Abstract
A complex relationship exists between autophagy and apoptosis, but the regulatory mechanisms underlying their interactions are largely unknown. We conducted a systematic study of Drosophila melanogaster cell death–related genes to determine their requirement in the regulation of starvation-induced autophagy. We discovered that six cell death genes—death caspase-1 (Dcp-1), hid, Bruce, Buffy, debcl, and p53—as well as Ras–Raf–mitogen activated protein kinase signaling pathway components had a role in autophagy regulation in D. melanogaster cultured cells. During D. melanogaster oogenesis, we found that autophagy is induced at two nutrient status checkpoints: germarium and mid-oogenesis. At these two stages, the effector caspase Dcp-1 and the inhibitor of apoptosis protein Bruce function to regulate both autophagy and starvation-induced cell death. Mutations in Atg1 and Atg7 resulted in reduced DNA fragmentation in degenerating midstage egg chambers but did not appear to affect nuclear condensation, which indicates that autophagy contributes in part to cell death in the ovary. Our study provides new insights into the molecular mechanisms that coordinately regulate autophagic and apoptotic events in vivo.
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved mechanism for the degradation of long-lived proteins and organelles. During autophagy, cytoplasmic components are sequestered into double membrane structures called autophagosomes, which then fuse with lysosomes to form autolysosomes, where degradation occurs (for review see Klionsky, 2007). Currently, there are 31 autophagy-related (Atg) genes in yeast, and 18 Atg proteins are essential for autophagosome formation (Mizushima, 2007). Most yeast Atg genes have orthologues in higher eukaryotes and encode proteins required for autophagy induction, autophagosome nucleation, expansion, and completion, and final retrieval of Atg protein complexes from mature autophagosomes (for review see Levine and Yuan, 2005).
Depending on the physiological and pathological conditions, autophagy has been shown to act as a pro-survival or pro-death mechanism in vertebrates (for reviews see Levine and Yuan, 2005; Maiuri et al., 2007). In the case of growth factor withdrawal, starvation, and neurodegeneration, autophagy has been shown to function in cell survival (Boya et al., 2005; Lum et al., 2005; Hara et al., 2006; Komatsu et al., 2006). In contrast, autophagy has been found to act as a cell death mechanism in derived cell lines where caspases or apoptotic regulators are impaired (Shimizu et al., 2004; Yu et al., 2004). The nature and perhaps level of the stress stimulus may also be important in determining whether autophagy promotes cell survival or cell death (Boya et al., 2005; Maiuri et al., 2007; Wang et al., 2008).
Overlaps between components in apoptosis and autophagic pathways have been described. Upstream signal transducers in apoptotic pathways, including TNF-related apoptosis-inducing ligand (TRAIL), TNF, Fas-associated protein with death domain (FADD), and death-associated protein kinase (DAPK), have been shown to play a role in autophagy regulation (Prins et al., 1998; Inbal et al., 2002; Mills et al., 2004; Thorburn et al., 2005), and two apoptotic inducers, including sphingolipid and ceramide, can activate autophagy in mammalian cells (Ghidoni et al., 1996; Scarlatti et al., 2004). In addition, two recent studies demonstrate physical and functional interactions between components of apoptosis and autophagy. First, the antiapoptosis protein, Bcl-2, suppresses autophagy through a direct interaction with Beclin 1, a protein required for autophagy (Pattingre et al., 2005). Second, Atg5, which is cleaved by calpain, associates with Bcl-XL, leading to cytochrome c release and caspase activation (Yousefi et al., 2006). Further examples and discussion of the connections between apoptosis and autophagy can be found in several recent reviews on this topic (Ferraro and Cecconi, 2007; Maiuri et al., 2007; Thorburn, 2008). The current findings indicate that there is a complex relationship between apoptosis and autophagy, but the regulatory mechanisms underlying the crosstalk between the two processes are still largely unknown.
Autophagy is observed in several Drosophila melanogaster tissues during development, and thus D. melanogaster is useful as a model to study autophagy in the context of a living organism. 14 D. melanogaster annotated genes share significant sequence identity with the yeast Atg genes, and, overall, eight D. melanogaster Atg homologues have already been shown to be required for autophagy function (Scott et al., 2004; Berry and Baehrecke, 2007). In addition, recent studies demonstrated the role of autophagy in D. melanogaster physiological cell death. Loss of Atg genes, including Atg1, Atg2, Atg3, Atg6, Atg7, Atg8, Atg12, and Atg18, inhibited proper degradation of salivary glands during development. Overexpression of Atg1 induced premature salivary gland cell death in a caspase-independent manner (Berry and Baehrecke, 2007). In contrast, caspase activity was required for Atg1-mediated apoptotic death in the fat body (Scott et al., 2007). Mutation of Atg7 resulted in an inhibition of DNA fragmentation in the midgut but led to an increase of DNA fragmentation in the adult D. melanogaster brain (Juhasz et al., 2007). Together, these results further suggest that the mechanistic role of autophagy in cell death and the interrelations between autophagy and apoptosis may be tissue and/or context dependent.
The adult D. melanogaster ovary contains 15–20 ovarioles comprised of developing egg chambers, which consist of 16 germ line cells (15 nurse cells and 1 oocyte) surrounded by a layer of somatic follicle cells. The germ line cells originate from stem cells that undergo mitosis to form 16-cell cysts in a specialized region called the germarium. In the late stage of oogenesis, the nurse cells support the development of the oocyte by transferring to it their cytoplasmic contents. After this “dumping” event, the nurse cells undergo cell death, and their remnants are engulfed by the surrounding follicle cells (King, 1970; Spradling, 1993). In addition to this late-stage developmental cell death, egg chambers can be induced to die at two earlier stages, during germarium formation (in region 2) and mid-oogenesis, by factors such as nutrient deprivation, chemical insults, and altered hormonal signaling (Drummond-Barbosa and Spradling, 2001; McCall, 2004). In some respects, cell death during D. melanogaster oogenesis is similar to the death of D. melanogaster larval salivary glands. Both nurse cells and salivary gland cells are large and polyploid, and the entire tissues undergo cell death simultaneously (McCall, 2004). Notably, morphological features of autophagy have been described during mid-oogenesis cell death in a related species, Drosophila virilis (Velentzas et al., 2007), which suggests that the cell death process in ovaries and salivary glands share additional similarities.
Previous studies have focused on characterizing the role of autophagy genes in cell death and determining the paradoxical functions of autophagy (pro-survival and pro-death) in various cell lines and organisms. However, a systematic approach that investigates the involvement of cell death genes in starvation-induced autophagy has not been conducted. Here, we present RNAi analyses to determine whether known cell death–related genes in D. melanogaster play a role in autophagy regulation in the lethal (2) malignant blood neoplasm (l(2)mbn) cell line. We also used D. melanogaster genetics to investigate a role for the effector caspase death caspase-1 (Dcp-1) and the inhibitor of apoptosis (IAP) family member Bruce in autophagy regulation in vivo during D. melanogaster oogenesis. Further, we examine the function of autophagy genes Atg7 and Atg1 in starvation-induced germ line cell death in the D. melanogaster ovary.
Results
The RNAi screening assay identifies known positive and negative regulators of starvation-induced autophagy in D. melanogaster l(2)mbn cells
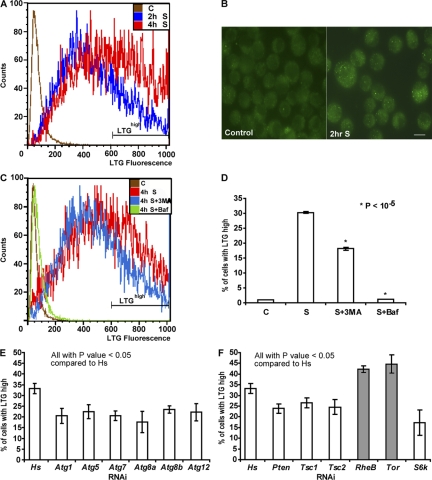
To quantify starvation-induced autophagy, we used a D. melanogaster tumorous larval hemocyte cell line, l(2)mbn (Ress et al., 2000), and used LysoTracker green (LTG) dye, which has been shown previously to label lysosomes and autolysosomes in D. melanogaster (Scott et al., 2004; Klionsky et al., 2007). Flow cytometry was used to acquire LTG fluorescence of individual cells. Under nutrient-full medium conditions, we detected a basal level of LTG labeling in l(2)mbn cells (Fig. 1 A). When cells were transferred into amino acid–deprived medium for 2 h, we observed a detectable increase in LTG labeling. After 4 h of amino acid deprivation, a further increase in the percentage of cells with high LTG fluorescence levels (LTGhigh population) was observed (Fig. 1 A). To confirm that autophagy is indeed up-regulated under nutrient-deprived conditions in l(2)mbn cells, we constructed a stable l(2)mbn cell line expressing mammalian microtubule-associated protein 1 light chain 3 (LC3)/Atg8 fused to GFP protein, a widely used marker for autophagy (Klionsky et al., 2007). During autophagy, LC3 conjugates to phosphatidylethanolamine, which then inserts into the autophagosomal membrane. Thus, localization of GFP-LC3 changes from a diffuse cytoplasmic pattern to a punctate autophagosomal membrane-bound pattern that can be monitored by microscopy (Mizushima et al., 2001; Klionsky et al., 2007). As expected, the percentage of cells with more than three GFP-LC3 puncta (GFP-LC3 positive) was increased from 9% (n = 216) in the nutrient-full condition to 32% (n = 200) in the nutrient-starved condition for 2 h (Fig. 1 B). Further, to confirm that LTG labeling correlates with autophagy levels in l(2)mbn cells, we used the pharmacological autophagy inhibitors 3-methyladenine (3MA) and bafilomycin A1 (Baf). 3MA blocks autophagy by inhibiting phosphoinositide 3-kinase (PI3K) activity (Seglen and Gordon, 1982). Baf is a specific inhibitor of lysosomal proton pumps and prevents the fusion of autophagosomes with lysosomes (Yamamoto et al., 1998). In l(2)mbn cells, both autophagy inhibitors significantly reduced LTG fluorescence levels after starvation treatment (Fig. 1, C and D). Consistently, the addition of 3MA also decreased the numbers of GFP-LC3–positive cells after starvation treatment. As expected, the addition of Baf, which is known to increase the numbers of autophagosomes by preventing their lysosomal degradation, resulted in increased GFP-LC3 puncta in starved cells (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200712091/DC1). These results indicate that our flow cytometry–based LTG assay is able to detect changes in the autophagy levels of l(2)mbn cells in response to starvation and autophagy-inhibiting drug treatments.
Figure 1.
Quantification of starvation-induced autophagy in D. melanogaster l(2)mbn cells. (A) Flow cytometry analysis of l(2)mbn cells starved for 2 h (2h S) or 4 h (4h S) showed an increase in LTG fluorescence levels (x axis) compared with control cells in full-nutrient medium (C). The gate shown on the histogram represents the LTGhigh population. (B) Representative images of GFP-LC3 puncta in control and 2-h starved l(2)mbn cells. An increase in GFP-LC3 puncta was observed in the starved cells (left). Bar, 10 μm. (C) Flow cytometry analysis of 4-h starved cells were incubated with 3MA (4h S + 3MA) and Baf (4h S + Baf). Both autophagy inhibitors reduced the LTG fluorescence levels compared with starved cells (4h S). Control cells in nutrient-full medium (C) are represented by the brown line. (D) Both autophagy inhibitors, 3MA and Baf, reduced the LTGhigh population significantly. (3MA, P = 0.00001; and Baf, P = 0.00006). (E) RNAi of representative Atg genes decreased the LTR fluorescence levels compared with control. (Atg1, P = 0.01; Atg5, P = 0.01; Atg7, P = 0.002; Atg8a, P = 0.008; Atg8b, P = 0.006; and Atg12, P = 0.02). (F) RNAi of all tested genes in the TOR–PI3K pathways had a statistically significant effect on LTG fluorescence levels. Known negative regulators of autophagy are shown with gray bars; positive regulators are shown with white bars. (Pten, P = 0.007; Tsc1, P = 0.027; Tsc2, P = 0.025; RheB, P = 0.005; Tor, P = 0.016; and S6k, P = 0.012). dsRNA corresponding to a human gene (Hs) was used as a negative control in E and F. Results represent the mean value ± SD from at least three independent experiments.
To further validate our flow cytometry–based LTG assay and determine its sensitivity, we used RNAi to specifically inhibit D. melanogaster autophagy genes. Currently, there are 14 D. melanogaster genes that share significant sequence identity with yeast Atg genes, and seven D. melanogaster Atg homologues have already been shown to be essential for starvation-induced autophagy in the larval fat body (Scott et al., 2004). In our assay, RNAi of 11 D. melanogaster Atg homologues individually resulted in a statistically significant reduction in the LTGhigh population after starvation treatment (Fig. 1 E and Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200712091/DC1). The effects on LTG staining by RNAi or 3MA were modest in size (e.g., 30–48% change relative to the Hs-RNAi–negative control in Fig. 1 E) but reproducible and statistically significant. The limited magnitude of the detectable effects may be due, at least in part, to the nature of this dye as an acidophilic probe that detects autolysosomes but also background lysosomal staining. As with any RNAi-based screen, incomplete target knockdown may also be a contributing factor. Comparison of our RNAi-mediated results with previous in vivo results from the D. melanogaster larval fat body (Table S1) indicates that l(2)mbn cells require the same autophagy genes as the fat body (Scott et al., 2004).
In higher eukaryotes, starvation-induced autophagy is suppressed by components of the insulin/class I PI3K and target of rapamycin (TOR) pathways (for reviews see Levine and Yuan, 2005; Klionsky, 2007; Maiuri et al., 2007). To determine whether PI3K and TOR pathways are required for starvation-induced autophagy in l(2)mbn cells, we designed double-stranded RNAs (dsRNAs) against several genes in these pathways. RNAi of Tor or RheB, negative regulators of autophagy, showed an increase in LTGhigh cells compared with Hs-dsRNA (negative control)-treated cells after starvation treatment (Fig. 1 F). In contrast, RNAi of Pten, Tsc1, Tsc2, and S6k, positive regulators of autophagy, showed a reduction in the LTGhigh population (Fig. 1 F). These results indicate that components of TOR and PI3K pathway are essential to regulate starvation-induced autophagy in D. melanogaster l(2)mbn cells. These results also demonstrate that our primary screening method, a flow cytometry–based LTG assay, is capable of detecting alterations induced by RNAi-mediated knockdown of positive and negative regulators of autophagy. Thus, this method can be used to identify novel components in starvation-induced autophagy. To ensure that the changes in LTG fluorescence levels were caused by alterations in autophagy, we used GFP-LC3 to track changes in autophagosome formation in cells. RNAi of Tor showed an increase in the numbers of GFP-LC3 positive cells after starvation treatment (Fig. 2 G). In contrast, reduction of Pten expression by RNAi resulted in a decrease in the number of the GFP-LC3–positive cells (Fig. 2 G). Together, these two methods allow us to monitor the dynamic steps, formation of autophagosomes (GFP-LC3), and autophagosome-lysosome fusion (LTG), during autophagy.
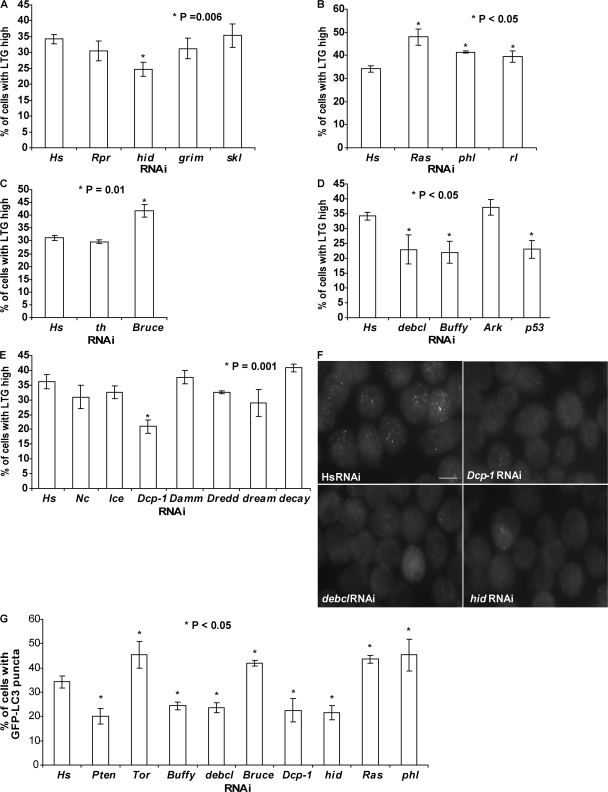
Figure 2.
Identification of known cell death–related genes in autophagy regulation in l(2)mbn cells using RNAi. (A) The percentage of LTGhigh cells was reduced by hid-RNAi (P = 0.006) but not by rpr, grim, and skl-RNAi. (B) Knockdown of Ras, phl, and rl expression by RNAi resulted in an increase in the percentage of LTGhigh cells. (Ras, P = 0.003; phl, P = 0.001; and rl, P = 0.028). (C) th-RNAi treatment (24 h) had no significant effect on LTG levels; in contrast, RNAi of Bruce resulted in an increase in LTG fluorescence levels (P = 0.01). (D) Reduction of debcl, Buffy, or p53 expression by RNAi resulted in a decrease in LTG fluorescence levels. (debcl, P = 0.018; Buffy, P = 0.006; and p53, P = 0.004). (E) RNAi of effector caspase Dcp-1 resulted in a significant decrease in the LTGhigh population (P = 0.001). (F) Representative images of GFP-LC3 puncta in cells treated with the indicated RNAi after a 2-h starvation treatment. Bar, 10 μm. (G) Quantification of cells with GFP-LC3 puncta after RNAi treatment. Cells with more than three GFP-LC3 punctate dots were considered to be GFP-LC3–positive cells. Cells treated with the RNAi indicated here all showed a significant difference (P < 0.05) in the percentage of GFP-LC3–positive cells compared with the human (Hs) RNAi control. (Pten, P = 0.006; Tor, P = 0.034; Buffy, P = 0.005; debcl, P = 0.003; Bruce, P = 0.003; Dcp-1, P = 0.007; hid, P = 0.002; Ras, P = 0.006; and phl, P = 0.050). Results represent the mean value ± SD from three independent experiments.
Identification of cell death–related genes that regulate starvation-induced autophagy in l(2)mbn cells
To better understand the relationship between autophagy and apoptosis, we investigated whether known cell death genes were required for starvation-induced autophagy in l(2)mbn cells. dsRNAs were designed against the D. melanogaster core cell death effectors, rpr, hid, grim and skl, and autophagy was evaluated by our flow cytometry–based LTG assay. Only dsRNA corresponding to hid but not rpr, grim, or skl showed an effect on autophagy by this assay. RNAi of hid decreased the percentage of LTGhigh cells after starvation treatment (Fig. 2 A). Previous studies showed that the Ras–Raf–MAPK pathway specifically inhibits the proapoptotic activity of hid (Bergmann et al., 1998). To determine whether the Ras–Raf–MAPK pathway also plays a regulatory role in autophagy in l(2)mbn cells, we designed dsRNAs to target these three components. RNAi of Ras, phl (also known as raf), or rl (also known as MAPK) all further enhanced the LTG fluorescence levels, suggesting that, like in apoptosis, they have an inhibitory role in autophagy regulation (Fig. 2 B). A second set of dsRNAs, nonoverlapping with the first set of dsRNAs, was designed to validate these new findings, and consistent results were observed (Fig. S1 B). In addition, GFP-LC3 was used to track changes in autophagosome formation in cells. RNAi of hid showed a decrease in the numbers of cells with GFP-LC3 puncta (Fig. 2, F and G), whereas RNAi of Ras, phl, or rl all resulted in a significant increase in the numbers of GFP-LC3–positive cells after starvation treatment (Fig. 2 G).
All RHG family members, Rpr, Hid, Grim, and Skl, bind to D. melanogaster IAP-1 (DIAP1) and inhibit its antiapoptotic activities (Hay et al., 2004). To test whether DIAP1 (encoded by th) is a putative downstream mediator of Hid-dependent autophagy in l(2)mbn cells, dsRNA was designed specifically to target th. We found that th-dsRNA–treated cells showed no difference in LTG fluorescence levels compared with Hs-dsRNA (negative control)-treated cells (Fig. 2 C). Interestingly, our data showed that reduced expression of Bruce, another IAP family member protein, further increased the LTG fluorescence levels after starvation treatment (confirmed using nonoverlapping dsRNAs; Fig. 2 C; see Fig. S1 C). RNAi of Bruce expression also resulted in an increase in GFP-LC3 puncta after starvation treatment (Fig. 2 G). These results suggest that Bruce, instead of DIAP1, could be the downstream target of Hid during starvation induced autophagy in l(2)mbn cells.
Next, we investigated whether the transducers of apoptotic signals, Ark, Buffy, and debcl, are required for starvation-induced autophagy. Reduced expression of Ark, the D. melanogaster homologue of mammalian Apaf-1, did not affect the LTG fluorescence levels (Fig. 2 D). RNAi of two Bcl-2 family members, Buffy or debcl, resulted in a decrease in the percentage of LTGhigh cells after starvation treatment (Fig. 2 D). Consistently, reduction of Buffy and debcl expression by RNAi decreased the percentage of GFP-LC3–positive cells after starvation treatment (Fig. 2, F and G). Reduced expression of Ark, Buffy, and debcl was determined using quantitative RT-PCR (QRT-PCR; Fig. S1 D). In addition, we reduced expression of the tumor suppressor p53 by RNAi and found that starvation-induced autophagy was inhibited (Fig. 2 D). Results were further confirmed using nonoverlapping dsRNAs (Fig. S1 C).
To investigate the requirement of caspases, the final effectors of apoptosis, in starvation-induced autophagy, we designed gene-specific dsRNAs corresponding to seven different D. melanogaster caspases. RNAi of just one caspase, Dcp-1, but not others resulted in a decrease in the percentage of LTGhigh cells after starvation treatment (Fig. 2 E). A second dsRNA against Dcp-1, nonoverlapping with the first dsRNA, yielded a similar result (Fig. S1 C). Reduction of Dcp-1 expression by RNAi was determined using QRT-PCR (Fig. S1 D). Consistent with the LTG derived data, RNAi-mediated knockdown of Dcp-1 resulted in a decrease in GFP-LC3–positive cells after starvation treatment (Fig. 2, F and G). These results indicate that Dcp-1 functions as a positive regulator of autophagy in D. melanogaster l(2)mbn cells.
Autophagy occurs in response to nutrient deprivation in germaria and midstage egg chambers in the D. melanogaster ovary
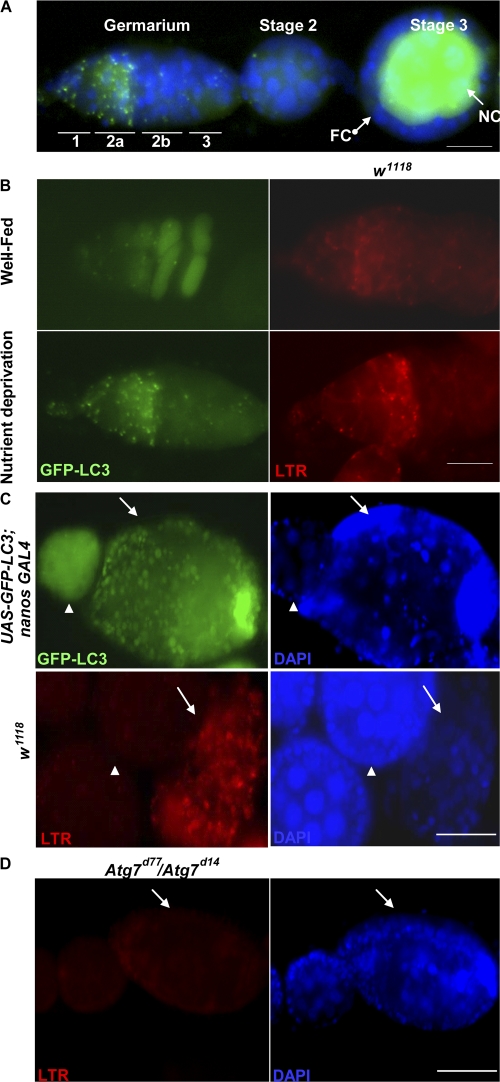
To further characterize the requirement of Dcp-1 and Bruce in autophagy regulation, we studied D. melanogaster oogenesis in vivo. We used a transgenic fly line that expresses a GFP-LC3 fusion protein under the control of the upstream activating sequence (UASp) promoter (Rusten et al., 2004). Coexpression of UASp-GFP-LC3 with the germ line–specific nanos-GAL4 driver resulted in detectable GFP-LC3 expression in the germ line (nurse cells and oocyte) cells but not in somatic (follicle) cells (Fig. 3 A; Rorth, 1998; Rusten et al., 2004). When UASp-GFP-LC3; nanos-GAL4 flies were subjected to nutrient deprivation, we observed numerous GFP-LC3 puncta in region 2 within the germarium (Fig. 3 B). In contrast, flies raised in the presence of yeast paste (Fig. 3 B, well-fed) had a diffuse GFP-LC3 pattern (Fig. 3 B). In addition, we found an increase in punctate LysoTracker red (LTR) staining in germaria of nutrient-deprived wild-type (w1118) flies compared with well-fed wild-type flies (Fig. 3 B and Table I). We also observed numerous GFP-LC3 puncta in nutrient-deprived degenerating stage 8 chambers, but a diffuse GFP-LC3 pattern was detected in healthy egg chambers (Fig. 3 C). Similarly, degenerating stage 8 egg chambers had numerous LTR-positive dots in the nurse cells, whereas healthy egg chambers had a low level of LTR staining (Fig. 3 C and Table II).In starved Atg7 mutants (Atg7d77/Atg7d14), there was a significant decrease in punctate LTR staining in region 2 of the germarium and in stage 8 degenerating egg chambers compared with flies with the genotype CG5335d30/Atg7d14, which were used previously as controls in Juhasz et al. (2007; Fig. 3 D and Tables I and II). These results indicated that nurse cells lacking the core autophagy regulator Atg7 failed to induce autophagy in response to nutrient deprivation. Overall, our observations showed that nutrient deprivation induces autophagy in region 2 germaria and in degenerating stage 8 egg chambers in D. melanogaster.
Figure 3.
Nutrient deprivation induces autophagy at region 2 within the germarium and in dying midstage egg chambers. (A) GFP-LC3 proteins were expressed in nurse cells (NC) but not in follicle cells (FC) by using the UASp/nanos Gal4 system. DAPI staining of nuclei is shown in blue. (B) UASp-GFP-LC3; nanos GAL4 flies were conditioned on yeast paste and had a diffuse GFP-LC3 pattern. Numerous GFP-LC3 puncta (green) at region 2 within germarium were observed in nutrient-deprived flies. Ovaries were stained with LTR in w1118 flies. Germarium of nutrient-deprived w1118 flies had an increase in punctate LTR staining (red) compared with well-fed germarium. (C) Degenerating stage 8 egg chambers (arrows) had numerous GFP-LC3 puncta (green) and an increase in LTR-positive dots (red) compared with healthy egg chambers (arrowheads). DAPI (blue) staining of nuclei is shown in the two panels on the right. (D) Degenerating stage 8 egg chambers (arrows) of nutrient-deprived Atg7 mutants (Atg7d77/Atg7d14) showed a dramatic decrease in LTR staining. DAPI staining of nuclei is shown in blue. Bars: (A and B) 20 μm; (C and D) 50 μm.
Table I. Quantification of autophagy in region 2 germaria.
| Genotype | Nutritional status | LTR positive | Number | Percentage of autophagy |
|---|---|---|---|---|
| w1118 | Fed | 8 | 30 | 27 |
| w1118 | Nutrient deprivation | 25 | 36 | 69 |
| Dcp-1Prev | Nutrient deprivation | 17 | 53 | 32 |
| BruceE81 | Fed | 67 | 116 | 58 |
| BruceE81/TM3 | Fed | 18 | 55 | 33 |
| BruceE16 | Fed | 35 | 63 | 56 |
| BruceE16/TM3 | Fed | 13 | 71 | 18 |
| Atg7d77/Atg7d14 | Nutrient deprivation | 14 | 65 | 22 |
| CG5335d30/Atg7d14 | Nutrient deprivation | 37 | 68 | 54 |
Numbers in the fourth column refer to the numbers of individual germariums scored in at least seven different animals.
Table II. Quantification of autophagy in stage 8 degenerating egg chambers.
| Genotype | Nutritional status | LTR positive | Number | Percentage of autophagy |
|---|---|---|---|---|
| w1118 | Nutrient deprivation | 29 | 40 | 73 |
| Dcp-1Prev | Nutrient deprivation | 8 | 54 | 15 |
| Atg7d77/Atg7d14 | Nutrient deprivation | 13 | 51 | 25 |
| CG5335d30/Atg7d14 | Nutrient deprivation | 7 | 14a | 50 |
| nanos-GAL4/UASp-fl-Dcp-1 | Fed | 62 | 74 | 84 |
| BruceE81 | Fed | 43 | 52 | 83 |
| BruceE81/TM3 | Fed | 0 | 0b | N/A |
Numbers in the fourth column refer to the number of individual degenerating stage 8 egg chambers scored in at least seven different animals.
n = 4 animals scored for this genotype.
No degenerating stage 8 egg chambers detected.
Dcp-1 and Bruce regulate autophagy in germaria and degenerating midstage egg chambers
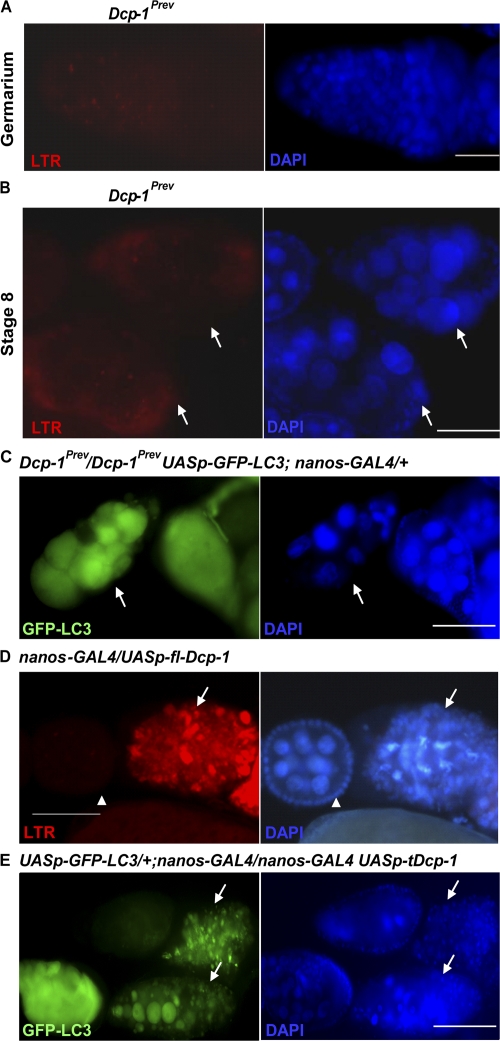
To determine whether Dcp-1 is required for autophagy at these two specific stages during oogenesis, we used the LTR staining in nutrient-deprived Dcp-1Prev mutant flies. We observed a decrease in punctate LTR staining in region 2 of the germarium and in stage 8 degenerating egg chambers (Fig. 4, A and B; and Tables I and II) compared with nutrient-deprived wild-type flies. Consistent results were observed using GFP-LC3. Degenerating stage 8 egg chambers in nutrient-deprived Dcp-1Prev mutants containing the GFP-LC3 transgene had a diffuse GFP-LC3 pattern instead of punctate GFP-LC3 structures (Fig. 4 C). Together, these results indicate that nurse cells lacking Dcp-1 function are severely impaired in the ability to induce autophagy in response to starvation.
Figure 4.
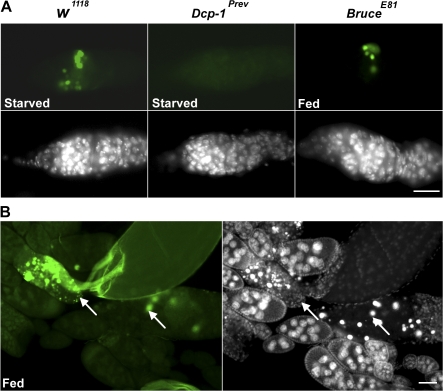
The effector caspase Dcp-1 is not only required for nutrient starvation–induced autophagy but is also sufficient for the induction of autophagy during D. melanogaster oogenesis. (A) Germaria of the nutrient-deprived Dcp-1Prev flies showed a dramatic decrease in LTR staining compared with nutrient-deprived wild-type flies shown in Fig. 3 B. DAPI staining of nuclei is shown in blue. (B) Degenerating stage 8 egg chambers (arrows) of nutrient-deprived Dcp-1Prev flies showed a dramatic decrease in LTR staining compared with nutrient-deprived wild-type flies shown in Fig. 3 C. (C) Lack of Dcp-1 function (UASp-GFP-LC3 Dcp-1Prev/Dcp-1Prev; nanos-GAL4/+) resulted in uniform diffuse staining of GFP-LC3 rather than the punctate pattern observed in wild-type degenerating stage 8 egg chambers shown in Fig 3 C. (D) Dying egg chambers (arrows) of NGT/+; nanos-GAL4/UASp-fl-Dcp-1 flies that were conditioned on yeast paste showed a significant increase in punctate LTR staining (red) compared with healthy egg chambers (arrowheads). (E) Expression of activated Dcp-1 (a truncated form) and GFP-LC3 in the germ line (UASp-GFP-LC3/+; nanos-GAL4/nanos-GAL4 UASp-tDcp-1) resulted in abundant degenerating stage 8 egg chambers (arrows) with numerous GFP-LC3 puncta (green). DAPI staining of nuclei is shown in blue. Bars: (A) 20 μm; (B–E) 50 μm.
To determine whether Dcp-1 was also sufficient to induce autophagy in vivo, we generated transgenic flies that express the full-length Dcp-1 (fl-Dcp-1) under the control of the UASp promoter. In the presence of a nutrient-rich food source, degenerating stage 8 egg chambers are observed only rarely in wild-type flies (Drummond-Barbosa and Spradling, 2001). However, under nutrient-rich conditions, we observed an abundance of degenerating stage 8 egg chambers in nanos-GAL4/UASp-fl-Dcp-1 flies with increased levels of punctate LTR staining (Fig. 4 D and Table II). Further, we expressed an activated form of Dcp-1 (missing the prodomain) and GFP-LC3 in the germ line using the UASp/nanos-GAL4 system and observed numerous degenerating stage 8 egg chambers with GFP-LC3 puncta (Fig. 4 E), which indicates that activity of effector caspase Dcp-1 is sufficient to induce autophagy during mid-oogenesis even under nutrient-rich conditions.
We identified the IAP protein Bruce as a negative regulator of autophagy in l(2)mbn cells. We next asked whether Bruce is able to inhibit autophagy during D. melanogaster oogenesis. We monitored the LTR staining in ovaries of BruceE81 flies that have a deletion in the baculoviral IAP repeat (BIR) domain, which binds to caspases (Arama et al., 2003). In the presence of a nutrient-rich food source, we observed an increase in punctate LTR staining in region 2 of the germarium in BruceE81 flies compared with controls (BruceE81/TM3; Fig. 5 A and Table I). Similarly, we observed numerous degenerating stage 8 egg chambers with increased levels of punctate LTR staining in BruceE81 flies, resembling overexpression of Dcp-1 (Fig. 5 B and Table II). In well-fed conditions, we observed no degenerating stage 8 egg chamber in control BruceE81/TM3 flies (n = 187 ovarioles; Table II). Punctate LTR staining was similarly observed in region 2 germaria (Table I) and degenerating stage 8 egg chambers (not depicted) in well-fed Bruce E16 flies that have a 10-kb deletion in the 3′ end of the Bruce gene sequence (Vernooy et al., 2002). Our results demonstrate that Bruce is normally required to inhibit autophagy under nutrient-rich conditions.
Figure 5.
Bruce suppresses autophagy at region 2 within germarium and in dying stage 8 egg chambers. (A) The germarium in well-fed BruceE81 flies showed an increase in LTR staining (red) compared with wild-type well-fed flies shown in Fig. 3 B (top right). DAPI staining (white) of nuclei is shown on the right. (B) In well-fed wild-type flies, mid-oogenesis nurse cell death is a rare event. Lack of Bruce function resulted in an increase in dying stage 8 egg chambers (arrows) in ovaries under well-fed conditions, and these degenerating stage 8 egg chambers had numerous LTR (red) punctate dots. DAPI staining (white) is shown on the right. Bars: (A) 20 μm; (B) 50 μm.
Dcp-1 and Bruce mutants have altered TUNEL staining in germaria and degenerating midstage egg chambers
Our previous work showed that nutrient-deprived Dcp-1 mutants (Dcp-1Prev) have defects in mid-oogenesis germ line cell death (Laundrie et al., 2003). To determine whether Dcp-1 is also required for germ line cell death in region 2 within the germarium, we used the TUNEL assay to detect levels of DNA fragmentation as an indication of cell death. We found that nutrient-deprived Dcp-1 mutants had decreased levels of TUNEL-positive cells in region 2 within the germarium compared with nutrient-deprived wild-type flies (Fig. 6 A and Table III), which indicates that Dcp-1 is also required for germarium stage cell death.
Figure 6.
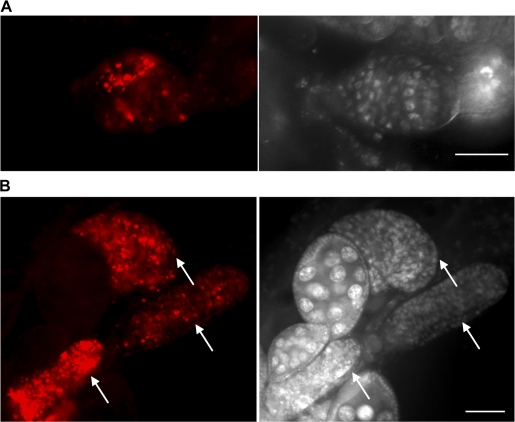
Dcp-1 is required for nutrient starvation–induced germarium cell death, and the IAP protein Bruce inhibits germarium and mid-oogenesis cell death. (A) Ovaries were stained with TUNEL (green) to detect DNA fragmentation. Clusters of cysts with TUNEL staining were observed in region 2 in nutrient-deprived w1118 files. In Dcp-1Prev flies, fewer TUNEL-positive cysts in region 2 were observed. Under well-fed conditions, numerous TUNEL-positive cysts were observed in BruceE81 flies. DAPI staining of nuclei is shown in white. (B) Numerous degenerating stage 8 egg chambers (arrows) with TUNEL-positive staining (green) were observed in well-fed BruceE81 flies. DAPI staining of nuclei (white) is shown on the right. Bars: (A) 20 μm; (B) 50 μm.
Table III. Quantification of cell death in region 2 germaria.
| Genotype | Nutritional status | TUNEL positive | Number | Percentage of TUNEL |
|---|---|---|---|---|
| w1118 | Fed | 9 | 50 | 18 |
| w1118 | Nutrient deprivation | 34 | 51 | 67 |
| Dcp-1Prev | Nutrient deprivation | 22 | 69 | 32 |
| Bruce E81 | Fed | 26 | 70 | 37 |
| BruceE81/TM3 | Fed | 12 | 69 | 17 |
| BruceE16 | Fed | 20 | 54 | 37 |
| BruceE16/TM3 | Fed | 14 | 80 | 18 |
| Atg7d77/Atg7d14 | Nutrient deprivation | 21 | 77 | 27 |
| CG5335d30/Atg7d14 | Nutrient deprivation | 95 | 202 | 47 |
| Atg1 GLC | Nutrient deprivation | 22 | 77 | 29 |
| Atg1Δ3D/TM3 | Nutrient deprivation | 53 | 64 | 83 |
Numbers in the fourth column refer to the number of individual germariums scored in at least seven different animals.
We also investigated the role of Bruce in cell death during oogenesis. In well-fed BruceE81 flies, we observed a degenerating ovary phenotype that has been shown previously in ovaries with partial loss of another IAP protein, DIAP1 (Xu et al., 2005). This ovary phenotype may be a consequence of excess cell death. Consistent with this possibility, we observed an increased number of cells with TUNEL-positive staining in region 2 within the germarium compared with controls (Fig. 6 A and Table III). Similar results were obtained with BruceE16 flies (Table III). Numerous TUNEL-positive dots were also observed in degenerating stage 8 egg chambers of BruceE81 well-fed flies (Fig. 6 B). These findings demonstrate that Bruce acts as an inhibitor of cell death in germaria and midstage egg chambers.
Autophagy contributes to cell death in nutrient-deprived ovaries
To assess the role of autophagy that we observed during the germarium and mid-oogenesis stages, we used the TUNEL assay to detect DNA fragmentation in two Atg gene mutants. Most Atg gene knockouts in D. melanogaster result in pupal or larval lethality; thus, we first analyzed the fully viable Atg7 mutant flies (Scott et al., 2004; Juhasz et al., 2007). We found that nutrient-deprived Atg7 mutants had reduced levels of TUNEL-positive cells in region 2 within the germarium compared with control flies (Table III). Further, degenerating stage 8 egg chambers in starved Atg7 mutants showed low or no TUNEL-positive staining compared with controls (Fig. 7, A and B; and Table IV). However, nuclear DNA condensation was still observed in the degenerating stage 8 egg chambers of starved Atg7 mutants (Fig. 7 B). To further investigate the role of autophagy in starvation-induced germ line cell death, we generated Atg1 germ line clones (GLCs), as mutations in Atg1 result in lethality at the pupal stage of development (Scott et al., 2004, 2007). Consistent with our Atg7 mutant observations, nutrient-deprived Atg1 GLC ovaries had decreased levels of TUNEL staining in both germaria and degenerating stage 8 egg chambers, which indicates a suppression of DNA fragmentation (Fig. 7, C and D; and Tables III and IV). Also consistent with Atg7, we observed nuclear DNA condensation in the Atg1 GLC degenerating stage 8 egg chambers (Fig. 7 D). Our results show that lack of autophagy results in a reduction of DNA fragmentation during starvation-induced cell death in the germaria and midstage egg chambers, which suggests that autophagy contributes to the cell death process at these stages.
Figure 7.
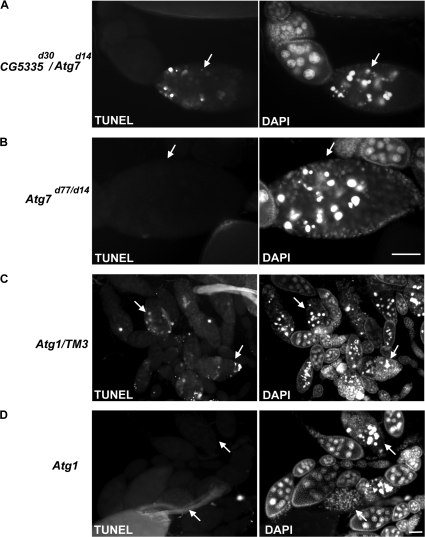
Lack of Atg7 or Atg1 function reduces DNA fragmentation during mid-oogenesis cell death. (A) TUNEL-positive staining was observed in dying stage 8 egg chambers (arrows) of starved control flies (CG5335d30/Atg7d14). DAPI staining of nuclei (white) is shown on the right. (B) In nutrient-deprived Atg7 mutants (Atg7d77/Atg7d14), degenerating stage 8 egg chambers (arrows) showed no or low levels of TUNEL staining. Nuclear DNA condensation, detected by DAPI, was still observed. (C) Dying stage 8 egg chambers (arrows) from nutrient-deprived control siblings (Atg1Δ3D/TM3) generated from the same cross in D had abundant TUNEL-positive staining. (D) In nutrient-deprived Atg1 GLCs, degenerating stage 8 egg chambers (arrows) showed no or low levels of TUNEL staining. Nuclear DNA condensation (DAPI, right) in degenerating egg chambers appeared to occur as in the controls. Bars, 50 μm.
Table IV. Quantification of cell death in stage 8 degenerating egg chambers.
| Genotype | Nutritional status | TUNEL positive | Number | Percentage of TUNEL |
|---|---|---|---|---|
| Atg7d77/Atg7d14 | Nutrient deprivation | 17 | 69 | 25 |
| CG5335d30/Atg7d14 | Nutrient deprivation | 39 | 62 | 63 |
| Atg1 GLC | Nutrient deprivation | 3 | 16 | 19 |
| Atg1Δ3D/TM3 | Nutrient deprivation | 19 | 37 | 51 |
Numbers in the fourth column refer to the number of individual degenerating stage 8 egg chambers scored in at least seven different animals.
Discussion
Key outstanding questions that need to be addressed are how autophagy and apoptosis pathways interact with each other, and whether common regulatory mechanisms exist between these two processes. We have shown here that six known cell death genes and the Ras–Raf–MAPK signaling pathway not only function in apoptosis but also act to regulate autophagy in D. melanogaster l(2)mbn cells. We cannot rule out the possibility that additional cell death genes that we screened may also function in autophagy but were not detected in our assay because of insufficient knockdown by RNAi, a long half-life of the corresponding proteins, and/or functional redundancy.
Consistent with our in vitro data, the involvement of Hid in autophagy regulation has been demonstrated in D. melanogaster. Overexpression of Hid induced autophagy in the fat body, larval epidermis, midgut, salivary gland, Malpighian tubules, and trachea epithelium (Juhasz and Sass, 2005). Further, expression of the constitutively active Ras form (RasV12), which has been shown to inhibit Hid activity in apoptosis (Bergmann et al., 1998), can also block Hid-induced autophagy (Juhasz and Sass, 2005). In D. melanogaster salivary glands, the Ras signaling pathway has also been shown to inhibit the autophagy process (Berry and Baehrecke, 2007). Based on our loss-of-function findings and these previous gain-of-function studies, we speculate that the Ras–Raf–MAPK pathway acts upstream to inhibit Hid activity in autophagy.
Poor nutrition has a dramatic effect on egg production in D. melanogaster. Flies fed on a protein-deprived diet showed an increase in cell death in germaria and midstage egg chambers (Drummond-Barbosa and Spradling, 2001). These two stages have been proposed to serve as nutrient status checkpoints where defective egg chambers are removed before the investment of energy into them. The molecular mechanisms of germarium cell death are still largely unknown, and Daughterless, a helix-loop-helix transcription factor, was the only known regulator involved in cell death of germaria (Smith et al., 2002). Nurse cell death during mid-oogenesis is also different from most developmental cell death in other D. melanogaster tissues because apoptotic regulators such as rpr, hid, or grim are not required for cell death in these cells (Peterson et al., 2007). However, the activity of caspases, particularly Dcp-1, was shown to be required for mid-oogenesis cell death (Laundrie et al., 2003; Baum et al., 2007). Our findings implicate several additional genes, Dcp-1, Bruce, Atg7, and Atg1, in nutrient deprivation–induced cell death in the germarium, as well as during mid-oogenesis.
Other forms of cell death, such as autophagic cell death, have been proposed previously to be involved in the elimination of defective egg chambers during mid-oogenesis. Known signaling pathways, including the insulin and ecdysone pathways, have been shown to be required not only for the survival of nurse cells in mid-oogenesis; they are also known to regulate the autophagy process, supporting the notion that autophagy plays a role in mid-oogenesis cell death (Drummond-Barbosa and Spradling, 2001; McCall, 2004). Features of autophagy were observed during D. virilis mid-oogenesis cell death as shown by monodansylcadaverine staining and transmission electron microscopy (Velentzas et al., 2007). Our results using GFP-LC3 and LTG demonstrate that autophagy occurs in degenerating midstage egg chambers and also in germaria of nutrient deprived D. melanogaster. We found that mutation of Atg7 results in a significant decrease of autophagy in dying mid-stage egg chambers and in germaria of starved flies, further supporting the presence of autophagy during these stages.
The role of autophagy in cell survival or cell death is still not well resolved and is likely to be context dependent. Our results show that autophagy contributes to the cell death process in the ovary. Loss of Atg7 or Atg1 activity in both dying midstage egg chambers and germaria leads to decreased TUNEL staining, which indicates a reduction in DNA fragmentation. Consistent results were observed previously in the larval midguts of Atg7 mutants, which also showed an inhibition of DNA fragmentation (Juhasz et al., 2007). Interestingly, lack of autophagy function does not appear to affect nuclear DNA condensation in nurse cells. Nurse cells in degenerating stage 8 egg chambers of starved Atg7 mutants or Atg1 GLCs appeared to still have condensed nuclei, as shown by DAPI staining (Fig. 7, B and D). Thus, based on Atg7 and Atg1 mutant analyses, autophagy contributes to DNA fragmentation but not all aspects of nurse cell death. Future studies are required to determine how autophagy is connected to known pathways leading to DNA fragmentation and chromatin condensation during cell death.
The IAP family member Bruce was shown previously to repress cell death in the D. melanogaster eye (Vernooy et al., 2002). Bruce was also shown to protect against excessive nuclear condensation and degeneration, perhaps by limiting excessive caspase activity, during sperm differentiation (Arama et al., 2003). Other IAP family members have been shown to bind caspases via a BIR domain and inhibit apoptosis (Riedl and Shi, 2004). The presence of a BIR domain in Bruce suggests that it may also have caspase-binding activity. We found that lack of Bruce function resulted in an increase in both LTR and TUNEL staining in germaria and degenerating midstage egg chambers. Thus, the Bruce mutant-degenerating phenotype in ovaries suggests that Bruce might function normally to restrain or limit caspase activity in this tissue. Because we found that Dcp-1 and Bruce are both required for the regulation of autophagy and DNA fragmentation in germaria and dying midstage egg chambers, it is possible that Bruce acts to bind and degrade Dcp-1 in nurse cells under nutrient-rich conditions. Future studies using epistasis and protein interaction analyses will be required to test this prediction. We cannot rule out the possibility that other IAP proteins, such as DIAP1, and other caspases also play a role during these stages. However, at least in response to starvation signals, Bruce and Dcp-1 play a nonredundant dual role in the regulation of autophagy and cell death in the ovary.
Numerous studies have linked caspase function to apoptosis, but recent findings indicate that caspases are also required for nonapoptotic processes including immunity and cell fate determination (for reviews see Kumar, 2004; Kuranaga and Miura, 2007). We have shown here that Dcp-1 is also required for starvation-induced autophagy. In the ovary, it appears that both apoptotic and autophagic events occur in the germaria and midstage egg chambers after nutrient deprivation. It is possible that Dcp-1 coordinates autophagy and apoptosis at these two nutrient status checkpoints to ensure elimination of defective egg chambers in the most efficient manner possible. Dcp-1 mutants exhibit intact nuclei in stage 8 defective egg chambers, which indicates a block in both DNA fragmentation and nuclear condensation, and further supports a dual regulatory role for Dcp-1 in mid-oogenesis cell death. Dcp-1 might function to induce autophagosome formation while coordinately acting upon alternate proteolytic targets to complete execution of apoptosis. Future studies to elucidate upstream regulators and downstream substrates of Dcp-1 in cells undergoing autophagy or apoptosis will help to establish the regulatory mechanisms governing the crosstalk between these two cellular processes. Given the multiple cellular effects associated with autophagy, our results also have important therapeutic implications for the use of modulators of caspase or IAP activity in the treatment of cancer and other diseases.
Materials and methods
Cell culture conditions
D. melanogaster l(2)mbn cells (provided by A. Dorn, Institute of Zoology, Johannes Gutenberg University, Mainz, Germany) were maintained in Schneider's D. melanogaster medium (Invitrogen) supplemented with 10% FBS in 25-cm2 suspension flasks (Sarstedt) at 25°C (Ress et al., 2000). All the experiments were performed 3 d after passage and the cells were discarded after 25 passages.
dsRNA synthesis
Individual PCR products containing coding sequences for the transcripts to be targeted were generated by RT-PCR using Superscript one-step RT-PCR kit with platinum taq (Invitrogen). Each primer used in the RT-PCR contained a 5′ T7 RNA polymerase-binding site (TAATACGACTCACTATAGG) followed by sequences specific for the targeted genes (Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200712091/DC1). For in vitro transcription reactions, 50 μl of each of the RT-PCR products was ethanol-precipitated and resuspended in 8 μl of nuclease-free water and then used as template. In vitro transcription reactions were performed using T7 RiboMax Express RNAi systems (Promega) according to the manufacturer's instructions. dsRNAs were ethanol-precipitated and resuspended in 50 μl of nuclease free water. A 5-μl aliquot of 1:100 dilution was analyzed by 1% agarose gel electrophoresis to determine the quality of dsRNA. The dsRNA was quantitated using the PicoGreen assay (Invitrogen) and adjusted to 200 ng/μl with nuclease-free water.
RNAi
66 μl of cells (106 cells/ml) in SFM922 medium were seeded into each well of a 96-well plate. 2 μg of dsRNA (20 μM) was added into each well. After 1 h of incubation at room temperature, the cells received Schneider's medium supplemented with 10% FBS to achieve a final volume of 200 μl. Cells were incubated for 72 h at 25°C.
Because RNAi of th triggered a massive amount of apoptosis at the standard incubation time of 72 h, we instead used a 24-h incubation period. After 24 h of th-dsRNA treatment, there was already a significant amount of apoptotic cells present, which indicates an efficient knockdown by RNAi, but a sufficient number of healthy cells (>10,000) remained for LTG analysis.
Flow cytometry–based LTG assay
For drug treatments, 10 mM 3MA or 0.1 μM Baf was added when nutrient-full medium was replaced with 2 mg/ml glucose in PBS. After a 4-h incubation at 25°C, cells were incubated for 20 min at room temperature with 50 nM LTG for quantification of autophagy levels and 2 μg/ml propidium iodide (PI) to eliminate dead cells. The cells were then analyzed using flow cytometry (FACSCalibur; Becton Dickinson). A minimum of 10,000 cells per sample was acquired for triplicate samples per experiment. LTG fluorescence levels of cells (excluding PI-positive cells) were analyzed using Flowjo software.
For RNAi experiments, the RNAi-treated cells, after a 72-h incubation, were transferred into a U-bottom 96-well plate and centrifuged at 800 rpm for 5 min. Nutrient-full medium was replaced with 2 mg/ml glucose/PBS with 20 μM dsRNA for a 4-h starvation treatment, then cells were labeled with LTG and PI for 20 min at room temperature and finally analyzed as described in the previous section.
GFP-LC3 detection
The p2ZOp2F-EGFP-LC3 plasmid was generated by restriction digestion of EGFP-LC3 from pUASP-EGFP-LC3 (Rusten et al., 2004) and cloning into the p2ZOp2F vector (a gift from T. Grigliatti, University of British Columbia, Vancouver, British Columbia, Canada; Hegedus et al., 1998). To create a stable cell line, D. melanogaster l(2)mbn cells were transfected with p2ZOp2F-EGFP-LC3 and selected for the presence of the construct using zeocin. The resulting p2ZOp2F-EGFP-LC3 stable (GFP-LC3) l(2)mbn cells were maintained in Schneider's D. melanogaster medium supplemented with 10% FBS and 10 μg/ml zeocin. 66 μl of these GFP-LC3 l(2)mbn cells (2.5 × 106 cells/ml) in SFM922 medium were seeded into each well of an 8-well CC2-coated chamber slide (LabTek). Cells were incubated with dsRNAs as described in the previous section, followed by a 4-h starvation treatment. Cells were fixed with 2% paraformaldehyde for 20 min and incubated with anti-GFP antibody (1:200; JL8; Clontech Laboratories, Inc.), followed by anti–mouse immunoglobulin Alexa 488 conjugates (Invitrogen). Cells were mounted with antifade reagent with DAPI at room temperature (SlowFade Gold; Invitrogen). Images were obtained using a 63× objective on a microscope (Axioplan2; all from Carl Zeiss, Inc.) and captured with a cooled mono 12-bit camera (QImaging) and Northern Eclipse image analysis software (Empix Imaging, Inc.). Cells with more than three GFP-LC3 punctate dots were considered to be GFP-LC3–positive cells. A minimum of 200 cells per sample were counted manually for triplicate samples per experiment.
Statistical analysis
Two-tailed student's t test (equal variances) was used to compare mean levels. n = 3. P < 0.05 was considered statistically significant.
Generation of transgenic flies
The UASp-full-length-Dcp-1 construct was generated by PCR amplification of the coding region from a Dcp-1 cDNA clone (Song et al., 1997) and cloning into the UASp vector (Rorth, 1998). Transgenic flies were generated using standard procedures. To express full-length Dcp-1 in the germ line, flies were crossed to NGT; nanosGAL4 flies (Cox and Spradling, 2003), and the resulting progeny were analyzed. To express truncated Dcp-1 and GFP-LC3 in the germ line, yw; nanosGAL4 UASp-tDcp-1 were crossed to UASp-GFP-LC3; nanos GAL4, and the resulting progeny were analyzed (Peterson et al., 2003).
Fly strains
w1118 was used as the wild-type stock. Other fly stocks used were as follows: Dcp-1Prev1 and UASp-GFP-LC3; nanos-GAL4 (from H. Stenmark, Norwegian Radium Hospital, Montebello, Oslo, Norway), BruceE81 (from H. Steller, the Rockefeller University, New York, NY), BruceE16 (from B.A. Hay, California Institute of Technology, Pasadena, CA), Atg7d77, Atg7d14, CG5335d30, and Atg1Δ3D (from T. Neufeld, University of Minnesota, Minneapolis, MN).
Generation of Atg 1 GLCs
To generate germ line clones, FRT2A was recombined onto the Atg1Δ3D chromosome. Correct recombinants were confirmed by failure to complement Df(3L)Bsc10 and Atg100305. Germ line clones were generated with the FLP/FRT/ovoD technique as described previously (Chou and Perrimon, 1996). Larvae of the genotype HSflp; ovoD FRT2A/Atg1Δ3D FRT2A were heat-shocked on days 4 and 5 for 1 h at 37°C.
LTR staining
For nutrient-deprivation experiments, flies were conditioned on yeast paste for 2 d and then placed in a dry vial with access to a 10% sucrose solution for 4–5 d (Peterson et al., 2003).
Ovaries were dissected in PBS and immediately transferred into PBS containing 0.8 μM LTR (Invitrogen) for 5 min at room temperature in the dark. The ovaries were then stained with 0.1 mg/ml DAPI for 30 s. The ovaries were washed three times with PBS and mounted with SlowFade antifade reagent at room temperature. Images were obtained using a 20 or 40× objective (Carl Zeiss, Inc.) on an Axioplan2 microscope and captured with a cooled mono 12-bit camera and Northern Eclipse image analysis software. Egg chambers with >10 LTR positive spots were considered to be LTR positive. Stage 8 degenerating egg chambers in w1118 flies were scored by the presence of condensed nurse cell nuclei. The degenerating egg chambers in Dcp-1Prev files were characterized by a disappearance of follicle cells and a persistence of nurse cell nuclei, as described previously (Laundrie et al., 2003).
For expression of Dcp-1 in ovaries, NGT/+; nanos-GAL4/UASp-fl-Dcp-1 flies were conditioned on wet yeast paste for 2–4 d and dissected in Ringers solution (Verheyen and Cooley, 1994). Ovaries were incubated with 50 μM LTR DND-99 in PBS for three minutes, washed three times for 5 min each in PBS, fixed for 10 min in 1:1 heptane/3.7% formaldehyde in Pipes buffer (0.1 M Pipes, 2 mM MgSO4, and 1 mM EGTA, pH 6.9), washed three times for 5 min in PBT (PBS + 0.1% Triton X-100), and mounted in antifade + 1.5 μg/ml Hoechst 33258. Egg chambers were viewed at room temperature using a UPlanFl 20×, 0.50 NA objective (Olympus) on a confocal microscope (BX50; Olympus). Images were captured using a Magnafire SP (S99810; Olympus; Hoechst) and Fluoview (LTR) cameras (Olympus). 1,4-diazabicyclo(2,2,2) octane (DABCO) was used as the imaging medium. Egg chambers with >10 LTR-positive spots were considered to be LTR positive. All figures were processed with Photoshop 7.0 (Adobe). Color was added to the LysoTracker image using ImageJ.
TUNEL assay
Ovaries were dissected in Schneider's D. melanogaster medium supplemented with 10% FBS. The ovaries were fixed in PBS containing 4% paraformaldehyde. The ovaries were then washed two times (5 min each) in PBS and permeabilized with 0.2% Triton X-100 for 5 min, followed by two washes in PBS. The TUNEL reaction was performed with the DeadEnd fluorometric TUNEL system (Promega). The ovaries were then stained with 0.1 mg/ml DAPI for 30 s at room temperature, mounted in SlowFade antifade reagent and observed under an Axioplan 2 microscope. Images were obtained using a 10, 20, or 40× objective (Carl Zeiss, Inc.) on an Axioplan2 microscope and captured with a cooled mono 12-bit camera and Northern Eclipse image analysis software.
QRT-PCR
Cells (∼2 × 105 cells in 600 μl) were incubated with dsRNAs as described (see “RNAi”) at 25°C for 72 h, followed by a 4-h starvation treatment. Cell cultures were transferred to RNase-free Eppendorf tubes (Ambion), and cells were pelleted at 1,000 rpm for 10 min. Cells were lysed in 1 ml Trizol (Invitrogen), and total RNA was extracted according to manufacturer's instructions. Isolated RNA was treated with RNase-free DNase, and 50 ng of total RNA was used in a 15-μl QRT-PCR reaction. QRT-PCR was performed using the one-step SYBR green RT-PCR reagent kit (Applied Biosystems) on a sequence detection system (7900; Applied Biosystems). Expression levels were calculated using the Comparative CT method with D. melanogaster rp49 as the reference. All samples were analyzed in triplicate. The knockdown efficiency was determined by comparing the fold change in expression between target dsRNA treated and untreated cells.
Online supplemental material
Fig. S1 includes representative images of GFP-LC3 puncta in l(2)mbn cells after starvation and treatment with autophagy inhibitors. Quantitative LTG data for additional nonoverlapping dsRNAs is also presented, along with assessment of target knockdown by QRT-PCR for selected genes. Table S1 shows a comparison of essential autophagy genes in the D. melanogaster larval fat body and l(2)mbn cells. Table S2 contains all primer sequences used for the preparation of dsRNAs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200712091/DC1.
Supplementary Material
Acknowledgments
We are grateful to A. Dorn for l(2)mbn cells, T.E. Rusten and H. Stenmark for the EGFP-LC3 plasmid and the UASp-GFP-LC3; nanos-GAL4 D. melanogaster strain, and T. Grigliatti for the p2ZOp2F plasmid. We thank B. Hay, T. Neufeld, and H. Steller for fly stocks. For technical assistance, we thank A.H. Tien, C. Helgason, J. Wilton, D. Mateos San Martin, E. Tanner, J. Chang, and B. Laundrie Stahl. We also thank M. Marra, M. Griffith, J. Halaschek-Wiener, and members of the Gorski laboratory for technical advice and helpful comments.
This work was supported by Canadian Institutes of Health Research grant MOP-78882 to S.M. Gorski. It was also supported by National Institutes of Health grant R01 GM60574 to K. McCall and the Summer Undergraduate Research Fellowship program at Boston University (to S.N. González Barbosa). S.N. González Barbosa was supported by the National Science Foundation–funded Northeast Alliance for Graduate Education and the Professoriate grant (NSF No. 0450339).
S.N. González Barbosa's present address is Rio Piedras Campus, University of Puerto Rico, San Juan 00931, Puerto Rico.
Abbreviations used in this paper: 3MA, 3-methyladenine; Atg, autophagy-related; Baf, bafilomycin A1; BIR, baculoviral IAP repeat; Dcp-1, death caspase-1; DIAP1, Drosophila melanogaster IAP protein-1; dsRNA, double-stranded RNA; fl-Dcp-1, full-length Dcp-1; GLC, germ line clone; IAP, inhibitor of apoptosis; l(2)mbn, lethal (2) malignant blood neoplasm; LC3, microtubule associated protein 1 light chain 3B; LTG, LysoTracker green; LTR, LysoTracker red; PI3K, phosphoinositide 3-kinase; QRT-PCR, quantitative RT-PCR; TOR, target of rapamycin; UASp, upstream activating sequence.
References
- Arama, E., J. Agapite, and H. Steller. 2003. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell. 4:687–697. [DOI] [PubMed] [Google Scholar]
- Baum, J.S., E. Arama, H. Steller, and K. McCall. 2007. The Drosophila caspases Strica and Dronc function redundantly in programmed cell death during oogenesis. Cell Death Differ. 14:1508–1517. [DOI] [PubMed] [Google Scholar]
- Bergmann, A., J. Agapite, K. McCall, and H. Steller. 1998. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 95:331–341. [DOI] [PubMed] [Google Scholar]
- Berry, D.L., and E.H. Baehrecke. 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 131:1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya, P., R.A. Gonzalez-Polo, N. Casares, J.L. Perfettini, P. Dessen, N. Larochette, D. Metivier, D. Meley, S. Souquere, T. Yoshimori, et al. 2005. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 25:1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, T.B., and N. Perrimon. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 144:1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, R.T., and A.C. Spradling. 2003. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development. 130:1579–1590. [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa, D., and A.C. Spradling. 2001. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev. Biol. 231:265–278. [DOI] [PubMed] [Google Scholar]
- Ferraro, E., and F. Cecconi. 2007. Autophagic and apoptotic response to stress signals in mammalian cells. Arch. Biochem. Biophys. 462:210–219. [DOI] [PubMed] [Google Scholar]
- Ghidoni, R., J.J. Houri, A. Giuliani, E. Ogier-Denis, E. Parolari, S. Botti, C. Bauvy, and P. Codogno. 1996. The metabolism of sphingo(glyco)lipids is correlated with the differentiation-dependent autophagic pathway in HT-29 cells. Eur. J. Biochem. 237:454–459. [DOI] [PubMed] [Google Scholar]
- Hara, T., K. Nakamura, M. Matsui, A. Yamamoto, Y. Nakahara, R. Suzuki-Migishima, M. Yokoyama, K. Mishima, I. Saito, H. Okano, and N. Mizushima. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. [DOI] [PubMed] [Google Scholar]
- Hay, B.A., J.R. Huh, and M. Guo. 2004. The genetics of cell death: approaches, insights and opportunities in Drosophila. Nat. Rev. Genet. 5:911–922. [DOI] [PubMed] [Google Scholar]
- Hegedus, D.D., T.A. Pfeifer, J. Hendry, D.A. Theilmann, and T.A. Grigliatti. 1998. A series of broad host range shuttle vectors for constitutive and inducible expression of heterologous proteins in insect cell lines. Gene. 207:241–249. [DOI] [PubMed] [Google Scholar]
- Inbal, B., S. Bialik, I. Sabanay, G. Shani, and A. Kimchi. 2002. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J. Cell Biol. 157:455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz, G., and M. Sass. 2005. Hid can induce, but is not required for autophagy in polyploid larval Drosophila tissues. Eur. J. Cell Biol. 84:491–502. [DOI] [PubMed] [Google Scholar]
- Juhasz, G., B. Erdi, M. Sass, and T.P. Neufeld. 2007. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 21:3061–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, R.C. 1970. Ovarian Development in Drosophila melanogaster. Academic Press, New York. 227 pp.
- Klionsky, D.J. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–987. [DOI] [PubMed] [Google Scholar]
- Klionsky, D.J., A.M. Cuervo, and P.O. Seglen. 2007. Methods for monitoring autophagy from yeast to human. Autophagy. 3:181–206. [DOI] [PubMed] [Google Scholar]
- Komatsu, M., S. Waguri, T. Chiba, S. Murata, J. Iwata, I. Tanida, T. Ueno, M. Koike, Y. Uchiyama, E. Kominami, and K. Tanaka. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884. [DOI] [PubMed] [Google Scholar]
- Kumar, S. 2004. Migrate, differentiate, proliferate, or die: pleiotropic functions of an apical “apoptotic caspase”. Sci. STKE. 2004:pe49. [DOI] [PubMed] [Google Scholar]
- Kuranaga, E., and M. Miura. 2007. Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 17:135–144. [DOI] [PubMed] [Google Scholar]
- Laundrie, B., J.S. Peterson, J.S. Baum, J.C. Chang, D. Fileppo, S.R. Thompson, and K. McCall. 2003. Germline cell death is inhibited by P-element insertions disrupting the dcp-1/pita nested gene pair in Drosophila. Genetics. 165:1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, B., and J. Yuan. 2005. Autophagy in cell death: an innocent convict? J. Clin. Invest. 115:2679–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum, J.J., D.E. Bauer, M. Kong, M.H. Harris, C. Li, T. Lindsten, and C.B. Thompson. 2005. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 120:237–248. [DOI] [PubMed] [Google Scholar]
- Maiuri, M.C., E. Zalckvar, A. Kimchi, and G. Kroemer. 2007. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8:741–752. [DOI] [PubMed] [Google Scholar]
- McCall, K. 2004. Eggs over easy: cell death in the Drosophila ovary. Dev. Biol. 274:3–14. [DOI] [PubMed] [Google Scholar]
- Mills, K.R., M. Reginato, J. Debnath, B. Queenan, and J.S. Brugge. 2004. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc. Natl. Acad. Sci. USA. 101:3438–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., A. Yamamoto, M. Hatano, Y. Kobayashi, Y. Kabeya, K. Suzuki, T. Tokuhisa, Y. Ohsumi, and T. Yoshimori. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre, S., A. Tassa, X. Qu, R. Garuti, X.H. Liang, N. Mizushima, M. Packer, M.D. Schneider, and B. Levine. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 122:927–939. [DOI] [PubMed] [Google Scholar]
- Peterson, J.S., M. Barkett, and K. McCall. 2003. Stage-specific regulation of caspase activity in Drosophila oogenesis. Dev. Biol. 260:113–123. [DOI] [PubMed] [Google Scholar]
- Peterson, J.S., B.P. Bass, D. Jue, A. Rodriguez, J.M. Abrams, and K. McCall. 2007. Noncanonical cell death pathways act during Drosophila oogenesis. Genesis. 45:396–404. [DOI] [PubMed] [Google Scholar]
- Prins, J.B., E.C. Ledgerwood, P. Ameloot, P. Vandenabeele, P.R. Faraco, N.A. Bright, S. O'Rahilly, and J.R. Bradley. 1998. Tumor necrosis factor-induced cytotoxicity is not related to rates of mitochondrial morphological abnormalities or autophagy-changes that can be mediated by TNFR-I or TNFR-II. Biosci. Rep. 18:329–340. [DOI] [PubMed] [Google Scholar]
- Ress, C., M. Holtmann, U. Maas, J. Sofsky, and A. Dorn. 2000. 20-Hydroxyecdysone-induced differentiation and apoptosis in the Drosophila cell line, l(2)mbn. Tissue Cell. 32:464–477. [DOI] [PubMed] [Google Scholar]
- Riedl, S.J., and Y. Shi. 2004. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 5:897–907. [DOI] [PubMed] [Google Scholar]
- Rorth, P. 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78:113–118. [DOI] [PubMed] [Google Scholar]
- Rusten, T.E., K. Lindmo, G. Juhasz, M. Sass, P.O. Seglen, A. Brech, and H. Stenmark. 2004. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell. 7:179–192. [DOI] [PubMed] [Google Scholar]
- Scarlatti, F., C. Bauvy, A. Ventruti, G. Sala, F. Cluzeaud, A. Vandewalle, R. Ghidoni, and P. Codogno. 2004. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J. Biol. Chem. 279:18384–18391. [DOI] [PubMed] [Google Scholar]
- Scott, R.C., O. Schuldiner, and T.P. Neufeld. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 7:167–178. [DOI] [PubMed] [Google Scholar]
- Scott, R.C., G. Juhasz, and T.P. Neufeld. 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen, P.O., and P.B. Gordon. 1982. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA. 79:1889–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, S., T. Kanaseki, N. Mizushima, T. Mizuta, S. Arakawa-Kobayashi, C.B. Thompson, and Y. Tsujimoto. 2004. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 6:1221–1228. [DOI] [PubMed] [Google Scholar]
- Smith, J.E. III, C.A. Cummings, and C. Cronmiller. 2002. Daughterless coordinates somatic cell proliferation, differentiation and germline cyst survival during follicle formation in Drosophila. Development. 129:3255–3267. [DOI] [PubMed] [Google Scholar]
- Song, Z., K. McCall, and H. Steller. 1997. DCP-1, a Drosophila cell death protease essential for development. Science. 275:536–540. [DOI] [PubMed] [Google Scholar]
- Spradling, A.C. 1993. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1558 pp.
- Thorburn, A. 2008. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis. 13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn, J., F. Moore, A. Rao, W.W. Barclay, L.R. Thomas, K.W. Grant, S.D. Cramer, and A. Thorburn. 2005. Selective inactivation of a Fas-associated death domain protein (FADD)-dependent apoptosis and autophagy pathway in immortal epithelial cells. Mol. Biol. Cell. 16:1189–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velentzas, A.D., I.P. Nezis, D.J. Stravopodis, I.S. Papassideri, and L.H. Margaritis. 2007. Mechanisms of programmed cell death during oogenesis in Drosophila virilis. Cell Tissue Res. 327:399–414. [DOI] [PubMed] [Google Scholar]
- Verheyen, E., and L. Cooley. 1994. Looking at oogenesis. Methods Cell Biol. 44:545–561. [PubMed] [Google Scholar]
- Vernooy, S.Y., V. Chow, J. Su, K. Verbrugghe, J. Yang, S. Cole, M.R. Olson, and B.A. Hay. 2002. Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not Hid-dependent cell death. Curr. Biol. 12:1164–1168. [DOI] [PubMed] [Google Scholar]
- Wang, Y., R. Singh, A.C. Massey, S.S. Kane, S. Kaushik, T. Grant, Y. Xiang, A.M. Cuervo, and M.J. Czaja. 2008. Loss of macroautophagy promotes or prevents fibroblast apoptosis depending on the death stimulus. J. Biol. Chem. 283:4766–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D., Y. Li, M. Arcaro, M. Lackey, and A. Bergmann. 2005. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 132:2125–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., Y. Tagawa, T. Yoshimori, Y. Moriyama, R. Masaki, and Y. Tashiro. 1998. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23:33–42. [DOI] [PubMed] [Google Scholar]
- Yousefi, S., R. Perozzo, I. Schmid, A. Ziemiecki, T. Schaffner, L. Scapozza, T. Brunner, and H.U. Simon. 2006. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 8:1124–1132. [DOI] [PubMed] [Google Scholar]
- Yu, L., A. Alva, H. Su, P. Dutt, E. Freundt, S. Welsh, E.H. Baehrecke, and M.J. Lenardo. 2004. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 304:1500–1502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.