Abstract
Lis1 and Ndel1 are essential for animal development. They interact directly with one another and with cytoplasmic dynein. The developing brain is especially sensitive to reduced Lis1 or Ndel1 levels, as both proteins influence spindle orientation, neural cell fate decisions, and neuronal migration. We report here that Lis1 and Ndel1 reduction in a mitotic cell line impairs prophase nuclear envelope (NE) invagination (PNEI). This dynein-dependent process facilitates NE breakdown (NEBD) and occurs before the establishment of the bipolar spindle. Ndel1 phosphorylation is important for this function, regulating binding to both Lis1 and dynein. Prophase cells in the ventricular zone (VZ) of embryonic day 13.5 Lis1+/− mouse brains show reduced PNEI, and the ratio of prophase to prometaphase cells is increased, suggesting an NEBD delay. Moreover, prophase cells in the VZ contain elevated levels of Ndel1 phosphorylated at a key cdk5 site. Our data suggest that a delay in NEBD in the VZ could contribute to developmental defects associated with Lis1–Ndel1 disruption.
Introduction
Lis1 haploinsufficiency causes a severe developmental brain disorder in humans, type 1 lissencephaly (Cardoso et al., 2002; Mei et al., 2008). Homozygous loss of Lis1 or its binding partner, Ndel1, is thought to be lethal, as is the case in mice (Hirotsune et al., 1998; Sasaki et al., 2005). The C terminus of Lis1 binds to a microtubule motor, cytoplasmic dynein (Reiner et al., 1993; Sasaki et al., 2000), whereas the N terminus contains a LisH homodimerization domain (Kim et al., 2004). Between these domains is a coiled-coil region that provides flexibility to the Lis1 dimer (Tarricone et al., 2004). Ndel1 contains an N-terminal coiled-coil domain that interacts with Lis1 and an unstructured C terminus possessing five Ser/Thr-Pro (S/T-P) sites targeted by Pro-directed kinases (Niethammer et al., 2000; Derewenda et al., 2007). Ndel1 also binds directly to dynein. In fact, Lis1 and Ndel1 are thought to associate with cytoplasmic dynein as a heterotetramer (Tarricone et al., 2004).
Cytoplasmic dynein is a multisubunit complex that contributes to processes requiring force generation. Dynein interacts with membranes through another multisubunit protein, dynactin, which also regulates dynein processivity (King and Schroer, 2000; Schroer, 2004). Although evidence that Lis1 and Ndel1 regulate dynein is accumulating, the precise mechanism is unclear. Lis1 modestly stimulates dynein's ATPase function in vitro (Mesngon et al., 2006), and there is evidence for functional interactions in several organisms (Vallee and Tsai, 2006). Reduction of Lis1 in developing rat neurons inhibited interkinetic nuclear oscillations in the ventricular zone (VZ), reduced mitosis, and perturbed the acquisition of a bipolar morphology in the subventricular/intermediate zones (Tsai et al., 2005). In mice, reduction of Lis1, Ndel1, or dynein heavy chain (DHC) disrupted the link between the centrosome and the nucleus in migrating neurons (Shu et al., 2004). Lis1 overexpression could partly compensate for Ndel1 reduction, but not vice versa. Neither protein was able to rescue DHC deficiency (Shu et al., 2004; Sasaki et al., 2005). More recently, Lis1 reduction was shown to alter spindle orientation in neuroepithelial stem cells so that the cellular cleavage plane was biased toward asymmetrical cell divisions and neurogenesis (Yingling et al., 2008). It was hypothesized that this resulted in the early depletion of a progenitor pool.
Two observations prompted us to explore another process that might be influenced by Lis1 pathways. The first was that dynein activity facilitated nuclear envelope (NE) breakdown (NEBD; Salina et al., 2002) by promoting prophase NE invagination (PNEI) in a microtubule-dependent manner, resulting in NE tearing. The second observation was a curious enrichment of Lis1 at the NE in prophase cells and in cells treated with nocodazole (Smith et al., 2000; Coquelle et al., 2002; Cockell et al., 2004). These observations suggest that Lis1 could function in neuroepithelial stem cells before its role in spindle orientation by contributing to dynein-dependent PNEI. We used in vivo and in vitro approaches to explore this possibility and found that both Lis1 and Ndel1 influence PNEI in cultured cell lines and in the developing mouse brain. Furthermore, we find that Ndel1 phosphorylation may be a key regulatory step and, as such, could fine tune the timing of NEBD.
Results and discussion
Reducing Lis1 expression in COS-7 cells reduces PNEI
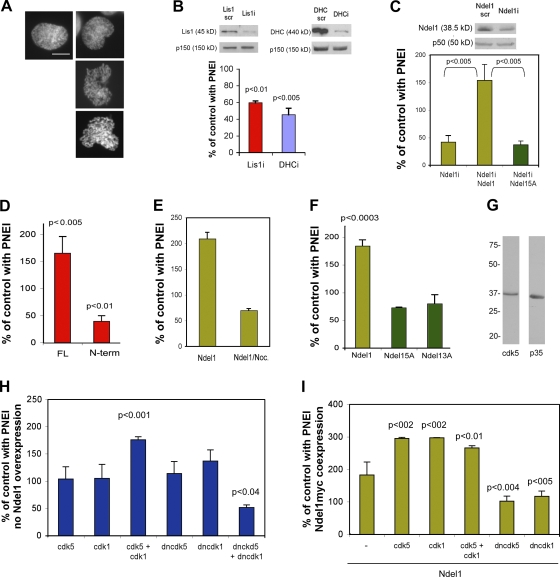
As expected, Lis1 and Ndel1 accumulated at the NE and within invaginations in prophase cells (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200803071/DC1). PNEI can be quantified by examining prophase nuclei labeled with Hoechst dye and determining the percentage with invaginations (Fig. 1 A). To determine whether Lis1 is important for PNEI, we transiently expressed a Lis1 short hairpin RNA (shRNA), which reduced Lis1 expression by 80% after 48 h (Fig. 1 B). An shRNA for DHC reduced DHC expression by 70%. Scrambled sequences were used as negative controls. Lis1 shRNA reduced PNEI by ∼40%, whereas DHC RNAi reduced PNEI by ∼50% (Fig. 1 B). The data support a model in which Lis1, like dynein, facilitates NEBD by promoting PNEI.
Figure 1.
Lis1 and Ndel1 contribute to PNEI. (A) COS-7 cell nuclei labeled with Hoechst. (left) A prophase nucleus before PNEI. (right) Nuclei with varying degrees of PNEI. (B, top) Lis1 shRNA (Lis1i) or dynein shRNA (DHCi) reduces protein expression by 48 h after transfection. (bottom) Relative to scrambled controls, the percentage of prophase cells with PNEI was reduced by Lis1 or DHC RNAi. (C) Similarly, Ndel1 RNAi reduced Ndel1 expression and PNEI. This was prevented by coexpression of Ndel1 but not Ndel15A, which has S/T→A mutations in five cdk sites. (D) Increased PNEI was caused by full-length Lis1 (FL), whereas a Lis1 fragment (N term) reduced PNEI. (E) Nocodazole treatment for 13 h after transfection prevents Ndel1-induced PNEI. (F) Neither Ndel15A nor Ndel13A (S/T→A mutations in the first three cdk sites) increased PNEI. (G) COS-7 cells express cdk5 and p35. (H) Cells were transfected with cdk5/p25, cdk1/cyclin B1, or dominant-negative kinases (dncdk5 and dncdk1). (I) The kinases described in H were cotransfected with Ndel1. Data are the mean ± SD for three cultures. Significance was determined by Student's t test. Error bars indicate ±SD. Bar, 10 μm.
Reducing Ndel1 expression in COS-7 cells reduces PNEI
Earlier studies suggested that Lis1 and Ndel1 cooperate to regulate dynein (Yan et al., 2003; Shu et al., 2004), and Ndel1 accumulates at the prophase NE (Fig. S1, E and F; and Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200803071/DC1). Therefore, we anticipated that reducing Ndel1 expression might have a similar effect on PNEI. The Ndel1 shRNA reduced Ndel1 protein expression by ∼50% in 48 h (Fig. 1 C, top) and also reduced PNEI by ∼50% (Fig. 1 C). The phenotype was rescued by coexpression of human myc-tagged Ndel1, so the level of expression of wild-type Ndel1 from the cytomegalovirus promoter was apparently high enough to overcome shRNA expression driven by the U6 promoter. Interestingly, the phenotype was not rescued by coexpression of an Ndel1 construct with mutations in five key cdk phosphorylation sites (Ndel15A; Fig. 1 C). Thus, Ndel1 phosphorylation could be a regulatory step in PNEI.
Increasing Lis1 or Ndel1 expression in COS-7 cells increases PNEI
We expected that transient overexpression of Lis1 or Ndel1 would block PNEI by disrupting normal stoichiometry of protein–protein interactions. Instead, we found that overexpression of Lis1 nearly doubled PNEI compared with controls (Fig. 1 D). This was dependent on intact microtubules, as the increase was blocked by nocodazole (unpublished data). A mutant construct that expresses an N-terminal fragment of Lis1 containing the LisH domain reduced PNEI by 50%, suggesting that Lis1 dimers are likely to be used during dynein-dependent PNEI. This fragment did not accumulate at the prophase NE (unpublished data), so the redistribution of Lis1 to the NE may also depend on homodimerization. Overexpression of Ndel1 was found to have a similar effect on PNEI (Fig. 1 E). This supports a model in which Ndel1 and Lis1 promote the microtubule-dependent invaginations attributed to dynein function (Beaudouin et al., 2002; Salina et al., 2002). Lis1 and Ndel1 expression may be elevated in prophase cells (Fig. S1, A and E), so transient overexpression of either protein may recapitulate or hasten a state conducive to PNEI.
S/T-P phosphorylation sites in Ndel1 are important for stimulation of PNEI
Interestingly, the phosphomutant Ndel15A did not induce PNEI. Normal PNEI was only modestly reduced by Ndel15A, so it did not significantly perturb endogenous Ndel1 function (Fig. 1 F). S197, T219, and S231 are targets for cdk5 (Niethammer et al., 2000), whereas cdk1 targets S242 and T245 (Yan et al., 2003). Because cdk5 mainly functions in postmitotic neurons, we expected that a construct in which S/T→A mutations were restricted to the cdk5 sites (Ndel13A) would retain its ability to stimulate PNEI. However, this was not the case (Fig. 1 F). Both cdk5 and the cdk5 activator, p35, were present at substantial levels in COS-7 cells (Fig. 1 G), raising the intriguing possibility that cdk5 activity is important during PNEI.
To further examine the role of phosphorylation, we increased or reduced cdk5 and cdk1 activity. The effect on PNEI was determined in cells with normal Ndel1 levels (Fig. 1 H) and in cells overexpressing Ndel1 (Fig. 1 I). Increasing cdk5 or cdk1 activity in the absence of Ndel1 overexpression did not alter PNEI. However, increasing the activity of both kinases almost doubled PNEI compared with controls. Furthermore, a significant inhibitory effect (50%) was observed when dominant-negative kinases (dncdk5 and dncdk1) were coexpressed. Neither dncdk5 nor dncdk1 was able to reduce PNEI when expressed individually, indicating that either (a) endogenous cdk5 or endogenous cdk1 is sufficient to allow normal PNEI to occur, or (b) the reagents incompletely inhibit their respective kinases. In fact, inhibiting cdk1 may actually increase PNEI (although this did not reach statistical significance), suggesting a complex relationship between the activity of this kinase and PNEI.
The aforementioned reagents produced somewhat different results when Ndel1 was also overexpressed. Cdk5 and cdk1 together enhanced PNEI over that stimulated by Ndel1. However, in this situation, the individual kinases were equally effective at stimulating PNEI. Expressing both dncdk5 and dncdk1 prevented PNEI induced by Ndel1 overexpression. However, PNEI was also inhibited when dncdk1 or dncdk5 was expressed individually, so each kinase alone is insufficient to fully phosphorylate ectopic Ndel1 at the appropriate sites to allow it to induce PNEI. Together, the data suggest that the activities of cdk5 and cdk1 can compensate for one another under some conditions. However, they also point to a dosage affect, i.e., the cell must have sufficient amounts of appropriately phosphorylated Ndel1 to carry out normal PNEI.
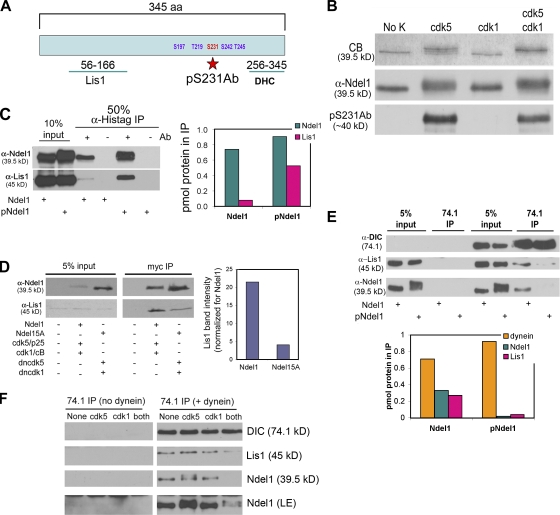
Ndel1 phosphorylation impacts Lis1 and dynein binding
Fig. 2 A shows a schematic of Ndel1 indicating the positions of cdk target residues. The pS231Ab antibody was generated against a peptide phosphorylated at S231, which is thought to be targeted by cdk5, not cdk1. To gain more insight into the role of Ndel1 phosphorylation, we turned to in vitro assays with purified proteins. We exposed Ndel1 to cdk5/p25 (p25 is a stable fragment of p35), cdk1/cyclin B, or both. Cdk5/p25 caused a distinct Ndel1 banding pattern in Coomassie-stained gels (Fig. 2 B). Cdk1/cyclin B produced a modest band shift. The band pattern observed when both kinases were included was distinct from that produced by cdk5 alone, indicating that both kinases target Ndel1 in vitro. pS231Ab recognized Ndel1 phosphorylated in vitro by cdk5 or by cdk5 and cdk1 together but not by cdk1 alone (Fig. 2 B). Yan et al. (2003) demonstrated that an Ndel1 construct with S/T→E mutations bound more Lis1. In support of this, we find that Ndel1 phosphorylated in vitro by both cdk5 and cdk1 pulled down approximately fivefold more Lis1 than unphosphorylated Ndel1 (Fig. 2 C). This was also true in coimmunoprecipitations (coIPs) of Ndel1 or Ndel15A coexpressed with active or dominant-negative kinases in COS-7 cells (Fig. 2 D). We anticipated that Ndel1 phosphorylation would have a similar impact on dynein binding but found instead that more unphosphorylated Ndel1 coimmunoprecipitated with purified bovine brain dynein (Fig. 2 E). Moreover, Ndel1 phosphorylation reduced the amount of Lis1 that coimmunoprecipitated with dynein, suggesting that phosphorylation not only strengthens Ndel1 binding to Lis1 but promotes the release of Lis1–pNdel1 complexes from dynein. Phosphorylation by either kinase alone was insufficient to reduce dynein binding (Fig. 2 F). Thus, although it is clear that Ndel1 phosphorylation impacts its interactions with the dynein motor machinery and PNEI, phosphorylation by individual kinases could have different biological consequences than phosphorylation by both kinases.
Figure 2.
Ndel1 phosphorylation alters Lis1 and dynein interactions. (A) Schematic of five S/T-P sites in Ndel1 showing the residue recognized by the pS231Ab antibody. (B) Ndel1 was exposed to GST-cdk5/p25, GST-cdk1/cyclin B, or both. Top, Coomassie staining; middle, Ndel1 antibody; bottom, pS231Ab. (C, left) Immunoprecipitations of unphosphorylated or phosphorylated Ndel1 (Ndel1 and pNdel1) incubated with purified Lis1. (right) Quantification is representative of three experiments. (D) COS-7 cells were transfected with Ndel1, cdk5/p25, and cdk1/cyclin B or with Ndel15A, dncdk5, and dncdk1. (left) Immunoprecipitations for Ndel1 constructs. (right) Quantification is representative of three experiments. (E) Dynein co-IPs. (top) Probed for DIC, Lis1, and Ndel1. (bottom) Quantification is representative of three experiments. (F) Dynein co-IP with Ndel1 phosphorylated by individual kinases. LE, longer exposure.
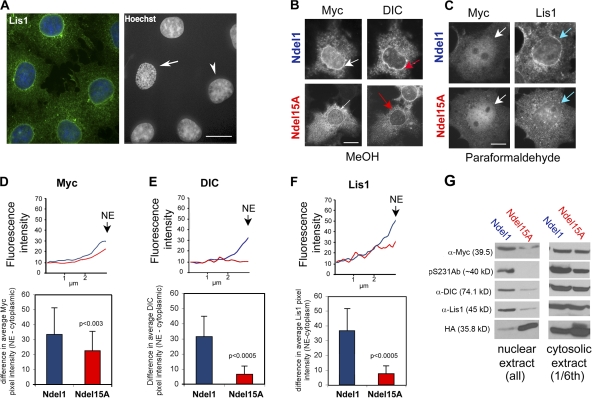
Accumulation of Lis1 and Ndel1 at the prophase NE can occur in the absence of microtubules but requires an intact dynactin complex
If dynein-dependent transport is important for relocation of Lis1 and Ndel1 to the NE, microtubule disruption should prevent their accumulation. However, prophase cells had typical NE enrichment of Lis1 3 h after exposure to nocodazole (Fig. 3 A) despite a complete loss of microtubules within minutes of drug addition. G2 cells also exhibited NE accumulation, and Ndel1 and dynein behaved similarly. A time course study indicated that nocodazole caused a transient increase in cells with Lis1–Ndel1–dynein enrichment at the NE and delayed NEBD (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200803071/DC1). A normal transient, microtubule-sparse state in G2 cells (Fig. S2 B) may be mimicked by microtubule disruption to allow accumulation of proteins at the NE.
Figure 3.
Ndel15A expression prevents accumulation of Lis1 and DIC at the NE in response to nocodazole. (A) 10-μM nocodazole delays NEBD in COS-7 cells and results in an increase in cells with Lis1 (green) at the NE. Arrow, prophase cell; arrowhead, G2 cell. (B) Ndel1 (top left) and DIC (top right) accumulate at the NE (red arrows) in a nocodazole-treated cell. Ndel15A (bottom left) but not DIC (bottom right) accumulates at the NE (white arrows) in a nocodazole-treated cell. Ndel1 constructs are myc tagged. (C) Lis1 at the NE was reduced by Ndel15A (bottom right). White arrows indicate accumulation of myc-tagged Ndel1 or Ndel15A at the NE. Blue arrows indicate the accumulation of Lis1 at the NE. (D–F) NE accumulation of Ndel1, DIC, and Lis1 was quantified (see Materials and methods). Statistical significance was determined by Student's t test. (G) COS-7 cells were cotransfected with Ndel1, cdk5, p25, HA-cdk1, and cyclin B (Ndel1) or Ndel15A, dncdk5, and HA-dncdk1 (Ndel15A) and exposed to nocodazole for 3 h. Nuclear and cytosolic extracts were examined by Western blotting. Error bars indicate ±SD. Bars, 10 μm.
Dynein's interaction with the NE is disrupted by overexpression of dynactin subunits (Salina et al., 2002). Dynactin colocalizes with Lis1 at the NE in nocodazole-treated cells (Fig. S2 D). Interestingly, overexpression of the p50 dynactin subunit prevented Lis1 redistribution to the NE in response to nocodazole (Fig. S2 E), indicating that Lis1 traffics to the NE with dynein. Lis1 colocalized at the NE with an ER marker (anti-protein disulfide isomerase; Fig. S2 C) but not with a Golgi or lysosomal marker (GM130; Lysotracker red; not depicted), so reorganization of the ER may contribute to accumulation of dynein machinery at the prophase NE.
Ndel1 phosphorylation influences the accumulation of Lis1 and dynein at the NE
When expressed in COS-7 cells, Ndel1 and, to a lesser extent, Ndel15A accumulated at the NE in response to nocodazole (Fig. 3 B). Comparing mean pixel intensities along lines ending at the NE confirmed this impression, as did comparison of pixel intensities around the NE and in the proximal cytoplasm (Fig. 3 D). Expression of Ndel15A substantially reduced dynein or Lis1 redistribution to the NE (Fig. 3, B–F). This was confirmed biochemically by analyzing Western blots of nuclear extracts from nocodazole-treated cells (Fig. 3 G). In this experiment, Ndel1 or Ndel15A was coexpressed with cdk5/cdk1 or dncdk5/dncdk1, respectively. Ponceau S and Coomassie staining of whole cell extract before nuclear isolation indicated that the samples had similar total protein levels (unpublished data). Substantially more Lis1 and dynein were present in nuclear extracts from cells expressing Ndel1. The pS231Ab labeled a band in this extract, but not in nuclear extract, from cells expressing Ndel15A. The pS231Ab band in the Ndel15A cytosolic extract may be endogenous Ndel1 that retained S231 phosphorylation in this short experiment (extracts were prepared 14 h after transfection).
The biochemical and immunofluorescence data indicate that Lis1 and dynein recruitment to the NE is reduced if Ndel1 cannot become phosphorylated by cdk5 and cdk1. This is consistent with the finding that Ndel1 phosphorylation increases its binding to Lis1. However, in vitro phosphorylation by both kinases reduced Ndel1 binding to dynein. Coupled with the finding that Ndel15A blocks recruitment to the NE and is unable to induce PNEI, a model is emerging in which recruitment to the NE and induction of PNEI may involve sequential phosphorylation of different sites and/or cycles of phosphorylation/dephosphorylation.
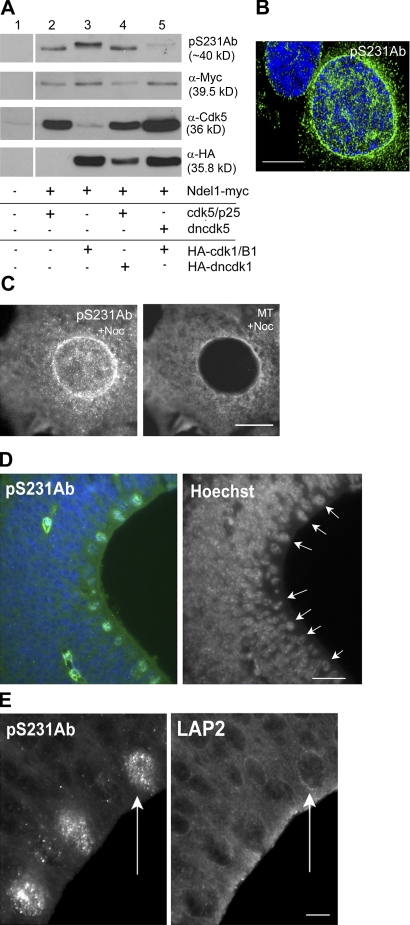
S231 is phosphorylated in prophase neural stem cells
Although both cdk5 and cdk1 activity in COS-7 cells induced Ndel1 phosphorylation at S231, inhibition of endogenous cdk5 (by dncdk5) blocked cdk1-stimulated S231 phosphorylation, whereas inhibition of endogenous cdk1 (by dncdk1) did not prevent cdk5-stimulated S231 phosphorylation (Fig. 4 A). One possible explanation for this is that phosphorylation of distinct sites by cdk1 increases S231 phosphorylation by cdk5 in the cell.
Figure 4.
Ndel1 phosphorylated at S231 is enriched at the prophase NE. (A) Western blot of COS-7 cells transfected as shown. Cdk5 and cdk1 activity increased pS231Ab signal (lanes 2 and 3). The presence of dncdk1 does not block phosphorylation of S231 by cdk5 (lane 4), but dncdk5 blocks S231 phosphorylation by cdk1 (lane 5). (B) A prophase NE labeled with pS231Ab (green). (C, right) pS231Ab labels the NE in cells treated with nocodazole. (left) Tubulin staining. (D, left) Parasagittal embryonic day 13.5 mouse embryo sections stained with pS231Ab (green) and Hoechst (blue). (right) Only the Hoechst is shown; arrows indicate mitotic cells at the ventricular surface. (E) pS231Ab (left) labels cells with intact LAP2 (right; arrows). Bars: 10 μm (B and C); 20 μm (D); 5 μm (E).
Several experiments indicated that pS231Ab could be used to label S231 phosphorylated by cdk5 in fixed cells (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200803071/DC1). p231 labeled the NE in prophase cells and in nocodazole-treated COS-7 cells (Fig. 4, B and C). Cdk5 function is critical during brain development, so we examined whether pS231Ab staining was present in neural stem cells in the developing VZ. Parasaggital sections of embryonic day 13.5 mouse brains were processed for pS231Ab immunofluorescence (Fig. 4 D). Mitotic cells identified by Hoechst staining and proximity to the ventricular surface were labeled by pS231Ab, suggesting that the timing of S231 phosphorylation is coordinated with mitotic entry. Some pS231Ab-labeled cells appeared to be in prophase based on a lamin-associated protein 2 (LAP2) pattern, so phosphorylation by cdk5 at S231 can occur at the appropriate time to contribute to PNEI and NEBD (Fig. 4 E).
Reduced PNEI and delayed NEBD in the VZ of developing Lis1+/− mice
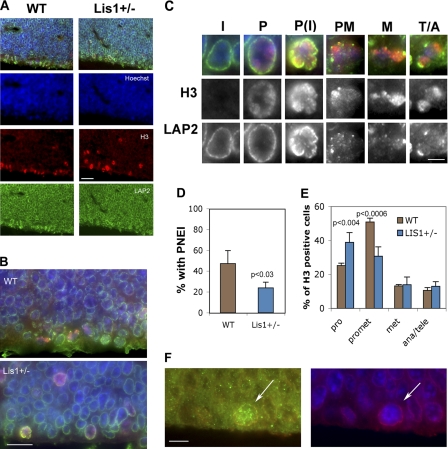
We were interested in whether Lis1 deficiency had an impact on PNEI or the timing of NEBD in the mouse VZ. To examine this, parasaggital sections of embryonic day 13.5 Lis1+/− mouse brains were immunostained for histone H3, which labels all stages of mitosis, and LAP2, which labels the NE (Fig. 5, A and B). As observed by others, mitotic cells were mispositioned in the Lis1+/− brains (Gambello et al., 2003). All H3-positive cells were analyzed in multiple sections from three animals of each genotype to determine whether there were differences in mitotic stage or in the number of prophase cells with invaginations. Mitotic cells were easily identified by H3 staining, and mitotic stage was easily determined by LAP2 staining and chromatin arrangement (Fig. 5 C). The percentage of H3-positive cells with intact NEs was significantly higher in Lis1+/− animals relative to wild-type littermates (prophase; Fig. 5 E). Of these, the percentage with clear PNEI was reduced in the mutant animals (Fig. 5 D), as was the percentage of prometaphase cells (Fig. 5 E), raising the possibility that NEBD is delayed in response to Lis1 deficiency. Upon examination of Lis1 immunofluorescence in the VZ, we found that prophase cells were more intensely labeled than surrounding cells, so Lis1 is present at the appropriate time to regulate dynein-dependent PNEI in the developing brain (Fig. 5 F).
Figure 5.
Embryonic day 13.5 Lis1+/− mouse brains have reduced PNEI and delayed NEBD. (A) Parasagittal sections through embryonic day 13.5 cortices from wild-type (WT) and Lis1+/− mice stained with LAP2 (green), H3 (red), and Hoechst (blue). (B) Enlargement of VZ. (C) Interphase (I) cells lack H3 staining and have intact NEs. Prophase (P) cells had an intact NE, sometimes with invaginations (PI). Prometaphase (PM), NE disrupted and chromosomes not aligned. Metaphase (M) and telophase/anaphase (T/A) cells were categorized by chromosome arrangement. (D) The percentage of prophase cells with PNEI was decreased in Lis1+/− animals. Three sections per animal were analyzed. Error bars indicate the mean ± SD for three animals per genotype. (E) Lis1+/− animals (blue) had more prophase (pro) and fewer prometaphase cells (promet) than wild-type littermates (brown). (F) A prophase cell in a wild-type embryonic day 13.5 brain stained for Lis1 (green) and LAP2 (red). (right) LAP2 (red) and Hoechst (blue). Arrows indicate prophase cells at the VZ. (D and E) Significance determined by Student's t test. Error bars indicate ±SD. Bars: 20 μm (A); 10 μm (B); 2.5 μm (C); 5 μm (F).
The simplest explanation of this Lis1+/− data is that Lis1 deficiency causes an NEBD delay in the VZ. Beaudouin et al. (2002) estimated that an NEBD delay was caused by microtubule disruption in living normal rat kidney (NRK) cells, which normally complete NEBD 10 min after the appearance of invaginations. After exposure to 1 μM nocodazole, the same process required 15 min. This represents a 5-min delay or a 50% increase in the duration of NEBD, which could have serious consequences in vivo. Our data do not allow us to estimate the duration of the delay caused by Lis1 deficiency. However, the finding that the proportion of mitotic cells with intact NEs was almost doubled in Lis1+/− embryos suggests a significant delay, which could conceivably alter cell fate. Neural stem cells in the cortical VZ are classified as neuroepithelial cells, radial glia (RG), or basal progenitors (BPs), and these appear to be progressively more restricted in their cell fate potential (Gotz and Huttner, 2005). During early development, NE and RG cells undergo mitosis at the apical surface nearest the ventricle. They divide symmetrically to expand their population or asymmetrically to produce BP cells or neurons. BP cells divide at the basal surface of the VZ (away from the ventricle) and are more restricted in their potential, primarily giving rise to neurons. There is evidence that G2 is of longer duration in BP cells, which are more likely to produce neurons, than in NE or RG cells (Calegari et al., 2005). Interestingly, Lis1 loss causes increased neurogenesis at the expense of the stem cell pool (Yingling et al., 2008). Although this has been correlated with altered cleavage plane orientation, our data raise the intriguing possibility that the defect originates even earlier in response to delayed NEBD, which could drive cells toward the BP fate and division within the sub-VZ. Our newly established link between Ndel1 phosphorylation and PNEI could allow precise control over the timing of NEBD by coordinated stimulation of cdk1 and cdk5 in response to intrinsic and extrinsic cues.
Materials and methods
Cell lines
NRK and green monkey kidney cells (COS-7) were used as indicated. All cell lines were maintained in DME supplemented with 10% fetal calf serum, l-glutamine, penicillin, and streptomycin.
Mouse brain sections
Pafah1b1-neo mice heterozygous for a null Lis1 allele were provided by T. Wynshaw-Boris (University of California, San Francisco, San Francisco, CA) and have been described previously (Hirotsune et al., 1998). We refer to these animals as Lis1+/− mice. Lis1+/− females impregnated by wild-type males were anesthetized 13.5 d after observing plugs, and embryos were dissected. Tails were genotyped at the time of harvesting. Embryonic brains were removed and drop fixed in 4% paraformaldehyde for 24–48 h. Fixed brains were dehydrated, embedded in paraffin, and sectioned into 8-μM-thick parasaggital sections.
Microscopy and immunofluorescence
COS-7 and NRK cells were plated onto 12-mm glass coverslips in 24-well plates. Coverslips were mounted on glass slides using ProLong Gold Antifade (Invitrogen). For Lis1, p150glued, and HA immunofluorescence, cells were fixed in 3% paraformaldehyde followed by permeabilization with 0.2% Triton X-100 for 10 min. For dynein intermediate chain (DIC) and Ndel1 immunofluorescence, cells were fixed in 100% ice-cold methanol or permeabilized for 30 s in 0.1% Triton X-100 before fixation in 3% paraformaldehyde. Cells were processed for immunofluorescence and observed using an inverted microscope (Axiovert 200; Carl Zeiss, Inc.) equipped with a Plan-Neo 100×/1.30 objective and a Plan-Apo 63×/1.40 objective. Immersion oil was used (Immersol 518F; Carl Zeiss, Inc.). Digital images were acquired with a charge-coupled device camera (AxioCam HR; Carl Zeiss, Inc.) linked to AxioVision software (version 4.2; Carl Zeiss, Inc.). Optical sections were deconvolved using AxioVision's combined iterative algorithm.
Mouse brains
Sections were stained on glass slides in a humidified chamber. Sections were dewaxed and permeabilized in 0.1% Triton X-100 for 10 min and incubated overnight in primary antibodies in PBS with 0.1% Tween 20, 10% normal goat serum, and 3% bovine serum albumin. FITC- or Texas red–conjugated secondary antibodies (MP Biomedicals) were used. Nuclei were visualized using Hoechst dye (33258; Sigma-Aldrich).
Antibodies
Lis1 and Ndel1 rabbit polyclonal antibodies have been extensively characterized (Niethammer et al., 2000; Smith et al., 2000). These specifically and selectively recognize Lis1 and Ndel1. The phosphoepitope-specific antibody to pNdel1 S231 has been described previously (Patzke et al., 2003). A keyhole limpet hemocyanin–coupled peptide, TENTFP(phosphoS)PKAIPC, was produced and injected into rabbits. Nonspecific antibodies were eliminated by affinity chromatography against unphosphorylated peptides. Phosphoepitope-specific antibodies in the flowthrough were purified on a phosphopeptide column. The cdk5 mouse monoclonal antibody (DC17) was provided by L.-H. Tsai (Massachusetts Institute of Technology, Cambridge, MA). The following antibodies were used (Santa Cruz Biotechnology, Inc.): DIC (74.1 mouse monoclonal), p35 (N19 rabbit polyclonal), histidine tag (Histag, mouse monoclonal), myc (9E10 mouse monoclonal), and HA (rabbit polyclonal and mouse monoclonal). The histone-H3 antibody was purchased from Millipore. Mouse monoclonal antibodies against p50, p150glued, and LAP2 were purchased from BD Biosciences. The anti–α-tubulin antibody and the anti-PDI mouse monoclonal antibodies were purchased from Sigma-Aldrich and Assay Designs, respectively.
Visualizing Ndel1-GFP in living cells
COS-7 cells were synchronized in G0 by growing at confluence for 3 d, replated at low density on 25-mm glass coverslips, and transfected with an expression construct designed to express full-length Ndel1 with an N-terminal EGFP tag. Control cultures were transfected with EGFP-only constructs. The next day, coverslips were transferred into fresh medium containing 25 mM Hepes, pH 7.4, in a water-heated custom-built microscope stage warmed to 37°C. Cells expressing a relatively low level of EGFP were selected for imaging using an inverted microscope (TE200; Nikon) and image acquisition software (DeltaVision; Applied Precision, LLC) coupled to an air-cooled charge-coupled device camera. Fluorescent images were acquired every 2 min for 90 min using a Plan-Apo 100×/1.6 oil immersion objective.
Constructs and transfections
Complementary hairpin sequences for Ndel1 (276–294 bp; GCAGGTCTCAGTGTTAGAA), DYNC1H1 (9,753–9,771 bp; GAAGGTCATGAGCCAAGAA), and Lis1 (1,062–1,080 bp; GAGTTGTGCTGATGACAAG) were synthesized and cloned into pSilencer under control of the U6 promoter (version 2.0; Ambion; Shu et al., 2004). To generate HA-FLLis1 and HA-NLis1, full-length murine, Lis1, and a Lis1 fragment encoding amino acids 1–87 were subcloned into a pCruzHA vector (Santa Cruz Biotechnology, Inc.). Myc-tagged Ndel1 constructs (Ndel1, Ndel1123, and Ndel15A), untagged Ndel1 point mutants (S231A, S242A, and T245A), Ndel1-GFP, cdk5, p25, and dncdk5 constructs have been described previously (Smith et al., 2000). Dncdk5 contains an R33T mutation (a putative ATP-binding site) and inhibits endogenous kinase activity by sequestering cdk5 activators p35/p25 and p39 (Nikolic et al., 1996). HA-tagged human cdk1, myc-tagged Xenopus laevis cyclin B1, and untagged human cyclin A2 expression vectors were obtained from Addgene. A dominant-negative cdk1 construct containing a mutation in A146, a conserved residue in protein kinases important for the phosphotransfer reaction (van den Heuvel and Harlow, 1993), was also obtained from Addgene. The dncdk5 and dncdk1 constructs are well characterized and have been used in hundreds of published papers from many laboratories. A p50 construct was provided by T.A. Schroer (The Johns Hopkins University, Baltimore, MD) and has been described previously (Quintyne et al., 1999). Cells were transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's directions. Transfection efficiencies were generally 90%.
PNEI analysis in COS-7 cells
Randomly cycling cells were transfected with the indicated constructs for the indicated times. Digital images of prophase cells were obtained from fixed cultures and analyzed for PNEI. The presence or absence of nuclear invaginations was readily observed after Hoechst staining. A nucleus was considered PNEI positive if invaginations were deeper than 2 μm. Kidney-shaped nuclei without clear invaginations were not considered PNEI positive. All prophase cells on each coverslip were included in the analysis. There were 30–50 of these per coverslip, and experiments were repeated three times, so on average, 90–150 prophase cells were analyzed. Our data may underestimate the effect of Lis1 and Ndel1 manipulation, as some untransfected cells may have been included in the counts.
Protein isolation and Ndel1 phosphorylation
Full-length human Ndel1 fused to a histidine tag was expressed in bacteria and purified using nickel–nitrilotriacetic acid chromatography (Invitrogen). Ndel1 was phosphorylated by incubation with GST-tagged cdk5/p25, cdk1/cyclin B1, or both (Cell Signaling Technology) according to the manufacturer's instructions. Full-length histidine-tagged Lis1 was purified from Sf9 insect cells and purified using nickel–nitrilotriacetic acid resin. The histidine tag was cleaved using tobacco etch virus protease. The purification and analysis of cytoplasmic dynein were isolated from bovine brains and were essentially performed as described previously (Mesngon et al., 2006).
Immunoprecipitation
For co-IP with DIC, equimolar Lis1 and histidine-tagged Ndel1 were incubated with and without dynein in BrB80 buffer (80 mM Pipes, 1 mM EGTA, and 1 mM MgCl2) at 37°C for 10 min. Proteins were exposed to a DIC mouse monoclonal antibody, 74.1, and conjugated to agarose beads (Santa Cruz Biotechnology, Inc.) for 1 h at 4°C. Precipitated proteins were analyzed by Western blotting of 10% SDS-PAGE gels. For co-IP with Ndel1, equimolar histidine-tagged Ndel1 and untagged Lis1 were incubated with or without the His-tag antibody for 1 h at 4°C. Protein A–Sepharose was added to each sample for 1 h at 4°C. After washing beads, precipitated proteins were analyzed by Western blotting. For co-IP with Ndel1 from COS-7 cells, cleared extracts from cells expressing either myc-tagged Ndel1 or myc-tagged Ndel15A were incubated with or without the 9E10 myc antibody or for 1 h at 4°C. Protein A–Sepharose was added to each sample for 1 h at 4°C. After washing beads, precipitated proteins were analyzed by Western blotting.
Nocodazole treatment
Cells were exposed to 10 μM nocodazole (EMD) for 3–5 h and processed as described in the Microscopy and immunofluorescence section. Co-IP was performed with anti–His tag antibody.
Preparation of crude nuclear fractions
Transfected COS-7 cells were exposed to 10 μM nocodazole for 3 h, incubated in 20 vol of cold hypotonic buffer (10 mM Hepes, pH 7.4, 2 mM MgCl2, 25 mM KCl, 1 mM DTT, and 1 mM PMSF) and 1× protease inhibitor cocktail (EMD) for 45 min, and lysed by Dounce homogenization. The lysate was centrifuged at 400 g through 12% sucrose. The supernatant (cytoplasmic fraction) was collected. Nuclei were washed in hypotonic buffer containing 8% sucrose and repelleted at 400 g for 5 min. Proteins in the crude nuclear pellet were extracted by incubation in 10 mM Hepes, pH 7.4, 2 mM MgCl2, 1 mM PMSF, and 7 M urea on ice for 30 min. This was cleared by centrifugation at 16,000 g for 10 min. 50% of the nuclear fraction from each set of transfected cultures was loaded onto an SDS-PAGE gel and immunoblotted to estimate the ratio of soluble to nuclear pools of endogenous DIC and Lis1 and exogenous Ndel1. Equal protein loading was confirmed by Ponceau S staining of the membrane and Coomassie blue staining of samples loaded on another gel.
Immunofluorescence intensity analysis
One set of COS-7 cell cultures was transfected with myc-tagged Ndel1, and a second set was transfected with myc-tagged Ndel15A. After 12 h, cells were exposed to nocodazole for 5 h and fixed and processed for Myc, Lis1, and DIC immunofluorescence. Multiple same-exposure three-channel digital images were acquired for each set of cultures (FITC for Lis1 and dynein, Texas red for Myc, and DAPI to label nuclei). Each image contained 5–10 transfected cells. Differences in fluorescence intensity at the NE were measured in two ways. (1) Pixel intensities were determined along 3-μm lines that ended within the NE region. The first pixel was set to an intensity of 10, and the remaining pixels were normalized accordingly. The mean intensities were calculated for 50 lines. (2) The mean of Myc pixel intensities of 10–30 points on the NE was compared with mean pixel intensities of 10–30 points in the cytoplasm near the NE. Pixel intensities were obtained using ImageJ software (National Institutes of Health).
Analysis of S231 phosphorylation by Western blotting
Transfected cells were lysed in 150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% SDS, and 50-mM Tris, pH 8. The protein concentrations were determined using protein assay concentrate (Bio-Rad Laboratories), and equal amounts of protein were separated on polyacrylamide SDS gels, transferred to polyvinylidene difluoride membranes, and immunoblotted with pS231Ab and other antibodies according to standard protocols. The blots were exposed to autoradiography film for a sufficient time to observe and minimize the saturation of bands.
Online supplemental material
Fig. S1 shows that Lis1 and Ndel1 accumulate at the prophase NE, where they colocalize with dynein and dynactin. Fig. S2 shows that dynactin integrity is important for Lis1 redistribution to the NE in response to nocodazole. Fig. S3 shows characterization of pS231Ab, a phosphospecific Ndel1 antibody. Video 1 shows Ndel1-GFP accumulating at the NE. Video 2 shows that GFP does not accumulate at the NE. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200803071/DC1.
Supplementary Material
Acknowledgments
Lis1+/− mice were provided by A. Wynshaw-Boris, and shRNA constructs were provided by L.-H. Tsai.
This work was supported in part through the Center for Colon Cancer Research at the University of South Carolina (National Institutes of Health grant 5P20RR017698-030001) and by a National Institutes of Health grant (1R01NS056314-01A2) to D. Smith.
Abbreviations used in this paper: BP, basal progenitor; co-IP, coimmunoprecipitation; DHC, dynein heavy chain; DIC, dynein intermediate chain; LAP2, lamin-associated protein 2; NE, nuclear envelope; NEBD, NE breakdown; NRK, normal rat kidney; PNEI, prophase NE invagination; RG, radial glia; shRNA, short hairpin RNA; S/T-P, Ser/Thr-Pro; VZ, ventricular zone.
References
- Beaudouin, J., D. Gerlich, N. Daigle, R. Eils, and J. Ellenberg. 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 108:83–96. [DOI] [PubMed] [Google Scholar]
- Calegari, F., W. Haubensak, C. Haffner, and W.B. Huttner. 2005. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J. Neurosci. 25:6533–6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso, C., R.J. Leventer, J.J. Dowling, H.L. Ward, J. Chung, K.S. Petras, J.A. Roseberry, A.M. Weiss, S. Das, C.L. Martin, et al. 2002. Clinical and molecular basis of classical lissencephaly: mutations in the LIS1 gene (PAFAH1B1). Hum. Mutat. 19:4–15. [DOI] [PubMed] [Google Scholar]
- Cockell, M.M., K. Baumer, and P. Gonczy. 2004. lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J. Cell Sci. 117:4571–4582. [DOI] [PubMed] [Google Scholar]
- Coquelle, F.M., M. Caspi, F.P. Cordelieres, J.P. Dompierre, D.L. Dujardin, C. Koifman, P. Martin, C.C. Hoogenraad, A. Akhmanova, N. Galjart, et al. 2002. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol. Cell. Biol. 22:3089–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda, U., C. Tarricone, W.C. Choi, D.R. Cooper, S. Lukasik, F. Perrina, A. Tripathy, M.H. Kim, D.S. Cafiso, A. Musacchio, and Z.S. Derewenda. 2007. The structure of the coiled-coil domain of ndel1 and the basis of its interaction with lis1, the causal protein of miller-dieker lissencephaly. Structure. 15:1467–1481. [DOI] [PubMed] [Google Scholar]
- Gambello, M.J., D.L. Darling, J. Yingling, T. Tanaka, J.G. Gleeson, and A. Wynshaw-Boris. 2003. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J. Neurosci. 23:1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz, M., and W.B. Huttner. 2005. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6:777–788. [DOI] [PubMed] [Google Scholar]
- Hirotsune, S., M.W. Fleck, M.J. Gambello, G.J. Bix, A. Chen, G.D. Clark, D.H. Ledbetter, C.J. McBain, and A. Wynshaw-Boris. 1998. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 19:333–339. [DOI] [PubMed] [Google Scholar]
- Kim, M.H., D.R. Cooper, A. Oleksy, Y. Devedjiev, U. Derewenda, O. Reiner, J. Otlewski, and Z.S. Derewenda. 2004. The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure. 12:987–998. [DOI] [PubMed] [Google Scholar]
- King, S.J., and T.A. Schroer. 2000. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2:20–24. [DOI] [PubMed] [Google Scholar]
- Mei, D., R. Lewis, E. Parrini, L.P. Lazarou, C. Marini, D.T. Pilz, and R. Guerrini. 2008. High frequency of genomic deletions–and a duplication–in the LIS1 gene in lissencephaly: implications for molecular diagnosis. J. Med. Genet. 45:355–361. [DOI] [PubMed] [Google Scholar]
- Mesngon, M.T., C. Tarricone, S. Hebbar, A.M. Guillotte, E.W. Schmitt, L. Lanier, A. Musacchio, S.J. King, and D.S. Smith. 2006. Regulation of cytoplasmic dynein ATPase by Lis1. J. Neurosci. 26:2132–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer, M., D.S. Smith, R. Ayala, J. Peng, J. Ko, M.S. Lee, M. Morabito, and L.H. Tsai. 2000. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 28:697–711. [DOI] [PubMed] [Google Scholar]
- Nikolic, M., H. Dudek, Y.T. Kwon, Y.F. Ramos, and L.H. Tsai. 1996. The cdk5/p35 kinase is essential for neurite outgrowth during neuronal differentiation. Genes Dev. 10:816–825. [DOI] [PubMed] [Google Scholar]
- Patzke, H., U. Maddineni, R. Ayala, M. Morabito, J. Volker, P. Dikkes, M.K. Ahlijanian, and L.H. Tsai. 2003. Partial rescue of the p35−/− brain phenotype by low expression of a neuronal-specific enolase p25 transgene. J. Neurosci. 23:2769–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne, N.J., S.R. Gill, D.M. Eckley, C.L. Crego, D.A. Compton, and T.A. Schroer. 1999. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147:321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner, O., R. Carrozzo, Y. Shen, M. Wehnert, F. Faustinella, W.B. Dobyns, C.T. Caskey, and D.H. Ledbetter. 1993. Isolation of a Miller-Dieker lissencephaly gene containing G protein β-subunit-like repeats. Nature. 364:717–721. [DOI] [PubMed] [Google Scholar]
- Salina, D., K. Bodoor, D.M. Eckley, T.A. Schroer, J.B. Rattner, and B. Burke. 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 108:97–107. [DOI] [PubMed] [Google Scholar]
- Sasaki, S., A. Shionoya, M. Ishida, M.J. Gambello, J. Yingling, A. Wynshaw-Boris, and S. Hirotsune. 2000. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 28:681–696. [DOI] [PubMed] [Google Scholar]
- Sasaki, S., D. Mori, K. Toyo-oka, A. Chen, L. Garrett-Beal, M. Muramatsu, S. Miyagawa, N. Hiraiwa, A. Yoshiki, A. Wynshaw-Boris, and S. Hirotsune. 2005. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol. Cell. Biol. 25:7812–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer, T.A. 2004. Dynactin. Annu. Rev. Cell Dev. Biol. 20:759–779. [DOI] [PubMed] [Google Scholar]
- Shu, T., R. Ayala, M.D. Nguyen, Z. Xie, J.G. Gleeson, and L.H. Tsai. 2004. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 44:263–277. [DOI] [PubMed] [Google Scholar]
- Smith, D.S., M. Niethammer, R. Ayala, Y. Zhou, M.J. Gambello, A. Wynshaw-Boris, and L.H. Tsai. 2000. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2:767–775. [DOI] [PubMed] [Google Scholar]
- Tarricone, C., F. Perrina, S. Monzani, L. Massimiliano, M.H. Kim, Z.S. Derewenda, S. Knapp, L.H. Tsai, and A. Musacchio. 2004. Coupling PAF signaling to dynein regulation: structure of LIS1 in complex with PAF-acetylhydrolase. Neuron. 44:809–821. [DOI] [PubMed] [Google Scholar]
- Tsai, J.W., Y. Chen, A.R. Kriegstein, and R.B. Vallee. 2005. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 170:935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee, R.B., and J.W. Tsai. 2006. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 20:1384–1393. [DOI] [PubMed] [Google Scholar]
- van den Heuvel, S., and E. Harlow. 1993. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 262:2050–2054. [DOI] [PubMed] [Google Scholar]
- Yan, X., F. Li, Y. Liang, Y. Shen, X. Zhao, Q. Huang, and X. Zhu. 2003. Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol. Cell. Biol. 23:1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling, J., Y.H. Youn, D. Darling, K. Toyo-Oka, T. Pramparo, S. Hirotsune, and A. Wynshaw-Boris. 2008. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 132:474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.