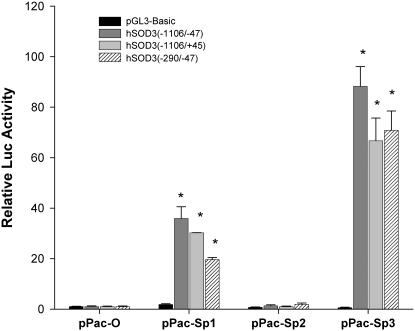
To further demonstrate in vivo that the Sp-family of transcription factors is involved in the transcriptional regulation of human EC-SOD, we co-transfected Drosophila SL2 cells with a luciferase reporter construct in the presence and absence of expression plasmids encoding Sp1, Sp2, and Sp3. We choose SL2 cells because they lack endogenous Sp proteins. Thus, activation of the EC-SOD promoter is dependent on exogenously introduced transcription factors. The activity of the pGL3-hSOD3(−1106/−47) construct was markedly stimulated in the presence of pPac-Sp1 and pPac-Sp3, whereas pPac-Sp2 had a minimal effect on reporter activity (Figure 4). The control pGL3-Basic construct was not activated by Sp-related transcription factors. Extension of the promoter sequence from −47 to +45 and deletion from −1106 to −290 did not affect the activation of promoter/reporter by Sp1 or Sp3. These results indicate that transcription factors Sp1 and Sp3 increase transcription from the endogenous human EC-SOD promoter.
Figure 4.
Activation of EC-SOD promoter by Sp1/Sp3 in vivo. Drosophila SL2 cells were transfected with 600 ng of promoter-reporter constructs depicted in the legend and 600 ng of pPac-O, pPac-Sp1, pPac-Sp2, or pPac-Sp3 plasmid as indicated. The luciferase activities were measured 24 hours later. Assays were repeated at least three times, and the mean ± SD for each value is indicated. Asterisks indicate the values of luciferase activity that are significantly different from those of control cells (P < 0.001).
Expression and Binding of Sp1/Sp3 to the EC-SOD Promoter In Vivo
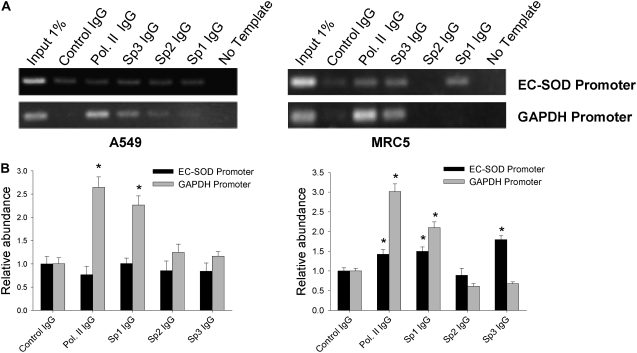
Our in vitro data suggested that Sp1 and Sp3 bind to their putative recognition sequence within the EC-SOD promoter in vitro, which in turn activates promoter/reporter activity in transient transfection experiments. To investigate the occupancy of EC-SOD promoter by Sp1/Sp3 in vivo, we performed chromatin immunoprecipitation (ChIP) assays in A549 and MRC5 cells. We detected Sp1/Sp3 binding only in MRC5 cells (Figure 5A, right panel), whereas no such binding was detected in A549 cells (Figure 5A, left panel). No enrichment of EC-SOD promoter fragments was detected in fractions precipitated with anti-Sp2 IgG in either cell line.
Figure 5.
Occupacy of EC-SOD promoter by Sp1/Sp3 and RNA polymerase II (Pol. II) in vivo. (A) Chromatin immunoprecipitation assay was used to analyze the interaction of RNA polymerase II, Sp3, Sp2, and Sp1 to the EC-SOD proximal promoter in A549 and MRC5 cells. Binding of RNA polymerase II to the GAPDH promoter was used as a positive control. Gel images are representative of at least three independent experiments. (B) Quantitative analysis of images presented in A. The intensity of fluorescence from amplified bands was compared and expressed as relative abundance of the protein at corresponding promoter. Asterisks indicate amplified bands whose intensity is significantly higher than the intensity of corresponding control IgG bands (P < 0.05). Bars represent ±SD.
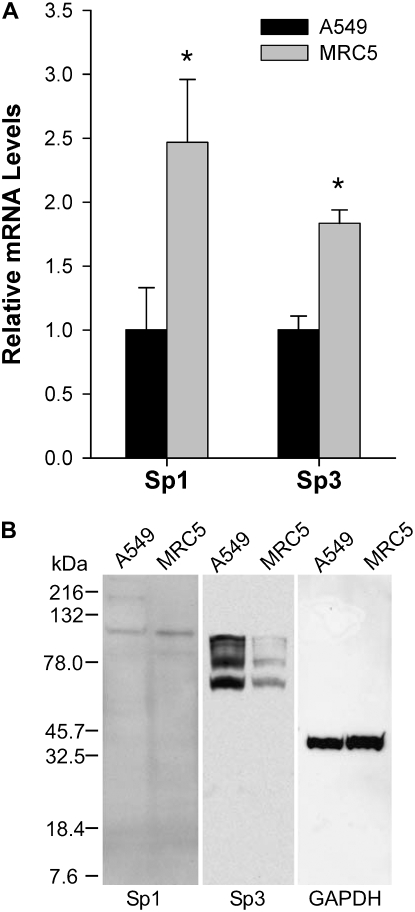
Because Sp1/Sp3 facilitated their activity through recruitment of RNA polymerase II, we anticipated detecting concomitant enrichment of RNA polymerase II. RNA polymerase II was enriched only in MRC5 lysates, whereas no enrichment of this protein was observed in A549 cells compared with the control IgG (Figure 5A). As a positive control, we used amplification of GAPDH promoter, which is expressed at high levels in both cell lines. Our ChIP assay showed marked enrichment of RNA polymerase II and Sp3 at the GAPDH promoter in A549 and MRC5 cells (Figure 5A). The higher occupancy of EC-SOD promoter by Sp1/Sp3 in MRC5 cells can be attributed to their higher expression in these cells. To test this hypothesis, we analyzed Sp1/Sp3 mRNA levels in two cell lines and found that both transcription factors showed slightly higher mRNA levels in MRC5 cells compared with A549 cells (Figure 6A). mRNA levels do not always correlate with corresponding protein levels. Western blot studies showed lower levels of Sp3 protein in MRC5 nuclear extract compared with A549 nuclear extract (Figure 6B). We were unable to detect Sp1 protein in the same nuclear extracts using the same antibodies as in the supershift experiments. Nevertheless, our data indicate that differences in the expression of Sp3 in these cells alone cannot ascribe the more than 2,000-fold higher EC-SOD mRNA levels in MRC5 cells. Moreover, these data indicate that in vivo occupancy of the EC-SOD promoter by Sp1/Sp3 and RNA polymerase II is observed only in MRC5 cells and correlates well with the levels of EC-SOD mRNA in these cells.
Figure 6.
Expression of Sp1 and Sp3 in A549 and MRC5 cells. (A) Relative mRNA levels of Sp1 and Sp3 in A549 and MRC5 cells. The mRNA levels were determined using quantitative RT-PCR and SYBR Green dye. Sp1 and Sp3 mRNA levels were normalized to GAPDH expression. Amplification was performed in triplicate, and SD is depicted as an error bar. Asterisks indicate the levels of mRNAs in MRC5 cells that are significantly different from those of A549 cells (P < 0.01). (B) Sp1 and Sp3 expression was assessed using Western blot. Nuclear extracts from A549 and MRC5 cells were separated on denaturing PAAG, transferred onto polyvinylidene difluoride membrane, and incubated with Sp1-, Sp3-, or GAPDH-specific IgG.
DISCUSSION
EC-SOD activity is essential for maintaining a balanced and physiologic concentration range of superoxide in extracellular spaces, which can indirectly regulate extracellular levels of other active and reactive biomolecules like nitric oxide and/or peroxynitrate. EC-SOD is expressed at high levels in lung, vasculature, and kidney. In the lung, EC-SOD is localized in close proximity to type II pneumonocytes and airway epithelial cells and around smooth muscle cells in the pulmonary vasculature and airways (13). Pulmonary fibroblasts and macrophages are an additional source of EC-SOD production in the lung (14). There is little known about specific transcriptional mechanisms that regulate basal expression of human EC-SOD and why this high level of expression occurs only in a limited number of specific cell types.
We have previously begun to identify the promoter region of the murine EC-SOD gene and have shown that EC-SOD transcription is dependent on proximal and distal cis-elements that interact with several transcription factors, including Ets, Kruppel, MZF, and Sp1/Sp3 (9). It seems to be the interaction between these transcription factors that accounts for, at least in part, its cell-specific expression (10). In the present study, we analyzed the transcriptional regulation of the human EC-SOD gene and found some similarities and differences compared with the murine EC-SOD gene. The analysis of transcription initiation sites for EC-SOD gene in a number of human tissues and cells revealed the presence of multiple transcription start sites located approximately 519 to 581 bp further downstream from the previously identified transcription start site (12). We cannot rule out the existence of previously identified longer RNA transcripts for human EC-SOD, taking into consideration that the previous site was determined by a different method that involved using primer extension analysis and RNA from human placenta. The discovery of multiple transcription start sites further downstream facilitated the analysis of DNA sequences located directly upstream of newly identified sites for transcriptional initiation. Using truncated promoter/reporter constructs, DNase I footprinting, and gel-shift assays, we identified a single Sp1-like binding site within this region. Simultaneous transient transfections of promoter/reporter constructs and the plasmids expressing Sp-like family of proteins into insect SL2 cells indicated that only Sp1 and Sp3 transcription factors were able to activate the human EC-SOD promoter. We found no evidence of Ets, MZF, or Kruppel-like transcription factors playing a role in human EC-SOD expression. These data share a high degree of similarity with the results we obtained for mouse EC-SOD promoter because both genes have the multiple transcriptional start sites, and their promoters are TATA-less and activated to a high degree by Sp1 and Sp3 (9).
Sp1 is the first discovered member of a growing family of transcription factors that exhibits similar structural features and binds to the GC-rich boxes and related motifs (for review, see Ref. 15). Sp1 is expressed ubiquitously and abundantly in almost all cell types and regulates the expression of a wide variety of metabolic enzymes, transcription factors, and cytoskeleton proteins. Developmental expression of Sp1 is critical because disruption of its gene causes embryonic growth retardation and death at the early gestational stages in mice (16). However, a lack of Sp1 expression can be compensated for, at least in part, by Sp3 or other Sp-family proteins. Sp1 and Sp3 bind to the same consensus binding sequences but might perform different physiologic functions. For example, studies with Sp3-null mice showed that during early development both proteins might have similar functions, but after their progression into late embryogenesis and postnatal period they become more specialized (17). It has been reported that three isoforms of Sp3 exist, and our Western blot experiment confirmed the presence of three distinct bands in A549 and MRC5 nuclear extracts (18). These isoforms have different functions and might act as activators or as repressors. Moreover, a transient transfection experiment in Drosophila SL2 cells and certain mammalian cells revealed that Sp3 can function as an activator or a repressor depending on the sequence composition and the organization of binding sites (19, 20). The dual function of Sp3 can be attributed to the presence of inhibitory domain located between the second glutamine-rich activation domain and the first zinc finger (20).
The electrophoretic mobility shift assay experiments performed with an excess of cold oligonucleotides indicated that another member of the Sp/Kruppel-like family of transcription factors early growth response factor (EGR)-1 might interact with the EC-SOD promoter, at least in in vitro settings. Egr-1 (also known as krox-24, NGFI-A, Zif-268, and TIS8) is a zinc-finger transcription factor that binds to GC-rich DNA sequences with the consensus binding sequence GCG(T/G)GGGCG (21). Thus, Egr-1 shares a similar consensus binding site with Sp1 (-GGGCGG-) and can potentially displace Sp1 from its binding site. Such cases were described for PDGF A-chain (22), CuZn superoxide dismutase (23), tissue factor (24), and others. Egr-1 stimulated transcription by interacting directly with components of the basal transcription apparatus or by recruitment of a coactivators, and its levels rapidly increased in response to a wide range of stimuli, including reactive oxygen species, angiotensin II, and shear stress (25). The binding site for Sp1/Sp3 in EC-SOD promoter (TGGGAGGGGCAGAG) does not resemble the consensus binding site for Sp1 or Egr-1, but it does share some degree of similarity. Although we cannot rule out the possibility that Egr-1 might play an important role in the activation of EC-SOD transcription in the lung cells, such a scenario is less favorable due to a lower affinity toward the protected region of Egr-1 compared with Sp1 (Figure 3B).
Experiments with mouse embryonic stem cells that have the Sp1 gene disrupted by homologous recombination provided data that a majority of genes that were documented to be regulated by Sp1 were not affected or were slightly down-regulated (16). To show that Sp1/Sp3 transcription factors bind to EC-SOD promoter in vivo, we performed chromatin immunoprecipitation assays in A549 and MRC5 cells. Sp1 and Sp3 were detected at their putative binding site in MRC5 cells but not in A549 cells. It has been shown that Sp1 can recruit the basal transcription machinery by directly interacting with TATA box binding protein (TBP) or other subunits of RNA polymerase II (26, 27). Our ChIP assay indicated that RNA polymerase II was recruited to the EC-SOD promoter only in MRC5 cells, which expressed EC-SOD at relatively high level. These data show a strong correlation between the occupancy of EC-SOD promoter by Sp1/Sp3 and RNA polymerase II in vivo and the expression of endogenous EC-SOD. A new question was raised about why Sp-like transcription factors do not interact with EC-SOD promoter in A549 cells even though their nuclear levels were comparable to their levels in MRC5 cells. Additional studies are required to elucidate the molecular mechanisms responsible for cell-specific differences in the EC-SOD promoter transcriptional machinery organization.
Acknowledgments
The authors acknowledge the support of the Duke University Tissue Culture Facility for providing the cell lines and reagents and the Duke University Sequencing Facility for help in sequencing of DNA clones. The authors thank Dr. J. Noti (Guthrie Research Institute) for providing the pPacO, pPacSp1, pPacSp2, and pPacSp3 plasmids.
This work was supported by National Institute of Health Grant HL64894 and HL074289 (R.J.F.).
Originally Published in Press as DOI: 10.1165/rcmb.2007-0378OC on February 28, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med 2002;33:337–349. [DOI] [PubMed] [Google Scholar]
- 2.Noor R, Mittal S, Iqbal J. Superoxide dismutase–applications and relevance to human diseases. Med Sci Monit 2002;8:RA210–RA215. [PubMed] [Google Scholar]
- 3.Fattman CL, Schaefer LM, Oury TD. Extracellular superoxide dismutase in biology and medicine. Free Radic Biol Med 2003;35:236–256. [DOI] [PubMed] [Google Scholar]
- 4.Folz RJ, Abushamaa AM, Suliman HB. Extracellular superoxide dismutase in the airways of transgenic mice reduces inflammation and attenuates lung toxicity following hyperoxia. J Clin Invest 1999;103:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed MN, Suliman HB, Folz RJ, Nozik-Grayck E, Golson ML, Mason SN, Auten RL. Extracellular superoxide dismutase protects lung development in hyperoxia-exposed newborn mice. Am J Respir Crit Care Med 2003;167:400–405. [DOI] [PubMed] [Google Scholar]
- 6.Rabbani ZN, Anscher MS, Folz RJ, Archer E, Huang H, Chen L, Golson ML, Samulski TS, Dewhirst MW, Vujaskovic Z. Overexpression of extracellular superoxide dismutase reduces acute radiation induced lung toxicity. BMC Cancer 2005;5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowler RP, Nicks M, Warnick K, Crapo JD. Role of extracellular superoxide dismutase in bleomycin-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2002;282:L719–L726. [DOI] [PubMed] [Google Scholar]
- 8.Fattman CL, Chang LY, Termin TA, Petersen L, Enghild JJ, Oury TD. Enhanced bleomycin-induced pulmonary damage in mice lacking extracellular superoxide dismutase. Free Radic Biol Med 2003;35:763–771. [DOI] [PubMed] [Google Scholar]
- 9.Zelko IN, Folz RJ. Sp1 and Sp3 transcription factors mediate trichostatin A-induced and basal expression of extracellular superoxide dismutase. Free Radic Biol Med 2004;37:1256–1271. [DOI] [PubMed] [Google Scholar]
- 10.Zelko IN, Folz RJ. Myeloid zinc finger (MZF)-like, kruppel-like and Ets families of transcription factors determine the cell-specific expression of mouse extracellular superoxide dismutase. Biochem J 2003;369:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrickson DJ, Fisher JH, Jones C, Ho YS. Regional localization of human extracellular superoxide dismutase gene to 4pter-q21. Genomics 1990;8:736–738. [DOI] [PubMed] [Google Scholar]
- 12.Folz RJ, Crapo JD. Extracellular superoxide dismutase (SOD3): tissue-specific expression, genomic characterization, and computer-assisted sequence analysis of the human EC SOD gene. Genomics 1994;22:162–171. [DOI] [PubMed] [Google Scholar]
- 13.Oury TD, Chang LY, Marklund SL, Day BJ, Crapo JD. Immunocytochemical localization of extracellular superoxide dismutase in human lung. Lab Invest 1994;70:889–898. [PubMed] [Google Scholar]
- 14.Su WY, Folz R, Chen JS, Crapo JD, Chang LY. Extracellular superoxide dismutase mrna expressions in the human lung by in situ hybridization. Am J Respir Cell Mol Biol 1997;16:162–170. [DOI] [PubMed] [Google Scholar]
- 15.Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol 2002;195:27–38. [DOI] [PubMed] [Google Scholar]
- 16.Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 1997;89:619–628. [DOI] [PubMed] [Google Scholar]
- 17.Bouwman P, Gollner H, Elsasser HP, Eckhoff G, Karis A, Grosveld F, Philipsen S, Suske G. Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J 2000;19:655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennett SB, Udvadia AJ, Horowitz JM. Sp3 encodes multiple proteins that differ in their capacity to stimulate or repress transcription. Nucleic Acids Res 1997;25:3110–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majello B, De Luca P, Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J Biol Chem 1997;272:4021–4026. [DOI] [PubMed] [Google Scholar]
- 20.Dennig J, Beato M, Suske G. An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J 1996;15:5659–5667. [PMC free article] [PubMed] [Google Scholar]
- 21.Gashler A, Sukhatme VP. Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol 1995;50:191–224. [DOI] [PubMed] [Google Scholar]
- 22.Silverman ES, Khachigian LM, Lindner V, Williams AJ, Collins T. Inducible pdgf a-chain transcription in smooth muscle cells is mediated by Egr-1 displacement of Sp1 and Sp3. Am J Physiol 1997;273:H1415–H1426. [DOI] [PubMed] [Google Scholar]
- 23.Minc E, de Coppet P, Masson P, Thiery L, Dutertre S, Amor-Gueret M, Jaulin C. The human copper-zinc superoxide dismutase gene (SOD1) proximal promoter is regulated by Sp1, Egr-1, and WT1 via non-canonical binding sites. J Biol Chem 1999;274:503–509. [DOI] [PubMed] [Google Scholar]
- 24.Cui MZ, Parry GC, Oeth P, Larson H, Smith M, Huang RP, Adamson ED, Mackman N. Transcriptional regulation of the tissue factor gene in human epithelial cells is mediated by Sp1 and EGR-1. J Biol Chem 1996;271:2731–2739. [DOI] [PubMed] [Google Scholar]
- 25.Silverman ES, Collins T. Pathways of Egr-1-mediated gene transcription in vascular biology. Am J Pathol 1999;154:665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emili A, Greenblatt J, Ingles CJ. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol Cell Biol 1994;14:1582–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang CM, Roeder RG. Cloning of an intrinsic human tfiid subunit that interacts with multiple transcriptional activators. Science 1995;267:531–536. [DOI] [PubMed] [Google Scholar]