Abstract
The four-subunit Ndc80 complex, comprised of Ndc80/Nuf2 and Spc24/Spc25 dimers, directly connects kinetochores to spindle microtubules. The complex is anchored to the kinetochore at the Spc24/25 end, and the Ndc80/Nuf2 dimer projects outward to bind to microtubules. Here, we use cryoelectron microscopy and helical image analysis to visualize the interaction of the Ndc80/Nuf2 dimer with microtubules. Our results, when combined with crystallography data, suggest that the globular domain of the Ndc80 subunit binds strongly at the interface between tubulin dimers and weakly at the adjacent intradimer interface along the protofilament axis. Such a binding mode, in which the Ndc80 complex interacts with sequential α/β-tubulin heterodimers, may be important for stabilizing kinetochore-bound microtubules. Additionally, we define the binding of the Ndc80 complex relative to microtubule polarity, which reveals that the microtubule interaction surface is at a considerable distance from the opposite kinetochore-anchored end; this binding geometry may facilitate polymerization and depolymerization at kinetochore-attached microtubule ends.
Introduction
Chromosome segregation requires dynamic interactions between kinetochores and spindle microtubules. Recent evidence has suggested that the core microtubule-binding site of the kinetochore is comprised of the KNL-1–Mis12 complex Ndc80 complex network (for review see Tanaka and Desai, 2008). This widely conserved set of interacting proteins is important for chromosome segregation in all systems analyzed. In vitro reconstitution experiments have indicated the presence of two distinct microtubule-binding activities within this set of proteins: the first in the Ndc80/Nuf2 heterodimer of the four-subunit Ndc80 complex (Cheeseman et al., 2006; Wei et al., 2007) and the second in KNL-1 (Cheeseman et al., 2006). Rotary shadowing EM, atomic force microscopy, and hydrodynamic studies of bacterially reconstituted Ndc80 complex have shown that this complex forms an ∼550-Å long rod-shaped structure with globular domains on both ends (Ciferri et al., 2005; Wei et al., 2005). The globular domains are formed by the N termini of Ndc80 and Nuf2 at one end and the N termini of Spc24 and Spc25 at the other end. Stretches of coiled coil immediately follow the N terminus of each of the four proteins in the complex. Antiparallel interaction of the coiled coils leads to the highly elongated shape of the heterotetrameric complex (Ciferri et al., 2005; Wei et al., 2005). Electron tomography of the kinetochore microtubule interface in cells has revealed a fibrous structure with multiple individual fibers contacting a single microtubule (Dong et al., 2007). These fibers may represent the Ndc80 complex in its native context at the kinetochore. Measurements in budding and fission yeast have indicated that five to eight Ndc80 complexes are present per kinetochore–microtubule attachment site (Joglekar et al., 2006, 2008). Collectively, these results suggest that multivalent associations between fibrous Ndc80 complexes extending out of the kinetochore and spindle microtubules are important for establishing dynamic kinetochore–microtubule interactions.
Structural studies of the globular domain of the human Ndc80 subunit have revealed that residues 81–196 comprise a calponin homology (CH) domain (Wei et al., 2007), a motif that was initially described in actin-binding proteins and more recently in the microtubule-binding proteins CLAMP and EB1 (Hayashi and Ikura, 2003; Dougherty et al., 2005; Slep and Vale, 2007). A recent crystal structure of an engineered, truncated Ndc80 complex revealed that the globular domain of Nuf2 also folds into a CH domain (Ciferri et al., 2008). The two CH domains of Ndc80 and Nuf2 are organized in an unusual arrangement relative to actin-binding tandem CH domain proteins (Ciferri et al., 2008). A basic 80–amino acid–long N-terminal region of Ndc80 is flexible, missing from the crystal structures, and critical for microtubule binding (Wei et al., 2007). This region is the major site of regulation on the Ndc80 complex by aurora B kinase (Cheeseman et al., 2006; DeLuca et al., 2006), which plays a central role in correcting improper chromosome–microtubule connections. Phosphorylation by aurora B reduces the affinity of the complex for microtubules (Cheeseman et al., 2006; Ciferri et al., 2008), and this is presumably part of the mechanism for aurora B–dependent removal of incorrect kinetochore–microtubule attachments in vivo. Consistent with this idea, mutation of aurora B target sites in the N-terminal region of Ndc80 leads to chromosome missegregation in human cells (DeLuca et al., 2006).
Negative stain EM of microtubules decorated with Ndc80 complexes has revealed that the complex binds to the lattice with the coiled-coil rods projecting away from the microtubule at variable angles (Cheeseman et al., 2006). Based on this geometry of binding, the microtubule-binding site of the Ndc80 complex is thought to reside in the globular CH domains of Ndc80 and Nuf2 and in the basic tail of Ndc80 (Wei et al., 2007; Ciferri et al., 2008). This is supported by the observations that the microtubule binding of the complex is localized to the Ndc80/Nuf2 heterodimer and does not require the Spc24/25 heterodimer (Cheeseman et al., 2006; Wei et al., 2007) and by mutational analysis of a conserved face of the paired CH domains (Ciferri et al., 2008). These prior studies established that the Ndc80 complex is centrally important in kinetochore–microtubule interactions and provided a framework for characterizing its microtubule-binding activity. Here, we directly visualize the footprint of the Ndc80/Nuf2 heterodimer on the microtubule lattice using cryo-EM. Additionally, we define the relationship between complex-binding geometry and microtubule polarity using centrosomal asters. The results reveal alternating strong and weak binding at every tubulin–tubulin interface along the microtubule protofilament and also indicate the presence of ordered regions that extend away from the microtubule. The binding geometry relative to the polymer's polarity is consistent with the requirement that kinetochore–microtubule attachments leave room for tubulin subunit addition and removal. We discuss these findings in light of crystallography data to suggest models for the interaction mechanism of the Ndc80 complex with microtubules.
Results and discussion
Ndc80/Nuf2 decoration of centrosomal asters
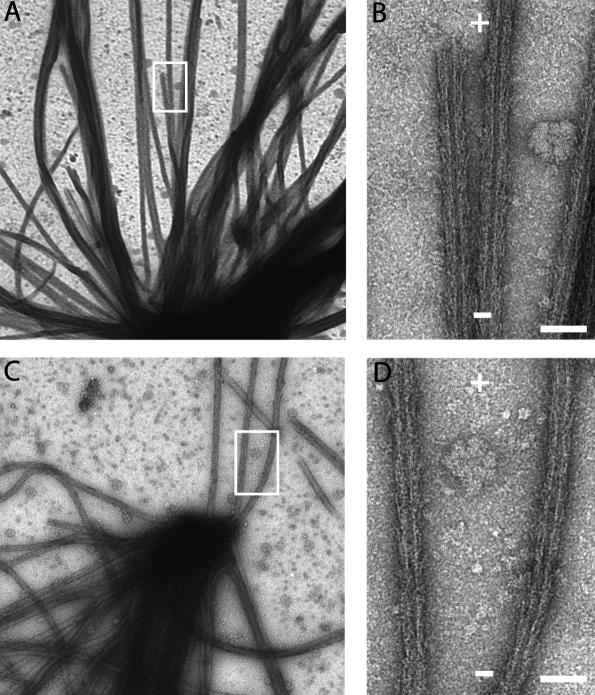
In the experiments described here, we used bacterially expressed Caenorhabditis elegans NDC-80/Nuf2HIM-10 heterodimers (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200804170/DC1; Cheeseman et al., 2006). We first analyzed the binding of Ndc80/Nuf2 relative to the polarity of the microtubule lattice. We grew microtubules of known polarity from centrosomes attached to EM grids and decorated these with the Ndc80/Nuf2 heterodimer. Examination of the negatively stained asters revealed that the arrowheads formed by the Ndc80/Nuf2 dimer point toward the centrosome (Fig. 1). Thus, the coiled-coil region of the heterodimer projects toward the plus end of the microtubule. In the heterotetrameric complex, this geometry of binding would force the Spc24/25 subunits to extend away from the microtubule plus end, where they likely bind to the kinetochore scaffold protein KNL-1 via their globular domains. This result is consistent with experiments comparing Spc24 and Ndc80/Hec1 localization at kinetochores in vertebrate cells using high resolution light microscopy (DeLuca et al., 2006). It is noteworthy that, given the length of the Ndc80 complex (∼550 Å), this binding geometry provides room at the microtubule plus end for polymerization and depolymerization reactions that are known to accompany chromosome movement in vivo.
Figure 1.
Orientation of the Ndc80/Nuf2 dimer relative to the polarity of the microtubule lattice. Negatively stained centrosome-nucleated microtubule asters decorated with the Ndc80/Nuf2 dimer (A and C). Boxed areas are magnified in B and D. + and − signs indicate microtubule polarity. Bars, 50 nm.
Ndc80/Nuf2 interaction with the microtubule
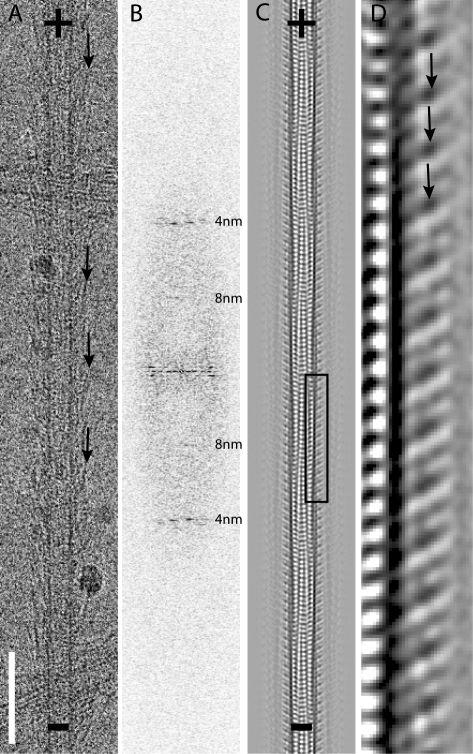
We next used EM and helical image analysis to investigate the interaction between the Ndc80 complex and the microtubule lattice. A preliminary examination of images of Ndc80/Nuf2 dimer–decorated microtubules (Fig. 2 A) revealed two significant features. First, diffraction patterns from fully decorated microtubules showed little evidence of 80-Å diffraction (Fig. 2 B). If each α/β-tubulin heterodimer bound a single Ndc80/Nuf2 dimer, as is the case for the kinesin motor domain, a strong 80-Å layer line would be present in the diffraction pattern. The absence of an 80-Å layer line indicates that Ndc80/Nuf2 binds with the same spacing as individual α- or β-tubulin monomers. Second, similar to previous observations in negative stain EM (Cheeseman et al., 2006), cryo-EM images of microtubules decorated with Ndc80/Nuf2 (Fig. 2 A) show relatively straight fibers (most likely the coiled-coil region) extending from the microtubule with a range of angles (∼20–60°). Because only the parts of Ndc80/Nuf2 that are rigidly attached to the lattice will be visible after averaging and image analysis, we could not visualize the entire heterotetrameric complex. Structural information associated with domains that are not reproducibly positioned with respect to the microtubule lattice, the variously angled fibers, is lost during averaging.
Figure 2.
Interaction of the Ndc80/Nuf2 dimer with the microtubule lattice. (A) An example of a vitrified microtubule decorated with the Ndc80/Nuf2 dimer. (B) A typical diffraction pattern (positions of 4- and 8-nm layer lines are marked). (C) A projection view of the three-dimensional map. (D) A magnified view of the boxed area in the three-dimensional map in C confirms the binding of the Ndc80/Nuf2 dimer to both α- and β-tubulin. Arrows point to the protruding density of the Ndc80/Nuf2 dimer in A and D. + and − signs indicate microtubule polarity. Bar, 85 nm.
To visualize the parts of Ndc80/Nuf2 firmly attached to the microtubule, we averaged data from 13 helical 15-protofilament Ndc80/Nuf2 dimer–decorated microtubules to obtain a three-dimensional map. A projection view of the map (Fig. 2, C and D) reveals alternating strong and weak (∼80-Å long) rod-shaped densities associated with each tubulin monomer along a protofilament. The characteristic moiré pattern in this projection can be used to infer microtubule polarity (Chretien et al., 1996). The rods make an angle of ∼60° with the microtubule surface, and, like the less well-ordered high radius coiled-coil regions, lean toward the plus end. A three-dimensional map calculated from five helical 16-protofilament microtubules, although of lower quality because of the limited number of images, confirmed the major features seen in the 15-protofilament map (unpublished data).
Features of the three-dimensional EM map
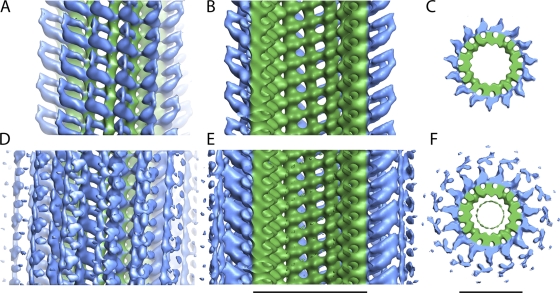
When the three-dimensional map is contoured such that only well-ordered regions are visualized (Fig. 3, A–C), densities that likely correspond to the N-terminal domains of the Ndc80/Nuf2 dimer and their connection to the tubulin protofilament are clearly apparent. A longitudinal slice through the map (Fig. 3 B) depicts two asymmetrical densities protruding orthogonally ∼80 Å from adjacent tubulin monomers at an angle of ∼60°. Most striking is an alternating pattern of strong and weak densities bridging sequential tubulin–tubulin interfaces along the protofilament axis. At this contour level, the coiled-coil rod of the Ndc80/Nuf2 dimer is not observed; the densities appear to merge ∼80 Å away from the microtubule surface. The lack of well-ordered density beyond this point suggests flexibility that is reflected in the variable angle of the rods observed on individual decorated microtubules.
Figure 3.
Surface representations of side and top views of the Ndc80/Nuf2 dimer–microtubule complex. The side views are oriented such that the plus end of the microtubule is at the top of the page. All views were colored using a cylinder radius that depicts the tubulin density as green and the Ndc80/Nuf2 dimer density as blue (Pettersen et al., 2004). Side and top views in A–C are contoured such that only the strongest, well-ordered density regions are visualized. Side and top views in D–F are contoured to show both strong and weak densities. Two sequential asymmetrical densities are clearly visible in the longitudinal slices, B and E, from the respective contoured views of the three-dimensional map, A and D. Top views of the slices are shown in C and F. Bars, 25 nm.
When we contoured the three-dimensional map to visualize less-ordered regions, weak densities at a significant distance (∼130 Å) from the microtubule surface were apparent (Fig. 3, D–F). To determine whether these weak densities were part of the map and not just noise, we generated projections of a raw image as well as for the three-dimensional map and used these to identify the radial positions of the density peaks (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200804170/DC1; Whittaker et al., 1995). We confirmed overlap between the radial distance of the density peaks found in the raw image and that of the final three-dimensional map, indicating that these weaker densities reflect ordered regions away from the microtubule surface. Microtubule binding of the Ndc80 complex exhibits cooperativity (Cheeseman et al., 2006; Ciferri et al., 2008), and a sequential “zippering”-type reaction that occurs off the microtubule axis by interactions between adjacent coiled coils may contribute to the cooperativity and be represented in the maps as the weak density at a higher radius. Although speculative, such a proposal should be testable using truncations and artificial dimerizing coiled coils in future work.
Interpretation of the strong densities associated with the microtubule surface in the three-dimensional EM map
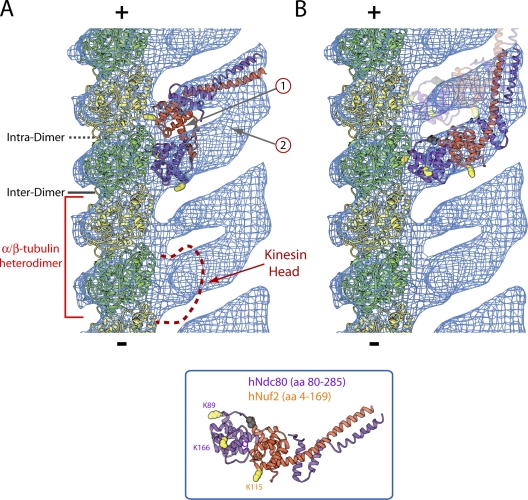
To help interpret the densities close to the microtubule surface, we docked a model of the high resolution polymerized form of the tubulin dimer into the electron density map (Fig. 4 A). This enabled observation of the contact points between the densities corresponding to the Ndc80/Nuf2 globular domains and the microtubule lattice. Given the resolution of the map, it is difficult to unambiguously distinguish between the intradimer and polymerization-dependent interdimer tubulin–tubulin interfaces along a protofilament. However, the best alignment between our map and a map of microtubule-bound kinesin-14 (Endres et al., 2006) indicates that the strong density binds near the interface between two different tubulin dimers (interdimer; Fig. 4 A) in a similar fashion to kinesins (Sosa et al., 1997; Highsmith et al., 2001), and the weaker density binds near the adjacent intradimer interface along the protofilament (the two densities are ∼35 Å apart; Fig. 4 A). Although it is possible that the alignment between maps is off register by one tubulin monomer, association of the strong density with the interdimer interface is supported by the lack of a significant affinity of the Ndc80 complex for unpolymerized tubulin dimer in vitro (unpublished data). In either scenario, unlike for kinesin, binding is observed near both intra- and intertubulin dimer interfaces (Fig. 4 A). This comparison with kinesin binding further suggests that it is unlikely for the Ndc80 complex and a kinesin family protein to simultaneously associate at the interdimer interface on a microtubule protofilament.
Figure 4.
Interaction site of the Ndc80/Nuf2 dimer on the microtubule lattice. (A and B) A model of a microtubule protofilament made from the tubulin dimer (PDB 1TUB) was docked into a single protofilament cut from the Ndc80/Nuf2 microtubule EM map. The crystal structure of an engineered, truncated Ndc80 complex was used for fitting into the remaining density in either a two-headed (A) or an alternating strong- or weak-binding (B) configuration. The red bracket demarcates an α/β-tubulin heterodimer based on the favored configuration in which the strong density overlaps the kinesin-binding site (red dashed line) at the interdimer interface. The + and − signs indicate microtubule polarity; β-tubulin of each α/β-tubulin heterodimer points toward the plus end. Lysines in Ndc80 and Nuf2, whose mutation to alanine or glutamate reduced microtubule-binding affinity (Ciferri et al., 2008), are indicated by yellow spheres; the black sphere marks the methionine preceding the first residue of Ndc80 (amino acid 80) in the crystal structure. For the two-headed model in A, the gray arrows labeled 1 and 2 highlight missing density and extra, unaccounted for density, respectively. For the strong or weak model in B, weak binding is schematized using light shading.
The asymmetry in densities along the protofilament axis observed in the three-dimensional map can be interpreted in two ways. First, the alternating pattern could represent strong binding of an Ndc80/Nuf2 dimer at a polymerization-dependent interdimer interface and weak binding to the adjacent intradimer interface along the protofilament axis. In this model, the observed pattern is caused by a difference in affinity at the sequential tubulin–tubulin interfaces. A second distinct model, which seems more favorable given the dual CH domain structure of the globular domains of Ndc80/Nuf2, is that the alternating densities represent a two-headed interaction of the Ndc80 complex along sequential tubulin–tubulin interfaces, with the strong density representing the N-terminal tail and CH domain of the Ndc80 subunit, and the weak density representing the Nuf2 CH domain. An electrostatic interaction between the basic tail of Ndc80 and the acidic C termini of α/β-tubulin is important for binding, as deletion of this tail (Wei et al., 2007) or subtilisin treatment of taxol-stabilized microtubules, which cleaves the acidic C termini of tubulin, compromises Ndc80 complex binding to microtubules in vitro (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200804170/DC1; Ciferri et al., 2008). Thus, examination of the densities close to the microtubule surface suggested two distinct mechanisms (alternating strong or weak binding versus a two-headed interaction) for how the Ndc80 complex associates with the microtubule lattice.
Discriminating models for interaction of the Ndc80 complex with microtubules by combining the three-dimensional EM map with crystallography data
We next attempted to fit the crystal structure of the Ndc80/Nuf2 globular regions (Ciferri et al., 2008) into the three-dimensional EM density map to discriminate between the two models for how the Ndc80 complex binds to microtubules. We did not obtain a fit compatible with the two-headed model, in which the conserved faces of the Ndc80 and Nuf2 CH domains are docked onto adjacent tubulin–tubulin interfaces (Fig. 4 A and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200804170/DC1). Specifically, significant predicted density from the interface between the two CH domains is not present in the three-dimensional map (Fig. 4 A, arrow 1), and significant density in the three-dimensional map is unaccounted for when the crystal structure is docked in this configuration (Fig. 4 A, arrow 2; and Video 1). In contrast, the crystal structure fits well into the strong density in the EM map with the Ndc80 CH domain docked close to the microtubule surface and the Nuf2 CH domain away from the lattice (Fig. 4 B and Video 2). This fit favors a model in which the binding interface for both the strong and weak densities is comprised exclusively by the N-terminal region of Ndc80, including the basic tail and CH domain, and the strong or weak densities arise from differing affinities for the sequential interfaces. It is possible that the weaker binding at the intradimer interface represents only an electrostatic interaction of the basic tail of Ndc80 with the acidic tail of tubulin, whereas the strong binding at the interdimer interface represents both an electrostatic interaction and a docking of the CH domain of Ndc80 onto specific features of the interdimer interface. A combination of electrostatic and a docking interaction has been previously observed with the kinesin-3 (Kif1A/Unc-104) subfamily (Okada and Hirokawa, 2000). The proposal for strong or weak binding with the geometry shown in Fig. 4 B contrasts with mutational analysis showing that altering the charge on the conserved surface of the Nuf2 CH domain reduces microtubule-binding affinity (Ciferri et al., 2008). One possible means to reconcile these observations is that the introduced mutations affect interactions between adjacent Ndc80 complexes and cooperativity of binding instead of direct contacts with the polymer surface.
In summary, a comparison of the crystal structure of the Ndc80/Nuf2 globular regions (lacking the basic tail) and the three-dimensional EM maps indicates that a two-headed mechanism of association with the microtubule lattice involving both the CH domains of Ndc80 and Nuf2 is unlikely. Instead, it appears that there is strong binding to the polymerization-dependent interdimer interface and weaker binding to the adjacent intradimer interface; the strong binding appears to be mediated by the Ndc80 subunit and may not involve direct contact of the Nuf2 CH domain with the protofilament surface.
Conclusion
Here, we provide a view of the Ndc80 complex's footprint on the microtubule lattice and define the relationship between the binding of this complex and microtubule polarity. The importance of this complex in chromosome segregation is well established, and it provides the evolutionarily ancient core microtubule attachment activity of the kinetochore. Our results indicate that the binding of the Ndc80 complex occurs at an angle relative to microtubule polarity such that if a complex were bound close to the end of a microtubule, there would be significant room (approximately five tubulin dimers long) for tubulin dimers to add and subtract at the end, assuming that the docking site on the kinetochore is restricted to the globular domains of Spc24 and Spc25. Three-dimensional maps of the tubulin–Ndc80 complex–binding interface are most consistent with the N-terminal region of Ndc80 binding strongly between two tubulin heterodimers and weakly at the interface of the same tubulin heterodimer. The CH domain of Nuf2 may contribute to binding cooperativity by interacting with an adjacent complex. The binding of the Ndc80 complex at an interdimer interface along the tubulin protofilament could stabilize microtubule attachment at the kinetochore and may also allow it to act as a coupler to dynamic microtubules. The presence of weaker densities distant from the microtubule surface suggests the presence of interactions away from the microtubule that may also contribute to the cooperativity of binding. These findings provide the basis for future studies of the interaction between the Ndc80 complex and microtubules.
Materials and methods
Microtubules polymerized in vitro are composed of variable numbers of protofilaments. Under the assembly conditions used here, 5–10% of the microtubules contain 15 and 16 protofilaments and can be identified in images by their diameter and by the characteristic moiré pattern resulting from the protofilament supertwist (Fig. 2 B). To identify the polarity of the microtubules decorated with the Ndc80/Nuf2 dimer head, we followed the method described previously by Hoenger and Milligan (1996). Centrosomes (provided by M. Moritz, University of California, San Francisco, San Francisco, CA) were adsorbed to carbon-coated electron microscope grids, a tubulin solution at a concentration of ∼1 mg/ml in BRB80 (80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA). 2 mM GTP and 0.25 mM taxol were incubated at 37°C in a humid chamber for 7 min. After aster formation (centrosomes with nucleated microtubules), the grids were washed with BRB80, incubated at room temperature with the Ndc80/Nuf2 dimer for 2 min, negatively stained with 1% uranyl acetate, and examined in the electron microscope.
Cryo-EM and image analysis
Microtubules were polymerized at 5 mg/ml in 80 mM Pipes, pH 6.8, 3 mM MgCl2, 16% (vol/vol) DMSO, and 2 mM GTP at 34°C for 30 min; 0.25 mM taxol was subsequently added, and incubation was continued for 30 min. The polymerized microtubules were left overnight at room temperature before use. Microtubules were diluted four to eight times with BRB80 before applying to plasma cleaned C-flat grids. The Ndc80/Nuf2 dimer (0.5 mg/ml in BRB80) was added to the microtubules by double blotting; the first 3-μl droplet of protein was partially blotted off before the second 3-μl of protein was added. The second protein droplet was incubated on the grid for 1–2 min before blotting and vitrifying in liquid ethane according to standard methods described previously (Dubochet et al., 1988) or by using a Vitrobot (FEI; humidity chamber set at 90%, and blot times between 3–4 s with an offset of −1). The specimens were examined with an electron microscope (CM200FEG; FEI) operating at 120 kV using a cryoholder (GATAN). Electron micrographs of the decorated microtubules preserved in vitreous ice were recorded in low dose conditions (<10 electrons/Å2) at a magnification of ∼38,000 and a nominal defocus between 1.5 and 2 μm. Electron micrographs were evaluated using an optical diffractometer to identify decorated microtubules with appropriate defocus and astigmatism. The selected micrographs were digitized on a scanner (Phodis SC; Carl Zeiss, Inc.) with a 7-μm pixel size. Groups of 3 × 3 pixels were averaged, giving a pixel size of 21 μm on the micrographs or 5.39 Å on the specimen.
Three-dimensional maps
Three-dimensional maps were calculated using Phoelix essentially as described previously (Whittaker et al., 1995). Surface representations of side and top views of the 15-protofilament map shown in Fig. 3 were produced with use of the Chimera software package (Resource for Biocomputing, Visualization, and Informatics; Pettersen et al., 2004). To obtain an accurate estimate of the length and angle at which the connected densities of Ndc80/Nuf2 dimer protrudes from the microtubule, we selected a section cut perpendicular to the three-dimensional map with clearly defined densities corresponding to the tubulin monomers and the connected densities. To determine the relationship between the Ndc80/Nuf2 dimer and the microtubule, we docked a model of the high resolution polymerized form of the tubulin dimer (PDB 1TUB) into the electron density as described previously by Nogales et al. (1999). To verify the positioning of kinesin versus the Ncd80/Nuf2 dimer, we overlapped a model of the crystal structure of Ncd-600k (PDB 1N6M) bound to the tubulin dimer (PDB 1TUB) as described previously by Endres et al. (2006) with our Ncd80/Nuf2 map (Fig. 4 A, red dashed line). To combine the EM densities with the crystal structure of a truncated, engineered Ndc80 complex (PDB 2VE7), we used the Ndc80/Nuf2 heterodimer (amino acids 80–285 of hNdc80 and 4–169 of hNuf2) to manually dock the structure into the densities in different configurations.
Online supplemental material
Fig. S1 shows schematics of the primary structures of C. elegans Ndc80 and Nuf2. Fig. S2 compares radial position of density peaks between the three-dimensional EM map and a raw image. Fig. S3 shows that subtilisin cleavage of microtubules significantly reduces binding of the Ndc80 complex. Video 1 shows a three-dimensional view of the EM density map into which the Ndc80/Nuf2 crystal structure has been placed in the two-headed configuration. Video 2 shows a three-dimensional view of the EM density map into which the Ndc80/Nuf2 crystal structure has been fitted into the strong density in a configuration in which the Nuf2 CH domain is off the microtubule lattice. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200804170/DC1.
Supplementary Material
Acknowledgments
We thank M. Moritz for providing centrosomes and A. Mulder (The Scripps Research Institute, La Jolla, CA) and J. Welburn (Whitehead Institute, Cambridge, MA) for comments on the manuscript.
Some of the work presented here was conducted at the National Resource for Automated Molecular Microscopy (supported by National Institutes of Health grant P41 RR-17573). Molecular graphics images were produced using the Chimera package from the University of California, San Francisco (supported by National Institutes of Health grant P41 RR-01081). This project was funded by National Institutes of Health grants to R.A. Milligan (GM 52468-13 and GM75820-03) and A. Desai (GM074215) and by funding from the Ludwig Institute for Cancer Research to A. Desai.
I.M. Cheeseman's present address is Whitehead Institute for Biomedical Research and Dept. of Biology, Massachusetts Institute of Technology, Cambridge, MA 02142.
Abbreviation used in this paper: CH, calponin homology.
References
- Cheeseman, I.M., J.S. Chappie, E.M. Wilson-Kubalek, and A. Desai. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. [DOI] [PubMed] [Google Scholar]
- Chretien, D., J.M. Kenney, S.D. Fuller, and R.H. Wade. 1996. Determination of microtubule polarity by cryo-electron microscopy. Structure. 4:1031–1040. [DOI] [PubMed] [Google Scholar]
- Ciferri, C., J. De Luca, S. Monzani, K.J. Ferrari, D. Ristic, C. Wyman, H. Stark, J. Kilmartin, E.D. Salmon, and A. Musacchio. 2005. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem. 280:29088–29095. [DOI] [PubMed] [Google Scholar]
- Ciferri, C., S. Pasqualato, E. Screpanti, G. Varetti, S. Santaguida, G. Dos Reis, A. Maiolica, J. Polka, J.G. De Luca, P. De Wulf, et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, J.G., W.E. Gall, C. Ciferri, D. Cimini, A. Musacchio, and E.D. Salmon. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. [DOI] [PubMed] [Google Scholar]
- Dong, Y., K.J. Vanden Beldt, X. Meng, A. Khodjakov, and B.F. McEwen. 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9:516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty, G.W., H.J. Adler, A. Rzadzinska, M. Gimona, Y. Tomita, M.C. Lattig, R.C. Merritt Jr., and B. Kachar. 2005. CLAMP, a novel microtubule-associated protein with EB-type calponin homology. Cell Motil. Cytoskeleton. 62:141–156. [DOI] [PubMed] [Google Scholar]
- Dubochet, J., M. Adrian, J.J. Chang, J.C. Homo, J. Lepault, A.W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21:129–228. [DOI] [PubMed] [Google Scholar]
- Endres, N.F., C. Yoshioka, R.A. Milligan, and R.D. Vale. 2006. A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Nature. 439:875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, I., and M. Ikura. 2003. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J. Biol. Chem. 278:36430–36434. [DOI] [PubMed] [Google Scholar]
- Highsmith, S., M. Thoene, E. Sablin, and K. Polosukhina. 2001. NCD activation of tubulin polymerization. Biophys. Chem. 92:127–139. [DOI] [PubMed] [Google Scholar]
- Hoenger, A., and R.A. Milligan. 1996. Polarity of 2-D and 3-D maps of tubulin sheets and motor-decorated sheets. J. Mol. Biol. 263:114–119. [DOI] [PubMed] [Google Scholar]
- Joglekar, A.P., D.C. Bouck, J.N. Molk, K.S. Bloom, and E.D. Salmon. 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8:581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar, A.P., D. Bouck, K. Finley, X. Liu, Y. Wan, J. Berman, X. He, E.D. Salmon, and K.S. Bloom. 2008. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 181:587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales, E., M. Whittaker, R.A. Milligan, and K.H. Downing. 1999. High-resolution model of the microtubule. Cell. 96:79–88. [DOI] [PubMed] [Google Scholar]
- Okada, Y., and N. Hirokawa. 2000. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc. Natl. Acad. Sci. USA. 97:640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F., T.D. Goddard, C.C. Huang, G.S. Couch, D.M. Greenblatt, E.C. Meng, and T.E. Ferrin. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Slep, K.C., and R.D. Vale. 2007. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol. Cell. 27:976–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa, H., D.P. Dias, A. Hoenger, M. Whittaker, E. Wilson-Kubalek, E. Sablin, R.J. Fletterick, R.D. Vale, and R.A. Milligan. 1997. A model for the microtubule-Ncd motor protein complex obtained by cryo-electron microscopy and image analysis. Cell. 90:217–224. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U., and A. Desai. 2008. Kinetochore-microtubule interactions: the means to the end. Curr. Opin. Cell Biol. 20:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, R.R., P.K. Sorger, and S.C. Harrison. 2005. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA. 102:5363–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, R.R., J. Al-Bassam, and S.C. Harrison. 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14:54–59. [DOI] [PubMed] [Google Scholar]
- Whittaker, M., B.O. Carragher, and R.A. Milligan. 1995. PHOELIX: a package for semi-automated helical reconstruction. Ultramicroscopy. 58:245–259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.