Abstract
RNA recombination occurs frequently during replication of tombusviruses and carmoviruses, which are related small plus-sense RNA viruses of plants. The most common recombinants generated by these viruses are either defective interfering (DI) RNAs or chimeric satellite RNAs, which are thought to be generated by template switching of the viral RNA-dependent RNA polymerase (RdRp) during the viral replication process. To test if RNA recombination is mediated by the viral RdRp, we used either a purified recombinant RdRp of Turnip crinkle carmovirus or a partially purified RdRp preparation of Cucumber necrosis tombusvirus. We demonstrated that these RdRp preparations generated RNA recombinants in vitro. The RdRp-driven template switching events occurred between either identical templates or two different RNA templates. The template containing a replication enhancer recombined more efficiently than templates containing artificial sequences. We also observed that AU-rich sequences promote recombination more efficiently than GC-rich sequences. Cloning and sequencing of the generated recombinants revealed that the junction sites were located frequently at the ends of the templates (end-to-end template switching). We also found several recombinants that were generated by template switching involving internal positions in the RNA templates. In contrast, RNA ligation-based RNA recombination was not detected in vitro. Demonstration of the ability of carmo- and tombusvirus RdRps to switch RNA templates in vitro supports the copy-choice models of RNA recombination and DI RNA formation for these viruses.
Viral RNA recombination, a process that joins together two noncontiguous RNA segments, is an especially powerful tool in virus evolution, since it can rapidly lead to dramatic changes in virus genomes by recombining or rearranging “battle-tested” (i.e., evolutionarily successful) sequences. Accordingly, the significant role of RNA recombination in emergence of new viruses or virus strains is well documented for numerous human, animal, plant, insect, fungal, and bacterial viruses (2, 4, 12, 17, 18, 20, 21, 27-31, 51, 57, 61, 62, 64, 65, 71). In addition to increasing sequence variability, RNA recombination can be an efficient tool for viruses to repair viral genomes, thus contributing to virus fitness (6, 14, 35, 36, 53, 66). In spite of its significance, our understanding of RNA recombination is incomplete. This is due to the complex nature of RNA recombination and the lack of tractable systems for mechanistic studies.
RNA recombination may also play a role in the formation of subviral RNAs that include defective interfering (DI) RNAs associated with many animal and plant viruses. DI RNAs are mainly derived from the parent (helper) virus via sequence deletion(s). The DI RNAs are deficient in replication and/or other functions, which makes them dependent on the helper virus for their survival and spread (50, 70). The best-known DI RNAs among plant viruses are those associated with tombusvirus infections. The tombusvirus DI RNAs are mosaic types that have two or three sequence deletions (70), resulting in 80 to 90% reduced genome size for DI RNAs when compared to the parental virus genome. Sequence deletions during DI RNA formation are thought to be the consequence of viral replicase jumping on the templates, and the deletions may occur in a stepwise manner (68, 70).
The most popular model of RNA recombination is the template-switching (copy-choice) mechanism, which predicts that the viral RNA-dependent RNA polymerase (RdRp) switches templates during cRNA synthesis (22, 26, 27, 33, 37). After the jump from the donor to the acceptor RNA, the RdRp is assumed to continue RNA synthesis on the acceptor template, using the nascent RNA as a primer. Experimental evidence supporting the template-switching mechanism has been obtained, for example, with poliovirus, bovine viral diarrhea virus, brome mosaic virus (BMV), cucumber mosaic virus, and bacteriophage Qβ (1, 5, 13, 25). In special cases observed with bacteriophage Qβ and poliovirus (11, 19), RNA breakage and ligation might also result in recombination. RNA recombinants generated via template switching or RNA breakage and ligation cannot be determined by sequence analysis of recombination sites. Mechanistic studies with purified protein and RNA components, however, are yielding valuable insights into the mechanism of RNA recombination (37).
Since the viral RdRp is thought to carry out viral recombination and replication, it is likely that RNA elements and protein factors involved in replication may also play roles in recombination. Previous in vivo and in vitro works revealed that replication of tombusviruses and carmoviruses (which are related viruses) is carried out by the replicase complex that includes two viral proteins and unknown host factors (58, 59). Functional replicase complexes have been partially purified from carmovirus-infected (60) and tombusvirus-infected (45) plants, which can synthesize complementary RNAs on added RNA templates. A functional viral RdRp has also been obtained after overexpression in Escherichia coli (52), which showed many of the features described for the plant-purified carmovirus replicase complex. Important cis-acting elements, including promoters and replication enhancers, have been defined for both viruses in vitro and in vivo (43, 46-49, 54, 55, 59). Some of these cis-acting elements have been shown to constitute recombination hot spots in vivo (8, 39, 59).
Previous in vivo RNA recombination studies with carmo- and tombusvirus have given insights into the mechanism of RNA recombination (8, 39, 67, 69) by supporting the template-switching (copy-choice) mechanism. The evidence includes the following: (i) at least one of the RNAs must have a replication signal, such as a promoter, for recombination to occur frequently (69); (ii) the recombination sites are frequently located at or close to known cis-acting replication elements, such as replication enhancers or promoters (8, 39); (iii) mutating these cis-acting replication elements can greatly affect the frequency of recombination; and (iv) the presence of nontemplated nucleotides at the recombination junction sites (39, 69). Although, based on the above facts, it is easier to explain recombinant formation by the RdRp-driven template switching, it is impossible to fully exclude the RNA ligation mechanism.
In this paper, using an in vitro assay with the recombinant carmovirus RdRp (Turnip crinkle virus [TCV]) and the partially purified tombusvirus RdRp (Cucumber necrosis virus [CNV]) from plants, we demonstrate that template switching occurs with high efficiency in the case of carmo- and tombusviruses in vitro. Characterization of the generated RNA recombinants revealed that the viral replication enhancer could promote template switching with higher efficiency than artificial sequences in both viral RdRp assays. Sequencing of the recombinants showed that most events occur at the ends of the templates, although internal events were also detected. RNA ligation has not been detected in these in vitro assays. Overall, these data firmly establish novel in vitro RNA recombination systems for carmo- and tombusviruses that should be useful in dissecting the factors facilitating viral RNA recombination.
MATERIALS AND METHODS
Recombinant TCV RdRp preparation.
The expression and purification of recombinant TCV RdRp was as described before (52). Briefly, p88C, which contains the C-terminal readthrough portion of full-length p88, was expressed as a fusion protein with the maltose-binding protein (MBP) domain in E. coli Epicurian BL21-CodonPlus (DE3)-RIL (Stratagene). Protein expression was induced as recommended by the supplier using isopropyl-β-d-thiogalactopyranoside. After 8 to 10 h of induction at 14°C, the cells were harvested, collected by centrifugation (3,000 × g for 5 min), resuspended, and sonicated as recommended by the supplier, except reducing the concentration of NaCl to 25 mM in the column buffer. The samples were then centrifuged again (15,000 rpm for 5 min), followed by affinity-based chromatography (amylose column from NEB) following the supplied procedure. After thorough washing with the column buffer, the proteins were eluted with maltose-containing column buffer (NEB). All steps were carried out on ice or in the cold room. The quality of the proteins obtained was checked by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (PAGE) analysis (52). The TCV RdRp studies were done with the MBP-p88C fusion proteins.
Plant inoculation and CNV RdRp preparation.
Nicotiana benthamiana plants were inoculated with CNV genomic RNA transcripts obtained by standard T7 RNA transcription using a SmaI-linearized clone of pK2/M5p20STOP for CNV (56). Construct pK2/M5p20STOP, which is a full-length T7 RNA polymerase-transcribable cDNA clone of CNV genomic RNA, was the generous gift of D'Ann Rochon. pK2/M5p20STOP contains a mutation within the p20 gene that eliminates the lethal necrosis induced by the wild-type CNV in N. benthamiana. CNV RdRp preparations were obtained from systemically infected leaves as described by Nagy and Pogany (45).
Preparation of DNA and RNA templates.
The DNA templates were generated with PCR using the templates and primers listed in Table 1. The RNA templates were generated from PCR products using in vitro transcription with T7 polymerase. The obtained RNAs were gel isolated from 5% denaturing PAGE to make sure that only the desired products were used for RdRp reactions (39). Labeling of RNA templates with biotin was done by using T7 RNA polymerase transcription in the presence of biotin-16-uridine-5′-triphosphate (1:5 ratio of biotin-16-UTP [Roche] and UTP). The biotinylated RNAs were gel isolated from 5% denaturing PAGE. The cPR21 RNA was chemically synthesized (Dharmacon Research).
TABLE 1.
Constructs and primers used for PCRs
| Construct | Template | Primer | Sequence of primer |
|---|---|---|---|
| Mot1/pr | TCV satC | P10 | GGGATAACTAAGGGTTTCATACGTTACTACATCCCAGACCCT |
| P11 | TAATACGACTCACTATAGGGCTGCCGCCGTTTTTGG | ||
| Mot1d10/pr | TCV satC | P12 | AAGGGTTTCATACGTTACTACATCCC |
| P10 | TAATACGACTCACTATAGGGCTGCCGCCGTTTTTGG | ||
| R3(−)/pr11 | DI-72 | 23 | GTAATACGACTCACTATAGGGACCCAACAAGAGTAACCTG |
| 253 | TTGGAAATTCTCCTTAGCGAGTAAGACAGACTC | ||
| AU1/pr11 | PN-R′+AU1 | 248 | GTAATACGACTCACTATAGGAGACCCTGTCCAGGTAG |
| 251 | TTGGAAATTCTCCTTGTGCTCGAGTTGGATCC | ||
| GC1/pr11 | PN-GC1 | 248 | GTAATACGACTCACTATAGGAGACCCTGTCCAGGTAG |
| 251 | TTGGAAATTCTCCTTGTGCTCGAGTTGGATCC | ||
| R3(−)/art | DI-72 | 116 | GTAATACGACTCACTATAGGACACCTAACTTTCGT |
| 112 | TCGTCTTATTGGACGAGATAGTCACTTGACTAC | ||
| AU1/art | PN-R′+AU1 | 248 | GTAATACGACTCACTATAGGAGACCCTGTCCAGGTAG |
| 249 | TCGTCTTATTGGACGAGTGCTCGAGTTGGATCC |
RdRp assay.
We used 0.5 μg of R3(−) RNA for the TCV or CNV RdRp reactions, while the other RNA templates were adjusted to the same molar amounts as R3(−). The RdRp reactions contained either 2 μl of recombinant TCV RdRp or 30 μl of CNV RdRp as described previously (45, 52). The concentrations of ribonucleotides were 0.2 mM ATP, CTP, and GTP; 0.02 mM UTP; and 0.3 μl of [32P]UTP (specific activity of 800 Ci/mmol from ICN) in the reaction mixture. The RdRp products were phenol-chloroform extracted and analyzed under complete denaturing conditions (i.e., 5% PAGE containing 40% formamide and 8 M urea, with the run performed at 70°C in the Bio-Rad DCode system apparatus).
To study the role of donor and acceptor templates (see Fig. 4), we modified the RdRp reaction to include biotin-labeled RNAs, which were produced with T7 RNA polymerase as described above. Biotinylated RNAs were as good templates as nonbiotinylated RNAs were in primer extension reactions with the TCV RdRp (data not shown). The biotin-labeled RNA templates were incubated for 3 h in the RdRp reactions, followed by the transfer of RdRp products to streptavidin-coated tubes (Roche) in a final volume of 200 μl (in 1× RdRp buffer). The tubes were incubated at 37°C for 30 min, and then we transferred the solution with unbound RNAs to new tubes for isopropanol precipitation. The tubes were further washed, first with 200 μl of RNase-free water (incubation at 85°C for 10 min) and then four times with a washing solution (10 mM Tris-HCl [pH 7.5], 200 mM LiCl, and 1 mM EDTA). Finally, the streptavidin-bound RNAs were recovered by adding a denaturing buffer (95% formamide and 10 mM EDTA; pH 8.0) and heating the tube to 85°C for 5 min (Dynabeads M-280 streptavidin manual, p. 156, Dynal), followed by rapid transfer of the denaturing buffer to new tubes and isopropanol precipitation. All the products were analyzed in 5% complete denaturing PAGE. Each experiment was repeated three times, and the quantification of gels was done using a PhosphorImager (45).
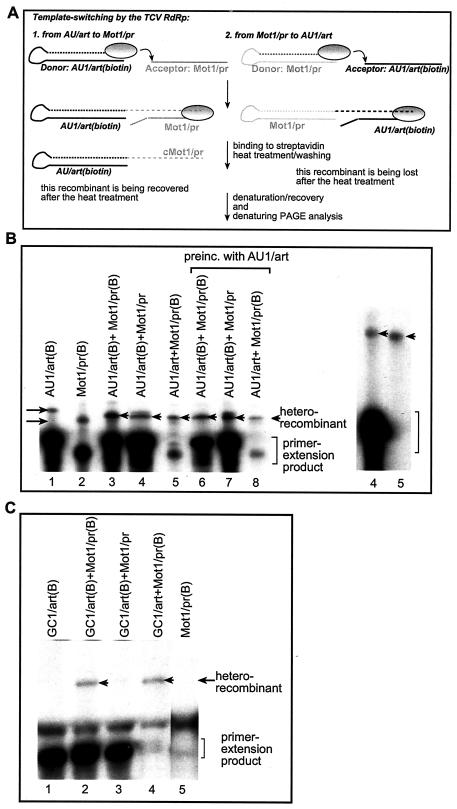
FIG. 4.
Determination of donor versus acceptor use by the TCV RdRp. (A) Schematic representation of the method used for selective purification of recombinant RdRp products. The template switching by the TCV RdRp (shown as an oval circle) is represented by an arrow. The newly synthesized, 32P-labeled RNA sequences are indicated by dotted lines. The two types of heterorecombinants are shown in the left and right panels, respectively. The left panel depicts the scenario when the biotin-labeled RNA template (AU1/art shown as an example) served as the donor RNA during the template-switching event, leading to the formation of a recombinant RNA that carries the label (due to initiation that takes place via 3′ primer extension). The biotin-labeled RdRp products (including the primer extension [data not shown] and the recombinants) were purified using streptavidin as described in Materials and Methods. Note that the heterorecombinants formed between Mot1/pr (donor; unlabeled with biotin) and AU1/art are not labeled with biotin and are lost during purification (as depicted in the right panel). (B) Analysis of the donor template selection by the recombinant TCV RdRp. The RdRp reaction was performed as described in the legend to Fig. 1, except that one of the templates was biotin labeled (indicated by the letter B after the name of the construct) before the reaction. After streptavidin-based purification, the RdRp products were analyzed on denaturing gels. The homorecombinants are depicted with arrows pointing rightward, while the heterorecombinants are marked with arrowheads. Lanes 6 to 8 include samples that were obtained by preincubating the RdRp reaction mixture with the AU1/art template (details are in Materials and Methods). Due to template competition, the primer extension products are lower in the two-template-containing experiments than in single-template-containing experiments. The gel on the right shows a longer run containing samples 4 and 5 to resolve the size differences between the recombinants (marked with arrowheads). (C) Inefficient use of a GC-rich template by the TCV RdRp during template switching. The names of the constructs used are shown above the lanes. See further details in panels A and B. Arrowheads point at the heterorecombinants.
RT-PCR and sequencing analysis.
To detect recombinants in the TCV RdRp assays, the recombinant RNA-containing bands were cut from denaturing gels and the RNAs were isolated as described earlier (39, 40). The use of the recombinant-sized, gel-isolated RdRp products was expected to eliminate the possibility of recombination during the reverse transcription-PCR (RT-PCR) analysis. The reverse primer (P11) for the RT reaction was designed to anneal to the 3′ end of the complementary Mot1/pr sequence, while the forward primer (251 [Table 1]) for PCR hybridized to the AU1/art as shown below in Fig. 3C. This primer set was used to detect template switching from AU1/art to Mot1/pr. To detect template switching from Mot1/pr to AU1/art, we used reverse primer 248 (Table 1), which anneals to the 3′ end of the complementary AU1/art sequence for RT and the forward primer (P10) (Table 1).
FIG. 3.
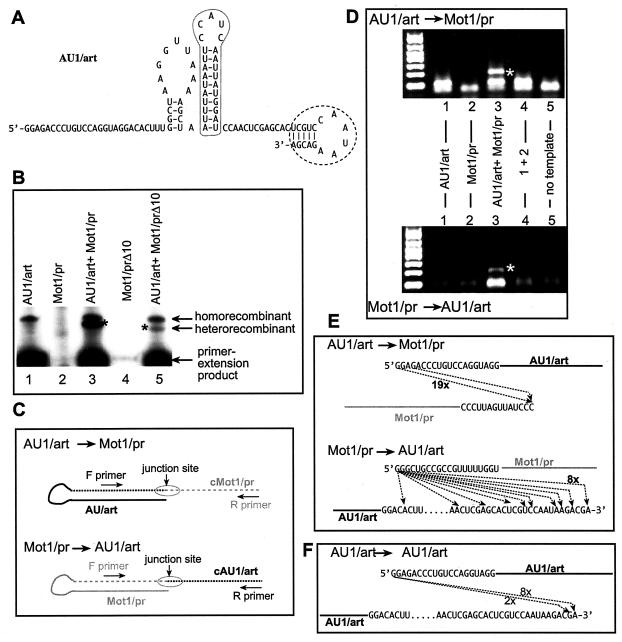
Increasing the efficiency of heterorecombinant formation by TCV RdRp. (A) A 5-bp primer sequence (named art5; encircled with dotted line) was used to replace the cPR11 sequence at the 3′ end of AU1 (Fig. 1) to increase the efficiency of primer extension by the TCV RdRp (10). (B) Denaturing gel analysis of the TCV RdRp products. The positions of the primer extension products and the putative homo- and heterorecombinants are marked on the right. The amount of Mot1/pr RNA was reduced to 0.1 μg to inhibit homorecombinant formation (Fig. 2). Construct Mot1/prΔ10 is derived from Mot1/pr (Fig. 1) by deletion of 10 nt from the 3′ end. (C) Schematic representation of the strategy used for the RT-PCR analysis of the heterorecombinants (see also panel D). Two different sets of primers were used to detect the heterorecombinants formed (depending on which template was used as a donor during the recombination events). Note that the dotted lines represent the newly synthesized RNA strands, which are complementary to the original templates (as indicated by the letter c in front of the names of the RNAs). (D) RT-PCR analysis of the putative recombinants. After the TCV RdRp reactions, the RdRp products migrating slower than the primer extension products were gel isolated and used for RT-PCR. The band representing heterorecombinants (lane 3) is marked with an asterisk. (E) Sequence analysis of the junction sites in the heterorecombinants. After the RT-PCR analysis, the bands representing the heterorecombinants were gel isolated and cloned in E. coli, and a representative number of clones was sequenced. Arrows indicate the template switching by the recombinant TCV RdRp from the donor template (top) to the acceptor template (bottom). The frequencies of clones with identical sequences are indicated by numbers next to the lines. (F) Sequences of the homorecombinants formed between identical AU1/art templates. A strategy similar to that shown in panel C was used for RT-PCR, cloning, and sequencing of homorecombinants.
To detect recombinants in the CNV RdRp assays, we cut the gel area above the visible primer extension products, followed by RNA isolation. The RT reaction included the reverse primer 23, which anneals to the 3′ end of complementary R3(−)/art, while the PCR mixture also contained the forward primer 251 to detect template switching from AU1/art to R3(−)/art. To detect template switch from R3(−)/art to AU1/art, we used reverse primer 248, which anneals to the 3′ end of the complementary AU1/art sequence for RT and the forward primer 18 (GTAATACGACTCACTATAGGAGAAAGCGAGTAAGACAG) for PCR. The obtained RT-PCR products were either gel isolated or directly cloned into pGEM-T Easy vector (Promega). A representative number of independent clones were sequenced using the M13/pUC19 reverse primer (Gibco BRL).
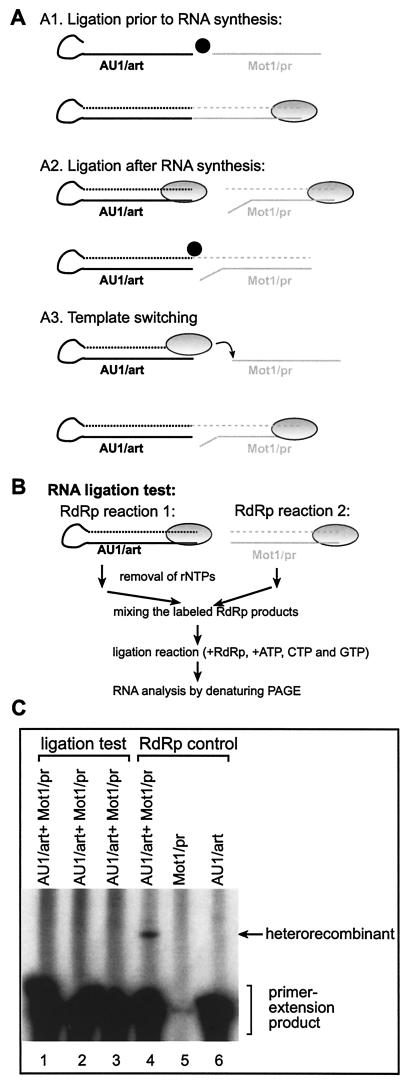
RdRp ligation assay.
First, standard TCV RdRp reactions (including [32P]UTP, see above) were performed with AU1/art and Mot1/pr templates separately, followed by removal of the free nucleotides by passing the reaction mixtures through Micro Bio-Spin columns (Bio-Rad). Then, the column-purified [32P]UTP-labeled RdRp products were mixed together (in a 1:1 ratio) along with additional TCV RdRp and three nucleotides (rATP, rCTP, and rGTP; 0.2 mM) followed by incubation for 2 h at 25°C. The RdRp products were then phenol-chloroform extracted, precipitated, and analyzed in 5% complete denaturing PAGE as described above.
RESULTS
Rationale.
Current models of RNA recombination in carmo- and tombusviruses predict that the viral RdRp jumps from the donor RNA to the acceptor RNA during viral RNA synthesis (37, 39, 70). Therefore, we tested the abilities of two viral RdRps, a recombinant TCV RdRp and the partially purified CNV RdRp, to switch templates in vitro. In these assays, the RdRp first has to initiate RNA synthesis on one of the templates (donor), followed by elongation and pausing or termination to generate the primer (which is the nascent strand) that is subsequently used to resume RNA synthesis on the second (acceptor) template. Since both TCV and CNV RdRps can initiate RNA synthesis on added templates (either using primer extension or de novo initiation [45, 52]), they were used in combinations with various RNA templates during this work.
Efficient template switching by the TCV RdRp in vitro.
To test template switching by the TCV RdRp in vitro, we used a previously characterized truncated version of the TCV RdRp protein lacking the N-terminal overlapping domain (termed p88C TCV RdRp) (52). This TCV RdRp protein was overexpressed in E. coli and affinity purified using an N-terminal MBP tag (52). The purified recombinant TCV RdRp is a highly active enzyme that can efficiently use various RNA templates in an in vitro assay. The RNA synthesis is initiated by either de novo initiation or by the more efficient 3′-terminal (primer) extension on the added RNA templates (52).
For the in vitro template-switching reactions, which contained the purified recombinant TCV RdRp and 32P-labeled UTP in addition to the unlabeled nucleotides, we selected four templates that are used with different efficiencies for cRNA synthesis by the recombinant TCV RdRp (Fig. 1A). One template (i.e., Mot1/pr [Fig. 1A]) contained the TCV satC replication enhancer (termed motif1-hairpin [39, 59]), which was shown to bind to the TCV RdRp with high affinity (39-41). The Mot1/pr template is used efficiently by the TCV RdRp for primer extension, which results in a major product in the RdRp reaction (Fig. 1B, lane 1). Interestingly, using the Mot1/pr template, the TCV RdRp makes additional products that are longer than the primer extension product (Fig. 1C). Based on their predicted sizes, we assumed that these long products could be recombinant RNAs. For example, the second-most-abundant RNA (marked as homorecombinant in Fig. 1C) is possibly formed by recombination between two identical Mot1/pr templates: one template serves as a donor, while the other RNA serves as an acceptor during the template-switching event. The three additional, longer RdRp products were estimated to be the result of two to four sequential template-switching events (depicted as multimeric homorecombinants in Fig. 1C). As expected, the amounts of these putative multimeric recombinants decreased as their sizes increased. These homorecombinants have not been characterized further.
FIG. 1.
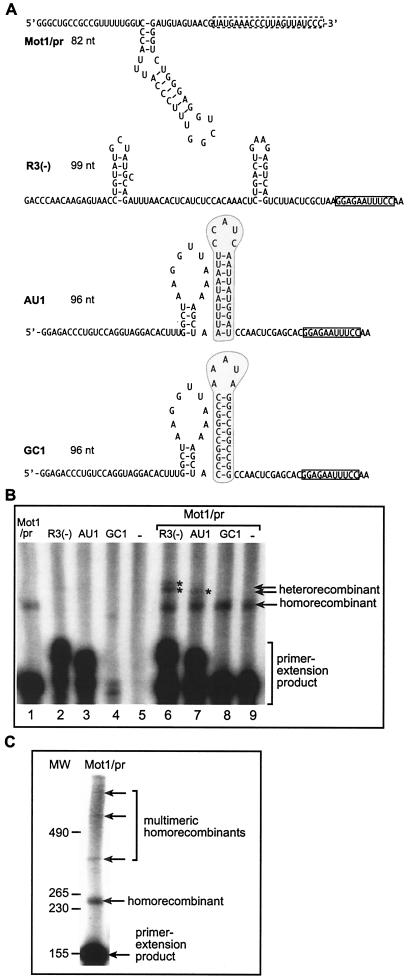
Template switching by the recombinant TCV RdRp in vitro. (A) Sequences and structures of RNA templates used in the TCV RdRp reactions. Construct Mot1/pr contains the minus-stranded motif1 hairpin replication enhancer (shown as the hairpin structure [39]) and the 3′-terminal plus-strand initiation sequence (boxed with dotted line) of TCV satC. Construct R3(−) contains the minus-stranded replication enhancer RIII(−) (49) and the minus-stranded 3′-terminal cPR11 promoter of the related TBSV (boxed) (47). Construct AU1 contains an artificial AU-rich sequence (encircled in gray box) (10), while the same-sized construct GC1 carries an artificial GC-rich sequence (shaded area) (10). In addition, constructs AU1 and GC1 contain the same 5′ and 3′ sequences (cPR11 is boxed); thus, these constructs are different only in the shaded regions. Note that the GC and AU regions differ not only in their sequences but also in the strength of the hairpin structures that could potentially influence initiation and/or recombination. (B) Detection of recombinants in an in vitro TCV RdRp assay. RNA templates were used in equal amounts (15 pmol/reaction mixture) under the conditions described in Materials and Methods. The RdRp products synthesized by in vitro transcription with the recombinant TCV RdRp were analyzed in a fully denaturing gel. The names of the RNA constructs used in the TCV RdRp reactions are shown above the lanes. The positions of heterorecombinants are marked with asterisks. Note that the combination of R3(−) and Mot1/pr resulted in two heterorecombinants (double band). The homorecombinants derived from the Mot1/pr template are also marked. Note that under the conditions used (reduced nucleotide concentration), the recombinant TCV RdRp performs 3′-terminal (primer) extensions for all four templates at various efficiencies. (C) TCV RdRp products obtained with the Mot1/pr template. The molecular size markers, which were obtained with T7 transcription, are shown on the left.
The second template used in this study was construct R3(−), which included the replication enhancer [termed region III(-) (49, 54, 55)] from a related tombusvirus (TBSV) which is also an efficient template for the TCV RdRp (Fig. 1B, lane 2). The third construct carried an artificial AU-rich sequence (construct AU1) (Fig. 1A) (10, 38), which can also be used efficiently by the recombinant TCV RdRp (Fig. 1B, lane 3). Both R3(−) and AU1 are used for primer extension by the TCV RdRp under the experimental conditions (reduced nucleotide concentration). Unlike Mot1/pr, R3(−) and AU1 did not support the accumulation of homorecombinants in detectable amounts (Fig. 1B, lanes 2 and 3), suggesting that these templates are either poor donors or poor acceptors. The fourth template in this study was an artificial GC-rich sequence (termed GC) (Fig. 1A) (10, 42), which is a poor template for the TCV RdRp in vitro (Fig. 1B, lane 4).
To test if the TCV RdRp can generate RNA recombinants in vitro, we added a mixture of two templates, such as Mot1/pr and a second template [either R3(−), AU1, or GC1] to the in vitro TCV RdRp assay. Note that Mot1/pr is a shorter template than the other RNAs in order to facilitate the discrimination among possible recombinants, which are formed either between two heterologous templates (called heterorecombinants) or between two identical templates (homorecombinants). Importantly, the RNA templates were gel purified prior to the TCV RdRp reaction to make sure that only the expected RNAs were present during the in vitro reactions. After the RdRp reactions, the products were separated by PAGE performed under fully denaturing conditions (see Materials and Methods). Long exposure of the gels revealed the accumulation of novel RdRp products (Fig. 1B) that were not generated in the control RdRp reactions, which contained only single templates. These novel RdRp products were easily detected in the combination of Mot1/pr and R3(−) (Fig. 1B, lane 6), and in a lesser amount in the combination of Mot1/pr and AU1 templates (Fig. 1B, lane 7), but they were not detected in the case of Mot1/pr and GC1 (Fig. 1B, lane 8). Based on the sizes of these novel products, they may be formed by template switching of the TCV RdRp from one template to the other during RNA synthesis.
The effect of template and enzyme concentration on template switching by the TCV RdRp.
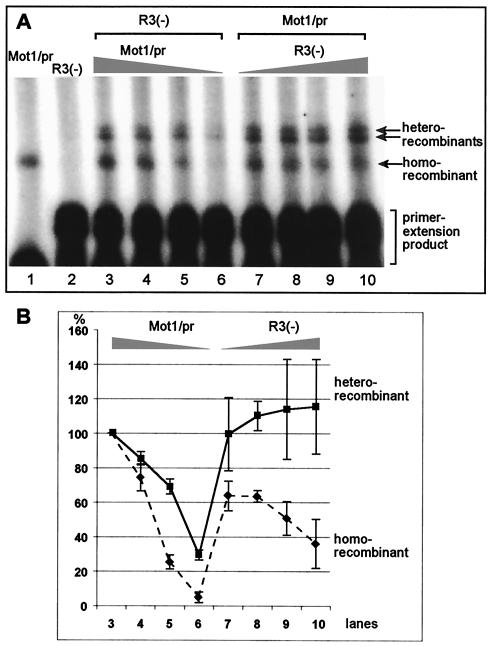
The observation that the putative homorecombinants were more abundant than the putative heterorecombinants suggested that the Mot1/pr RNA might be used more efficiently than either R3(−) or AU1 RNAs during template-switching events (based on Fig. 1B). Therefore, to increase the chance of recombination events that take place between R3(−) RNA and Mot1/pr RNA, we changed the amounts of Mot1/pr and R3(−) RNAs in the TCV RdRp reactions. Reduction of the Mot1/pr RNA by 10-fold, while leaving the amount of R3(−) RNA unchanged, decreased the overall efficiency of homorecombinant formation by ∼95% (Fig. 2, lanes 3 to 6). Interestingly, the efficiency of heterorecombinant formation was reduced to a lesser extent (∼70%) than that of homorecombinant formation (Fig. 2B, lanes 3 to 6). In contrast, increasing the amount of R3(−) RNA in the TCV RdRp reaction mixture by fourfold, while leaving the concentration of Mot1/pr RNA unchanged, increased the amount of heterorecombinants by ∼20% and decreased the amount of homorecombinants by ∼60% (Fig. 2B, lanes 7 to 10). Overall, the observation that the ratio of homorecombinants versus heterorecombinants depends on the relative concentrations of the two parental RNAs is consistent with their recombinant nature.
FIG. 2.
Effect of template concentrations on recombinant formation. (A) All in vitro TCV RdRp reactions were done as described in the legend to Fig. 1. Samples in lanes 1 and 2 contained single templates, while the RdRp reactions included two templates in lanes 3 to 10 as shown. The amount of templates used in the RdRp reaction mixtures was 0.5 μg of R3(−) in lanes 3 to 6, while the relative molar ratios between R3(−) and Mot1/pr were 1:1, 1:0.7, 1:0.3, and 1:0.1. The samples in lanes 7 to 10 contained 0.5 μg of Mot1/pr, while the relative molar ratios between Mot1/pr and R3(−) were 1:1.5, 1:2, 1:3, and 1:4, respectively. (B) The relative amounts of heterorecombinants and homorecombinants based on quantification of three independent experiments (such as shown in panel A) are shown. The relative amounts of heterorecombinants and homorecombinants are shown as a percentage in comparison with the amounts of heterorecombinants and homorecombinants obtained with R3(−) and Mot1/pr (the template ratio of 1:1 in lane 3 was chosen as 100%).
The effect of concentration of the recombinant TCV RdRp on the frequency of template-switching events was tested by using various amounts of the enzyme in the RdRp reaction that contained Mot1/pr and AU1 RNAs as templates (data not shown). We found that increasing the amount of the TCV RdRp by 10-fold in the RdRp assay increased the level of RNA synthesis by 4-fold (not shown). Similarly, we observed that the amount of recombinants was increased by ∼4-fold, but changing the concentration of the TCV RdRp did not change the ratio of hetero- versus homorecombinants (data not shown). This suggests that increasing the amount of TCV RdRp in the reaction mixture enhances RNA synthesis and the frequency of template switching.
Efficient template switching by the TCV RdRp on templates with artificial 3′ ends.
To further increase the efficiency of recombination events, we tested the in vitro recombination activity of the AU1 sequence carrying an artificial 3′ sequence (termed art5) (Fig. 3A), which is known to increase primer extension (10). Testing the AU1/art construct alone in the TCV RdRp reaction revealed that it generated homorecombinants efficiently (Fig. 3B, lane 1). This observation confirmed that AU1/art is more efficient in recombination than AU1, which did not generate homorecombinants in detectable amounts (Fig. 1B, lane 3). Interestingly, introduction of the artificial art5 sequence at the 3′ end of R3(−) (Fig. 1A) also increased the recombination frequency between identical R3(−)/art RNAs (results not shown). These observations demonstrate that the art5-containing AU1 and R3(−) RNAs are better templates for recombination studies with the TCV RdRp than the original R3(−) and AU1 templates (Fig. 1A).
When the construct AU1/art was tested in combination with Mot1/pr RNA (the latter was applied in reduced concentration to inhibit recombination between two identical Mot1/pr RNAs [Fig. 3B, lane 2]), then we observed the generation of both homorecombinants (between two identical AU1/art RNAs) and heterorecombinants (Fig. 3B, lane 3). To demonstrate that the sizes of the heterorecombinants depend on the two parent templates, we changed the size of Mot1/pr by deleting 10 nucleotides (nt) from the 3′ end (construct Mot1/prΔ10) and used it together with AU1/art RNA in the TCV RdRp reaction. As expected, the novel heterorecombinants became slightly smaller than that obtained with the longer Mot1/pr RNA (Fig. 3B, compare lanes 3 and 5). Since the putative heterorecombinants were only observed in the two template-containing TCV RdRp reactions, this observation further supports that their formation depends on template-switching events.
Determination of junction sites in the recombinant RNAs.
To obtain evidence that the heterorecombinants are indeed formed between the two templates during the in vitro RdRp reaction, we isolated the putative heterorecombinant products from denaturing gels, followed by RT-PCR analysis, cloning, and sequencing (Fig. 3C). Two sets of primers for RT-PCR were designed to detect either AU1/art-to-Mot1/pr or Mot1/pr-to-AU1/art template switches as shown schematically in Fig. 3C. The expected recombinant-sized RT-PCR products were detected for both combinations of primer pairs (top and bottom gels in Fig. 3D, lane 3), suggesting that both templates served as donors as well as acceptors during the template-switching events. To exclude that the obtained RT-PCR products are artifacts of RT-PCR amplification, we performed the following three control reactions. The first two sets of control RT-PCRs included RdRp samples obtained with single templates (AU1/art or Mot1/pr alone [Fig. 3D, lanes 1 and 2]). To obtain these control samples, the RdRp products from the single-template-containing RdRp reactions were run on the same denaturing gels as the double-template-containing samples. The corresponding areas of the gel, which contained the heterorecombinants in the double-template-containing samples (Fig. 3B, lane 3), were cut in case of single-template-containing samples (Fig. 3B, lanes 1 to 2), and the RdRp products were recovered from the gel. The obtained RdRp products were then used in RT-PCRs using the same primer sets as shown in Fig. 3D. For the third control reaction, we mixed the above gel-isolated RdRp products from single-template-containing RdRp reactions (Fig. 3D, lanes 1 and 2) prior to the RT-PCR. Yet, no RT-PCR products similar to the heterorecombinants were observed in these control experiments (Fig. 3D, lanes 1, 2, and 4). Overall, the lack of recombinant-sized RT-PCR products in the control experiments makes it unlikely that RT-PCR was responsible for the generation of the heterorecombinants observed in mixed-template RdRp assays (Fig. 3D, lane 3).
Sequencing of the cloned RT-PCR products of the heterorecombinants demonstrated that most of the recombinants (73%) were the result of end-to-end template switching (Fig. 3E). The remaining portion of the recombinants was due to end-to-internal or internal-to-end switches. The recombinants also contained nontemplated nucleotides at the junctions (data not shown).
We also RT-PCR amplified (results not shown), cloned, and sequenced gel-isolated homorecombinants generated with the AU1/art construct in the TCV RdRp reaction (Fig. 3B, lane 1). Similar to the heterorecombinants, most of the homorecombinants also contained extra nucleotides at the junction sites and were the result of end-to-end template switching (Fig. 3F). The overall similarity between the homo- and heterorecombinants suggests that they were generated by the same template-switching mechanism.
Selection of the donor versus the acceptor RNAs by the recombinant TCV RdRp during in vitro template-switching events.
The RT-PCR analysis of heterorecombinants (Fig. 3) indicated that both Mot1/pr and AU1/art RNAs could serve as donors as well as acceptors during the template-switching events. To test quantitatively which of the two RNAs are favored to serve more frequently as donors during the template-switching events, we have developed a method to purify the recombinant RNAs based on labeling the donor templates with biotin prior to the RdRp reaction. This method is based on the fact that RNA synthesis is initiated by the recombinant TCV RdRp via primer extension (Fig. 1) (52). Thus, the recombinant RNA is expected to be linked covalently to the donor template but not to the acceptor template, as shown schematically in Fig. 4A. In addition, the RdRp products were labeled with [32P]UTP during the RdRp reactions. After the RdRp reaction, the biotin-labeled RdRp products were isolated using streptavidin-coated tubes (see Materials and Methods). As schematically shown in Fig. 4A, when one of the templates is biotin labeled, then those recombinants should be detected after the streptavidin-based purification step that are formed using the biotin-labeled template as the donor RNA (Fig. 4A). In contrast, those recombinants that were obtained with nonbiotinylated donor RNA should not be detected after streptavidin-based purification (Fig. 4A, right panel). When both templates are biotin labeled, then we should detect both types of recombinants (we call this total recombination).
Based on the above strategy to capture biotinylated recombinant RNAs, we compared donor versus acceptor RNA selection by the recombinant TCV RdRp. These studies revealed that the designed biotin-streptavidin-based purification (Fig. 4A) indeed led to the recovery of primer extension and homorecombination products in single-template-containing samples (Fig. 4B, lanes 1 and 2). The two-template-containing RdRp reactions also resulted in heterorecombinant-sized products after the biotin-streptavidin-based purification step (Fig. 4B, lanes 3 to 8). Comparison of the amounts of heterorecombinants obtained with doubly biotin-labeled templates (100% total recombination; Fig. 4B, lane 3) and singly labeled templates revealed that the AU1/art RNA (Fig. 4B, lane 4) was used as a donor in ∼60%, while Mot1/pr RNA (Fig. 4B, lane 5) was used as a donor in ∼40% of total heterorecombinants (Fig. 4B, lane 3) under the conditions used (i.e., reduced concentration of Mot1/pr to favor heterorecombination [Fig. 2]).
The observation that the sum of recombination with single biotin-labeled templates (60 and 40% of recombination frequency) (Fig. 4B, lanes 4 and 5) was comparable with the amount of recombinants obtained with the doubly biotin-labeled templates (100% in lane 3) suggests the radioactive bands in Fig. 4B are not derived from contamination left behind due to incomplete washing of the tubes after binding of the biotin to streptavidin. Another piece of evidence against possible contamination is the loss of the abundant primer extension product obtained from the non-biotin-labeled AU1/art template (Fig. 4B, lane 5). In contrast, the band representing the heterorecombinant formed between AU1/art and Mot1/pr (biotin labeled) was easily detectable after the purification step (lane 4). In addition, a longer run of the heterorecombinants in denaturing gels revealed that the heterorecombinants recovered after the biotin-streptavidin-based purification were different in size, depending on the donor RNA (Fig. 4B, right panel, lanes 4 and 5; see the slight difference in migration of the heterorecombinants after purification). This is consistent with the difference in size between the two templates (Fig. 4A; note that the donor sequence is represented twice in the recombinants due to primer extension). The last piece of evidence against the role of contamination was obtained with a construct (GC1/art; Fig. 4C) that contains a GC-rich hairpin (Fig. 1A) in place of the AU-rich hairpin in AU1/art (Fig. 3A). The remaining sequences were the same in AU1/art and GC1/art (Fig. 3A). The use of Mot1/pr in combination with the GC1/art RNA resulted in a small amount of heterorecombinants (fivefold less than that observed with AU1/art [data not shown]) when both Mot1/pr and GC1/art RNAs were biotin labeled (i.e., Fig. 4C, lane 2, total amount of heterorecombinants). When GC1/art was the only biotin-labeled RNA in the RdRp reaction, then we could not detect heterorecombinants after purification. This suggests that GC1/art RNA is not used as a donor RNA by the recombinant TCV RdRp at a detectable level under the experimental conditions used. In contrast, biotin labeling of Mot1/pr RNA resulted in as much heterorecombinants (Fig. 4C, lane 4) as that shown for the total heterorecombinants (lane 2), suggesting that the donor RNA was Mot1/pr in ∼100% of template-switching events (Fig. 4C), while the GC1/art RNA was used, albeit inefficiently, only as an acceptor RNA during the template-switching events. A more detailed analysis of the role of various sequences will be the scope of future work. Importantly, the lack of heterorecombinants in the sample containing biotin-labeled GC1/art and unlabeled Mot1/pr after the purification excludes the possibility of contamination with the non-biotin-labeled heterorecombinant formed between Mot1/pr (donor) and GC1/art (acceptor) in the same RdRp reaction (Fig. 4A). Based on the above observations, we conclude that the developed biotin-streptavidin-based purification is suitable for selective purification of heterorecombinants that share the same donor template.
To test what factors, in addition to the previously tested template concentration effect (Fig. 2), might influence the selection of templates as donors, we preincubated the biotinylated AU1/art RNA with the recombinant TCV RdRp in the presence of only three nucleotides (ATP, CTP, and GTP) for 10 min. This step was expected to allow the RdRp to bind to the AU1/art template, thus possibly increasing its chance of being used as a donor RNA. After the preincubation step, the Mot1/pr template was added to the RdRp reaction together with the missing nucleotide, followed by incubation to allow the RdRp to finish RNA synthesis and to switch template. As expected, the above conditions increased the use of the AU1/art template as donor by 30% (Fig. 4B, lanes 6 and 7) when compared to the above experiments in which the two templates were added and incubated simultaneously in the RdRp assay (Fig. 4B, lanes 3 to 5). Preincubation of the RdRp reaction with unlabeled AU1/art under the same conditions followed by addition of biotin-labeled Mot1/pr RNA resulted in reduction of the use of Mot1/pr to 20% as a donor in the template-switching reactions (Fig. 4B, lane 8). This observation confirmed that preincubation of the RdRp with a template could enhance the chance for that template to serve as a donor during the template-switching events.
Evidence supporting template switching and excluding RNA ligation as the mechanism of recombinant formation in vitro.
The above experiments indicated that the likely mechanism of recombinant formation in vitro is the RdRp-driven template-switching (copy-choice) mechanism. In contrast, the simple RNA breakage-ligation model (11) (Fig. 5A), which involves breakage and ligation of the input single-stranded templates prior to cRNA synthesis by the viral RdRp, is unlikely to contribute to the formation of RNA recombinants. This is because this type of RNA breakage and ligation would lead to the generation of recombinant templates, which results in RdRp products that are close to double and/or four times the size of the input templates (depending on de novo or primer extension mode of initiation by the RdRp) (Fig. 5A, model A1). However, we have not detected these recombinant RNAs in the above experiments (Fig. 1 to 4 and data not shown). Instead, the observed recombinant RNAs were approximately 3 times larger than the original templates (Fig. 3C and E). Therefore, it is unlikely that this simple RNA breakage-ligation (occurring prior to RNA synthesis) mechanism would contribute significantly to recombinant RNA formation in vitro.
FIG. 5.
Testing of whether the recombinants are generated by template switching or RNA ligation. (A) Schematic representation of the three most likely mechanisms that might lead to formation of recombinants shown in Fig. 1 to 4. (A1) If RNA ligation (due to RNA ligase activity, shown as a black circle) occurs between the two heterologous templates prior to RNA synthesis, then the RNA product synthesized by the RdRp (gray oval circle) would be approximately double or four times larger than the input templates (if initiation takes place via de novo initiation [data not shown] or primer extension [as indicated in the figure]). Since the observed recombinants were approximately three times larger than the input templates in the above experiments (Fig. 1 to 4), this mechanism cannot explain the observed recombinants in the in vitro system and it was not tested further. Dotted lines represent the newly synthesized labeled RNAs. (A2) If RNA ligation between the heterologous RNAs occurs after the RdRp completes RNA synthesis on the input templates, then, depending on ligation involving one strand (as shown) or two strands (data not shown), recombinants either three or four times larger than the input RNAs are generated. Note that RNA synthesis by the TCV RdRp must initiate de novo on the template in the right panel in order to generate a substrate for RNA ligation that could generate the recombinants observed in this work (Fig. 3C to E). Since this mechanism could potentially generate the correct-sized recombinants, we tested this possibility as described below for panel B. (A3) Recombinants may also be generated by template switching of the RdRp as shown, resulting in the correct-sized recombinants. (B) Schematic description of the procedure used to test RNA ligation in vitro. See details in Materials and Methods. Note that the RdRp products were labeled with [32P]UTP independently in reactions 1 and 2 (top). (C) Denaturing PAGE analysis of the RdRp products after the RNA ligation test. Samples in lanes 1 to 3 were prepared as shown in panel B, while samples in lanes 4 to 6 were obtained as for Fig. 3B. The ligation reactions were performed in the presence of 10 mM ATP (lane 1), 10 mM ATP, CTP, and GTP (lane 2), or no ribonucleotides (lane 3). The expected heterorecombination product is marked with an arrow.
A more complex RNA breakage-ligation model (Fig. 5A, model A2), however, cannot be ruled out without further experiments. For example, the RNA breakage and ligation might occur after the RNA synthesis being completed by the RdRp. In this model, the RdRp first performs RNA synthesis on the two templates independently and simultaneously. After termination of RNA synthesis by the RdRp, a putative RNA ligase might ligate the strands of the RdRp products to generate recombinants. If the two newly synthesized RNA strands were ligated together (as shown schematically in Fig. 5A, model A2), then RNA ligation would lead to recombinants that are similar to those described in Fig. 1 to 4.
To test if the above RNA breakage-ligation mechanism (Fig. 5A, model A2) could generate recombinants in our RdRp assay, first we made two independent TCV RdRp reactions, each reaction containing only one template (see Fig. 5B for a schematic description of the assay). This allowed for the 32P labeling of the RdRp products but excluded the possibility of RNA recombination (i.e., formation of heterorecombinants). After the separate RdRp reactions were completed, we removed the nucleotides by pushing the RdRp products through size-exclusion columns. Then, we mixed the 32P-labeled RdRp products from the two RdRp reactions and added the recombinant TCV RdRp, the reaction buffer, and three of the four ribonucleotides (see Materials and Methods). The idea is that RNA ligation, which might be mediated by either a contaminating ligase or the RdRp itself, should be possible under these conditions, while extensive RNA synthesis and template switching by the RdRp should not be possible, due to the absence of a ribonucleotide. These RNA ligation experiments failed to detect RNA recombinants (i.e., heterorecombinants), while the expected recombinants were readily detected in the control RdRp reactions that contained both Mot1/pr and AU1 templates simultaneously in the presence of all four ribonucleotides (Fig. 5C, compare lanes 1 to 3 versus 4). Based on these data, we conclude that the RNA breakage-ligation mechanism is unlikely to contribute to the formation of recombinants observed in vitro under the conditions used.
RNA recombination driven by the CNV RdRp purified from plants.
To obtain evidence that the CNV RdRp is capable of template switching in vitro, we used a partially purified CNV RdRp preparation obtained from plants. This preparation contains both the RdRp protein (i.e., p92) and the small replicase protein, p33 (J. Pogany and P. D. Nagy, unpublished results). The above CNV RdRp is a functional polymerase under the same conditions as the recombinant TCV RdRp (45, 52).
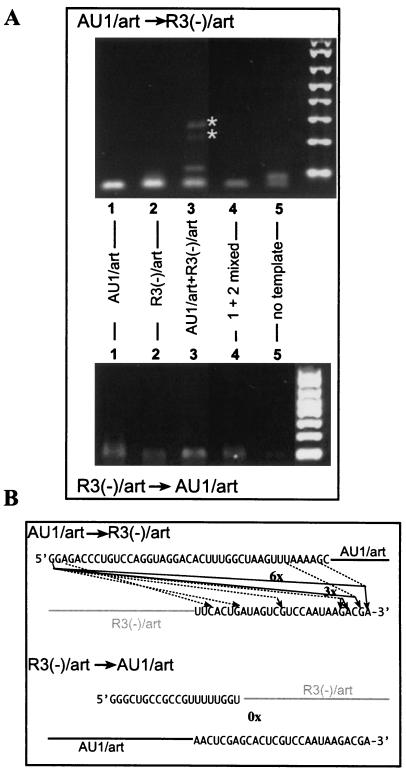
To test template switching by the CNV RdRp, we selected templates that we used in the above recombination assays with the recombinant TCV RdRp. Namely, constructs AU1/art (Fig. 3A) and R3(−)/art, which contains the tombusvirus-derived replication enhancer (49, 55), and the artificial art5 sequence (Fig. 3A and reference 10), which promotes efficient primer extension in vitro (10). We found that CNV RdRp reactions containing standard or reduced amounts of ribonucleotides and the two templates did not result in heterorecombinant-sized RNAs at detectable levels (data not shown), while the primer extension products were easily detectable (10). Similarly, denaturing gel analysis of single-template-containing CNV RdRp reactions did not reveal formation of homorecombinants in detectable amounts (data not shown and reference 10). This suggests that recombinant formation with the CNV RdRp is inefficient with these two templates under the conditions used. However, RT-PCR analysis of the gel-isolated RdRp products obtained from the portion of denaturing gels that contained heterorecombinants in the TCV RdRp reactions revealed recombinant-sized products only from CNV RdRp reactions that contained both templates (Fig. 6A, lane 3). Interestingly, the recombinant RT-PCR products were observed only with the primer set that detects AU1/art-to-R3(−)/art recombinants (Fig. 6A, top gel). We suggest that the recombinants were generated by the CNV RdRp, since mixing the gel-isolated CNV RdRp products from single-template-containing reactions did not result in recombinant-sized RT-PCR products (Fig. 6A, lane 4). This excludes the possibility that RT-PCR was responsible for generating the recombinant-sized products.
FIG. 6.
RT-PCR and sequence analysis of the template-switching products obtained with a partially purified CNV RdRp preparation. (A) Agarose gel electrophoresis of the RT-PCR products obtained from CNV RdRp reactions. The names of the templates used for the RdRp reactions are shown above the lanes. The sequences of the templates are the same as in Fig. 1 and 3 [except R3(−) carried the art5 sequence at the 3′ end (Fig. 3A)]. To obtain the RNA samples for the RT-PCR analysis, we cut a portion of the denaturing gel that should contain the recombinant-sized products (larger than the primer extension products) and used it for RNA isolation. Sample 4 contained the RdRp products from samples 1 and 2 mixed prior to the RT-PCR (control reaction). The RT-PCR method used is similar to that described for Fig. 3C and D, except that the primer sets used were different (see Materials and Methods). Asterisks depict the predicted-sized heterorecombination products. Note that the bottom gel does not show any recombinant-sized product, suggesting that the recombination is inefficient from R3(−)/art to AU1/art. (B) Sequence analysis of the heterorecombinants after RT-PCR and cloning. Those recombinants found only once are depicted with dotted lines, while recombinants with the same junctions are depicted with solid lines with the numbers indicating their frequencies in the cloned library. Many clones contained extra, nontemplated nucleotides at the junctions (data not shown).
Cloning and sequencing of the RT-PCR products obtained from the above CNV RdRp reactions (Fig. 6A, lane 3) revealed that the CNV RdRp frequently generates end-to-end recombinants (6 out of 15). In addition, we also observed that template switching to internal locations is also common with the CNV RdRp (Fig. 6B).
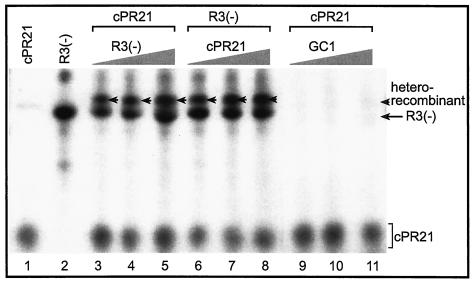
Since the CNV RdRp obtained from plants is capable of efficient de novo initiation (45, 47, 48), we also tested RNA recombination with templates that promote de novo initiation. The two templates chosen were (i) construct R3(−) (Fig. 1A), with the region III(-) replication enhancer sequence and a minimal tombusvirus promoter (cPR11) (47); and (ii) the 21-nt-long cPR21, which contains the tombusvirus extended complementary (plus-strand initiation) promoter sequence (47). Both templates were capable of supporting de novo initiation by the CNV RdRp as shown in Fig. 7 (lanes 1 and 2). The presence of both templates in the CNV RdRp reaction, however, resulted in the appearance of a novel, recombinant-sized band (Fig. 7, lane 3). Increasing the amounts of R3(−) (lanes 3 to 5) and cPR21 (lanes 6 to 8) separately in the CNV RdRp reactions increased the amounts of recombinant-sized RdRp products, indicating that the recombination events are dependent on the concentration of the templates in the reaction mixture. Testing an additional template, namely GC1 (Fig. 1A), which is similar to the AU1 template except that it contains a GC-rich hairpin, in combination with cPR21 revealed that GC1 is a poor template for recombination (Fig. 7, lanes 9 to 11), even in high concentrations. This suggests that the CNV RdRp has the ability to select among the templates during the template-switching events (compare Fig. 6 and 7). RT-PCR analysis of the recombinants from the above experiments (Fig. 7) failed in spite of our extensive efforts, probably due to the difficulty in annealing the primer complementary to the cPR21 sequence in the recombinants.
FIG. 7.
A denaturing PAGE analysis of heterorecombinants generated with the CNV RdRp preparation. A chemically synthesized 21-mer RNA, called cPR21, which represents the very 3′-terminal promoter sequence in the minus-stranded TBSV (47), is used in combination with two other RNA templates (Fig. 1) as shown. The amounts of RNAs used for the CNV RdRp reactions were as follows: cPR21 and R3(−) were 300 and 45 pmol in lanes 1 and 2, respectively. The samples in lanes 3 to 5 contained 300 pmol of cPR21 and increasing amounts of R3(−) (15, 45, and 75 pmol, respectively); in lanes 6 to 8, the amount of R3(−) was fixed (45 pmol), while that of cPR21 increased progressively (100, 300, and 500 pmol, respectively). The samples in lanes 9 to 11 contained 300 pmol of cPR21 and increasing amounts of GC1 (15, 45, and 75 pmol, respectively). The heterorecombinants are marked with arrowheads. The long arrow indicates the de novo initiation products of R3(−). The GC1 RNA resulted in an almost-undetectable amount of de novo and recombinant-sized products. Note that the CNV RdRp preparation initiates de novo on these templates.
DISCUSSION
Development of efficient recombination assays with the recombinant TCV RdRp and the plant-purified CNV RdRp extends previous in vitro works that were based on primer extension on templates (10, 39-41). The previous primer extension studies contributed to our understanding of the so-called late steps in recombination (37), which might be similar to the recombination steps occurring during and/or after template switching, such as binding of the RdRp to the acceptor template and initiation (resumption) of RNA synthesis on the acceptor template via a primer. In contrast, the recombinant assays presented in this work also included the donor as well as the acceptor RNAs. This made the present recombination assays more complex than the previous primer extension assay, since the present tests also include initiation (via either primer extension or de novo), elongation, pausing or termination, and template switching (37). Nevertheless, we observed many similarities among the results. For example, templates containing the replication enhancers were the best templates, while an AU-rich sequence was better template than a GC-rich sequence in both primer extension and template-switching reactions (10). In addition, the recombinants isolated in this work (Fig. 3 and 6) did not contain extensive sequence similarities at the junction sites, and the primer extension studies revealed that the TCV and CNV RdRps can extend on primers with no or only limited base-pairing between the primer sequence and the acceptor template (10, 39-41, 59, 70).
The template-switching mechanism of recombination versus RNA breakage and ligation.
Previous in vitro primer extension studies with partially purified TCV and CNV RdRp preparations supported the template-switching mechanism of RNA recombination for these viruses (10, 39-41). In this work, we provide further evidence supporting the template-switching mechanism, while our data do not support the role of an RNA breakage-ligation mechanism in the recombination events in vitro. For example, a simple RNA breakage-ligation mechanism, which might occur between the input templates prior to RNA synthesis by the RdRp, would generate recombinants that are different in size and sequence from those observed in this work in vitro (Fig. 5A, model A1). Moreover, we found no evidence that a more complex version of the RNA breakage-ligation mechanism, which would occur after the RNA synthesis was accomplished by the RdRp (Fig. 5A, model A2), could lead to the expected recombinants obtained under conditions which should favor ligation but that interfere with template switching (Fig. 5). Although we cannot completely rule out the contribution of RNA breakage and ligation to recombination events, we propose that template switching by the TCV and CNV RdRps is the mechanism leading to recombinant formation.
Similarities between in vivo and in vitro recombination experiments.
Although the above experiments with the recombinant TCV RdRp strongly supported the template-switching model, it is important to analyze if similar mechanism might also operate in vivo. This possibility is supported by the following observations: (i) the replication enhancer-containing RNA templates are more efficient in the in vitro template-switching reactions for both the TCV RdRp (i.e., the motif1-hairpin in Mot1/pr [Fig. 1B]) and the CNV RdRp [region III(-) sequence in R3(−) (Fig. 7)] than other templates lacking the replication enhancers. Accordingly, the motif1-hairpin replication enhancer was also found to facilitate RNA recombination in vivo between two satellite RNAs associated with TCV (8) and region III sequence constitute a recombination hot spot in TBSV-associated DI RNAs in plant protoplasts (Z. Panaviene and P. D. Nagy, unpublished results). (ii) RNA recombinants with junctions at the end or near to the end of the templates are frequently isolated both in vitro (Fig. 3 and 6) and in vivo in the case of subviral RNAs that are associated with carmo- and tombusvirus infections (7, 8, 16, 69). (iii) The recombination sites can include internal positions in the templates (8, 69). (iv) There is no absolute need for the presence of sequence identity or similarity between the templates for RNA recombination to occur in vitro or in vivo (true for both carmo- or tombusviruses) (37). (v) The recombination sites frequently contain extra, nontemplated nucleotides in in vitro and in vivo recombinants (7, 8, 69). These common features support the model that RNA recombination events are mediated by the same RdRp-driven template-switching mechanism both in vivo (8, 69) and in vitro (this work).
Differences between in vivo and in vitro recombination experiments.
In spite of the similar observations, there are also notable differences between the in vitro and in vivo conditions and the obtained data. These include the following: first, the motif1-hairpin replication enhancer of satC (a chimeric satellite RNA associated with TCV) is proposed to serve primarily as an acceptor in recombination in vivo (8), while templates containing the motif1-hairpin replication enhancer (construct Mot1/pr [Fig. 1]) can be used both as a donor and an acceptor in the in vitro assay (Fig. 4). This difference in template use in recombination may come from the differences in the conditions between the in vivo and in vitro assays. For example, in the in vitro assay, the templates were added at the same time (Fig. 1), which could favor the binding of template containing the motif1-hairpin replication enhancer to the TCV RdRp when compared with other templates that bind less efficiently than the motif1-hairpin replication enhancer (8, 39-41). Therefore, it is likely that the TCV RdRp frequently has the opportunity to use the motif1-hairpin replication enhancer-containing template as a donor in vitro. It is not surprising that the motif1-hairpin replication enhancer can also facilitate the use of the template as an acceptor, since the motif1-hairpin may facilitate the efficient binding to the TCV RdRp (which possibly carries the primer) during the “jumping events.” On the contrary, the in vivo recombination assay was based on recombination that included a mutated satC RNA which did not replicate at a detectable level (7, 8). In contrast, the donor RNA was an efficiently replicating satD RNA (another satellite RNA that is related to the 5′ half of the satC genome) (8). Therefore, the more-abundant satD RNA might have a better chance to bind to the TCV RdRp and serve as a donor than the mutated low-abundant satC. Accordingly, we found that preincubation of the less-efficient AU1/art template with the recombinant TCV RdRp or reduction of the amount of Mot1/pr (the motif1 hairpin-containing template), which facilitated the binding of AU1 to the RdRp prior to the addition of the more efficient Mot1/pr template, enhanced the use of AU1/art as a donor RNA (Fig. 2 and 4B).
The second difference between the in vivo and in vitro recombination assays is that we have used shorter templates in vitro than those used in vivo, which were full-length or close to full-length sized. The use of long templates would reduce the sensitivity of the in vitro assay and decreasing resolution of the RdRp products in gels, while our goal was to visualize the recombination products directly in the gels (Fig. 1 and 7). The advantage of direct visualization of recombinant products in the gels is that it allows for isolation of the RdRp products from the gel prior to their analysis with RT-PCR. This approach prevented the generation of RT-PCR artifacts (Fig. 3). It is likely that long templates could be used for RNA recombination by the carmo- and tombusvirus RdRp's in vitro, generating recombinants that are basically similar to the recombinants obtained in this work. Indeed, recombinants with full-length poliovirus RNAs were obtained in vitro, although RT-PCR was needed to detect these recombinants (63). It is also interesting that Kim and Kao (25) observed the highest frequency of template switching when using short RNA templates of 8 to 15 nt in length. The reason for the enhanced template-switching activity of the RdRp on short templates is that the RdRp is operating in “initiation mode” on the short templates (24). In contrast, the longer-than-15-nt templates used in this work should favor the RdRp carrying out most of the RNA synthesis in “elongation mode,” which is a significantly more stable association between the template and the RdRp (24). This is possibly the reason that we observed multimeric recombinants with lower frequencies than that described for the RdRp of bovine viral diarrhea virus (25). It is also highly likely that different viral RdRps may support template-switching with different efficiencies as demonstrated for the BMV and cucumber mosaic virus RdRps in vitro (13, 15, 25, 34).
The third difference between the in vitro and in vivo assays is that RNA transcription was initiated by primer extension on the templates in our in vitro recombinant TCV RdRp assay (Fig. 1). In contrast, initiation of RNA synthesis occurs de novo (without a primer) in the TCV-infected plants. However, the difference in the mode of initiation should not affect the actual mechanism of template switching, since the RdRp is predicted to jump template not during initiation but rather during elongation or during pausing or termination (37). Irrespective of the mode of initiation, the RdRp should use the primer derived from transcription on the donor RNA for resuming synthesis on the acceptor RNA. Accordingly, we observed that the CNV RdRp generated RNA recombinants regardless of the mode of initiation (i.e., primer extension in Fig. 6 and de novo initiation in Fig. 7). The major effect of the mode of initiation on RNA recombination is predicted to be indirect, and it would affect the actual frequency of the template-switching events. For example, efficient RNA initiation is expected to lead to the generation of ample primers (nascent RNA products), while inefficient initiation should lead to small amounts of primer. Larger amounts of primers are predicted to increase the chance and thus the frequency of recombination events. Indeed, the CNV RdRp, which favors de novo initiation over primer extension, supported generation of recombinants more efficiently when promoter sequences (to promote de novo initiation) were present in the templates than when in the presence of self-priming sequences in the templates (compare Fig. 6 and 7). The opposite is true for the recombinant TCV RdRp, which favored primer extension even in the presence of promoter sequences (Fig. 1). Based on these observations, we suggest that the primary effect of initiation is on the amount of putative primers generated. We cannot completely exclude other alternative models, such as the absence of additional factors in our in vitro assays that might affect the precision of RNA synthesis in vivo.
The fourth major difference between the in vitro and in vivo recombination experiments is the existence of postrecombinational selection in the in vivo systems. This model of selection for the best-fit recombinants predicts that the best-adapted (the most efficiently replicating) recombinants will be overrepresented in the recombination pool, since they will be amplified to a greater extent than the poorly adapted recombinants. Competition among different recombinants is indeed a major factor which affects interpretation of in vivo results, as demonstrated for tombusviruses, BMV, and coronaviruses (3, 32, 44, 66, 67). It is important that the effect of the selection pressure on recombinant accumulation can be highly variable from one viral or host system to another (9), as demonstrated for TCV-associated satellite RNAs, which showed remarkable variations among the recombinants recovered from plants (7). In contrast to the possible major effect of selection pressure on recombinant RNA accumulation in vivo, the role of selection pressure in the in vitro RdRp assays is predicted to be insignificant. This is because the detected recombinant RNAs in the gels are expected to be the original recombinant RNAs (we do not have evidence supporting postrecombinational replication in vitro [data not shown]). Therefore, we believe that the in vitro-generated recombinants might represent the recombination pool fairly. This is expected to be a major advantage for studying the mechanism of RNA recombination in vitro (23, 25).
In summary, our results firmly establish the carmo- and tombusvirus recombination assays for in vitro analysis of recombinants. This will help future studies on dissecting RNA elements and protein factors which could affect recombination events in these viruses.
Acknowledgments
We thank Judit Pogany and K. S. Rajendran for their critical comments.
This work was supported by the National Science Foundation (MCB0078152) and by the National Institutes of Health (to P.D.N.).
Footnotes
This study is publication no. 03-12-043 of the Kentucky Agricultural Experiment Station.
REFERENCES
- 1.Arnold, J. J., and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 274:2706-2716. [DOI] [PubMed] [Google Scholar]
- 2.Ayllon, M. A., C. Lopez, J. Navas-Castillo, M. Mawassi, W. O. Dawson, J. Guerri, R. Flores, and P. Moreno. 1999. New defective RNAs from citrus tristeza virus: evidence for a replicase-driven template switching mechanism in their generation. J. Gen. Virol. 80:817-821. [DOI] [PubMed] [Google Scholar]
- 3.Banner, L. R., and M. M. Lai. 1991. Random nature of coronavirus RNA recombination in the absence of selection pressure. Virology 185:441-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becher, P., M. Orlich, and H. J. Thiel. 2001. RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease. J. Virol. 75:6256-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biebricher, C. K., and R. Luce. 1992. In vitro recombination and terminal elongation of RNA by Q beta replicase. EMBO J. 11:5129-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borja, M., T. Rubio, H. B. Scholthof, and A. O. Jackson. 1999. Restoration of wild-type virus by double recombination of tombusvirus mutants with a host transgene. Mol. Plant-Microbe Interact. 12:153-162. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter, C. D., J. W. Oh, C. Zhang, and A. E. Simon. 1995. Involvement of a stem-loop structure in the location of junction sites in viral RNA recombination. J. Mol. Biol. 245:608-622. [DOI] [PubMed] [Google Scholar]
- 8.Cascone, P. J., T. F. Haydar, and A. E. Simon. 1993. Sequences and structures required for recombination between virus-associated RNAs. Science 260:801-805. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y. C., M. Borja, H. B. Scholthof, A. O. Jackson, and T. J. Morris. 1995. Host effects and sequences essential for accumulation of defective interfering RNAs of cucumber necrosis and tomato bushy stunt tombusviruses. Virology 210:41-53. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, C. P., J. Pogany, and P. D. Nagy. 2002. Mechanism of DI RNA formation in tombusviruses: dissecting the requirement for primer extension by the tombusvirus RNA-dependent RNA polymerase in vitro. Virology 304:460-473. [DOI] [PubMed] [Google Scholar]
- 11.Chetverin, A. B., H. V. Chetverina, A. A. Demidenko, and V. I. Ugarov. 1997. Nonhomologous RNA recombination in a cell-free system: evidence for a transesterification mechanism guided by secondary structure. Cell 88:503-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desport, M., M. E. Collins, and J. Brownlie. 1998. Genome instability in BVDV: an examination of the sequence and structural influences on RNA recombination. Virology 246:352-361. [DOI] [PubMed] [Google Scholar]
- 13.Dzianott, A., N. Rauffer-Bruyere, and J. J. Bujarski. 2001. Studies on functional interaction between brome mosaic virus replicase proteins during RNA recombination, using combined mutants in vivo and in vitro. Virology 289:137-149. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Cuartero, B., J. Burgyan, M. A. Aranda, K. Salanki, E. Moriones, and F. Garcia-Arenal. 1994. Increase in the relative fitness of a plant virus RNA associated with its recombinant nature. Virology 203:373-377. [DOI] [PubMed] [Google Scholar]
- 15.Figlerowicz, M., P. D. Nagy, and J. J. Bujarski. 1997. A mutation in the putative RNA polymerase gene inhibits nonhomologous, but not homologous, genetic recombination in an RNA virus. Proc. Natl. Acad. Sci. USA 94:2073-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finnen, R. L., and D. M. Rochon. 1995. Characterization and biological activity of DI RNA dimers formed during cucumber necrosis virus coinfections. Virology 207:282-286. [DOI] [PubMed] [Google Scholar]
- 17.Furuya, T., T. B. Macnaughton, N. La Monica, and M. M. Lai. 1993. Natural evolution of coronavirus defective-interfering RNA involves RNA recombination. Virology 194:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbs, A. 1987. Molecular evolution of viruses; “trees,” “clocks” and “modules.” J. Cell Sci. Suppl. 7:319-337. [DOI] [PubMed] [Google Scholar]
- 19.Gmyl, A. P., E. V. Belousov, S. V. Maslova, E. V. Khitrina, A. B. Chetverin, and V. I. Agol. 1999. Nonreplicative RNA recombination in poliovirus. J. Virol. 73:8958-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajjou, M., K. R. Hill, S. V. Subramaniam, J. Y. Hu, and R. Raju. 1996. Nonhomologous RNA-RNA recombination events at the 3′ nontranslated region of the Sindbis virus genome: hot spots and utilization of nonviral sequences. J. Virol. 70:5153-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, K. R., M. Hajjou, J. Y. Hu, and R. Raju. 1997. RNA-RNA recombination in Sindbis virus: roles of the 3′ conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching. J. Virol. 71:2693-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvis, T. C., and K. Kirkegaard. 1991. The polymerase in its labyrinth: mechanisms and implications of RNA recombination. Trends Genet. 7:186-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarvis, T. C., and K. Kirkegaard. 1992. Poliovirus RNA recombination: mechanistic studies in the absence of selection. EMBO J. 11:3135-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 25.Kim, M. J., and C. Kao. 2001. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc. Natl. Acad. Sci. USA 98:4972-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirkegaard, K., and D. Baltimore. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai, M. M. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 56:61-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y., and L. A. Ball. 1993. Nonhomologous RNA recombination during negative-strand synthesis of flock house virus RNA. J. Virol. 67:3854-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao, C. L., and M. M. Lai. 1992. RNA recombination in a coronavirus: recombination between viral genomic RNA and transfected RNA fragments. J. Virol. 66:6117-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mindich, L., X. Qiao, S. Onodera, P. Gottlieb, and J. Strassman. 1992. Heterologous recombination in the double-stranded RNA bacteriophage phi 6. J. Virol. 66:2605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molenkamp, R., B. C. Rozier, S. Greve, W. J. Spaan, and E. J. Snijder. 2001. Characterization of an arterivirus defective interfering RNA. Replication and homologous recombination. Adv. Exp. Med. Biol. 494:519-525. [DOI] [PubMed] [Google Scholar]
- 32.Nagy, P. D., and J. J. Bujarski. 1992. Genetic recombination in brome mosaic virus: effect of sequence and replication of RNA on accumulation of recombinants. J. Virol. 66:6824-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy, P. D., and J. J. Bujarski. 1993. Targeting the site of RNA-RNA recombination in brome mosaic virus with antisense sequences. Proc. Natl. Acad. Sci. USA 90:6390-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagy, P. D., A. Dzianott, P. Ahlquist, and J. J. Bujarski. 1995. Mutations in the helicase-like domain of protein 1a alter the sites of RNA-RNA recombination in brome mosaic virus. J. Virol. 69:2547-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy, P. D., and J. J. Bujarski. 1995. Efficient system of homologous RNA recombination in brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J. Virol. 69:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy, P. D., and J. J. Bujarski. 1996. Homologous RNA recombination in brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J. Virol. 70:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagy, P. D., and A. E. Simon. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 38.Nagy, P. D., and J. J. Bujarski. 1997. Engineering of homologous recombination hot spots with AU-rich sequences in brome mosaic virus. J. Virol. 71:3799-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagy, P. D., C. Zhang, and A. E. Simon. 1998. Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J. 17:2392-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagy, P. D., and A. E. Simon. 1998. In vitro characterization of late steps of RNA recombination in turnip crinkle virus. I. Role of motif1-hairpin structure. Virology 249:379-392. [DOI] [PubMed] [Google Scholar]
- 41.Nagy, P. D., and A. E. Simon. 1998. In vitro characterization of late steps of RNA recombination in turnip crinkle virus. II. The role of the priming stem and flanking sequences. Virology 249:393-405. [DOI] [PubMed] [Google Scholar]
- 42.Nagy, P. D., and J. J. Bujarski. 1998. Silencing homologous RNA recombination hot spots with GC-rich sequences in brome mosaic virus. J. Virol. 72:1122-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagy, P. D., J. Pogany, and A. E. Simon. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 18:5653-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagy, P. D., C. Ogiela, and J. J. Bujarski. 1999. Mapping sequences active in homologous RNA recombination in brome mosaic virus: prediction of recombination hot spots. Virology 254:92-104. [DOI] [PubMed] [Google Scholar]
- 45.Nagy, P. D., and J. Pogany. 2000. Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology 276:279-288. [DOI] [PubMed] [Google Scholar]
- 46.Nagy, P. D., J. Pogany, and A. E. Simon. 2001. In vivo and in vitro characterization of an RNA replication enhancer in a satellite RNA associated with turnip crinkle virus. Virology 288:315-324. [DOI] [PubMed] [Google Scholar]
- 47.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Analysis of minimal promoter sequences for plus-strand synthesis by the Cucumber necrosis virus RNA-dependent RNA polymerase. Virology 296:263-274. [DOI] [PubMed] [Google Scholar]
- 48.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Internal initiation by the cucumber necrosis virus RNA-dependent RNA polymerase is facilitated by promoter-like sequences. Virology 296:275-287. [DOI] [PubMed] [Google Scholar]
- 49.Panavas, T., and P. D. Nagy. 2003. The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis. J. Virol. 77:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrault, J. 1981. Origin and replication of defective interfering particles. Curr. Top. Microbiol. Immunol. 93:151-207. [DOI] [PubMed] [Google Scholar]
- 51.Qiao, X., J. Qiao, and L. Mindich. 1997. An in vitro system for the investigation of heterologous RNA recombination. Virology 227:103-110. [DOI] [PubMed] [Google Scholar]
- 52.Rajendran, K. S., J. Pogany, and P. D. Nagy. 2002. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 76:1707-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao, A. L., and T. C. Hall. 1993. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J. Virol. 67:969-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ray, D., and K. A. White. 1999. Enhancer-like properties of an RNA element that modulates tombusvirus RNA accumulation. Virology 256:162-171. [DOI] [PubMed] [Google Scholar]
- 55.Ray, D., and K. A. White. 2003. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol. 77:245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rochon, D. M. 1991. Rapid de novo generation of defective interfering RNA by cucumber necrosis virus mutants that do not express the 20-kDa nonstructural protein. Proc. Natl. Acad. Sci. USA 88:11153-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlesinger, S., and B. G. Weiss. 1994. Recombination between Sindbis virus RNAs. Arch. Virol. Suppl. 9:213-220. [DOI] [PubMed] [Google Scholar]
- 58.Scholthof, K. B., H. B. Scholthof, and A. O. Jackson. 1995. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology 208:365-369. [DOI] [PubMed] [Google Scholar]
- 59.Simon, A. E. 1999. Replication, recombination, and symptom-modulation properties of the satellite RNAs of turnip crinkle virus. Curr. Top. Microbiol. Immunol. 239:19-36. [DOI] [PubMed] [Google Scholar]
- 60.Song, C., and A. E. Simon. 1994. RNA-dependent RNA polymerase from plants infected with turnip crinkle virus can transcribe (+)- and (−)-strands of virus-associated RNAs. Proc. Natl. Acad. Sci. USA 91:8792-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strauss, J. H., and E. G. Strauss. 1988. Evolution of RNA viruses. Annu. Rev. Microbiol. 42:657-683. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki, Y., T. Gojobori, and O. Nakagomi. 1998. Intragenic recombinations in rotaviruses. FEBS Lett. 427:183-187. [DOI] [PubMed] [Google Scholar]
- 63.Tang, R. S., D. J. Barton, J. B. Flanegan, and K. Kirkegaard. 1997. Poliovirus RNA recombination in cell-free extracts. RNA 3:624-633. [PMC free article] [PubMed] [Google Scholar]
- 64.Walter, J. E., J. Briggs, M. L. Guerrero, D. O. Matson, L. K. Pickering, G. Ruiz-Palacios, T. Berke, and D. K. Mitchell. 2001. Molecular characterization of a novel recombinant strain of human astrovirus associated with gastroenteritis in children. Arch. Virol. 146:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiss, B. G., and S. Schlesinger. 1991. Recombination between Sindbis virus RNAs. J. Virol. 65:4017-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White, K. A., and T. J. Morris. 1994. Enhanced competitiveness of tomato bushy stunt virus defective interfering RNAs by segment duplication or nucleotide insertion. J. Virol. 68:6092-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White, K. A., and T. J. Morris. 1994. Recombination between defective tombusvirus RNAs generates functional hybrid genomes. Proc. Natl. Acad. Sci. USA 91:3642-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.White, K. A., and T. J. Morris. 1994. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 68:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.White, K. A., and T. J. Morris. 1995. RNA determinants of junction site selection in RNA virus recombinants and defective interfering RNAs. RNA 1:1029-1040. [PMC free article] [PubMed] [Google Scholar]
- 70.White, K. A., and T. J. Morris. 1999. Defective and defective interfering RNAs of monopartite plus-strand RNA plant viruses. Curr. Top. Microbiol. Immunol. 239:1-17. [DOI] [PubMed] [Google Scholar]
- 71.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80:2535-2543. [DOI] [PubMed] [Google Scholar]