Abstract
At the FASEB summer research conference on “Arf Family GTPases”, held in Il Ciocco, Italy in June, 2007, it became evident to researchers that our understanding of the family of Arf GTPase activating proteins (ArfGAPs) has grown exponentially in recent years. A common nomenclature for these genes and proteins will facilitate discovery of biological functions and possible connections to pathogenesis. Nearly 100 researchers were contacted to generate a consensus nomenclature for human ArfGAPs. This article describes the resulting consensus nomenclature and provides a brief description of each of the 10 subfamilies of 31 human genes encoding proteins containing the ArfGAP domain.
Introduction
Regulatory GTPases act as molecular switches that can rapidly interconvert between two conformational states, depending on whether GTP or GDP is bound. The cellular actions of GTPases are typically initiated by GTP binding, promoted by guanine nucleotide exchange factors (GEFs), and terminated by GTP hydrolysis that is facilitated by GTPase-activating proteins (GAPs). Within the Ras superfamily (Wennerberg et al., 2005), comprised of >150 GTPases, each family (Ras, Rho, Rab, Arf, and Ran) uses distinct sets of GEFs and GAPs to regulate signaling. Though originally envisioned as simple “off switches” in Arf signaling, the ArfGAPs have also emerged as effectors and key components in the assembly of nanomachines with complex signaling potential (Gillingham and Munro, 2007; Inoue and Randazzo, 2007).
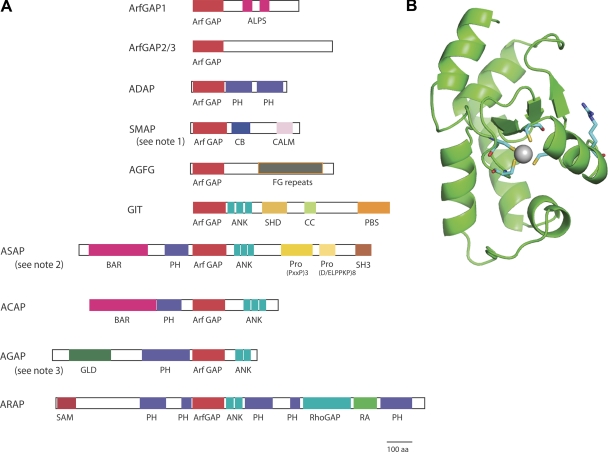
ArfGAPs are a family of proteins containing a characteristic module, the ArfGAP domain, which was first identified in rat ArfGAP1 as the domain responsible for stimulation of GTP hydrolysis on Arf1 (Cukierman et al., 1995). ArfGAP domains are ancient and highly conserved since the earliest eukaryotes. Five ArfGAPs have been identified in the yeast Saccharomyces cerevisiae and shown to display a combination of redundant and unique functions. Mammalian cells express an array of ArfGAPs ranging from relatively small proteins resembling those found in yeast to the large, multi-domain ArfGAPs that are proposed to function as scaffolds for cell signaling (Fig. 1 A).
Figure 1.
Domain organization of human ArfGAP subfamilies and structure of the ArfGAP domain. (A) Representative domain structures of each human ArfGAP subfamily are depicted and are drawn to scale. Abbreviations are: ALPS, ArfGAP1 lipid-packing sensor; ArfGAP, ArfGAP domain; ANK, ankyrin repeat; BAR, Bin/Amphiphysin/Rvs; CALM, CALM binding domain; CB, clathrin-box; CC, coiled-coil; FG repeats, multiple copies of the XXFG motif; GLD, GTP-binding protein-like domain; PBS, Paxillin binding site; PH, pleckstrin homology domain; Pro(PxxP)3, cluster of three Proline-rich (PxxP) motifs; Pro(D/ELPPKP)8, eight tandem Proline-rich (D/ELPPKP) motifs; RA, Ras association motif; RhoGAP, RhoGAP domain; SAM, sterile α-motif; SH3, Src homology 3 domain; SHD, Spa-homology domain. Notes: (1) SMAP2 has CALM BD, but SMAP1 does not. (2) ASAP1 contains the indicated Pro-rich domains; ASAP2 and ASAP3 lack the Pro (D/ELPPKP) repeat and ASAP3 does not have an SH3 domain. (3) AGAP2 has a splice variant with three N-terminal PxxP motifs, called PIKE-L. (B) The structure of the isolated ArfGAP domain of human ArfGAP1 (residues 6–120) is displayed with the backbone shown in green with secondary structures indicated. The side chains of only the conserved arginine (Arg50, on the right) and the four zinc finger cysteines (center; Cys22, 25, 42, and 45) are displayed along with the coordinated Zn2+ (gray sphere). This image was generated using PyMol.
Structure, mechanism, and specificity
ArfGAP domains are ∼130 amino acids in length and were originally defined as the minimal fragment possessing ArfGAP activity (Cukierman et al., 1995). They contain a characteristic C4-type zinc finger motif and a conserved arginine that is required for activity, within a particular spacing (CX2CX16CX2CX4R). The zinc finger has an architectural rather than catalytic role (Fig. 1 B) (Goldberg, 1999). The invariant arginine was proposed to serve in a catalytic “arginine finger” mechanism, similar to that found in GAPs for other GTPases, including Ras and Rho (Scheffzek et al., 1998), and is highly exposed to solvent in the crystal structure (Fig. 1 B). However, the potential for other binding partners (e.g., coatomer; Goldberg 1999) or other domains within some ArfGAPs (e.g., PH domains) serving supportive or regulatory roles in GAP-stimulated hydrolysis has also been demonstrated. For example, the ArfGAP activity of ASAP1 is dependent on the PH domain and is sensitive to PI(4,5)P2, and that of GITs is stimulated by PIP3.
ArfGAPs display various degrees of specificity for individual members of the Arf family both in vitro and in live cells. However, these data are not easy to interpret due to uncertainties in colocalization of the ArfGAP and Arfs in cells, incomplete knowledge of the importance of coregulators (lipids or other proteins), and because some ArfGAPs use their GAP domain to bind Arf without promoting GTP hydrolysis. For the most part (see below) ArfGAPs are active on one or more of the “true” Arfs (Arf1-6) but not on the Arf-like (Arl) or Sar proteins, which use distinct families of GAPs. Arf GAP activity has been demonstrated in vitro for at least one member of each subfamily, with the exception of the ADAPs, which appear to lack in vitro GAP activity. However, overexpression of ADAP1 reduces activated Arf6 (but not Arf1) levels, and diminishes cortical actin and stress fibers, consistent with function as an Arf6 GAP.
Although ArfGAPs had been thought to distinguish between Arfs and Arls, Gcs1p (a yeast orthologue of mammalian ArfGAP1) was reported to function as GAP for both yeast Arf1p and Arl1p (Liu et al., 2005). Conversely, Arl2 GAP isolated from bovine testis lacks the ArfGAP domain but exhibits GAP activity toward Arfs (Bowzard et al., 2007). The latter finding suggests that the “canonical” ArfGAPs, those containing the ArfGAP domain, may not represent the entire repertoire of proteins capable of stimulating GTP hydrolysis by Arf proteins.
Consensus nomenclature of the ArfGAP family of human genes and encoded proteins
Like most gene families today, gene/protein discovery and the accompanying nomenclature of ArfGAPs has come from a number of different laboratories and techniques and over a span of more than a decade. We have developed a consensus nomenclature for the human ArfGAPs that is based upon the phylogeny of the ArfGAP domains and the domain organization of the proteins, and updated or clarified the acronyms in use to more accurately describe the current understanding of each protein. Table I lists the consensus nomenclature for the human genes, previously published gene symbols and names, database identifiers, chromosome locations, and accession numbers. These gene names should also be used for the encoded proteins, with appropriate indications when more than one splice variant is known to exist. The 31 predicted human ArfGAPs have been classified into 10 subfamilies, based on sequence similarities of their ArfGAP domains, and supported by the conservation of the domain architecture within each subfamily (see Fig. 1 A). The acquisition of additional domains throughout eukaryotic evolution has contributed to the acquisition of new functions by different ArfGAPs. A brief summary of each subfamily is presented below.
Table I. Consensus names for human ArfGAPs, with additional information.
| Subfamily | New consensus gene symbols |
Previous HGNC gene symbols |
Literature or database aliases/synonyms |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name #1 | Name #2 | Name #3 | Name #4 | KIAA# | NCBI gene ID |
Human gene location |
Accession number (length) | |||
| ARFGAP1 | ARFGAP1 | ARFGAP1 | ArfGAP1 | Arf1GAP | Arf GAP | MGC39924 | 55738 | 20q13.33 | NM_018209; NP_060679 (406aa) | |
| ARFGAP2 | ARFGAP2 | ARFGAP2 | ArfGAP2 | ZNF289/Zfp289 | FLJ14576 | FLJ26000 | 84364 | 11p11.2-p11.12 | NM_032389; NP_115765 (521aa) | |
| ARFGAP3 | ARFGAP3 | ArfGAP3 | ArfGAP1 | 26286 | 22q13.2-q13.3 | NM_014570; NP_055385 (516aa) | ||||
| ADAP | ADAP1 | CENTA1 | Centaurin (alpha1) |
PIPBP | p42IP4 | GCS1L | 11033 | 7p22.3 | NM_006869; NP_006860 (374aa) | |
| ADAP2 | CENTA2 | Centaurin (beta) |
Cent. Alpha2 | cent-b | HSA272195 | 55803 | 17q11.2 | NM_018404; NP_060874 (381aa) | ||
| SMAP | SMAP1 | SMAP1 | SMAP-1 | FLJ13159 | FLJ42245 | 60682 | 6q13 | NM_001044305; NP_068759 (440aa) | ||
| SMAP2 | SMAP2 | SMAP1L | RP1-228H13.3 | SMAP2 | 64744 | 1p35.3-p34.1 | NM_022733; NP_073570 (429aa) | |||
| AGFG | AGFG1 | HRB | HRB1 | RIP | HIV-1 Rev BP | RAB | 3267 | 2q36.3 | NM_004504; NP_004495 (562aa) | |
| AGFG2 | HRBL | HRB2 | RAB-R/HRB1 | HIV-1 Rev BP-L | 3268 | 7q22.1 | NM_006076; NP_006067 (481aa) | |||
| GIT | GIT1 | GIT1 | Git1 | Cat1 | p95APP1 | 28964 | 17p11.2 | NM_014030; NP_054749 (761aa) | ||
| GIT2 | GIT2 | Git2 | Cat2 | p95APP2 | p95PKL | KIAA0148 | 9815 | 12q24.1 | NM_057169; NP_476510 (759aa) | |
| ASAP | ASAP1 | DDEF1 | AMAP1 | DEF1/DDEF1 | PAG2/SHAG1 | Cent. Beta4 | KIAA1249 | 50807 | 8q24.1-q24.2 | NM_018482; NP_060952 (1129aa) |
| ASAP2 | DDEF2 | AMAP2 | PAP/DDEF2 | SHAG2/PAG3 | Cent. Beta3 | KIAA0400 | 8853 | 2p24 | NM_003887: NP_003878 (1006aa) | |
| ASAP3 | DDEFL1 | DDEFL1 | UPLC1 | ACAP4 | Cent. Beta6 | 55616 | 1p36.12 | NM_017707; NP_060177 (903aa) | ||
| ACAP | ACAP1 | CENTB1 | ACAP1 | Cent. Beta1 | KIAA0050 | 9744 | 17p13.1 | NM_014716; NP_055531 (740aa) | ||
| ACAP2 | CENTB2 | ACAP2 | Cent. Beta2 | KIAA0041 | 23527 | 3q29 | NM_012287; NP_036419 (778aa) | |||
| ACAP3 | CENTB5 | ACAP3 | Cent. Beta5 | KIAA1716 | 116983 | 1p36 | NM_030649; NP_085152 (759aa) | |||
| AGAP | AGAP1 | CENTG2 | AGAP1 | Cent. Gamma2 | GGAP1 | MGC71657 | KIAA1099 | 116987 | 2q37 | NM_014914; NP_055729 (804aa) |
| AGAP2 | CENTG1 | AGAP2 | Cent. Gamma1 | GGAP2/PIKE | FLJ16430 | KIAA0167 | 116986 | 12q14.1 |
NM_014770; NP_055585 (836aa); AAM97540 (1192aa) |
|
| AGAP3 | CENTG3 | AGAP3 | Cent. Gamma3 | MRIP1/CRAG | FLJ16146 | 116988 | 7q36.1 | NM_031946; NP_114152 (911aa) | ||
| AGAP4 | CTGLF1 | CTGLF1 | Centaurin gamma-like family 1 | MRIP2 | 119016 | 10q11.21 | NM_133446; NP_597703 (663aa) | |||
| AGAP5 | CTGLF2 | CTGLF2 | Centaurin gamma-like family 2 | 729092 | 10q22.2 | XM_001132588; XP_001132588 (686aa) | ||||
| AGAP6 | CTGLF3 | CTGLF3 | Centaurin gamma-like family 3 | 414189 | 10q11.23 | NM_001077665; NP_001071133 (686aa) | ||||
| AGAP7 | CTGLF4 | CTGLF4 | Centaurin gamma-like family 4 | 653268 | 10q11.23 | NM_001077685; NP_001071153 (663aa) | ||||
| AGAP8 | CTGLF5 | CTGLF5 | Centaurin gamma-like family 5 | 728404 | 10q11.23 | NM_001077686; NP_001071154 (663aa) | ||||
| AGAP9 | CTGLF6 | CTGLF6 | Centaurin gamma-like family 6 | FLJ00312 | 642517 | 10q11.22 | NM_001077686 (663aa) | |||
| AGAP10 | CTGLF7 | CTGLF7 | Centaurin gamma-like family 7 | 728127 | 10q11.22 | XR_015281 (655aa) | ||||
| AGAP11 | KIAA1975 | Similar to MRIP2 | KIAA1975 | 119385 | 10q23.2 | NP_597704 (550aa) | ||||
| ARAP | ARAP1 | CENTD2 | ARAP1 | Cent. Delta2 | KIAA0782 | 116985 | 11q13.4 | NM_001040118; NP_001035207 (1450aa) | ||
| ARAP2 | CENTD1 | ARAP2 | Cent. Delta1 | FLJ13675 | PARX | KIAA0580 | 116984 | 4p14 | NM_015230; NP_056045 (1704aa) | |
| ARAP3 | CENTD3 | ARAP3 | Cent. Delta3 | FLJ21065 | DRAG1 | 64411 | 5q31.3 | NM_022481; NP_071926 (1544aa) | ||
List of consensus acronyms: ArfGAP = ADP-ribosylation factor GTPase activating proteins. ACAP = ArfGAP with coiled-coil, ankyrin repeat, and PH domains. ADAP = ArfGAP with dual PH domains. AGAP = ArfGAP with GTPase domain, ankyrin repeat, and PH domain. AGFG = ArfGAP with FG repeats. ARAP = ArfGAP with Rho GAP domain, ankyrin repeats and PH domain. ASAP = ArfGAP with SH3 domain, ankyrin repeat, and PH domain. GIT = G protein receptor kinase (GRK) interacting ArfGAP. SMAP = Small ArfGAP.
List of proposed discontinued acronyms: APP = ArfGAP putative, PIX1-interacting, paxillin binding protein. CAT = Cool-associated tyrosine phosphorylated protein. CTGLF = Centaurin gamma-like family. DEF = differentiation-enhancing factor. HRB = HIV Rev binding protein. PAG = paxillin associated Arf GAP. PAP = Pyk2 associated protein. PKL = paxillin-kinase linker. RAB = HIV Rev-associated binding protein. RIP = HIV Rev-interacting protein.
ArfGAP1 subfamily
This founder of the ArfGAP family is also the smallest member, at ∼45 kD. ArfGAP1 shuttles between cytosol and the Golgi, where it is involved in regulating the COPI mechanism of membrane traffic. The region of ArfGAP1 C-terminal to the ArfGAP domain is predicted to be largely unstructured. This region contains two stretches, termed ALPS motifs, that have the propensity in vitro to fold into amphipathic α-helices upon interaction with membranes that are highly curved or contain loosely packed lipids (Bigay et al., 2005; Mesmin et al., 2007), and are required for Golgi targeting of the protein in vivo (Levi et al., 2008). The ALPS motifs are predicted to orient the protein on the membrane and to contribute substantially to regulation of its activity in cells. ArfGAP activity is also regulated by the COPI coat complex (Goldberg, 1999). While most studies have focused on the role of ArfGAP1 in the COPI system, ArfGAP1 also interacts with components of clathrin coated carriers (including clathrin, AP-1, and AP-2), although the functional consequences of these interactions remains to be established.
ArfGAP2 subfamily
ArfGAPs 1–3 are predicted to have arisen from a common ancestor (e.g., a single gene is present in G. lamblia) with an early split into distinctive ArfGAP1 and ArfGAP2 subfamilies and later duplications leading to the ArfGAP2/ArfGAP3 divergence. Human ArfGAP2 and 3 are closely related proteins (58% identity) with little similarity to ArfGAP1 outside the catalytic domain. ArfGAP1 and ArfGAP2/3 display functional interplay, as indicated by synthetic lethality observed between these ArfGAPs in HeLa cells (Frigerio et al., 2007), and in S. cerevisiae (Gcs1 and Glo3; Poon et al., 1999). ArfGAP2/3 lack ALPS motifs but are found on Golgi membranes as a result of strong interactions with the COPI coat (Watson et al., 2004; Frigerio et al., 2007). Like ArfGAP1, the GAP activities of ArfGAP2/3 are stimulated by coatomer (unpublished data). How each of these ArfGAPs contributes to the COPI mechanism remains unclear.
ADAP subfamily
ADAP1 (ArfGAP with dual PH domains; previously centaurin α1, p42(IP4), or PIP3BP) was identified as a high affinity PI(3,4,5)P3 and Ins(1,3,4,5)P4 binding protein. ADAP1 is proposed to function as an Arf6 GAP that regulates the actin cytoskeleton, membrane traffic, and neuronal differentiation (Thacker et al., 2004; Venkateswarlu et al., 2004). ADAP1 localizes to dendrites, spines, and synapses of developing and adult neurons and can impact traffic of regulated secretory vesicles in neuronal cells. Studies of ADAP2 have not been reported.
SMAP subfamily
SMAPs have been implicated as regulators of endocytosis and oncogenesis (Tanabe et al., 2006). Human SMAPs are ∼50 kD and lack other defined domains, thus the acronym small ArfGAP protein. The two human SMAP proteins share 47% identity overall, which rises to 83% identity in the ArfGAP domain. SMAPs bind to clathrin heavy chain via the clathrin binding motif (LLGLD) as well as the clathrin assembly protein, CALM (Natsume et al., 2006). SMAP1 is cytosolic but is recruited to membranes where it regulates constitutive endocytosis. SMAP2 is more stably bound to endosomes and is involved in the retrograde transport of TGN46 from early endosomes to the TGN (Natsume et al., 2006).
AGFG subfamily
AGFG1 has been reported to be an essential HIV Rev cofactor, based on experiments that suggest that the ArfGAP domain mediates the release of Rev-directed HIV-1 RNAs from the perinuclear region (Sanchez-Velar et al., 2004). The Rev–AGFG1 interaction is indirect and possibly bridged by the nuclear export receptor CRM1. The AGFG subfamily arose very early in eukaryotes, with a predicted progenitor in G. lamblia and representatives in molds, plants, fish, and mammals. AGFG1 contains 10 phenylalanine-glycine (FG) repeats, reminiscent of those found in nucleoporins. Thus, the AGFGs are ArfGAPs with FG repeats. Much less information is available on AGFG2.
GIT subfamily
GIT genes arose in animals and were duplicated in vertebrates. GIT2 expression is nearly ubiquitous, whereas GIT1 appears absent from many major cell types (muscle, hepatoctyes, pneumocytes, adipocytes) but is especially prominent in endothelial cells (Schmalzigaug et al., 2007). Unlike other ArfGAPs, GITs tightly associate with a specific partner, the PIX/Cool proteins, to form oligomeric complexes (Premont et al., 2004). PIX/Cool proteins are GEFs for Rac1 and Cdc42 GTPases. GIT/PIX complexes function as scaffolds for a variety of signaling enzymes (including G protein receptor kinases, p21 activated kinases, focal adhesion kinases, MEK/Erk, and phospholipase Cγ) and are recruited to distinct cellular locations via specific partners (e.g., focal adhesions via paxillin or integrinα4, synapses via piccolo or liprin-α, and plasma membranes via scribble) (Hoefen and Berk, 2006). GIT/PIX complexes function as sites of signal integration from multiple GTPase inputs, a feature shared by the ARAPs (see below).
ASAP subfamily
The ASAPs are associated with plasma membrane specializations (e.g., focal adhesions and invadopodia), and regulate aspects of endocytic traffic and actin remodeling (Nie and Randazzo, 2006; Inoue and Randazzo, 2007; Randazzo et al., 2007). ASAPs are found exclusively in animals, and have been duplicated in vertebrates, with three genes in humans that share multiple domains (see Fig. 1 A) but differ in their C termini. ASAP1 has tandem repeats of [D/ELPPKP] and an SH3 domain, ASAP2 has an SH3 domain but no E/DLPPKP repeats, and ASAP3 has neither of these motifs. These differences at the C termini have led to confusion in nomenclature, but phylogenetic analysis of the ArfGAP domains supports the conclusion that these three ASAPs form a distinct group.
ASAP1 was identified as a Src-binding protein but also binds CrkL, focal adhesion kinase, CD2AP, and CIN85 (Inoue and Randazzo, 2007). ASAP2 binds pyk2, a focal adhesion kinase, whereas ASAP3 associates with focal adhesions and regulates stress fibers (Ha et al., 2008).
AGAP subfamily
AGAPs are present in animals only and there are 11 human genes predicted to encode AGAP-type proteins, arising from amplifications in regions of human chromosome 10q, with a number of pseudogenes predicted in this same region. AGAPs possess a GTP-binding domain, reported to directly bind and activate Akt and other Ras effectors (Ye and Snyder, 2004). AGAP2 has multiple splice variants with one possessing an SH3 binding motif N-terminal to the GTP-binding protein-like domain. AGAP1 and AGAP2 have been the most extensively characterized members of this group. They function in the endocytic system; AGAP1 working with AP-3 and AGAP2 with AP-1 (Nie and Randazzo, 2006).
ACAP subfamily
ACAPs regulate Arf6-dependent actin remodeling and endocytosis and receptor tyrosine kinase-dependent cell movement (Inoue and Randazzo, 2007). ACAP1 functions as part of an Arf6-regulated clathrin coat (Li et al., 2007). ACAPs are found in Dictyostelium and metazoans, with vertebrate duplications resulting in three human genes. ACAP is an acronym for ArfGAP with coiled coil, ankyrin repeat, and PH domains, though the coiled coil domain was later identified as a BAR domain.
ARAP subfamily
The presence of ArfGAP, RhoGAP, ankyrin repeats, and Ras association domains in ARAPs intimates that they are important coordinators of two or more GTPase signaling pathways. Although the domain structures of the three ARAPs are similar, the proteins are distinct in functions and cellular locations with individual ARAPs showing different Arf, Rho, and Ras binding specificities. Pathways affected by different family members include signaling through EGF receptor, focal adhesion dynamics, and lamellipodia formation (Inoue and Randazzo, 2007; Randazzo et al., 2007). ARAPs are specific to chordates, with three human genes.
Summary
ArfGAPs play critical roles in secretory and endocytic membrane traffic as well as actin remodeling. Other ArfGAPs serve as scaffolds for cell signaling, allowing them to mediate cross-talk between members of different GTPase families. Future studies aimed at dissecting ArfGAP functions and at identifying sources of specificity and regulation in their actions are certain to provide important insights into the role of ArfGAPs in cell physiology and in human pathologies. These outcomes should be facilitated by rapid adoption of the consensus nomenclature described herein.
Acknowledgments
The authors thank Jonathan Goldberg (Memorial Sloan-Kettering Cancer Center) for providing coordinates of his ArfGAP structure and Eric Ortlund (Emory University) for help in the preparation of Fig. 1 B. We also acknowledge the many contributions to the field that we were not able to specifically cite due to limits on the number of citations.
This work was performed with support from extramural grants (GM67226, GM68029, R.A. Kahn; National Institutes of Health GM59989, DA016347, R.T. Premont; AI-043208, M.L. Zapp) and that from the Intramural Program (P.A. Randazzo, H. Inoue) of the National Institutes of Health, Department of Health and Human Services as well as the American Heart Association (0655454U, R.T. Premont), and a grant from the Israel Science Foundation 448/04 (D. Cassel).
Abbreviations used in this paper: ArfGAP, Arf GTPase activating protein; Arl, Arf-like; FG, phenylalanine-glycine; GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor.
References
- Bigay, J., J.F. Casella, G. Drin, B. Mesmin, and B. Antonny. 2005. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 24:2244–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowzard, J.B., D. Cheng, J. Peng, and R.A. Kahn. 2007. ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J. Biol. Chem. 282:17568–17580. [DOI] [PubMed] [Google Scholar]
- Cukierman, E., I. Huber, M. Rotman, and D. Cassel. 1995. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 270:1999–2002. [DOI] [PubMed] [Google Scholar]
- Frigerio, G., N. Grimsey, M. Dale, I. Majoul, and R. Duden. 2007. Two human ARFGAPs associated with COP-I-coated vesicles. Traffic. 8:1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham, A.K., and S. Munro. 2007. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23:579–611. [DOI] [PubMed] [Google Scholar]
- Goldberg, J. 1999. Structural and functional analysis of the ARF1-ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell. 96:893–902. [DOI] [PubMed] [Google Scholar]
- Ha, V.L., S. Bharti, H. Inoue, W.C. Vass, F. Campa, Z. Nie, A. de Gramont, Y. Ward, and P.A. Randazzo. 2008. ASAP3 is a focal adhesion-associated Arf GAP that functions in cell migration and invasion. J. Biol. Chem. 283:14915–14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefen, R.J., and B.C. Berk. 2006. The multifunctional GIT family of proteins. J. Cell Sci. 119:1469–1475. [DOI] [PubMed] [Google Scholar]
- Inoue, H., and P.A. Randazzo. 2007. Arf GAPs and their interacting proteins. Traffic. 8:1465–1475. [DOI] [PubMed] [Google Scholar]
- Levi, S., M. Rawet, L. Kliouchnikov, A. Parnis, and D. Cassel. 2008. Topology of amphipathic motifs mediating Golgi localization in ArfGAP1 and its splice isoforms. J. Biol. Chem. 283:8564–8572. [DOI] [PubMed] [Google Scholar]
- Li, J., P.J. Peters, M. Bai, J. Dai, E. Bos, T. Kirchhausen, K.V. Kandror, and V.W. Hsu. 2007. An ACAP1-containing clathrin coat complex for endocytic recycling. J. Cell Biol. 178:453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.W., C.F. Huang, K.B. Huang, and F.J. Lee. 2005. Role for Gcs1p in regulation of Arl1p at trans-Golgi compartments. Mol. Biol. Cell. 16:4024–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin, B., G. Drin, S. Levi, M. Rawet, D. Cassel, J. Bigay, and B. Antonny. 2007. Two lipid-packing sensor motifs contribute to the sensitivity of ArfGAP1 to membrane curvature. Biochemistry. 46:1779–1790. [DOI] [PubMed] [Google Scholar]
- Natsume, W., K. Tanabe, S. Kon, N. Yoshida, T. Watanabe, T. Torii, and M. Satake. 2006. SMAP2, a novel ARF GTPase-activating protein, interacts with clathrin and clathrin assembly protein and functions on the AP-1-positive early endosome/trans-Golgi network. Mol. Biol. Cell. 17:2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, Z., and P.A. Randazzo. 2006. Arf GAPs and membrane traffic. J. Cell Sci. 119:1203–1211. [DOI] [PubMed] [Google Scholar]
- Poon, P.P., D. Cassel, A. Spang, M. Rotman, E. Pick, R.A. Singer, and G.C. Johnston. 1999. Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 18:555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premont, R.T., S.J. Perry, R. Schmalzigaug, J.T. Roseman, Y. Xing, and A. Claing. 2004. The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell. Signal. 16:1001–1011. [DOI] [PubMed] [Google Scholar]
- Randazzo, P.A., H. Inoue, and S. Bharti. 2007. Arf GAPs as regulators of the actin cytoskeleton. Biol. Cell. 99:583–600. [DOI] [PubMed] [Google Scholar]
- Sanchez-Velar, N., E.B. Udofia, Z. Yu, and M.L. Zapp. 2004. hRIP, a cellular cofactor for Rev function, promotes release of HIV RNAs from the perinuclear region. Genes Dev. 18:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek, K., M.R. Ahmadian, and A. Wittinghofer. 1998. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci. 23:257–262. [DOI] [PubMed] [Google Scholar]
- Schmalzigaug, R., H. Phee, C.E. Davidson, A. Weiss, and R.T. Premont. 2007. Differential expression of the ARF GAP genes GIT1 and GIT2 in mouse tissues. J. Histochem. Cytochem. 55:1039–1048. [DOI] [PubMed] [Google Scholar]
- Tanabe, K., S. Kon, W. Natsume, T. Torii, T. Watanabe, and M. Satake. 2006. Involvement of a novel ADP-ribosylation factor GTPase-activating protein, SMAP, in membrane trafficking: implications in cancer cell biology. Cancer Sci. 97:801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker, E., B. Kearns, C. Chapman, J. Hammond, A. Howell, and A. Theibert. 2004. The arf6 GAP centaurin alpha-1 is a neuronal actin-binding protein which also functions via GAP-independent activity to regulate the actin cytoskeleton. Eur. J. Cell Biol. 83:541–554. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu, K., K.G. Brandom, and J.L. Lawrence. 2004. Centaurin-alpha1 is an in vivo phosphatidylinositol 3,4,5-trisphosphate-dependent GTPase-activating protein for ARF6 that is involved in actin cytoskeleton organization. J. Biol. Chem. 279:6205–6208. [DOI] [PubMed] [Google Scholar]
- Watson, P.J., G. Frigerio, B.M. Collins, R. Duden, and D.J. Owen. 2004. Gamma-COP appendage domain—structure and function. Traffic. 5:79–88. [DOI] [PubMed] [Google Scholar]
- Wennerberg, K., K.L. Rossman, and C.J. Der. 2005. The Ras superfamily at a glance. J. Cell Sci. 118:843–846. [DOI] [PubMed] [Google Scholar]
- Ye, K., and S.H. Snyder. 2004. PIKE GTPase: a novel mediator of phosphoinositide signaling. J. Cell Sci. 117:155–161. [DOI] [PubMed] [Google Scholar]