Abstract
The study of bound-state conformations of ligands interacting with proteins is important to the understanding of protein function and the design of drugs that alter function. Traditionally, transferred nuclear Overhauser effects (trNOEs), measured from NMR spectra of ligands in rapid exchange between bound and free states, have been used in these studies, owing to the inherent heavy weighting of bound state data in the averaged ligand signals. In principle, residual dipolar couplings (RDCs) provide a useful complement to NOE data in that they provide orientational constraints as opposed to distance constraints, but use in ligand binding applications has been limited due to the absence of heavy weighting of bound-state data. A widely applicable approach to increasing the weighting of bound state data in averaged RDCs measured on ligands is presented. The approach rests on association of a His-tagged protein with a Ni-chelate carrying lipid inserted into the lipid-bilayer-like alignment media used in the acquisition of RDCs. The approach is validated through the observation of bound-state RDCs for the disaccharide, lactose, bound to the carbohydrate recognition domain of the mammalian lectin, Galectin-3
Introduction
Optimization of lead compounds in drug discovery can often be facilitated with direct information on the geometry of the bound compound.1,2. Knowledge of the bound geometry of native ligands can also be of use in understanding the molecular origin of binding specificity. For cases in which ligands are in rapid exchange, NMR methods, and specifically transferred Nuclear Overhauser Effects (trNOEs), have proven especially useful1,3. TrNOEs are applicable to large proteins under rather dilute conditions, typically with the ligand in at least ten fold molar excess. They report preferentially on the bound state of the ligand because the efficiency of the magnetization transfer producing the NOE is proportional to the molecular weight of the complex (at least in the large complex limit). When used properly trNOEs provide distance constraints between pairs of protons on ligands that lie less than 5Å apart4. However, it is not always possible to observe trNOEs between sufficient numbers of such pairs to accurately define bound geometries of ligands. It is also difficult to observe protein to ligand NOEs needed in orienting the ligand relative to the protein in its binding site. In what follows we present a new method for acquiring complementary data that overcomes some of the limitations of trNOEs, namely transferred residual dipolar couplings (trRDCs).
Residual dipolar couplings (RDCs) can often provide data that describe the relative geometry of remote parts of ligands as well as the orientation of a ligand relative to a protein. There have been many applications of RDCs to the determination of ligand structures free in solution5,6, and while there have been a few applications of RDCs to the determination of bound ligand geometry, there is a problem in that, unlike trNOEs, RDCs from the bound ligand do not normally dominate the observed average. Recently, we presented one approach to enhancing the RDC contribution of bound ligands7. By adding a short alkyl chain to the protein’s amino terminus, the protein and any bound ligand become more strongly associated with media used in many RDC measurements, and contributions of the bound state to measured RDCs increase. However, this approach is not generally applicable because of the nature of required protein modifications. In what follows, we present a more widely applicable method for enhancing the bound ligand contribution that rests simply on the presence of a His-tag commonly used in protein expression and purification.
Measurement of RDCs in solution requires weak alignment in order to keep dipolar couplings from averaging to zero. The requisite level of order is most often achieved through use of nematic liquid crystalline media exhibiting no specific protein or ligand interactions. The dissolved macromolecule and ligand adopt a preferential degree of order (a few tenths of a percent) through weak (often collisional) interactions with the medium. The problem is that these interactions often depend on size-independent properties, such as anisotropy of shape. Hence, measured RDCs of ligands can be nearly equally weighted by populations of bound and free species. To circumvent this problem we introduce an association with the medium that is specific for the protein as opposed to the ligand. In particular we exploit association of a poly-histidine-tag (His-tag) added to the protein terminus with a His-tag binding lipid inserted into a lipid bilayer-like alignment medium. Alignment by direct association can produce order approaching a few percent as opposed to tenths of a percent, greatly accentuating bound-state data for exchanging ligands.
His-tag addition is a very common modification to protein constructs and is routinely used for affinity purification. Purification proceeds through ionic interactions of poly-Histidine residues with chelated nickel bound to an immobile resin. Release of the bound protein is often through competitive binding of imidazole or alteration of ionic interactions by changing pH. More recently, in order to provide a chelate that can reside on membrane-like surfaces, the same Ni chelate (Ni-NTA) used in purification columns has been covalently attached to fatty acyl chains9 (DOGS-NTA-Ni, 1,2-dioleoyl-sn-glycero-3-{[N(5-amino-1-carboxypentyl) iminodiacetic acid]succinyl} (nickel salt) ), and used in conjunction with alignment media10. This chelate is now commercially available, and has, in addition, allowed for the immobilization of his-tagged proteins on lipid surfaces for two-dimensional protein crystallization9,11. Here, incorporation of nickel-chelating lipids into the bicelle-like liquid crystal media used for sample alignment in RDC measurements12 will be shown to provide enhanced alignment of a His-tagged protein, and hence, enhanced RDCs measured for bound ligands.
The carbohydrate-binding protein, Galectin-3, was chosen as a system to validate the proposed method. Galectin-3 is a mammalian S-type lectin with an affinity for galactose terminated oligosaccharides13. It has been implicated in the inhibition of apoptosis, progression of cancer, and mediation of inflammation through interaction with a variety of carbohydrates14,15. Hence, the origin of its binding specificity for various carbohydrate ligands is of considerable interest. It has a crystal structure with the ligand N-acetyl lactosamine in the binding site13 as well as well as NMR structural data on lactose in the binding site16. Here we illustrate the determination of the bound geometry for the simple disaccharide, α-lactose, using the His-tagged Galectin-3 carbohydrate binding domain and the methods described.
Experimental Section
Sample preparation
cDNA encoding the carbohydrate recognition domain (CRD) of Galectin-3 (residues 114-250) was cloned from an existing full length human Galectin-3 gene (pET-3c) into the Escherichia coli expression vector pET-23a containing a C-terminal 6 × His-tag. Proper cloning was verified by DNA sequencing. BL21 (DE3) codon plus cells were transformed with the expression plasmid for Galectin-3-CRD (pET-23a) in 50ml of LB rich media containing 100ug/ml ampicillin (Amp) and incubated at 37°C overnight. Cells were collected, resuspended, and added to 1L M9/Amp media, containing 1g/L 15NH4Cl and 4g/L glucose. Protein expression was initiated by the addition of IPTG to 0.5mM (at OD600 of 1.0) and incubated for an additional 2-4hrs. Cells were resuspended in 10ml Phosphate buffer (75 mM KH2PO4, 75mM NaCl, 2mM EDTA, 1mM DTT, 5mM NaN3 pH 6.8 at room temperature), broken using sonication (8min, 10s pulse and 20s pause), and subsequently clarified by centrifugation (100k × g, 1hr). Passage over a 25 ml lactosyl-agarose affinity column (Sigma) equilibrated in phosphate buffer efficiently extracted Galectin-3-CRD from crude cell lysate. Elution with 300mM lactose yielded 25-40 mg pure Galectin-3-CRD as assessed by SDS-PAGE. To verify His-tag presence and accessibility, the Galectin-3-CRD was passed over a 25 ml nickel affinity column (Amersham) and eluted by the addition 300mM imidazole. Dialysis against phosphate buffer (4°C, overnight) effectively removed residual imidazole. NMR samples were made to be 0.3mM 15N His-tagged Galectin-3-CRD in 75mM KH2PO4, 75mM NaCl, 5mM NaN3 (pH=6.8).
Nickel chelated bicelle preparation
A bicelle-like alignment medium, based on mixtures of hexanol and the alkyl-polyethylene glycol detergent, C12E5, (pentaethylene glycol monododecyl ether)(Sigma, St Louis MO) was chosen for RDC measurement 20. The DOGS-NTA nickel salt is hydrophobic and by itself insoluble in water. However, in the presence of C12E5 or hexanol it is readily incorporated into the alignment medium. The nickel-chelated C12E5 medium was made in the samefashion as previously described by Ruckert et al17, with DOGS-NTA-Ni being added directly to the appropriate volume of hexanol prior to mixing with lipids. C12E5 was mixed thoroughly with the buffer. Hexanol/DOGS-NTA-Ni was added in 2–3 uL increments vortexing after each addition. Alternatively, it is possible to mix the desired amount of DOGS-NTA ammonium salt with C12E5 sincethe ammonium salt is soluble in the presence of C12E5. Nickel chloride is then added to a ratio of 1:1with respect to DOGS-NTA ammonium and vortexed overnight to complete complexation. The alignment of this medium was checked by measuring the deuterium splitting at 25°C on a spectrometer operating at 600MHz for protons. A 12–14Hz deuterium splitting was obtained in 3% C12E5/hexanol/0.5mM DOGS-NTA nickel salt.
NMR spectroscopy
All other NMR spectra were recorded with a Varian Inova 800MHz spectrometer equipped with a cryogenic triple resonance probe. One-dimensional 1H-15N HSQC spectra were collected to monitor the association of His-tag Galectin-3-CRD with DOGS-NTA-Ni. Spectra were recorded at 298K using 1000 scans on samples containing 0.3mM Galectin-3-CRD in 3% C12E5, 0.3mM Galectin-3-CRD in 3% C12E5/0.5mM DOGS-NTA, and 0.3mM Galectin-3-CRD in 3% C12E5/0.5mM DOGS-NTA-Ni.
1H-15N-coupled gradient HSQC experiments were used to collect RDCs from protein resonances run under isotropic (298K) and partially aligned (298K) conditions on 3% C12E5/0.5mM DOGS-NTA-Ni samples. Data collection typically included 1024 t2 points and 256 t1 points, subsequently extended with linear prediction and zero filling to 2048 and 1024 points respectively. Differences in the splittings under aligned and isotropic conditions (298K) yielded residual dipolar couplings.
The sensitivity-enhanced 1H-13C coupled HSQC (natural abundance) experiment18 was used to observe RDCs emanating from ligand resonances. Spectra were recorded under isotropic (298K) and partially aligned (298K) conditions with samples containing either 6mM free lactose, 6mM lactose in 3% C12E5/0.5mM DOGS-NTA-Ni, or 6mM lactose with 0.3mM His Galectin-3-CRD in 3% C12E5/0.5mM DOGS-NTA-Ni. Data collection included 1024 t2 points and 512 t1 points with 16 scans each for free lactose and lactose in media. Due to the line broadening observed on membrane association in the presence of protein 64 acquisitions per t1 point were acquired in samples containing lactose with His-Galectin-3-CRD in 3% C12E5/hexanol/0.5mM DOGS-NTA-Ni.
Intensity-based constant-time 1H-1H COSY (CT-COSY) experiments 19 were run to obtain the remaining 1H-1H lactose couplings needed for order tensor analysis. As above, spectra were recorded under isotropic (298K) and partially aligned (298K) conditions with samples containing either 6mMfree lactose, 6mM lactose in 3% C12E5/hexanol/0.5mM DOGS-NTA-Ni, and 6mM lactose with 0.3mM His Galectin-3-CRD in 3% C12E5/hexanol/0.5mM DOGS-NTA-Ni. Constant time periods were arrayed from 0.1ms to 0.42ms with time increments of 0.04ms (free lactose and lactose in media) or 0.018ms to0.153ms with time increments of 0.015ms (lactose with His-Galectin-3-CRD in 3% C12E5/hexanol/0.5mM DOGS-NTA-Ni).
Data processing, including extraction of residual dipolar couplings for the protein, was performed with the nmrPipe/nmrDraw package20. RDC measurement used the frequency-domain least squares module (NLinLS) found in NMRDraw, resulting in measured couplings having estimated errors of 1 Hz (20 % of the overall range). For precise measurement of ligand couplings in our strongly aligned system a Bayesian fitting program, XRAMBO, was used to extract the couplings for each C-H pair21. Methods such as these have allowed for measurement of ligand RDCs with estimated errors less than 0.5 Hz. Intensity-based CT-COSY spectra were processed and dipolar couplings extracted as described in the literature19 All experimental dipolar couplings were analyzed using the REDCAT analysis package to provide a principal order frame and order parameters22.
Results
His-tag addition to the Galectin-3 Carbohydrate Recognition Domain (CRD)
The C-terminal domain of Galectin-3 (residues 114-250) comprises the carbohydrate recognition domain (CRD) and carries the galactose-specific binding site. Due to the spatial proximity of the ligand-binding site to the Galectin-3-CRD N-terminus as observed in the crystal structure13, the His-tag was introduced onto the C-terminal end. This was done in order to minimize paramagnetic broadening effects of the Nickel ion on resonances from the bound ligand and to prevent masking of the oligosaccharide-binding site by His-tag-Nickel-chelate interactions.
In order to address the potential structural alteration resulting from His-tag addition,1H-15N Heteronuclear Single Quantum Coherence (HSQC) spectra were recorded on His-tagged and non-His-tagged samples. HSQC spectra are commonly used to report on the structural characteristics of soluble macromolecules isotopically enriched in 15N23 Positions of amide cross-peaks are sensitive to the local electronic environment, and changes in cross-peak position would be expected to reflect any changes in local structure. Overlaying the 1H-15N HSQC spectra for the Galectin-3-CRD and the 6×His-tag Galectin-3-CRD, showed little change in observed peak positions.
Nickel-Chelated Lipid Alignment and Stability
An ability to create media containing theNi-chelate (DOGS-NTA) that align in a magnetic field, are homogeneous, and are stable over time, is key to the collection of quality RDC data. DOGS-NTA was added to a commonly used pentaethyleneglycol monododecyl ether medium (C12E5/hexanol bicelles)17 at a ratio of approximately 0.5mM DOGS-NTA to 3%(m/m) lipid. Splitting of the deuterium resonance from 10% 2H2O included in the medium was used to monitor alignment, homogeneity and stability. A 3% C12E5/hexanol preparationshowed splittings of 12-14Hz with no residual isotropic peak. Splittings remained within this range for more than 14 days. Moreover, the addition of Galectin-3-CRD to levels of 0.3mM did not substantially change these splittings. This behavior is very similar to that of undoped 3% C12E5/hexanol bicelles.
Association of the Target Protein with Ni chelate Media
Association of a His-Tagged protein with DOGS-NTA-Ni doped bicelles is easily demonstrated. One-dimensional versions of HSQC spectra provide a good indicator of association. In isotropic solution each amide 1H15-N pair gives rise to a sharp line. With membrane association we expect correlation times to rise and numerous through-space dipolar couplings to appear; both will contribute to increased line widths and loss of signal. Changes in the signal-to-noise ratio in spectra with and without the chelating metal can therefore be indicative of selective lipid anchoring of His-tagged macromolecules in solution.
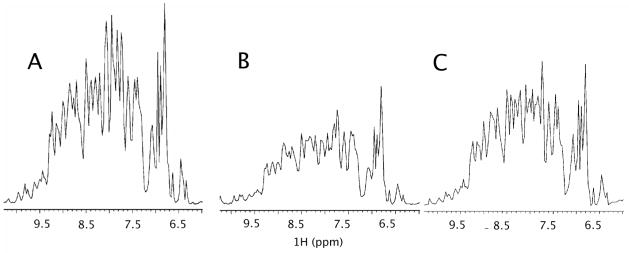
Figure 1 shows a 1D H-15N HSQC spectrum collected on a 0.3mM sample of Galectin-3-CRD (pH 7.4) in a 3% C12E5/hexanol bicelle preparation doped with (A) 0.5mM DOGS-NTA ammonium salt (no chelating metal) and (B) 0.5mM DOGS-NTA-Ni. There is obviously a reduction in intensity of resonances in the presence of the Ni-loaded DOGS-NTA. However, the widths of the residual lines are only slightly broader than in the absence of activated chelator. Also attempts to collect RDC data from coupled HSQC spectra, using the residual protein lines, resulted in spectra with 1H-15N splittings identical to those for protein in bicelles without DOGS-NTA-Ni. The most likely interpretation is that protein is in slow exchange between bound and free states. Resonances from the bound protein are broadened beyond detection and only resonances from free protein are detected. Since RDCs will be averaged over all states, this will have little consequence on interpretability of transferred RDCs of ligands, as long as there is sufficient enhancement of some protein population to allow that population to dominate the measured average.
Fig 1.
1D HSQC spectra of 1H-15N His-tagged-Galectin-3-CRD. (A) 0.3mM Galectin-3 (pH 7.4) in a 3% C12E5/Hexanol bicelle preparation doped with 0.5mM DOGS-NTA ammonium salt (no chelating metal); (B) 0.3mM Galectin-3 (pH 7.4) in a 3% C12E5/Hexanol bicelle preparation doped with 0.5mM DOGS-NTA-Ni; (C) 0.3mM G0.5mM DOGS-NTA-Ni.
Since we expect the ligand to be in fast exchange, not only RDCs, but line widths, will be averaged over bound and free forms. This can result in excessive line broadening of ligand resonances, and this can inhibit observation of RDCs, if the fraction of bound ligand is too high. While one could simple raise the ligand to protein ratio to restore an acceptable line width, it is also convenient to be able to reduce, after sample preparation, the fraction of protein associated with the alignment medium. This can be done by addition of competitors (imidazole when compatible with the protein under study) or by adjustment of pH. The effect of pH is shown in Figure 1C. It is known that pH affects His-tag – nickel association due to the altered protonation state of the histidine residues. This results in a reduced affinity as the pH is decreased. At low pH (6.8, Figure 1C) signal-to-noise levels are substantially higher than those observed at higher pH values (7.4, Figure 1B), suggesting a decrease in lipid-anchored protein. As described below, the smaller fraction of associated protein results in observable resonances and relatively large RDC values for the ligand.
Measurement of ligand RDCs
Lactose (Galβ1-4Glc) was chosen as a ligand for validation of our proposed method for the measurement of transferred RDCs. 1H-13C RDCs, as well as a few 1H-1HRDCs, were measured at a 20:1 ratio of ligand to protein, or 6 mM lactose in the presence of 0.3mMGalectin-3-CRD in pH 6.8, 3% C12E5/0.5mM DOGS-NTA. At this relatively high concentration of lactose it is possible to measure one-bond 1H-13C RDCs at natural abundance. RDCs were measured by taking the difference between 1H-13C splittings in the indirect dimensions of 1H-13C coupled HSQCs taken under isotropic conditions (no alignment media) and aligned conditions (DOSGS-NTA doped media with and without Ni). In select cases, 1H-1H couplings were measured using a 1H-1H CT COSY experiment19. All measured RDCs are summarized in Table 1.
Table 1.
RDCs measurements on lactose bound to Galectin-3.
| Free | DOGS-NTA Ammonium | DOGS-NTA Nickel salt | ||||
|---|---|---|---|---|---|---|
| RDC(Hz) | RDC(Hz) | RDC(Hz) | RDCbound(Hz)a | RDCcal(Hz)b | ||
| Glc(α) | C1-H1 | −4 | −3.9 | −4.5 | −14 | −13 |
| C2-H2 | 2.7 | 2.5 | 0.7 | −37 | −21 | |
| C3-H3 | 3.7 | 3.4 | 2.9 | −12 | −22 | |
| C5-H5 | 3.9 | 3.2 | 2.8 | −18 | −26 | |
| H1-H2 | −0.1 | −1.5 | −29 | −24 | ||
| Gal | C1-H1 | 1.7 | 1.5 | −2.6 | −84 | −86 |
| C2-H2 | 1.9 | 2.1 | −1.8 | −72 | −70 | |
| C3-H3 | 2.4 | 1.8 | −2.3 | −91 | −96 | |
| C4-H4 | 0.1 | 1 | 1.4 | 26 | 23 | |
| C5-H5 | 2.8 | 2.4 | −1.8 | −89 | −88 | |
| H1-H2 | 0.2 | 0.9 | 15 | 13 | ||
|
| ||||||
| Order tensor | Sxx | Syy | Szz | |||
|
| ||||||
| Free lactose | −1.1E-04 | −1.7E-04 | 1.8E-04 | |||
|
| ||||||
| Bound -lactose | −2.8E-04 | −2.9E-03 | 3.2E-03 | |||
RDCfor the bound state have been extracted using a dissociation constant of 1mM.
RDCs have been back-calculated using the lactose geometry found in the crystal structure, 1ULC.
As expected, measured RDCs appear virtually unchanged when comparing those from the ligand in the absence of protein with those from the ligand in the presence of Galectin 3-CRD and uncharged DOGS-NTA (columns 1 and 2, Table 1). Given a dissociation constant of 1mM for the lactose –Galectin-3 interactions we would expect about 5 % of the ligand to be bound under the conditions of our experiment. Even if the level of order for the protein were twice as large as that for the free ligand, changes would be less than 1Hz. It would obviously be very difficult to extract RDCs for the bound state from the measured differences.
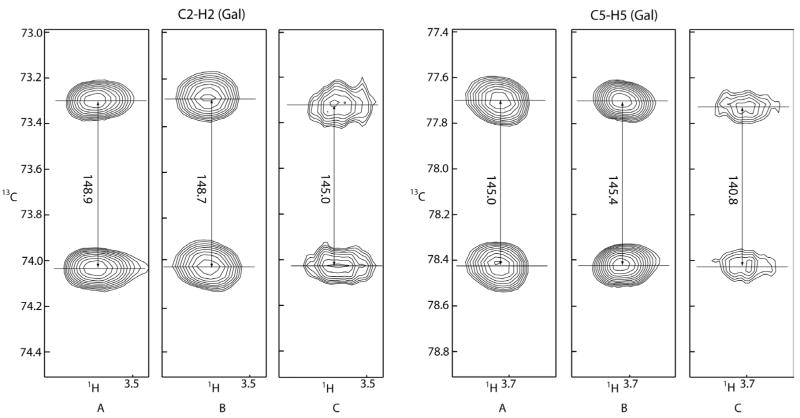
Charging of DOGS-NTA with Ni (column 3, Table 1) allows for an increase in the difference between observed RDCs for the free-state and the protein complexed state to nearly 5Hz. As an illustration of the increase, splittings of C2-H2 and C5-H5 of the galactose residue of α-lactose are shown in Figure 2. Changes in splittings in the presence and absence of protein using media lacking the Ni required for enhanced protein alignment (Figures 2A and 2B) are almost imperceptible. A substantial change in splitting can, however, be observed in Fig 2C when the protein is associated with the Ni-charged medium (lactose with His-tagged Galectin-3-CRD and DOGS-NTA nickel chelated lipid). As expected for a more strongly oriented molecule, the cross peaks in Figure 2C are broadened and show partially resolved 1H-1H splittings. While RDCs are somewhat harder to measure, the fact that they are more strongly influenced by the protein-associated ligand, makes data on the bound state easier to extract and interpret.
Fig 2.
Splittings of C2-H2 and C5-H5 of the lactose galactose residue. (A) 6mM lactose, 0.3mM Galectin-3 (pH 6.8) in 3% C12E5/Hexanol bicelle preparation doped with 0.5mM DOGS-NTA ammonium salt (no chelating metal); (B) 6mM lactose (pH 6.8) in 3% C12E5/Hexanol bicelle preparationdopedwith0.5mM DOGS-NTA-Ni; (C) 6mM lactose, 0.3mM Galectin-3 (pH 6.8) in 3% C12E5/Hexanol bicelle preparation doped with 0.5mM DOGS-NTA-Ni.
RDCs for the bound state can be extracted using the known binding constant for lactose (1mM)13 and equating a simple population weighted average of RDCs in the free state (column 1) and RDCs of the bound state to measured RDCs (column 3). The results are given in column 4. The magnitude of order observed in the bound state can be estimated by comparing the maximum splitting to splittings expected for an immobile 1H-13C bond directed perpendicular to the magnetic field (~32,500 Hz). The level of order would be 2.8 × 10−3 The level of order in free and bound states can be more accurately determined by solving for order matrix parameters using the free-state lactose geometry (phi, psi = 50, −116 respectively) simulated with Glycam force field24,25 and the data on free lactose, or the bound lactose geometry found in the crystal structure (PDB id 1ULC, phi, psi = 48, −95 respectively) and the data on bound lactos26. The resulting principal order matrix parameters are Sxx = −1.1E-05, Syy = −1.7E-04, Szz = 1.8E-04 and Sxx =− 2.8E-4, Syy = −2.9E-03, Szz = 3.2E-03, respectively. This suggests an averaged increase in order of a factor of 15 due to the association of a fraction of the Galectin-3-CRD with the alignment medium.
One can also test for consistency of bound RDCs with the actual geometry of lactose found in the crystal structure. Once an order tensor and the principal order frame are known, back-calculation of RDCs, and examination of the correlation between observed couplings and those calculated from the model, can give an indication of successful sampling of the bound state conformation. The bound-state RDCs were back-calculated using the REDCAT program27 and results are given in column 5 of table 1. Very good agreement between the experimental and back-calculated bound-state RDCs is seen except for Glc C2-H2. The resonances from this site are partially obscured by background from the C12E5 medium and errors in measurement are large. The RMSD of measured RDCs from back-calculated RDCs, excluding C2-H2, is 5Hz. We can also attempt back calculation using the solution structure in which the ligand geometry is slightly different. The agreement is not as good yielding an RMSD of 8Hz. While these differences are not large, the better agreement in the former case suggests consistency with the geometry found in the crystal structure.
Discussion
RDCs measured from rapidly exchanging systems, while information rich, have historically been underutilized because of an inability to accurately extract bound state contributions from their measured averages. Here, we have shown that incorporation of target proteins containing poly-histidine tags into lipid bilayer–like preparations doped with DOGS-NTA-Ni can result in sufficient enhancement of bound state couplings to facilitate extraction. We have validated the approach by demonstrating the extraction of a set of bound-state RDCs for lactose bound to the Galectin-3-CRD, and showing that they are quite consistent with the bound state geometry of lactose found in a Galectin-3-CRD crystal structure13.
While it would be advantageous to measure RDCs and derive alignment parameters for the protein itself, signal-to-noise levels of resonances from the protein were dramatically reduced on association with the alignment media. One explanation is that the protein is in slow exchange on and off the bicelle with the bound form giving resonances too broad to detect; resonances seen from the fraction free in solution, therefore, do not carry information on protein alignment. This does not invalidate information on conformations of bound ligands. In fact, most transferred NOE data are obtained on proteins too large to characterize by NMR. Transferred RDC data on ligands for proteins that are unable to be observed directly is equally valuable.
In the present case, in which there is a crystal structure for the complex, it is possible to obtain a representation of protein orientation indirectly from dipolar couplings emanating from bound lactose. When order parameters in the principal order frame are derived for the bound ligand, a rotation matrix, needed to bring protein coordinates into the principal frame, also results. The model produced by moving the protein to the principal frame agrees well with one having the His-tag pointing towards the membrane surface. Such a model could not be derived in cases where appropriate protein structures did not already exist. However, in the present case, it does support the view that interactions of the His-tag with a lipid-anchored chelate are responsible for the enhanced level of order.
In addition to our own studies27, there have been a few other attempts to accentuate RDC contributions of the bound ligand by relying on orientational properties specifically originating in the proteins. For example, the large magnetic anisotropy of ordered peptide fibrils28, ordered protein assemblies, or proteins with paramagnetic centers have been used30,31. However, these approaches are only possible in special cases where magnetic anisotropies exist naturally or where proteins can be engineered to contain highly anisotropic metal centers. When measured RDCs are the result of exchange with excess free ligand, measured RDCs are also very small in many of these cases.
There may be some limitations to application of the current method as well. For example, in cases where hydrophobic ligands are investigated, their tendency to independently associate with bilayer-like alignment media can cause problems. However, there are very pronounced advantages in terms of broad applicability. The widespread incorporation of His-tags in proteins prepared for biophysical characterization means that there will be many proteins to which the methodology is directly applicable. We expect many applications to the determination of bound state geometries of rapidly exchanging protein ligands in the future.
Acknowledgments
This work was supported by grant number GM33225 from the National Institutes of Health.
References
- 1.Moore JM. Biopolymers. 1999;51(3):221–243. doi: 10.1002/(SICI)1097-0282(1999)51:3<221::AID-BIP5>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 2.Homans SW. Angew Chem Int Ed Engl. 2004;43(3):290–300. doi: 10.1002/anie.200300581. [DOI] [PubMed] [Google Scholar]
- 3.Jiménez-Barbero J, Peters T. Nmr Spectroscopy of Glycoconjugates. Wiley-Vch; Weinheim, Germany: 2002. [Google Scholar]
- 4.Tugarinov V, Hwang PM, Kay LE. Annu Rev Biochem. 2004;73:107–146. doi: 10.1146/annurev.biochem.73.011303.074004. [DOI] [PubMed] [Google Scholar]
- 5.Tian F, Al-Hashimi HM, Craighead JL, Prestegard JH. J Am Chem Soc. 2001;123(3):485–492. doi: 10.1021/ja002900l. [DOI] [PubMed] [Google Scholar]
- 6.Martin-Pastor M, Canales A, Corzana F, Asensio JL, Jimenez-Barbero J. J Am Chem Soc. 2005;127(10):3589–3595. doi: 10.1021/ja043445m. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu H, Donohue-Rolfe A, Homans SW. J Am Chem Soc. 1999;121(24):5815–5816. [Google Scholar]
- 8.Jain NU, Noble S, Prestegard JH. J Mol Biol. 2003;328(2):451–462. doi: 10.1016/s0022-2836(03)00268-7. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt L, Dietrich C, Tampe R. J Am Chem Soc. 1994;116(19):8485–8491. [Google Scholar]
- 10.Prosser RS, Bryant H, Bryant RG, Vold RR. J Magn Reson. 1999;141(2):256–260. doi: 10.1006/jmre.1999.1855. [DOI] [PubMed] [Google Scholar]
- 11.Kubalek EW, Legrice SFJ, Brown PO. J Struct Biol. 1994;113(2):117–123. doi: 10.1006/jsbi.1994.1039. [DOI] [PubMed] [Google Scholar]
- 12.Sanders CR, Hare BJ, Howard KP, Prestegard JH. Progress in Nuclear Magnetic Resonance Spectroscopy. 1994;26:421–444. [Google Scholar]
- 13.Seetharaman J, Kanigsberg A, Slaaby R, Leffler H, Barondes SH, Rini JM. J Biol Chem. 1998;273(21):13047–13052. doi: 10.1074/jbc.273.21.13047. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura M, Inufusa H, Adachi T, Aga M, Kurimoto M, Nakatani Y, Wakano T, Nakajima A, Hida JI, Miyake M, Shindo K, Yasutomi M. Int J Oncol. 1999;15(1):143–148. doi: 10.3892/ijo.15.1.143. [DOI] [PubMed] [Google Scholar]
- 15.Almkvist J, Dahlgren C, Leffler H, Karlsson A. J Immunol. 2002;168(8):4034–4041. doi: 10.4049/jimmunol.168.8.4034. [DOI] [PubMed] [Google Scholar]
- 16.Umemoto K, Leffler H, Venot A, Valafar H, Prestegard JH. Biochemistry. 2003;42(13):3688–3695. doi: 10.1021/bi026671m. [DOI] [PubMed] [Google Scholar]
- 17.Ruckert M, Otting G. J Am Chem Soc. 2000;122(32):7793–7797. [Google Scholar]
- 18.Schleucher J, Schwendinger M, Sattler M, Schmidt P, Schedletzky O, Glaser SJ, Sorensen OW, Griesinger C. J Biomol NMR. 1994;4(2):301–306. doi: 10.1007/BF00175254. [DOI] [PubMed] [Google Scholar]
- 19.Tian F, Bolon PJ, Prestegard JH. J Am Chem Soc. 1999;121(33):7712–7713. [Google Scholar]
- 20.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Andrec M, Prestegard JH. J Magn Reson. 1998;130(2):217–232. doi: 10.1006/jmre.1997.1304. [DOI] [PubMed] [Google Scholar]
- 22.Valafar H, Prestegard JH. J Magn Reson. 2004;167(2):228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Wuthrich K. Nmr of Proteins and Nucleic Acids. Wiley-Interscience; New York, Ny USA: 1986. [Google Scholar]
- 24.Woods RJ, Dwek RA, Edge CJ, Fraserreid B. J Phys Chem. 1995;99(11):3832–3846. [Google Scholar]
- 25.Kirschner KN, Woods RJ. Proc Natl Acad Sci U S A. 2001;98(19):10541–10545. doi: 10.1073/pnas.191362798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. J Magn Reson. 1999;138(2):334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang TD, Leffler H, Prestegard JH. Protein Sci. 2006;15(7):1780–1790. doi: 10.1110/ps.051994306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen ZJ, Reif B. J Biomol NMR. 2004;29(4):525–530. doi: 10.1023/B:JNMR.0000034353.98902.4f. [DOI] [PubMed] [Google Scholar]
- 29.Koenig BW, Mitchell DC, Konig S, Grzesiek S, Litman BJ, Bax A. J Biomol NMR. 2000;16(2):121–125. doi: 10.1023/a:1008378523816. [DOI] [PubMed] [Google Scholar]
- 30.Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C. J Biomol NMR. 2004;29(3):339–349. doi: 10.1023/B:JNMR.0000032611.72827.de. [DOI] [PubMed] [Google Scholar]
- 31.Feeney J, Birdsall B, Bradbury AF, Biekofsky RR, Bayley PM. J Biomol NMR. 2001;21(1):41–4. doi: 10.1023/a:1011924017938. [DOI] [PubMed] [Google Scholar]