Abstract
Genes on different chromosomes can be spatially associated in the nucleus in several transcriptional and regulatory situations; however, the functional significance of such associations remains unclear. Using human erythropoiesis as a model, we show that five cotranscribed genes, which are found on four different chromosomes, associate with each other at significant but variable frequencies. Those genes most frequently in association lie in decondensed stretches of chromatin. By replacing the mouse α-globin gene cluster in situ with its human counterpart, we demonstrate a direct effect of the regional chromatin environment on the frequency of association, whereas nascent transcription from the human α-globin gene appears unaffected. We see no evidence that cotranscribed erythroid genes associate at shared transcription foci, but we do see stochastic clustering of active genes around common nuclear SC35-enriched speckles (hence the apparent nonrandom association between genes). Thus, association between active genes may result from their location on decondensed chromatin that enables clustering around common nuclear speckles.
Introduction
The genome is organized in a nonrandom fashion, both three dimensionally within the nucleus and with regard to linear gene distribution (for reviews see Cremer et al., 2004; Hurst et al., 2004), and it has been proposed that this organization may play a role in the modulation of transcription (for review see Fraser and Bickmore, 2007; Misteli, 2007). Gene-dense domains that are widely expressed show nonrandom clustering along the linear length of the genome and also tend to be positioned more toward the interior of the nucleus in a manner that is broadly independent of variation in gene expression between tissues (Gilbert et al., 2004; Sproul et al., 2005; Gierman et al., 2007; Goetze et al., 2007). Chromatin structure shows a relationship to linear gene distribution, with gene-dense or highly expressing regions being more decondensed than areas of gene paucity or low expression (Gilbert et al., 2004; Gierman et al., 2007). Upon activation, gene-dense clusters or even individual genes can sometimes, but not always, be seen looping out from their respective chromosome territories on decondensed stretches of chromatin (Volpi et al., 2000; Mahy et al., 2002; Williams et al., 2002; Ragoczy et al., 2003; Chambeyron and Bickmore, 2004; Brown et al., 2006; Morey et al., 2007). The arrangement of particular chromatin domains within the arena of the nucleus show not only preferred radial positionings as described above but can also show favored relative locations in some situations. Changes in relative positioning can be seen during cell differentiation and between cell types (Kuroda et al., 2004; Parada et al., 2004; Kosak et al., 2007), and specific genomic regions show nonrandom spatial proximity (or association) in certain situations.
Associations have been reported both in cis and in trans when certain genes are active (Thompson et al., 2003; Osborne et al., 2004, 2007; Simonis et al., 2006), and trans-association of specific genes has also been described in several situations in which there is a binary outcome (e.g., X inactivation, genomic imprinting, and before selective activation), therefore implying a regulatory role for spatial association (LaSalle and Lalande, 1996; Spilianakis et al., 2005; Bacher et al., 2006; Lomvardas et al., 2006; Xu et al., 2006; Augui et al., 2007; Takizawa et al., 2008). However, other studies have suggested that such gene convergences, which occur in only a proportion of nuclei, may not have a functional basis (Brown et al., 2006; Simonis et al., 2006; Fuss et al., 2007; Nishizumi et al., 2007; Teller et al., 2007; Palstra et al., 2008). These observations pose several key questions. Is spatial association a common occurrence between active genes? If so, what is the basis of this phenomenon? Is it a requirement for properly regulated gene transcription, or does it result from transcriptional activity within the region interrogated (de Laat, 2007)? These questions have important relevance for interpreting the outcome of any procedures involving nuclear disruption, such as somatic cloning, stem cell differentiation, and gene therapy (Misteli, 2007).
We have used human primary erythroid differentiation as a model where the differential expression of erythroid genes allows us to investigate possible roles that nuclear organization may play in gene transcription. Previously using this system, we looked at the positioning of globin genes during erythroid differentation as they switch on and transcribe heavily over the course of a few days in primary erythroblasts. We found that trans-associations (both inter-allelic and inter-genic) between α- and β-globin genes did increase with gene activity. However, we concluded that such associations were not required for coordinate regulation of globin transcription because they only occurred in a percentage of active genes, there was no evidence of increased association before transcription, and the frequency of associations was very different between the human α- and β-globin genes and between the globin genes in mouse and human erythroblasts (Brown et al., 2006). The aim of our present study is to understand how widespread gene association is during erythropoiesis and to gain insight into the basis of association between active genes. We look at the levels of association of five genes that are cotranscribed during erythropoiesis, we investigate the role played by chromatin environment on the marked variation in association frequency displayed by different genes, we examine the roles of nuclear foci in bringing genes together in trans, and, finally, we present a model to explain the basis of gene association.
Results
Gene proximity is a common but variable feature of active erythroid genes
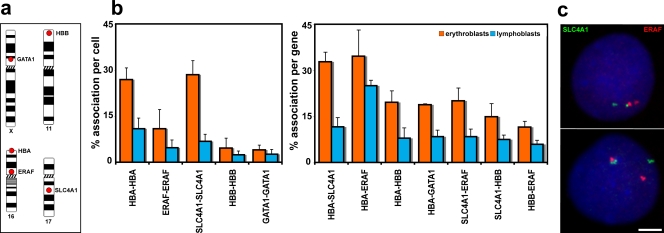
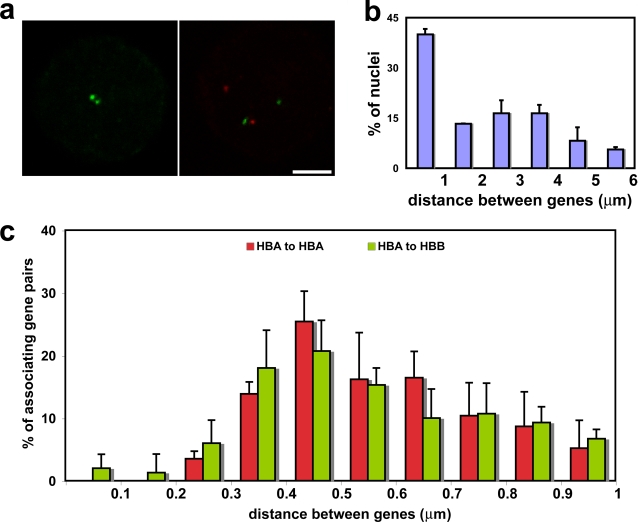
Red blood cells differentiate from proerythroblasts in the bone marrow over just a few days, and, during this time, many erythroid-restricted genes switch on and are highly transcribed (see Hembase at http://hembase.niddk.nih.gov). We compared nuclear organization in primary human intermediate erythroblasts where erythroid genes are active with that in human lymphoblasts, which have a similar mean nuclear volume but in which the erythroid-specific genes are silent. We analyzed five genes known to be highly up-regulated during erythropoiesis (Brown et al., 2006; Keller et al., 2006): α-globin (HBA), β-globin (HBB), erythrocyte membrane protein band 3 (SLC4A1), erythroid-associated factor (ERAF), and globin transcription factor 1 (GATA1) located on four different chromosomes (Fig. 1 a). We then scored the positions of these genes with respect to one another in intermediate erythroblasts (active) and lymphoblasts (inactive). We found that trans-associations (defined as closer than 1 μm) were indeed a feature of these active genes in a proportion of nuclei (Fig. 1, b and c; and Table I). The numbers of associations scored for these erythroid genes were significantly increased in erythroblast compared with lymphoblast nuclei (Table I), with >30% of loci involved for some genes. An exception was association between the GATA1 genes on the X chromosomes, which is to be expected because only one allele is active in each cell. As an additional control, we looked at the positioning of a third clone from chromosome (HSA) 16 located 2.5 Mb from ERAF but which does not cover any erythroid-specific genes (RP11-264B17). Inter-allelic associations scored for this clone were just 5% in both erythroblasts and lymphoblasts, indicating that it is unlikely that the general positioning of this chromosome in erythroblasts would increase levels of association.
Figure 1.
Associations between genes up-regulated in erythroblasts. (a) Chromosomal locations of the five genes analyzed. (b) Degree of associations between erythroid genes detected by DNA-FISH to intermediate erythroblasts and lymphoblasts. Allelic associations are depicted on the left, and inter-genic associations are depicted on the right. Allelic associations involving the HBA, SLC4A1, and ERAF genes are the most common in erythroblasts and demonstrate highly significant increases over the levels scored in lymphoblasts. The HBB and GATA1 genes are more frequently involved in inter-genic than allelic associations in erythroblasts. The scores for inter-genic associations in the right-hand chart represent the percent association per gene, not per cell, because many cells had both SLC4A1 genes involved in associations. GATA1 was scored only in female cells. Numbers of nuclei analyzed and individual p-values are given in Table I. Values represent the mean ± SD (error bars) from 2–22 independent experiments. These patterns of association mirrored those obtained by 3D hybridizations for α- and β-globin (Brown et al., 2006) and for SLC4A1 and ERAF (not depicted), although slightly higher values of association were obtained by 3D FISH. (c) Typical trans-associations between SLC4A1 (green) and ERAF (red) detected by DNA-FISH. Bar, 4 μm.
Table I. Associations between genes up-regulated in intermediate erythroblasts.
| DNA-FISH | Numbers of nuclei scored for associations between genes |
Fisher's exact test p-value |
|
|---|---|---|---|
| Erythroblast | Lymphoblast | ||
| HBA-HBA | 800 | 2,200 | 4.91 × 10−25 |
| ERAF-ERAF | 1,100 | 1,500 | 2.64 × 10−09 |
| SLC4A1-SLC4A1 | 600 | 1,600 | 1.79 × 10−37 |
| HBB-HBB | 1,900 | 1,300 | 0.0011 |
| GATA1-GATA1 | 1,100 | 1,200 | 0.056 |
| HBA-SLC4A1 | 400 | 300 | 5.47 × 10−17 |
| HBA-ERAF | 400 | 400 | 0.0015 |
| HBA-HBB | 700 | 400 | 5.52 × 10−11 |
| HBA-GATA1 | 200 | 400 | 3.62 × 10−06 |
| SLC4A1-ERAF | 500 | 500 | 9.96 × 10−10 |
| SLC4A1-HBB | 400 | 400 | 4.79 × 10−05 |
| HBB-ERAF | 400 | 400 | 0.00042 |
P-values for Fisher's exact test of differences between associations scored in erythroblasts and lymphoblasts (Fig. 1). A p-value of 0.004 is required to achieve significance after a Bonferroni correction for multiplicity.
Importantly, what became clear from our analysis was that there is a marked variation in the frequency with which different erythroid genes are involved in associations. The allelic association scores (Fig. 1 b, left) showed that the α-globin and SLC4A1 genes on chromosomes 16 and 17 have much higher levels of association than, for example, β-globin on HSA11. What could be the basis for the striking variation in levels of association between different genes?
The gene most frequently involved in both allelic and inter-genic associations was SLC4A1, whereas β-globin was much less frequently involved (Fig. 1 b). This could not be related to levels of transcription of the individual genes because β-globin is very active in intermediate erythroblasts, with nascent transcripts at 77% of loci (Brown et al., 2006). One potential contributor to this variability in association is radial position in the nucleus because allelic associations at least would be less likely for genes near the nuclear periphery. However, radial position would not explain the different levels of association observed between erythroblasts and lymphoblasts because β-globin genes are found near the nuclear periphery in both cell types (Brown et al., 2006). Another potential contributor is the nature of the surrounding chromatin environment. Chromatin is organized into different domains of expression (Caron et al., 2001; Lercher et al., 2002; Versteeg et al., 2003), which has been related to differing levels of decondensation (Gilbert et al., 2004; Sproul et al., 2005; Gierman et al., 2007; Goetze et al., 2007) and the frequency with which genes may sit away from their chromosome territories (Mahy et al., 2002; Brown et al., 2006). Therefore, we compared the immediate chromatin environments around SLC4A1 and β-globin.
SLC4A1 and β-globin genes sit in very different chromatin environments.
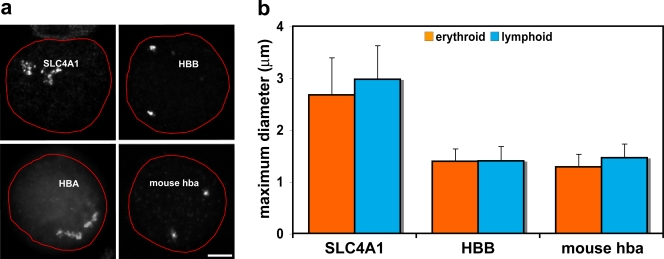
We established bacterial artificial chromosome (BAC) probe pools covering 2 Mb centered around SLC4A1 on HSA17 and β-globin on HSA11 and examined the appearance of these contigs by FISH when hybridized to erythroblast and lymphoblast nuclei. The SLC4A1 region was decondensed in around two thirds of cases in both cell types and frequently appeared as an expanded cloud of signal (decondensed in erythroblasts, 62%, n = 105; decondensed in lymphoblasts, 71%, n = 50; Fig. 2 a). In contrast, the FISH signal for the β-globin region on HSA11 was most often condensed (decondensed in erythroblasts, 24%, n = 100; decondensed in lymphoblasts, 21%, n = 100; Fig. 2 a) and was never observed decondensed to the same degree as seen for the SLC4A1 region. To quantify the degree of decondensation, we measured the diameter of each contig signal at its widest point and found significant differences between values for SLC4A1 and β-globin in both erythroblasts (P = 2.51 × 10−29) and lymphoblasts (P = 9.87 × 10−26; n = 88/54 for SLC4A1 and n = 50/50 for β-globin in erythroblasts/lymphoblasts; Fig. 2 b).
Figure 2.
Chromosomal environments differ between erythroid-specific genes. (a) Representative intermediate erythroblast nuclei after DNA-FISH of four 2-Mb BAC contigs. The nuclear periphery is outlined in red. The contig centered on the SLC4A1 gene on HSA17 shows marked decondensation of the chromatin region. The contig centered on the HBB gene on HSA11 does not exhibit the same levels of decondensation. The contig covering the mouse α-globin genes on MMU11 again does not display the levels of decondensation seen around SLC4A1. The contig covering the terminal 2 Mb of 16p13.3, surrounding the human α-globin genes, has been published previously (Brown et al., 2006) and is included here to demonstrate the marked difference in chromatin decondensation around the human and mouse α-globin genes. Note that the telomeric chromatin around HBA is more linearly extended than that around interstitial SLC4A1. (b) Maximum diameter measurements of BAC contig FISH signals in erythroblast and lymphoblast nuclei for SLC4A1, HBB, and mouse α-globin. These measurements minimize differences in signal size because they cannot take account of folding in the discontinuous SLC4A1 signal, yet there are significant differences between values for SLC4A1 and HBB and also for SLC4A1 and mouse α-globin. Values represent the mean ± SD (error bars) of measurements taken from one to two hybridizations for each contig probe set and cell type. Bar, 3 μm.
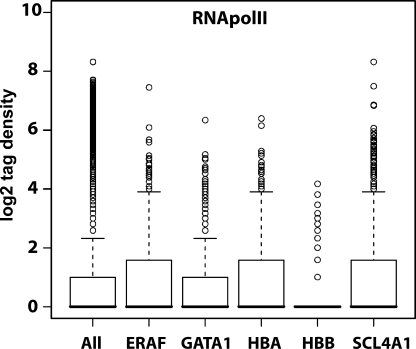
This analysis indicates that SLC4A1 sits in a region of chromatin that is frequently decondensed, like the human α-globin genes in 16p13.3 (Fig. 2 a; Mahy et al., 2002; Brown et al., 2006). We compared further aspects of the chromatin environment surrounding the five human erythroid up-regulated genes together with the murine α-globin (mouse hba) region on mouse chromosome (MMU) 11 because we previously observed much less association involving the α-globin genes in mouse than in human erythroblasts (Brown et al., 2006). Looking at the 2-Mb region surrounding each of the genes, both the proportion of the region occupied by full-length transcripts, i.e., coding density, and the number of CpG islands reflect the levels of gene association (Table II). Furthermore, we analyzed the density of RNA polymerase II (RNApolII) across the five human regions using high resolution chromatin immunoprecipitation and sequencing profiles (there is a caveat that these data were obtained from CD4+ T cells; therefore, RNApolII distribution could be different in the lymphoblasts and erythroblasts studied here; Barski et al., 2007; Fig. 3). The highly expressed β-globin gene sits in a region of comparatively low RNApolII density when compared with SLC4A1, which links transcriptional activity of the region to levels of decondensation, as has been previously observed (Goetze et al., 2007).
Table II. Characteristics of 2-Mb chromatin environments surrounding erythroid up-regulated genes.
| Gene | Chromosome | 2-Mb coordinates | Number of genes |
Coding density |
Number of CpG islands | GC content |
|---|---|---|---|---|---|---|
| % | % | |||||
| HBA | HSA 16 | 1-2000000 | 114 | 11.7 | 256 | 57.25 ± 22.7 |
| ERAF | HSA 16 | 30446680-32446680 | 55 | 6.6 | 72 | 49.07 ± 23.7 |
| SLC4A1 | HSA 17 | 38691896-40691896 | 61 | 8 | 81 | 45.49 ± 23.6 |
| GATA1 | HSA X | 47529906-49529906 | 62 | 5.8 | 60 | 45.39 ± 23.1 |
| HBB | HSA 11 | 4204075-6204075 | 84 | 4.9 | 1 | 38.5 ± 21.7 |
| hba | MMU 11 | 31184083-33184083 | 26 | 3 | NA | 45.3 ± 21.5 |
GC, guanine-cytosine; NA, not applicable. Genes counted have Ensembl gene IDs, and the coding densities of the full-length transcripts are based on data from the Ensembl genome browser. All other data were obtained from the University of California Santa Cruz genome browser. CpG islands scored are >200 bp with >50% guanine-cytosine content and >0.6 observed/expected ratio. Guanine-cytosine percent content is scanned in five base windows, and SDs are given.
Figure 3.
Different levels of transcriptional activity surround erythroid up-regulated genes. Box plots showing the distribution of RNApolII across 2-Mb regions surrounding five erythroid up-regulated genes together with a box plot for the whole genome (All) for comparison. ERAF, HBA, and SLC4A1 all have higher than average densities of RNApolII bound to their surrounding chromatin regions. Data were extracted from high resolution maps for the genome-wide distribution of RNApolII in CD4+ T cells using the Solexa 1G sequencing technology (Barski et al., 2007).
Mouse α-globin genes lie in a region of condensed chromatin.
A prediction of this work would be that the region around the murine α-globin genes, which associate to a much lower degree than either human α-globin or SLC4A1, would also be more condensed than the SLC4A1 region. Therefore, we hybridized a 2-Mb contig of labeled mouse BAC probes centered around the α-globin genes to adult mouse splenic intermediate erythroblasts and activated B lymphocytes. This contig FISH signal was most frequently condensed in a manner similar to the human β-globin region (Fig. 2 a), and the maximum diameter was significantly less than the human SLC4A1 region in both erythroid (P = 4.72 × 10−32) and lymphoid cell types (P = 1.10−24; n = 48/50 erythroid/lymphoid; Fig. 2 b).
We have demonstrated that genes showing higher levels of trans-association are found in decondensed regions of chromatin. Levels of association are also directly reflective of other regional chromatin features, including coding density, number of CpG islands, and RNApolII density. A principal determinant for the marked variation in association between active genes may therefore be the chromatin environment in which a gene is embedded.
Manipulation of chromatin environment affects human α-globin gene positioning
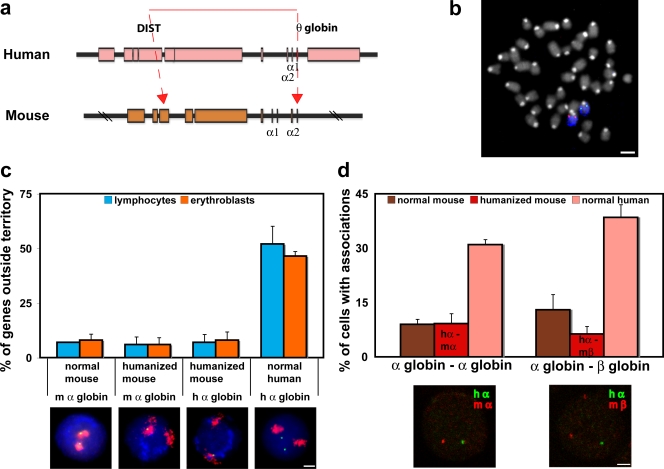
To test the effect of the chromatin environment more directly, we analyzed a mouse in which the human α-globin genes have been transferred to the mouse α-globin chromatin environment. This was achieved in mouse embryonic stem cells in which the entire 117-kb human α-globin locus, including all known regulatory elements, replaced the mouse α-globin locus (Wallace et al., 2007; Fig. 4 a). With germline transmission, the heterozygous humanized mouse carries one normal MMU11 with the endogenous mouse α-globin locus and one MMU11 with the human α-globin locus. This human sequence responds to normal murine developmental control and is expressed appropriately during terminal erythroid differentiation. Nascent transcription from the human α-globin gene in the “humanized” mouse is as high (active in 61% of cells) as from the endogenous mouse locus (active in 64% of cells), as detected by RNA-FISH to heterozygous humanized mouse acetylphenylhydrazine-treated spleen cells (n = 200). The heterozygous mouse allows us to contemporaneously follow the positionings of both human and mouse α-globin loci in primary mouse erythroblasts (Fig. 4 b).
Figure 4.
Positioning of human globin genes in a mouse chromatin environment. (a) Map of the α-globin genes and surrounding regulatory regions on HSA16 and MMU11. Red lines denote the region of human 16p13.3 that has been inserted into the corresponding region of the mouse α-globin locus in mouse embryonic stem cells through heterotypic lox sites in the DIST (rhomboid 5 homologue 1 [Drosophila] [RHBDF1]) and θ-globin genes. For a full explanation, see Wallace et al., 2007. (b) Heterozygous “humanized” α-globin mouse. Metaphase spread from spleen cells showing both MMU11 (blue), one with the endogenous mouse α-globin gene locus (red) and one with the human α-globin locus (green). (c) Position of α-globin genes with respect to chromosome territory. Human and mouse α-globin FISH signals (green) were scored as within or outside the MMU11 territory delineated by whole chromosome FISH paint (red). Nuclei are stained with DAPI (blue). In activated lymphocytes and erythroblasts (n = 300–550) from the heterozygous humanized mouse, endogenous mouse α-globin genes sat mainly at the MMU11 territory (n = 300), with the human α-globin genes behaving similarly in both cell types (n = 500–550). The normal mouse also shows the α-globin genes sitting mainly at their territory in both cell types (n = 100–150) and contrasts with the normal human situation in which ∼50% of the α-globin genes sit outside the HSA16 territory in both cell types (n = 200–500). (d) Association between globin genes in normal and humanized mouse erythroblasts. In normal mouse (n = 150–200), α-globin genes and α–β-globin genes associate with similar low frequencies. In normal human (n = 200), α-globin genes and α–β-globin genes associate much more frequently; however, in the humanized mouse (n = 300–500), both the human and the endogenous mouse α-globin genes and the human α-globin and mouse β-globin genes show association levels similar to that seen in the normal mouse. Therefore, in the humanized mouse, the human α-globin locus associates with other globin genes (hα–mα and hα–mβ) much less than in the normal human chromatin environment. Representative hybridizations to the humanized mouse are shown. (c and d) Values represent means ± SD (error bars) from two to five independent experiments. Bars, 4 μm.
We measured gene positioning in two ways: the frequency with which genes sit outside of their chromosome territory and the frequency with which genes associate. The human α-globin genes frequently sit outside the normal HSA16 territory on a decondensed terminal piece of chromatin, irrespective of transcriptional status; however, this does not happen to the same degree in the normal mouse (Mahy et al., 2002; Brown et al., 2006). We found that in the humanized mouse, both the endogenous mouse α-globin genes and the human α-globin genes behaved like those of the normal mouse in activated lymphocytes and in intermediate erythroblasts (Fig. 4 c). The degree of association between globin genes in intermediate erythroblasts from the heterozygous humanized mouse was also at the level of the normal mouse and much less than in human erythroblasts (Fig. 4 d).
Thus, the human α-globin locus in the mouse environment behaves just like the normal endogenous mouse α-globin genes and not like normal human α-globin genes by both measures of gene positioning; i.e., the chromatin region is not decondensed, and association is reduced. These findings show that the variability in levels of association between genes is principally affected by regional chromatin environment.
The role of transcription factories in globin gene proximity
What could be the underlying drivers bringing active genes into association? We considered whether a regulatory requirement for genes to relocate to specific transcription factories might be involved, as has been proposed for active murine erythroid genes (Osborne et al., 2004). When bromo-UTP (BrUTP) is incorporated into nascent transcripts, punctate sites of transcription, termed transcription factories, can be visualized (Iborra et al., 1996). These factories have been measured at ∼50 nm in diameter in several cell types by electron microscopy (Iborra and Cook, 1998; Iborra et al., 1998). If coordinately regulated genes do need to share a transcription factory, we would expect to find evidence at the human α- and β-globin genes because these genes need to be transcribed in roughly equal quantities per cell. Therefore, we examined the spatial relationship between transcription foci and active human globin genes, where we have already demonstrated an increase in proximity related to transcription (Brown et al., 2006).
Distance between associating globin genes exceeds the mean transcription factory diameter.
The distance between the two homologous active α-globin signals in erythroblast nuclei, as detected by RNA-FISH, ranged from 0.24 to 5.57 μm (n = 195), of which 40% were within a micrometer of each other and were classed as associating (Fig. 5, a and b). In a more detailed analysis of the spatial distribution below 1 μm, the mean distance apart of α–α-associated signals (n = 173) was 571 nm (Fig. 5 c). The distance between α- and β-globin signals ranged from coincident to 11.29 μm (n = 190; not depicted), and the mean distance between associating signals below 1 μm was very similar to α–α at 542 nm (n = 150; Fig. 5, a and c). The mean distances apart for α–α and α–β gene associations are therefore both just over 500 nm, which is much larger than the mean diameter of transcription foci at ∼50 nm.
Figure 5.
Associating genes are not commonly coincident. Measurements between globin RNA-FISH signals in intermediate erythroblasts. (a) Typical α–α (green-green) and α–β (green-red) associations. (b) Measurements between α-globin signals showing a bimodal distribution; a similar distribution was seen for α–β distances (not depicted). Values represent means ± SD (error bars) from two independent experiments. (c) Distances between associating α–α signals (HBA–HBA) and α–β signals (HBA–HBB) separated by <1 μm. Both have a mean separation of ∼0.5 μm. Values represent means ± SD from four independent experiments for both α–α associations and α–β associations. Bar, 4 μm.
Estimate of transcription factory number in intermediate erythroblasts.
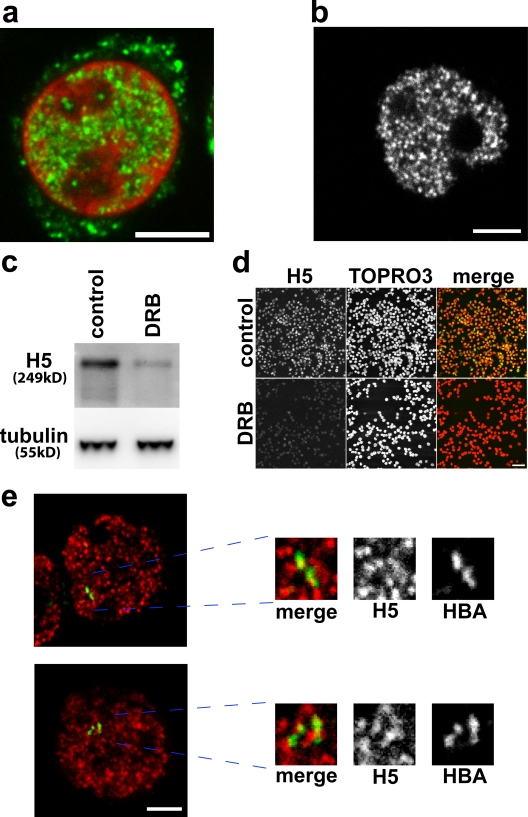
To understand the relationship of transcription factories to associating gene pairs, we used incorporation of BrUTP into active sites of transcription to give an indication of the number of transcription factories in a given cell type (Fay et al., 1997). This approach may underrepresent the true number of transcription foci when compared with the counting of gold particles by electron microscopy (Iborra et al., 1998; Pombo et al., 1999; Iborra, 2002), which indicates up to 8,000 foci in a HeLa cell nucleus, but certainly will not overestimate. BrUTP was detected by immunofluorescence (Fig. 6 a), and a mean value for the number of transcription foci per human intermediate erythroblast nucleus was calculated (see Materials and methods) as between 1,138 ± 264 and 1,897 ± 440 foci, depending on the value of optical section assumed (400 or 700 nm).
Figure 6.
Transcription in intermediate erythroblasts. (a) Incorporation of BrUTP into active sites of transcription, showing a typical distribution of transcription foci (green) in a single confocal slice of a TOPRO3-stained nucleus (red). (b–d) H5 antibody detects the active elongating form of RNApolII. (b) Typical H5 immunofluorescence staining. (c and d) Incubation of erythroblasts with DRB inhibits elongation and results in reduced H5 signal by immunoblotting (c) and by immunofluorescence (d). (e) Associating globin genes (α–α associations and α–β associations) do not share common foci of RNApolII. The image shows a typical RNA immuno-FISH detection of α-globin (green) together with H5 antibody to RNApolII transcription foci (red). Associating globin signals are magnified to show that they sit at different transcription foci. Bars: (a, b, and e) 4 μm; (d) 40 μm.
Active, associating globin genes do not commonly share transcription factories.
To define the location of associating active genes with respect to transcription factories, we used RNA immuno-FISH, detecting the transcripts of specific genes together with an antibody specific for active phosphorylated RNApolII. RNApolII exists in the nucleus in at least three forms: freely diffusible, initiating (phosphorylated at serine 5 on the carboxy-terminal domain [CTD]), and the active elongating form (phosphorylated at CTD serine 2). The latter is recognized by an antibody, H5 (Phatnani and Greenleaf, 2006), that has a very similar staining pattern to BrUTP in erythroblasts (Fig. 6 b). Using 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside (DRB) to block transcript elongation by inhibiting RNApolII phosphorylation at CTD serine 2 (Marshall et al., 1996; Trimborn et al., 1999), H5 levels were substantially reduced in erythroblasts, as demonstrated by both immunoblotting (Fig. 6 c) and immunofluorescence (Fig. 6 d). This indicates that the H5 antibody detects sites of elongation and distinguishes those sites (corresponding to sites of BrUTP incorporation; Pombo et al., 1999) from sites of nonproductive binding of RNApolII (Hieda et al., 2005).
We scored active, associating globin FISH signals in erythroblasts with respect to the foci of transcription detected using the H5 antibody by RNA immuno-FISH (Fig. 6 e). For α-globin, 95% (n = 56) of associating active genes were sitting at different transcription foci, and 5% were at the same focus. For α–β associations, the figures were 73% (n = 63) at different transcription foci and 27% at the same focus. This analysis indicates that association between active globin genes in human erythroblasts is not caused by a requirement to share a common transcription factory.
Positioning of multiple active genes
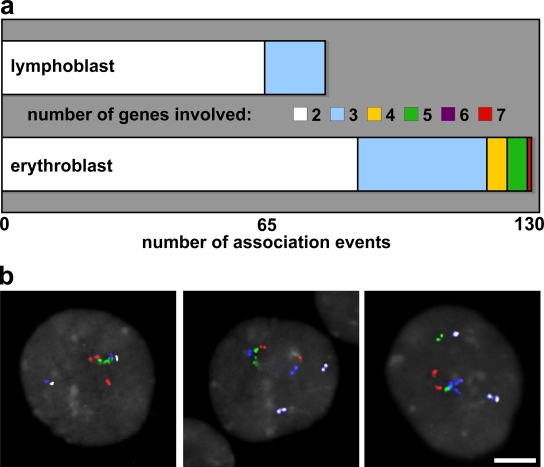
We have demonstrated that associating globin genes do not commonly share transcription factories, but the numbers of trans-associations we scored between active genes in erythroblasts imply that, in some instances, several of the genes analyzed must be clustered together. Using multicolor FISH (M-FISH) hybridization to simultaneously detect four erythroid genes (i.e., eight loci), we scored the number of genes associating and the number of association clusters per nucleus in 90 nuclei of both erythroblasts and lymphoblasts. A maximum number of three of the possible eight hybridization signals in each nucleus clustered in association in lymphoblasts, whereas up to seven gene signals were found to be clustered in erythroblasts (Fig. 7 a). The number of nuclei with more than one association cluster was also greater in erythroblasts than lymphoblasts (47 vs. 13%). Critically, the genes were not all concentrating at a single point, as might be expected if they were sharing transcription foci. Instead, they were grouped in a more curvilinear manner, where each gene was not necessarily in close proximity with all other genes in the cluster (Fig. 7 b). Therefore, we considered another explanation of the gene configurations in erythroblasts.
Figure 7.
Positioning of multiple up-regulated genes. (a) M-FISH detection of concurrent associations between genes HBA, SLC4A1, ERAF, and HBB scored in erythroblast and lymphoblast nuclei. Up to seven FISH signals were found together in nuclear association in intermediate erythroblasts compared with a maximum of three in lymphoblasts. (b) M-FISH detection of HBA (red), SLC4A1 (green), ERAF (blue), and HBB (white) within erythroblast nuclei (gray). Associating signals are not grouped tightly around a single point but in a curvilinear manner. Panels (left to right) show groups of five, four, and three genes. Bar, 4 μm.
Associating genes contact common nuclear speckles
Nuclear speckles are aggregations of splicing-related factors that form after each cell division and during the course of interphase (Misteli et al., 1997; Lamond and Spector, 2003; Prasanth et al., 2003). Certain active genes have been described as sitting at the surface of nuclear speckles (Nielsen et al., 2002; Shopland et al., 2003; Moen et al., 2004), and we have shown previously in intermediate erythroblasts that the majority of associating human globin genes contact the same nuclear speckle (Brown et al., 2006). We speculated that the nucleation of splicing factors into speckles either after cell division or during interphase may bring active genes into association.
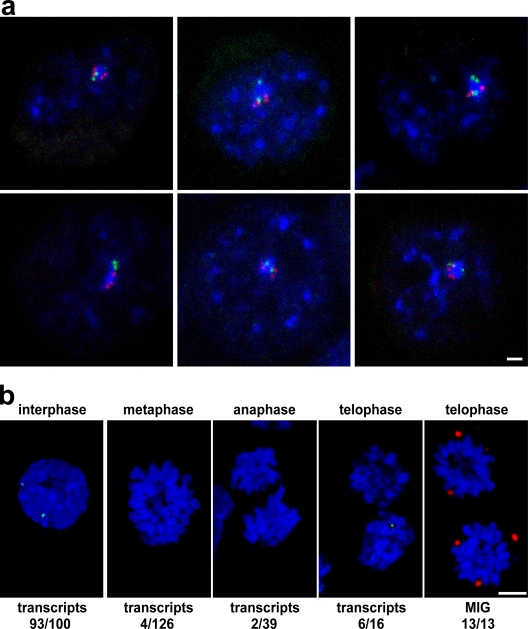
We looked at the contacts made by associating genes with nuclear speckles. Detection of HBA, HBB, or SLC4A1 (in a pairwise fashion) with SC35-enriched splicing factor speckles by DNA immuno-FISH in erythroblasts showed that almost all associating HBA/SLC4A1 signals were in contact with a nuclear speckle, and 92% contacted the same speckle (n = 104). These included not just pairs of associating genes but groups of three (HBA/HBA/SLC4A1 or HBA/SLC4A1/SLC4A1) and even all four genes (Fig. 8 a). Similar results were scored for HBA/HBB and HBB/SLC4A1 associations (93%, n = 58 and 91%, n = 22, respectively). In contrast, the locations of the HBA/SLC4A1 associations scored in lymphoblasts (n = 50) showed that only 28% shared the same speckle, and 50% of the associations were not in contact with any speckle compared with only 1% in erythroid cells. These data show that the vast majority of associations between erythroid genes in erythroblasts, where the genes are active, involve the same speckle. Conversely, in lymphoblasts, where the genes are not active, the same associations do not commonly involve nuclear speckles; i.e., they may represent random juxtapositions of two genes constrained to the inner core of the nucleus.
Figure 8.
A role for nuclear speckles in interphase association of active genes. (a) Immuno-FISH of HBA (green) and SLC4A1 (red) with SC35-enriched nuclear speckles (blue) in intermediate erythroblasts showing that all four genes can be found in association and contacting the same speckle. (b) HBB is active before formation of nuclear speckles after mitosis. RNA-FISH for HBB detects transcripts (green) in interphase intermediate erythroblast nuclei but also in late telophase figures, when the SC35 protein (red) is still present in MIGs. DNA is stained with DAPI (blue). Bars: (a) 1 μm; (b) 4 μm.
If indeed it is the nucleation of splicing factors into speckles after each cell division or during interphase that pulls active genes into close proximity, one would expect that frequently transcribed genes like the globins would be active in a proportion of daughter nuclei before speckle formation. During mitosis, splicing factors accumulate in the cytosol in mitotic interchromatin granules (MIGs) and then migrate into daughter nuclei in late telophase to reform into nuclear speckles (Prasanth et al., 2003). We divided mitotic figures of erythroblasts into three groups: metaphase, anaphase, and telophase (after cytokinesis but before the chromosome structure has fully broken down). In this last group, we found by immunofluorescence that SC35 splicing factors were still sitting in MIGs in 13/13 telophase pairs (Fig. 8 b). We found little evidence of β-globin transcription by RNA-FISH during metaphase or anaphase but scored β-globin nascent transcripts in 6/16 telophase figures (Fig. 8 b). This analysis shows that it would be temporally possible for speckle formation to influence the nuclear positioning of active genes in late telophase.
Discussion
The positioning of genomic regions can be nonrandom within the three-dimensional context of the nucleus; however, the importance this plays in their regulation is still unclear (Fraser and Bickmore, 2007; Misteli, 2007). Although certain regions may have preferred locations, these are not absolute, and several factors seem to interplay to affect their probabilistic positioning (Pederson, 2004; Misteli, 2007).
Chromatin environment affects levels of association
In this study, we find that erythroid up-regulated genes in humans demonstrate significantly higher levels of association when active. However, association levels are strikingly variable. The levels of associations involving SLC4A1 can be as high as 33% when active, whereas association levels for β-globin are generally much lower. β-Globin sits peripherally in the nucleus (Brown et al., 2006), which makes serendipitous allelic associations less likely. Nevertheless, ERAF and α-globin associate allelically to markedly different degrees despite being just 31 Mb apart on the same chromosome; therefore, their specific chromatin environments must play a part. We already know that human α-globin, which also exhibits high levels of association, lies in a dramatically decondensed stretch of terminal chromatin on 16p, regardless of transcriptional status (Brown et al., 2006). The mobility allowed by location in such a decondensed chromatin stretch, along with the preferred internal location of human α-globin on HSA16 (unpublished data), may explain the high levels of association involving the human α-globin gene in comparison with the human β-globin gene. Looking at the 2 Mb surrounding the frequently associating SLC4A1 gene, we find that it too lies in a decondensed chromatin environment. Indeed, Shopland et al. (2003) observed the chromosome band in which SLC4A1 resides (17q21) to be highly extended in fibroblasts. However, in contrast, the 2-Mb region around the β-globin genes appears grossly condensed, even in erythroblasts in which β-globin is highly active. Gene density alone cannot explain the differing chromatin condensation because the β-globin region is more gene rich than the SLC4A1 region. However, we note that the coding density is higher where there is more decondensation, and other factors reflective of decondensation levels include CpG island density and RNApolII density. One can imagine that all of these factors could impinge on local chromatin condensation, with regions of more densely packed genes frequently engaged by transcriptional machinery being much less condensed than areas where genes are sparser and less frequently transcribed. Collectively, one can envisage that a gene that occupies a more internal nuclear address and sits in a decondensed, mobile chromatin region may show more marked levels of association than a gene that is peripherally located on a constrained stretch of chromatin.
Like the human β-globin genes, the mouse α-globin genes show less marked levels of association than their human counterparts (Brown et al., 2006) despite their location on MMU11, which occupies the most internal positioning of the mouse chromosomes (Mayer et al., 2005). In this study, we see that, like human β-globin, the 2-Mb chromatin region around the mouse α-globin genes is condensed regardless of transcriptional status. The profound effect of chromatin environment on gene positioning is demonstrated when the mouse α-globin locus is replaced with the human counterpart (Wallace et al., 2007). The human α-globin locus then adopts the endogenous mouse α-globin gene positioning, yet the normal developmental program of the human α-globin locus is maintained. Interestingly, Morey et al. (2008) have demonstrated that the decondensation of a region can be uncoupled from transcriptional activity. Our findings imply that the degree of association is principally affected by regional chromatin environment and also that severely reduced levels of association do not interfere with the transcriptional program. This last point is most clearly demonstrated by the fact that normal α-globin genes associate much more frequently in human than in mouse erythroblasts yet are expressed optimally in both (Brown et al., 2006). Indeed, deletion of the human α-globin genes from one chromosome does not affect levels of expression from the other α-globin locus or from the two β-globin loci, which are normally transcribed in equal amounts to α-globin (Weatherall and Clegg, 2001). 4C data also suggests that trans-interactions detected by that method reflect chromatin context rather than specific functional relationships (Simonis et al., 2006; Palstra et al., 2008).
Gene positioning and transcription factories
Active murine erythroid genes, both in cis and in trans, have been described as preferentially sharing transcription factories (as defined using the 4H8 antibody against RNApolII; Osborne et al., 2004). Furthermore, it has been proposed that genes may be dynamically relocated to preassembled transcription factories rather than the transcriptional machinery assembling around a gene (Osborne et al., 2004, 2007). Transcription factories are punctate sites of transcription, and the definition is necessarily a functional one because they are the visualization of BrUTP incorporation into nascent transcripts (Iborra et al., 1996). They contain on average eight active molecules of RNApolII, which raises the possibility that more than one gene may share a factory (Iborra, 2002). Using BrUTP incorporation, we estimated that the number of transcription factories per human intermediate erythroblast is >1,500. This is in line with previous studies (Fay et al., 1997; Jackson et al., 1998) and is many more than that detected using the 4H8 antibody to RNApolII in mouse erythroblasts (Osborne et al., 2004). The discrepancy between the number of transcription foci reported in mouse versus human erythroblasts may represent a species difference but could also highlight the need for the use of BrUTP to estimate the transcription factory numbers. The high number of transcription factories we see does not uphold the idea, in human erythroblasts at least, that their limiting number would necessitate the sharing of this resource between physically distant transcribing sites (Osborne et al., 2004).
Recent work modeling RNApolII dynamics in vivo estimated that only around 9% of initiated RNApolII proceeds to the elongation phase of transcription (Darzacq et al., 2007), highlighting the importance of selecting an antibody best suited to detecting the elongating form of RNApolII. We and others (Palstra et al., 2008) demonstrate that the antibody H5 principally detects the elongating form of RNApolII. In contrast, the 4H8 antibody detects foci that remain in murine fetal liver cells when transcription is abolished by DRB treatment or by heat shock (Mitchell and Fraser, 2008). In this study, we find the mean distance between active human globin genes (α–α and α–β) to be ∼500 nm, and, given the 50-nm size of transcription factories, it is suggestive that associating genes lie most frequently at different factories. We confirmed this by looking at associating active globin genes in combination with H5 staining, and we indeed see that associating pairs of human globin genes do not commonly share transcription factories.
Association of genomic elements: common themes
Although we show that association at common transcription factories is not frequent, it is clear that transcriptional activity of the gene itself and of the region in which it lies are important factors in the association levels we observe. We found that multiple active genes can group together in the erythroid nucleus, suggesting that some aspect involved in the expression or processing of active regions may be involved in their convergence. Shopland et al. (2003) have shown that coordinately expressed genes can be found at the same nuclear speckles when active, although the choice of speckle is random, and that gene-rich regions contact nuclear speckles more frequently than gene-poor regions. We postulated that genes found in proximity may be associating at such splicing factor–enriched nuclear speckles. This was lent credence by the curvilinear nature (as opposed to tight congregation) of the gene clusters, as if converging around a large nuclear body. This hypothesis was supported by the multiple gene associations we found at SC35-enriched nuclear speckles in erythroid cells. We show that it is temporally possible for globin genes to begin transcription at the very beginning of the cell cycle, just before the renucleation of splicing factors, which are known to nucleate at early transcribing genes (Prasanth et al., 2003). Therefore, erythroid genes active at the beginning of the cell cycle and on decondensed mobile stretches of chromatin may be brought into association by the nucleation of splicing factors into speckles (Fig. 9). The same must be true of speckles forming during the course of interphase (Misteli et al., 1997). Transcripts from genes sited at the surface of nuclear speckles have been shown to traffic through those structures and to be modified there before export from the nucleus (Johnson et al., 2000; Smith et al., 2007). It is unclear at present whether there is any posttranscriptional advantage for allelic or coregulated genes to converge at the same speckle.
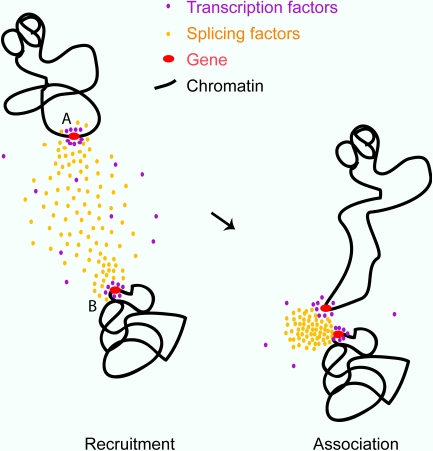
Figure 9.
A model for involvement of splicing factor aggregations in the stochastic association of transcriptionally active genes. Genes in the process of transcriptional elongation accumulate splicing-related factors (left). Nuclear speckles are known to form as concentrations of such factors in early G1 and randomly during interphase. This process of nucleation may pull more mobile chromatin (around gene A) into association with other active genes (gene B; right).
In addition to the association of active genes (Osborne et al., 2004, 2007; Brown et al., 2006), convergences have also been noted in several situations that could be thought of as regulatory; e.g., genomic imprinting (LaSalle and Lalande, 1996; Ling et al., 2006), before selective activation (Spilianakis et al., 2005; Lomvardas et al., 2006; Takizawa et al., 2008), or before inactivation (Bacher et al., 2006; Xu et al., 2006). The functional significance of these associations, however, remains unclear (Fuss et al., 2007; Nishizumi et al., 2007; Teller et al., 2007). Could there be common ground linking the associations seen between active genes and those in regulatory situations? One possibility is unrecognized transcriptional activity in the region surrounding the gene in question (de Laat, 2007). For example, it has been postulated that transcription of noncoding RNAs may be generally important in the regulation of genomic imprinting and in the establishment of X inactivation (Pauler et al., 2007; Yang and Kuroda, 2007). Another possibility is that gene regions are converging at other nuclear sites, for example the nucleolus (Thompson et al., 2003; Teller et al., 2007). Finally, self-interacting proteins such as the diversely functioning protein CTCF (CCCTC-binding factor) may have a role to play (Pant et al., 2004; Wallace and Felsenfeld, 2007; Filippova, 2008). Association of the imprinted maternal Igf2/H19 allele on MMU7 and the paternal Wsb1/Nf1 locus on MMU11 and pairing of the X chromosomes both involve CTCF, with knockdown of CTCF in both cases abrogating association (Ling et al., 2006; Xu et al., 2007). Interestingly, CTCF has been shown to recruit RNApolII to CTCF sites by interaction with the CTD with a possible role in inter-genic transcription (Chernukhin et al., 2007).
Summary
Within a model of erythroid differentiation, we find that the tendency to associate, which varies between genes, is directly dependent on the surrounding chromatin environment. We find that spatial associations are a frequent occurrence between some active genes but that this is not a result of shared transcription factories. Finally, we demonstrate that multiple active genes may be found together around nuclear speckles. We have already shown for the globin genes that association is not a functional necessity for transcription (Brown et al., 2006). We now suggest that active genes, particularly those on stretches of decondensed chromatin, can be brought into close proximity by the nucleation of multiple splicing-associated proteins and small nuclear RNPs into nuclear speckles (Misteli et al., 1997; Lamond and Spector, 2003; Prasanth et al., 2003). We propose that whole groups of coexpressed genes will occasionally be found around the same speckle but that these clusters have a probabilistic basis. Our findings contribute to the growing body of evidence that the central processes of transcription, replication, and repair are all self-organized within the nucleus (Misteli, 2007).
Materials and methods
Cell culture
Human intermediate erythroblasts and lymphoblasts were prepared as previously described (Brown et al., 2006). Mouse intermediate erythroblasts and activated B lymphocytes were obtained from mouse spleen as previously described (Brown et al., 2006) under license from the Oxford University Local Ethical Review Process.
Probes and nick-translation labeling
Probes used for detection of human and mouse globin genes by RNA-FISH and DNA-FISH were previously described (Brown et al., 2006). Additional FISH probes used were human BAC clones BAC RP11-1137J13 (GATA1), BAC RP11-194H10 (ERAF), BAC RP11-264B17 (control probe for HSA16), and BAC RP11-976E1 (SLC4A1 and biotinylated whole chromosome paints for HSA16, HSA17, and MMU11 [Cambio Biosys]). The mouse α-globin 2-Mb contig consisted of nine BAC clones (RP23-31D18, RP23-475A8, RP24-212L20, RP23-414P21, RP23-322D7, RP23-395O6, RP23-94F11, RP23-413O13, and RP23-323L8); the human SLC4A1 2-Mb contig consisted of eight BAC probes (RP11-100E5, RP11-392O1, RP11-261A9, RP11-810P3, RP11-812N9, RP11-1072C15, RP11-454D13, and RP11-403G3); and the human β-globin consisted of 14 BAC probes (RP11-437G21, RP11-54I6, RP11-648B16, RP11-3P24, RP11-534K22, RP11-1205H24, RP11-910P5, RP11-454O22, RP11-54F1, RP11-622B13, Rp11-957H15, RP11-451K18, RP11-290F24, and RP11-175111). All BACs were obtained from the BACPAC Resources Center (Children's Hospital Oakland Research Institute).
FISH probes were labeled by nick translation as previously described (Smith and Higgs, 1999). Probes for M-FISH were directly or indirectly labeled by nick translation using Cy3-dUTP (GE Healthcare), Cy5-dUTP (GE Healthcare), AlexaFluor488–5-dUTP (Invitrogen), and biotin–16-dUTP (Roche).
2D DNA-FISH
DNA-FISH was performed as described previously with minor changes (Brown et al., 2006). In brief, methanol–acetic acid–fixed cells were treated with 100 μg/ml RNase for 30 min at 37°C, washed in 2× SSC, and dehydrated. Cells were denatured in 70% formamide at 70°C for 5 min and dehydrated. Labeled probes (100 ng each) were denatured in hybridization mix at 90°C for 5 min and were preannealed at 37°C for 20 min. Where whole chromosome paints were used for each slide, 100 ng of denatured labeled probe in 5 μl was combined with 10 μl of denatured chromosome paint and applied to the slide. Slides were hybridized overnight at 42°C. After hybridization, slides were washed four times in 50% formamide at 45°C for 3 min, four times in 2× SSC at 45°C for 3 min, and four times in 0.1× SSC at 60°C for 3 min. Slides were blocked in 3% BSA, biotin was detected with streptavidin Cy3 (Jackson ImmunoResearch Laboratories), and digoxygenin was detected with sheep antidigoxigenin FITC (Roche) followed by rabbit anti–sheep FITC (Vector Laboratories).
3D DNA-FISH
3D DNA-FISH was performed as described previously (Brown et al., 2006). In brief, cells were fixed in 4% PFA for 15 min and permeabilized in 1% Triton X-100 for 10 min. Cells were treated with 100 μg/ml RNase for 1 h at 37°C, denatured in 3.5 N HCl for 20 min, and neutralized in ice-cold PBS. Probes were prepared as in the previous section, and slides were hybridized overnight at 37°C. Cells were washed two times in 2× SSC at 37°C, once in 1× SSC at RT, and blocked in 3% BSA. Probes were detected as in the previous section.
M-FISH
M-FISH was performed as previously described (Brown et al., 2006) on methanol–acetic acid–fixed erythroblasts and lymphoblasts. Multiple BAC probes were labeled as follows: ERAF-Cy3, SLC4A1-AlexaFluor488, HBB-biotin, and HBA-Cy5. After hybridization and washing, the biotin-labeled probe was detected using streptavidin Cy3.5 (GE Healthcare). We were only able to undertake this analysis by DNA-FISH, not RNA-FISH, because of technical constraints. Therefore, we cannot be certain that all genes scored as associating are active. However, we compared the levels of association between intermediate erythroblasts, where they are very active, and lymphoblasts, where they are not active. Furthermore, we already know that association levels between individual pairs of the four genes tested are higher in erythroblasts than lymphoblasts.
Immuno-FISH
Immuno-FISH was performed as described previously with minor changes (Brown et al., 2006). In brief, cells were fixed in 4% PFA and permeabilized in 0.5% Triton X-100. Nonspecific sites were blocked using 10% FCS. Antibodies were prepared in blocking solution as follows: IgM mouse antiphosphorylated CTDser2 RNApolII (H5; Covance) at 1:100 and mouse anti-SC35 (Sigma-Aldrich) at 1:500. Secondary antibodies used were anti–mouse IgM Cy3 or Cy5 (Jackson ImmunoResearch Laboratories) and horse anti–mouse Texas red (Vector Laboratories). Cells were treated with 100 μg/ml RNase for 1 h at 37°C, denatured in 3.5 N HCl for 20 min, and neutralized in ice-cold PBS. Probes were prepared as in the 2D DNA-FISH section, and slides were hybridized overnight at 37°C. Cells were washed twice in 2× SSC at 37°C, once in 1× SSC at RT, and blocked in 3% BSA. Probes were detected as in the 2D DNA-FISH section.
RNA-FISH and RNA immuno-FISH
RNA-FISH and RNA immuno-FISH were performed as described previously (Brown et al., 2006).
Immunofluorescence
Immunofluorescence was performed as described previously with minor changes (Brown et al., 2006). In brief, cells were fixed in 4% PFA and permeabilized in 1% Triton X-100. Nonspecific sites were blocked using 10% FCS. Antibodies were prepared in blocking solution as follows: IgM mouse antiphosphorylated CTDser2 RNApolII (H5; Covance) at 1:100 and anti-IgM Cy3 (Jackson ImmunoResearch Laboratories) at 1:100.
BrUTP incorporation
BrUTP was incorporated into nascent transcripts as previously described (Iborra et al., 2004). To calculate an average figure for transcription foci in erythroblast nuclei, we scored the number of foci in the confocal slice with maximum diameter from 54 nuclei and calculated the density per unit of surface area. The mean diameter of intermediate erythroblasts was taken from 200 nuclei, and the density per μm3 (ρ) was then calculated with Abercrombie's formula (Iborra et al., 1996) assuming an optical section thickness of 400–700 nm.
Immunoblotting
Protein extracts were prepared by harvesting intermediate erythroblasts in Laemmli buffer. Total denatured protein extracts were separated by SDS-PAGE on a 3–8% Tris-acetate gradient gel (Invitrogen) and transferred to Immobilon-PSQ polyvinylidene difluoride membrane (Millipore). Nonspecific binding sites were blocked using 5% powdered milk and 0.1% Tween-20 in PBS (PBS-T) for 1 h. The membrane was then incubated for 1 h with primary antibodies (made in PBS-T) against phosphorylated CTDser2 RNApolII (H5; Covance) at 1:1,000 and β-tubulin (Sigma-Aldrich) at 1:5,000 and was washed in PBS-T for 30 min. The membrane was incubated for 1 h with the HRP-conjugated secondary antibodies anti–mouse IgM HRP and anti–mouse IgG HRP at 1:1,000 (Novus Biologicals) and was washed for 30 min in PBS-T. Blots were washed in PBS-T before detection of the HRP signal with ECL detection reagent (GE Healthcare).
Imaging and distance measurements
All cells were mounted in Vectashield (Vector Laboratories) with 1 μg/ml DAPI (Roche) counterstain. 2D M-FISH analysis was performed using a microscope (BX60; Olympus) equipped with Perceptive Scientific International MacProbe version 4.3 image analysis package (Applied Imaging) and a SenSys KAF1400 charge-coupled device camera (Photometrics). A narrow band-pass filter set (Chroma Technology Corp.) suitable for M-FISH analysis was used to discriminate fluorochromes. Chromosome territories were imaged on the same system. All other fluorescent analysis was undertaken with a Radiance 2000 confocal system (Bio-Rad Laboratories) on a microscope (BX51; Olympus). All images were taken through a 100× NA 1.3 UPlanF1 oil immersion objective (Olympus) at RT using Lasersharp software (Bio-Rad Laboratories). Contrast-stretch and γ adjustments were made using Photoshop (Adobe) only for display in figures. Laserpix software (Bio-Rad Laboratories) was used for distance measurements between FISH signals. The diameters of contig FISH signals were measured with Easivision software (Soft Imaging Systems) as the extent outer max value: the maximum diameter through particle center from outer border to outer border. Immunoblots were imaged with Universal Hood II (Bio-Rad Laboratories), and analysis was performed using Quantity One software (Bio-Rad Laboratories).
Statistical analysis
P-values have been calculated either by t test with two-tailed distributions of unequal variance and α level of 0.05 or by Fisher's two-tailed exact test for categorical data with an α level of 0.004 after a Bonferroni correction for multiplicity (Table I). All error bars denote SDs.
Acknowledgments
We thank Sue Butler, Jackie Sloane-Stanley, Jackie Sharpe, and Kathryn Robson for provision of cells.
This work was supported by the Medical Research Council.
Abbreviations used in this paper: BAC, bacterial artificial chromosome; BrUTP, bromo-UTP; CTD, carboxy-terminal domain; DRB, 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside; ERAF, erythroid-associated factor; M-FISH, multicolor FISH; MIG, mitotic interchromatin granule; RNApolII, RNA polymerase II.
References
- Augui, S., G.J. Filion, S. Huart, E. Nora, M. Guggiari, M. Maresca, A.F. Stewart, and E. Heard. 2007. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 318:1632–1636. [DOI] [PubMed] [Google Scholar]
- Bacher, C.P., M. Guggiari, B. Brors, S. Augui, P. Clerc, P. Avner, R. Eils, and E. Heard. 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8:293–299. [DOI] [PubMed] [Google Scholar]
- Barski, A., S. Cuddapah, K. Cui, T.Y. Roh, D.E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell. 129:823–837. [DOI] [PubMed] [Google Scholar]
- Brown, J.M., J. Leach, J.E. Reittie, A. Atzberger, J. Lee-Prudhoe, W.G. Wood, D.R. Higgs, F.J. Iborra, and V.J. Buckle. 2006. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 172:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, H., B. van Schaik, M. van der Mee, F. Baas, G. Riggins, P. van Sluis, M.C. Hermus, R. van Asperen, K. Boon, P.A. Voute, et al. 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 291:1289–1292. [DOI] [PubMed] [Google Scholar]
- Chambeyron, S., and W.A. Bickmore. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18:1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernukhin, I., S. Shamsuddin, S.Y. Kang, R. Bergstrom, Y.W. Kwon, W. Yu, J. Whitehead, R. Mukhopadhyay, F. Docquier, D. Farrar, et al. 2007. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol. Cell. Biol. 27:1631–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer, T., K. Kupper, S. Dietzel, and S. Fakan. 2004. Higher order chromatin architecture in the cell nucleus: on the way from structure to function. Biol. Cell. 96:555–567. [DOI] [PubMed] [Google Scholar]
- Darzacq, X., Y. Shav-Tal, V. de Turris, Y. Brody, S.M. Shenoy, R.D. Phair, and R.H. Singer. 2007. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14:796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat, W. 2007. Long-range DNA contacts: romance in the nucleus? Curr. Opin. Cell Biol. 19:317–320. [DOI] [PubMed] [Google Scholar]
- Fay, F.S., K.L. Taneja, S. Shenoy, L. Lifshitz, and R.H. Singer. 1997. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A). Exp. Cell Res. 231:27–37. [DOI] [PubMed] [Google Scholar]
- Filippova, G.N. 2008. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 80:337–360. [DOI] [PubMed] [Google Scholar]
- Fraser, P., and W. Bickmore. 2007. Nuclear organization of the genome and the potential for gene regulation. Nature. 447:413–417. [DOI] [PubMed] [Google Scholar]
- Fuss, S.H., M. Omura, and P. Mombaerts. 2007. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 130:373–384. [DOI] [PubMed] [Google Scholar]
- Gierman, H.J., M.H. Indemans, J. Koster, S. Goetze, J. Seppen, D. Geerts, R. van Driel, and R. Versteeg. 2007. Domain-wide regulation of gene expression in the human genome. Genome Res. 17:1286–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N., S. Boyle, H. Fiegler, K. Woodfine, N.P. Carter, and W.A. Bickmore. 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 118:555–566. [DOI] [PubMed] [Google Scholar]
- Goetze, S., J. Mateos-Langerak, H.J. Gierman, W. de Leeuw, O. Giromus, M.H. Indemans, J. Koster, V. Ondrej, R. Versteeg, and R. van Driel. 2007. The three-dimensional structure of human interphase chromosomes is related to the transcriptome map. Mol. Cell. Biol. 27:4475–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieda, M., H. Winstanley, P. Maini, F.J. Iborra, and P.R. Cook. 2005. Different populations of RNA polymerase II in living mammalian cells. Chromosome Res. 13:135–144. [DOI] [PubMed] [Google Scholar]
- Hurst, L.D., C. Pal, and M. Lercher. 2004. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5:299–310. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J. 2002. The path that RNA takes from the nucleus to the cytoplasm: a trip with some surprises. Histochem. Cell Biol. 118:95–103. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., and P.R. Cook. 1998. The size of sites containing SR proteins in human nuclei. Problems associated with characterizing small structures by immunogold labeling. J. Histochem. Cytochem. 46:985–992. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., A. Pombo, D. Jackson, and P. Cook. 1996. Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. J. Cell Sci. 109:1427–1436. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., D. Jackson, and P. Cook. 1998. The path of transcripts from extra-nucleolar synthetic sites to nuclear pores: transcripts in transit are concentrated in discrete structures containing SR proteins. J. Cell Sci. 111:2269–2282. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., A.E. Escargueil, K.Y. Kwek, A. Akoulitchev, and P.R. Cook. 2004. Molecular cross-talk between the transcription, translation, and nonsense-mediated decay machineries. J. Cell Sci. 117:899–906. [DOI] [PubMed] [Google Scholar]
- Jackson, D.A., F.J. Iborra, E.M. Manders, and P.R. Cook. 1998. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell. 9:1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C., D. Primorac, M. McKinstry, J. McNeil, D. Rowe, and J.B. Lawrence. 2000. Tracking COL1A1 RNA in osteogenesis imperfecta. splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J. Cell Biol. 150:417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, M.A., S. Addya, R. Vadigepalli, B. Banini, K. Delgrosso, H. Huang, and S. Surrey. 2006. Transcriptional regulatory network analysis of developing human erythroid progenitors reveals patterns of coregulation and potential transcriptional regulators. Physiol. Genomics. 28:114–128. [DOI] [PubMed] [Google Scholar]
- Kosak, S.T., D. Scalzo, S.V. Alworth, F. Li, S. Palmer, T. Enver, J.S. Lee, and M. Groudine. 2007. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS Biol. 5:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, M., H. Tanabe, K. Yoshida, K. Oikawa, A. Saito, T. Kiyuna, H. Mizusawa, and K. Mukai. 2004. Alteration of chromosome positioning during adipocyte differentiation. J. Cell Sci. 117:5897–5903. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I., and D.L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4:605–612. [DOI] [PubMed] [Google Scholar]
- LaSalle, J.M., and M. Lalande. 1996. Homologous association of oppositely imprinted chromosomal domains. Science. 272:725–728. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., A.O. Urrutia, and L.D. Hurst. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31:180–183. [DOI] [PubMed] [Google Scholar]
- Ling, J.Q., T. Li, J.F. Hu, T.H. Vu, H.L. Chen, X.W. Qiu, A.M. Cherry, and A.R. Hoffman. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 312:269–272. [DOI] [PubMed] [Google Scholar]
- Lomvardas, S., G. Barnea, D.J. Pisapia, M. Mendelsohn, J. Kirkland, and R. Axel. 2006. Interchromosomal interactions and olfactory receptor choice. Cell. 126:403–413. [DOI] [PubMed] [Google Scholar]
- Mahy, N.L., P.E. Perry, and W.A. Bickmore. 2002. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159:753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, N.F., J. Peng, Z. Xie, and D.H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176–27183. [DOI] [PubMed] [Google Scholar]
- Mayer, R., A. Brero, J. von Hase, T. Schroeder, T. Cremer, and S. Dietzel. 2005. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. 2007. Beyond the sequence: cellular organization of genome function. Cell. 128:787–800. [DOI] [PubMed] [Google Scholar]
- Misteli, T., J.F. Caceres, and D.L. Spector. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 387:523–527. [DOI] [PubMed] [Google Scholar]
- Mitchell, J.A., and P. Fraser. 2008. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 22:20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen, P.T. Jr., C.V. Johnson, M. Byron, L.S. Shopland, I.L. de la Serna, A.N. Imbalzano, and J.B. Lawrence. 2004. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol. Biol. Cell. 15:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey, C., N.R. Da Silva, P. Perry, and W.A. Bickmore. 2007. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 134:909–919. [DOI] [PubMed] [Google Scholar]
- Morey, C., N.R. Da Silva, M. Kmita, D. Duboule, and W.A. Bickmore. 2008. Ectopic nuclear reorganisation driven by a Hoxb1 transgene transposed into Hoxd. J. Cell Sci. 121:571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, J.A., L.D. Hudson, and R.C. Armstrong. 2002. Nuclear organization in differentiating oligodendrocytes. J. Cell Sci. 115:4071–4079. [DOI] [PubMed] [Google Scholar]
- Nishizumi, H., K. Kumasaka, N. Inoue, A. Nakashima, and H. Sakano. 2007. Deletion of the core-H region in mice abolishes the expression of three proximal odorant receptor genes in cis. Proc. Natl. Acad. Sci. USA. 104:20067–20072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, C.S., L. Chakalova, K. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065–1071. [DOI] [PubMed] [Google Scholar]
- Osborne, C.S., L. Chakalova, J.A. Mitchell, A. Horton, A.L. Wood, D.J. Bolland, A.E. Corcoran, and P. Fraser. 2007. Myc dynamically and preferentially relocates to a transcription factory occupied by igh. PLoS Biol. 5:e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra, R.J., M. Simonis, P. Klous, E. Brasset, B. Eijkelkamp, and W. de Laat. 2008. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS ONE. 3:e1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant, V., S. Kurukuti, E. Pugacheva, S. Shamsuddin, P. Mariano, R. Renkawitz, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 24:3497–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada, L.A., P.G. McQueen, and T. Misteli. 2004. Tissue-specific spatial organization of genomes. Genome Biol. 5:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauler, F.M., M.V. Koerner, and D.P. Barlow. 2007. Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet. 23:284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T. 2004. The spatial organization of the genome in mammalian cells. Curr. Opin. Genet. Dev. 14:203–209. [DOI] [PubMed] [Google Scholar]
- Phatnani, H.P., and A.L. Greenleaf. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20:2922–2936. [DOI] [PubMed] [Google Scholar]
- Pombo, A., D.A. Jackson, M. Hollinshead, Z. Wang, R.G. Roeder, and P.R. Cook. 1999. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 18:2241–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth, K.V., P. Sacco-Bubulya, S. Prasanth, and D. Spector. 2003. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell. 14:1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy, T., A. Telling, T. Sawado, M. Groudine, and S.T. Kosak. 2003. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 11:513–525. [DOI] [PubMed] [Google Scholar]
- Shopland, L.S., C.V. Johnson, M. Byron, J. McNeil, and J.B. Lawrence. 2003. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J. Cell Biol. 162:981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis, M., P. Klous, E. Splinter, Y. Moshkin, R. Willemsen, E. de Wit, B. van Steensel, and W. de Laat. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38:1348–1354. [DOI] [PubMed] [Google Scholar]
- Smith, K.P., M. Byron, C. Johnson, Y. Xing, and J.B. Lawrence. 2007. Defining early steps in mRNA transport: mutant mRNA in myotonic dystrophy type I is blocked at entry into SC-35 domains. J. Cell Biol. 178:951–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, Z.E., and D.R. Higgs. 1999. The pattern of replication at a human telomeric region (16p13.3): its relationship to chromosome structure and gene expression. Hum. Mol. Genet. 8:1373–1386. [DOI] [PubMed] [Google Scholar]
- Spilianakis, C.G., M.D. Lalioti, T. Town, G.R. Lee, and R.A. Flavell. 2005. Interchromosomal associations between alternatively expressed loci. Nature. 435:637–645. [DOI] [PubMed] [Google Scholar]
- Sproul, D., N. Gilbert, and W.A. Bickmore. 2005. The role of chromatin structure in regulating the expression of clustered genes. Nat. Rev. Genet. 6:775–781. [DOI] [PubMed] [Google Scholar]
- Takizawa, T., P.R. Gudla, L. Guo, S. Lockett, and T. Misteli. 2008. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 22:489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teller, K., I. Solovei, K. Buiting, B. Horsthemke, and T. Cremer. 2007. Maintenance of imprinting and nuclear architecture in cycling cells. Proc. Natl. Acad. Sci. USA. 104:14970–14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, M., R.A. Haeusler, P.D. Good, and D.R. Engelke. 2003. Nucleolar clustering of dispersed tRNA genes. Science. 302:1399–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn, T., J. Gribnau, F. Grosveld, and P. Fraser. 1999. Mechanisms of developmental control of transcription in the murine alpha- and beta-globin loci. Genes Dev. 13:112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg, R., B.D. van Schaik, M.F. van Batenburg, M. Roos, R. Monajemi, H. Caron, H.J. Bussemaker, and A.H. van Kampen. 2003. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 13:1998–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi, E.V., E. Chevret, T. Jones, R. Vatcheva, J. Williamson, S. Beck, R.D. Campbell, M. Goldsworthy, S.H. Powis, J. Ragoussis, et al. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565–1576. [DOI] [PubMed] [Google Scholar]
- Wallace, H.A., F. Marques-Kranc, M. Richardson, F. Luna-Crespo, J.A. Sharpe, J. Hughes, W.G. Wood, D.R. Higgs, and A.J. Smith. 2007. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell. 128:197–209. [DOI] [PubMed] [Google Scholar]
- Wallace, J.A., and G. Felsenfeld. 2007. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17:400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherall, D.J., and J.B. Clegg. 2001. The Thalassaemia Syndromes. Fourth edition. Blackwell Science, Oxford. 846 pp.
- Williams, R.R., S. Broad, D. Sheer, and J. Ragoussis. 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272:163–175. [DOI] [PubMed] [Google Scholar]
- Xu, N., C.L. Tsai, and J.T. Lee. 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 311:1149–1152. [DOI] [PubMed] [Google Scholar]
- Xu, N., M.E. Donohoe, S.S. Silva, and J.T. Lee. 2007. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat. Genet. 39:1390–1396. [DOI] [PubMed] [Google Scholar]
- Yang, P.K., and M.I. Kuroda. 2007. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell. 128:777–786. [DOI] [PubMed] [Google Scholar]