Abstract
We have begun an analysis of the functional architecture of the ICP0 promoter in neurons in vivo with the ultimate goal of determining how this gene is regulated during reactivation in vivo. Promoter/reporter mutants in which the Escherichia coli beta-galactosidase (β-Gal) gene was driven by various permutations of the ICP0 promoter were employed to permit the analysis of promoter function without the added complications that would arise due to inappropriate regulation of ICP0 protein levels. A whole-ganglion immunohistochemical staining procedure (N. M. Sawtell, J. Virol. 77:4127-4138, 2003) was used for direct comparisons of the expression of the promoter/reporter gene to expression of the native protein in the same cell. In this way, the expression of the putative wild-type promoter could be validated and results for mutant promoters could be compared to expression of the native gene. We found that a DNA fragment from bp −562 through the methionine start codon of the ICP0 gene contained all sequences required for properly regulated ICP0 expression in diverse cell types (including sensory neurons of the trigeminal ganglia [TG]) in vitro and in vivo, as indicated by colocalization of ICP0 and β-Gal. Truncation of the ICP0 promoter to bp −145 or −129 resulted in the loss of immediate-early (α) kinetics. The truncated promoters expressed high levels of the reporter gene with leaky late (γ1) kinetics in vitro and in some cell types in vivo. Unexpectedly, the truncated promoters did not express in TG neurons. Thus, TAATGARAT or other sequences upstream of bp −145 in the ICP0 promoter are required for basal expression of ICP0 in neurons but are not required for basal expression in other cells in vivo. There was a >95% concordance between reporter and native protein expression detected with the 562-bp promoter in neurons during the acute stage. However, this was not the case during reactivation from latency in vivo, as nearly twice as many neurons contained detectable β-Gal as contained detectable ICP0. This same 562-bp promoter/reporter cassette, when placed in the context of a latency-associated transcript (LAT) null mutant, resulted in >95% concordance of expression of β-Gal and ICP0 during reactivation in vivo. These last results strongly suggest that there is a posttranscriptional constraint on the expression of ICP0 protein during reactivation from latency and that this constraint is mediated by LAT.
Despite the extensive repression of herpes simplex virus type 1 (HSV-1) gene transcription during latency, a variety of stressful stimuli can result in the reactivation of the latent genome and reentry into lytic-phase transcription (71). How the quiescent genome exits latency is not yet known. It has been suggested that ICP0 is uniquely important for the initiation of viral reactivation (reviewed in reference 19). ICP0 is a major viral transcriptional activator important for the expression of viral genes of diverse kinetic classes (8-10, 12, 60). In addition, this protein functions to promote viral mRNA translation and regulates levels of cellular proteins through interaction with the ubiquitin-proteosome pathway (18, 20, 45, 49). These properties suggest that ICP0 can regulate both viral and host protein levels through transcriptional, translational, and posttranslational mechanisms. In a tissue culture model of latent infection, ICP0 was implicated in the efficient establishment of latency (74). ICP0-expressing adenovirus vectors that lack all other HSV genes have been shown to induce viral replication in quiescently infected tissue cultures (28, 75). Further, a viral mutant lacking functional ICP0 but containing all other viral genes did not induce viral replication in similar cultures, suggesting a specific role for ICP0 (28). However, adenovirus vectors expressing ICP4 and VP16 can also induce viral replication in primary cultures derived from latently infected ganglia (26).
In animal models ICP0 mutant strains can establish latent infections and can reactivate in cocultivation assays, but the levels of efficiency for both of these processes are greatly reduced compared to that seen with the wild-type strain (7, 10, 27, 33, 37). One problem with these studies is that ICP0 null mutants establish latency inefficiently, and it is therefore difficult to distinguish between the importance of this gene for establishing latency and its importance for initiating reactivation. Recently, Halford and Schaffer infected immunosuppressed mice with ICP0 null mutants in an effort to increase the establishment of latency by the null mutants. Those authors concluded (on the basis of the total amount of HSV DNA detected in ganglia by PCR and the number of explant cultures in which virus reactivation was detected) that ICP0 is required for the efficient induction of reactivation from latency in vitro (27). Thus, the evidence suggesting that ICP0 is important for the induction of reactivation seems at first compelling and yet this conclusion may still be premature. Halford et al. did not include experiments that would distinguish between a specific requirement for ICP0 for the induction of reactivation and its requirement for the modulation of some unmeasured but important parameter (such as the actual number of neurons in which latency is established or the range and mean of the number of viral genomes within individual latently infected neurons) (51, 54, 57) for the establishment of latency already known to affect reactivation frequency. Indeed, it is possible that the requirement for ICP0 in such models is simply a requirement for amplification of reactivated virus to detectable levels within the explant cultures.
One strategy alternate and complementary to the use of ICP0 null mutants for deducing a direct role for this gene in latency is the examination of its promoter function in neurons in vivo. If ICP0 does initiate the reactivation process, as has been hypothesized, its promoter must have evolved to contain regulatory sequences that sense specific cellular environments and direct appropriate viral transcriptional activity in response to signals transduced in neurons by stress. The promoter of ICP0 is complex, containing elements that bind both cellular and viral encoded transcriptional factors (1, 11, 15, 22, 46). Interestingly, deletion of most of the ICP0 promoter, including the sequences that confer immediate-early (IE) gene kinetics, had no detected effect on the ability of the virus to reactivate following explant of ganglia into culture (11). This result suggests either that the ICP0 promoter does not contain specialized reactivation trigger-sensitive sequences or that the physiologic changes induced in ganglia by explantation into cultures obviate the need for appropriately regulated ICP0 during reactivation.
Here we report an analysis of the functional architecture of the ICP0 promoter in neurons in vivo. Using promoter/reporter viruses in which the Escherichia coli beta-galactosidase (β-Gal) gene is driven by various permutations of the ICP0 promoter, we made the following observations. First, basal ICP0 promoter activity is dependent on sequences upstream of −145 bp in neurons during the acute stage of infection. The upstream region is not required for basal expression in other cell types in vivo, a finding consistent with prior results seen with cell cultures (46). Second, we provide evidence for a posttranscriptional restraint on the expression of ICP0 protein in neurons during the early stages of reactivation from latency. The latter results emphasize the importance of investigating viral promoter function in neurons in vivo during the various stages of infection.
MATERIALS AND METHODS
Cells and viruses.
The wild-type HSV-1 strain 17syn+ was originally obtained from J. Subak-Sharpe of the MRC Virology Unit in Glasgow, Scotland, and was plaque purified as previously described (68, 70). Virus stocks were generated by routine propagation in rabbit skin cell (RSC) monolayers as previously described (68, 70).
Construction of viral mutants.
All viral DNA sequences were derived from the isolate of strain 17syn+ described above. The methods for inserting promoter/reporter genes cassettes into the glycoprotein C (gC) locus were previously detailed (55, 65). Briefly, to limit promoter occlusion or interference effects, promoter/reporter cassettes were inserted into the gC gene at the XbaI site at bp 97,669 on the viral genome in an orientation opposite to that of gC. For insertion of promoter reporters into the gJ locus, a modification of the plasmid pHD5 was generated. PHD5 contains the gJ locus (into which a polylinker was inserted) and flanking regions of HSV-1 strain SC16 (36) and was a kind gift of S. Efstathiou, Cambridge, United Kingdom. The polylinker present in the gJ gene of pHD5 was modified to replace the unique EcoRV site with an XbaI site and was transferred into the gJ gene of strain 17syn+ at the SacI site at bp 137,941 to generate the plasmid gJinPucXba. Reporter/promoter constructs were inserted into XbaI site in the polylinker in the gJ gene in gJinPucXba in an orientation opposite to that of gJ to limit any potential promoter occlusion or interference effects. The plasmid sequences were recombined into the HSV-1 genome as previously described in detail (63, 64, 66, 70). A blue plaque assay was utilized for the screening of the progeny of single plaques for the presence of the appropriate promoter/reporter constructs first, and purified virus stocks were employed as a source of DNA for restriction enzyme length polymorphism analysis as previously detailed (63, 64, 66, 70). All restriction enzyme site and base pair numbering is referred to according to the corresponding positions in the published HSV-1 sequence of strain 17syn+ (43, 47), as present in GenBank (NID g1944536).
Inoculation of mice.
Male, outbred, Swiss Webster mice (Harlan Laboratories) were used throughout these studies. Animals were housed in American Association for Laboratory Animal Care-approved quarters, with unlimited access to food and water. Mice were anesthetized by intraperitoneal injection of sodium pentobarbital at a dosage of 50 mg/kg of body weight. Both corneas and/or both sides of the snout were scarified, and a total inoculum of 2 × 105 PFU of 17syn+ or the mutant isolates was applied as previously described (55, 56, 66).
In vitro and in vivo acute replication kinetics.
Single and multistep replication kinetic analysis was performed on slightly subconfluent RSC or SK-N-SH monolayers following infection at a high multiplicity of infection (MOI) (10 PFU/cell) or a low MOI (0.01 PFU/cell). Cells and media were harvested at 4, 8, 12, 18, and 24 h postinfection (p.i.) (high MOI) or 4, 24, 48, and 72 h p.i. (low MOI) and subjected to three cycles of freezing and thawing. Virus titers were determined on RSC monolayers as previously described (68, 69).
Groups of 45 mice (225 mice total) were inoculated with each of the five viruses analyzed as described above. At the indicated times p.i., animals were sacrificed and the appropriate tissues were removed, snap frozen, and stored at −80°C. These tissues were homogenized in 1 ml of MEM, clarified at 5,000 × g for 5 min, and assayed on RSC monolayers for infectious virus titer (68, 69). The remaining mice were maintained for at least 40 days p.i. and employed in latency studies as described below.
Reactivation of latent HSV-1 by hyperthermic stress (HS) in vivo.
To determine the reactivation potential of the mutants in vivo, infected mice were subjected to a transient hyperthermia induction procedure as previously described (50, 51, 53, 54, 56, 57, 66, 67). Briefly, mice were inoculated as detailed above and maintained for at least 40 days. Animals were then subjected to 10 min of hyperthermia at 43°C, and pairs of trigeminal ganglia (TG) were removed from sacrificed animals at 22 h posttreatment, homogenized, and plated on RSC monolayers to detect infectious virus as previously described. DNA was purified from positive cultures, and the genomic structure of the reactivating virus was confirmed by Southern blot restriction enzyme fragment length polymorphism analysis as described above and as previously described (50, 51, 53, 54, 57, 66, 67). In all cases the reactivated virus had a genomic structure indistinguishable from that of the infecting strain or mutant (data not shown).
Analysis of mRNA expression in vitro.
RSC monolayers were infected at an MOI of 5 or 10 PFU per cell as indicated in the text. At 2, 4, and 6 h p.i., RNA was isolated using Ultraspec RNA (Biotecx Laboratories) according to the manufacturer's protocol. A total of 10 μg of RNA was glyoxylated, electrophoresed, transferred to nylon membranes (GeneScreen), and probed as described below (66). To control for gel loading artifacts, all blots were simultaneously probed (using a probe containing nucleotides −18 to +186) for the highly conserved L32 ribosomal protein (16).
The blots were incubated for 2 h at 50°C in prehybridization mix, a 32P-labeled probe specific for either the second exon of ICP0 (bp 122709 to 123030) or the E. coli β-Gal gene was then added, and the blots were incubated at 50°C for an additional 24 h as previously described (66). The blots were washed in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 1% sodium dodecyl sulfate at 65°C and exposed to a storage phosphor screen (Molecular Dynamics). Plates were analyzed using a STORM 860 phosphor imaging system and ImageQuant software.
For determination of IE gene kinetics, RSC were plated in 60-mm-diameter culture plates (Falcon) for 16 h. Medium was removed from the monolayers and replaced with medium with or without 50 μg of cycloheximide/ml for incubation for 30 min at 37°C. Following this preincubation, the cells were infected for 1 h at 37°C at an MOI of 10. Cycloheximide at 50 μg/ml was added to the virus inoculum on the pretreated cells. At the times p.i. indicated in the text, total RNA was isolated with Ultraspec (Biotecx) and analyzed as described above. To distinguish between early or late gene expression patterns, viral DNA synthesis was inhibited with phosphonoacetic acid (PAA) or acyclovir (ACV). Cells were pretreated with 500 μg/ml of PAA for 60 min, infected in the presence of PAA as described above, and maintained in PAA containing medium. At the times indicated in the text, total RNA was harvested and analyzed as described above. Similar experiments were performed with ACV at 100 μM.
Histochemical and immunohistochemical staining of whole ganglia.
Immunohistochemistry assays on whole ganglia were carried out with modifications of a procedure previously reported by Luque et al. (42). This method is detailed and has been validated as a method for the detection of reactivating neurons (52). In brief, mice were euthanized and TG were rapidly removed and placed in 0.5% paraformaldehyde for 2 h, rinsed in PBS twice for 15 min, and placed in X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) buffer containing 100 μg of X-Gal/ml for 5 h at 37°C (55). Following rinsing in phosphate-buffered saline (PBS), ganglia were postfixed overnight in methanol containing 20% dimethyl sulfoxide (DMSO). Ganglia were then incubated for 1 h in a solution of methanol containing 20% DMSO and 10% H2O2. Following two 15-min rinses in 100% methanol, ganglia were stored overnight in methanol at −70°C. Ganglia were allowed to equilibrate at room temperature for 15 min, rinsed twice in PBS, and incubated for 2 h at 37°C in PBS containing 3.6 mg of β-d+-glucose (Sigma)/ml, 100 μg of glucose oxidase (Sigma)/ml, and 130 μg of sodium azide (Mallinckrodt)/ml (42).
Incubation with primary antibody.
Ganglia were rinsed twice in PBS and incubated overnight at 37°C in the primary antibodies (rabbit anti-HSV-1 and rabbit anti-HSV-2 [Accurate] diluted 1:3,000, rabbit anti-VP16 [Clonetech] diluted 1:500, and rabbit anti-ICP0 [Zymed] diluted 1:500) in PBS containing 2% bovine serum albumin, 5% DMSO, and 5% normal horse serum.
Incubation with horseradish peroxidase conjugate.
Following rinsing for 5 h (which included five changes of PBS), ganglia were incubated overnight at room temperature in a 1:500 dilution of anti-rabbit horseradish peroxidase conjugate (Vector) in PBS containing 2% bovine serum albumin, 5% DMSO, and 5% normal horse serum. Tissue was again rinsed in PBS (including five changes of PBS over a period of 5 h) followed by a final rinse in 0.05 M Tris-Cl (pH 8.2).
Color development.
Ganglia were then incubated in a solution containing 250 μg of diaminobenzidine (Aldrich)/ml and 0.004% H202 in 0.1 M Tris (pH 8.2). The reaction was carefully monitored by visualizing color development and stopped by rinsing in distilled water. Rinsed ganglia were cleared in glycerol and mounted between two glass slides.
RESULTS
Construction and characterization of promoter/reporter viruses.
Transcription of ICP0 (as an IE gene) during lytic infection is induced by the interaction of viral and host proteins with a cis-acting promoter sequence known as a TAATGARAT sequence (reviewed in reference 73). Several additional host cell-encoded transcription factors have been shown to affect basal IE gene promoter activity in transient transfection assays of cultures of various cell lines (13, 32, 34, 40, 46) (Fig. 1). The importance of these particular factors in the context of viral infection in vivo, however, has not been tested.
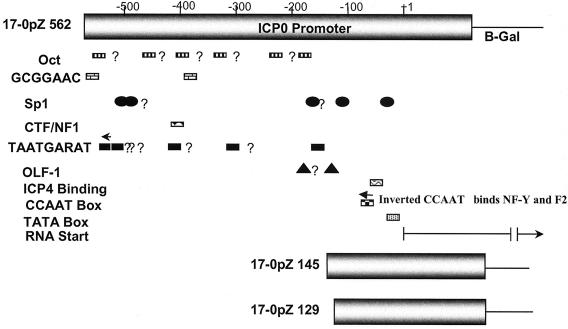
FIG. 1.
A schematic representation of the ICP0 promoter/reporter gene constructs used to generate reporter mutants. Binding sites for factors shown to influence ICP0 promoter function in transient transfection assays in tissue culture cells are indicated. Question marks denote potential TAATGRAT sequences, Octa protein binding sites (Oct), or factor binding sequences similar to those shown to be functional in a more proximal promoter region in vitro.
As a first step toward analyzing ICP0 promoter function in vivo, we sought to distinguish between the importance of cis sequences that confer IE gene expression (e.g., TAATGARATs) and that of other known promoter elements. The overall strategy employed is shown schematically in Fig. 1. Sequences from bp −562 of the ICP0 promoter through the entire 89-bp 5′ untranslated region of the ICP0 mRNA were utilized as the full-length promoter. These sequences contain all known promoter elements implicated in ICP0 transcriptional regulation in cell cultures as well as six additional upstream TAATGARAT-like sequences that might be functional in vivo. The 562-bp promoter was first fused to the β-Gal gene to generate a protein in which the methionine translation start of ICP0 was fused in frame to the β-Gal open reading frame and then inserted into the gC locus of HSV-1 strain 17syn+. This mutant was designated 17-0pZ562gC (17, HSV-1 strain 17syn+; 0pZ, 0 promoter driving LacZ; 562, length of promoter in base pairs; gC, site of insertion). To control for potential effects of the genomic context, this 562-bp promoter β-Gal construct was also inserted into the gJ locus to generate two independently derived mutants. These mutants were designated 17-0pZ562gJ-1 and 17-0pZ562gJ-14. ICP0 promoters were truncated at the 5′ end at bp −145 and bp −129 and employed to generate two additional mutants designated 17-0pZ145gC and 17-0pZ129gC, respectively.
The purpose of these truncations was to remove all elements known to induce IE gene expression (including all potential TAATGARATs) while preserving other important promoter elements, including the CCAAT box (shown by O'Rourke and O'Hare to be of paramount importance for basal promoter activity) (46), the ICP4 binding site, the TATA box, and the 5′ untranslated region (Fig. 1). The genomic structures of the mutants were confirmed by restriction enzyme fragment length polymorphism analysis, utilizing various restriction endonucleases and probes specific for β-Gal or various regions of the HSV-1 genome as previously described (data not shown) (65, 66).
Replication kinetics in vitro.
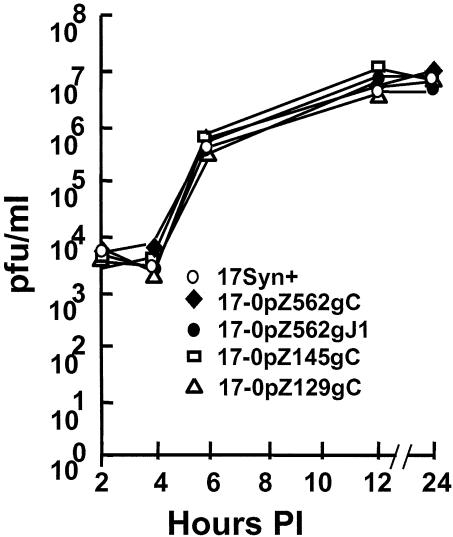
Other null mutants at the gC and gJ loci have been shown to replicate normally in cell cultures (2, 39, 61). Therefore, any generalized replication defects in our mutants may indicate the presence of unknown second-site mutations that could conceivably affect ICP0 promoter function. Replication kinetic experiments performed in RSC and SK-N-SH cell monolayers at high (10 PFU/cell; Fig. 2) and low (0.01 PFU/cell; not shown) MOIs did not reveal any generalized replication defects. There was no evidence of any unselected second-site mutations that affected viral replication in these cell types.
FIG. 2.
Single-step viral replication kinetics in rabbit RSC monolayers. Cultures were infected with the indicated viruses at an MOI of 10. At the times indicated, triplicate samples were harvested, frozen and thawed three times, and analyzed for virus content.
Kinetics of reporter mRNA transcription.
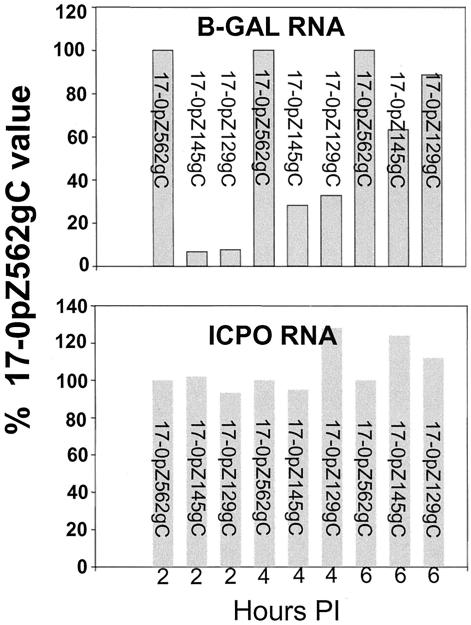
Expression from the inserted promoter in infected tissue cultures was directly compared to that of the native gene promoter at the transcriptional level (Fig. 3). Mutants containing the 562-bp promoter expressed β-Gal mRNA with kinetics similar to that of the ICP0 transcript and reached maximal levels by 2 h p.i. At all time points tested, the ratios of ICP0/β-Gal signal were similar in RNAs isolated from 17-0pZ562gC-infected RSCs (Fig. 3) and SK-N-SH neuroblastoma cells (data not shown). Similar findings were obtained with both independent isolates of 17-0pZ562gJ (data not shown). In contrast, the expression of β-Gal mRNA by mutants 17-0pZ145gC and 17-0pZ129gC was considerably delayed compared to that of ICP0. Significant levels of β-Gal mRNA were not detected until 4 h p.i., and expression levels were still reduced at 6 h p.i. At 2 h p.i., the ICP0/β-Gal mRNA ratio was more than 20-fold higher in cultures infected with the truncated promoter mutants than in cultures infected with either 17-0pZ562gC or 17-0pZ562gJ. By these criteria, expression of the 562-bp promoter was similar to that of the native ICP0 gene and the truncated promoters appeared to be expressed with delayed early or late kinetics.
FIG. 3.
Expression of ICP0 or β-Gal mRNA at 2 h p.i. RSC monolayers were infected at an MOI of 5, and total cellular RNA was harvested from triplicate cultures at 2, 4, and 6 h p.i. Total RNA was electrophoresed, blotted, transferred, and hybridized to probes specific for β-Gal, ICP0, or the cellular transcript L32, which was employed as a loading control. The signals were quantified with a PhosphorImager using ImageQuant software and normalized to the L32 control signal. The bar graphs represent the percentages of normalized signal relative to the 17-0pZ562gC signal, which was set to 100%.
Transcription in the absence of protein production or viral DNA replication.
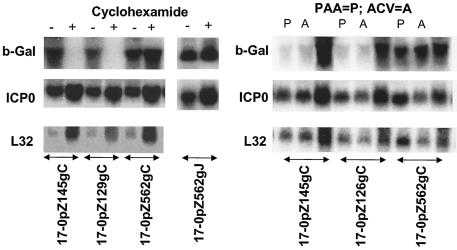
Cycloheximide blocks protein synthesis, and only IE viral mRNAs are expressed in the presence of this inhibitor (29). As anticipated, strains 17-0pZ562gC and 17-0pZ562gJ produced both ICP0 and β-Gal mRNAs in the presence of cycloheximide (Fig. 4). In cultures infected with 17-0pZ145gC or 17-0pZ129gC the production of ICP0 mRNA appeared similar to that seen with the wild-type strain, but no detectable β-Gal mRNA was produced in the presence of cycloheximide. Thus, expression of the 562-bp promoter in the presence of the protein synthesis inhibitor cycloheximide was similar to that of an IE gene but the truncated promoters were not expressed.
FIG. 4.
RNA transcription in the absence of viral protein production or viral DNA replication. RSC monolayers were infected at an MOI of 5. (Left panels) Cultures were maintained for 16 h in the presence of cycloheximide. A total of 10 μg of total RNA was loaded per lane, and the blots were analyzed for ICP0 or β-Gal mRNA. Signals were normalized to L32, a cellular ribosomal gene employed as a loading control. (Right panels) Infected cultures were maintained for 6 h in the presence or absence of PAA (500 μg/ml) or ACV (100 μM). RNA was isolated and processed as described in the Fig. 3 legend.
Early viral genes are expressed to high levels prior to viral DNA replication and delayed early genes are expressed at low levels in the absence of DNA replication, while expression of true late genes requires DNA synthesis (73). In all samples from cells infected with mutants expressing the 562-bp ICP0 promoter/reporter in the presence of either of two viral DNA synthesis inhibitors (PAA and ACV), the ratios of ICP0 to β-Gal signal were similar. The levels of β-Gal mRNA in cells infected with either strain 17-0pZ145gC or 17-0pZ129gC were severely reduced in the presence of either drug, however, although positive hybridization signals were still detectable (Fig. 4). The inhibition of viral DNA synthesis resulted in high ratios of ICP0 to β-Gal signal (>50:1) compared to the ratio seen in the absence of DNA replication inhibitors (approximately 0.5:1). By these criteria, the truncated ICP0 promoters were expressed with leaky late (γ1) kinetics.
Analysis of the promoter/reporter viruses in vivo.
In vivo, the lack of synchrony of replication and the diversity of infected cell types added a level of complexity that required alternative strategies for evaluation of promoter reporter function. In this study, we adopted the following criteria as indicative of the presence of a promoter/reporter that accurately reflects the endogenous gene's promoter in a particular in vivo context. First, during acute infection the cells that show evidence of expression of the reporter β-Gal should also express the protein from the native locus. Second, reporter gene expression levels from two different loci (both the gC and gJ loci) should be equivalent during the acute phase of infection, demonstrating that the context in which the promoter resides is not a dominant factor for expression. Finally, the kinetics of repression of the promoter/reporter gene (that is, the time p.i. that the promoter is shut off in neurons) should be similar to the kinetics of repression of the native promoter. Meeting these criteria would indicate that the function of the test promoter in vivo accurately reflected expression of the native promoter and strongly suggest that meaningful data on promoter function in vivo could be obtained.
Replication kinetics in vivo.
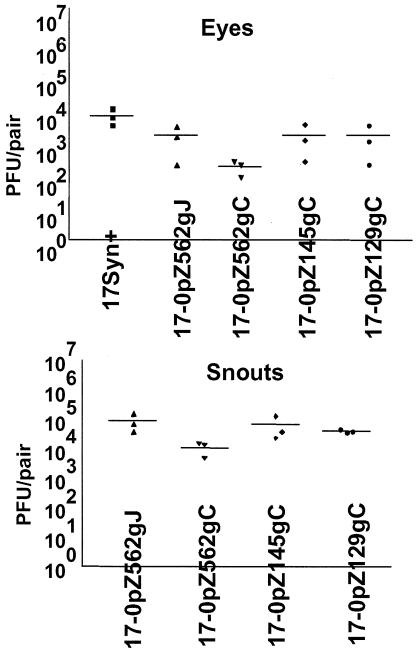
Groups of animals were inoculated with the promoter/reporter viruses or the parental strain 17syn+ on corneas and snouts, and eyes and snouts were examined for virus content on day 4 p.i. As can be seen in Fig. 5, all viruses replicated efficiently in peripheral tissues. In this experiment the replication of mutant strain 170pZ562gC was reduced somewhat from that of the other isolates, but in other experiments this was not the case. This experiment demonstrated that all tested mutants were capable of efficient replication at the surface, and as is shown below (Fig. 6), all four promoter/reporter mutants reached the TG and expressed viral proteins in neurons efficiently at 4 days p.i.
FIG. 5.
Virus replication in vivo. Mice were infected via the cornea and snout with the total of 2 × 105 PFU of mutant or wild-type viruses. On day 4 p.i., tissues were analyzed for virus content. Each point on the graph represents the amount of virus obtained from an individual mouse. The horizontal bars indicate the means of the results for the corresponding groups.
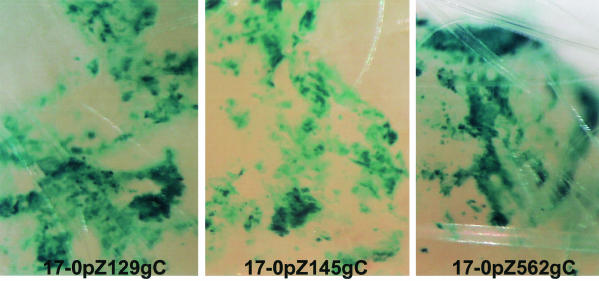
FIG. 6.
Expression of β-Gal in mouse snouts in vivo. Mice were infected with 105 PFU of the promoter reporter mutants, and at 4 days p.i. tissues were processed for the detection of β-Gal. Snout tissue from the area of inoculation was cleared in xylene and photographed under a dissecting microscope. The presence of a blue precipitate indicates reporter gene activity.
Comparison of the general distribution of β-Gal and lytic viral protein expression in tissues in vivo.
Additional mice infected as described above were processed for the detection of either HSV proteins or β-Gal activity at day 4 p.i. To facilitate these comparative studies, a modification of procedures developed to immunohistochemically stain whole-mouse embryos (42) was employed to stain whole-mouse tissues for the expression of viral proteins (see Materials and Methods). Peripheral tissues: As anticipated from previous studies on the distribution of HSV proteins during the acute stage of infection (Sawtell and Thompson [55], Simmons et al. [59a], Balan et al. [2], and Sawtell [52]) detectable evidence of viral protein and β-Gal activity was found in numerous cells in the cornea and snouts of these mice. All of the mutants exhibited similar general distribution patterns of β-Gal expression, and these patterns were similar to the pattern of HSV protein expression (data not shown). Representative samples of β-Gal expression from mouse snouts are shown in Fig. 6. This experiment demonstrates that all of the promoter/reporter constructs were expressed efficiently in peripheral mouse tissues in vivo. The expression of the truncated promoters most likely represents the basal expression with leaky late kinetics detected in cell cultures.
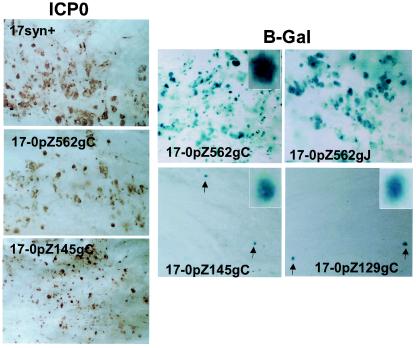
In ganglia infected with the 17-0pZ562gJ or 17-0pZ562gC (β-Gal expressed from the 562-bp ICP0 promoter) mutant strain, the distribution pattern of β-Gal activity was similar to that of viral proteins (Fig. 7; compare brown staining for viral protein in the left panels with blue staining for β-Gal in the right panels). Quite unexpectedly, this concordance of β-Gal activity and viral protein expression was not found with TG infected with the truncated promoter/reporter mutants 17-0pZ145gC and 17-0pZ129gC. Although there was abundant expression of viral proteins (similar to that observed for TG infected with the 562-bp promoter/reporter mutants) (Fig. 7), β-Gal-positive cells were very rare. The ratio of β-Gal-positive/viral protein-positive neural cells was at least 100-fold reduced compared to that of the 562-bp promoter mutants. The basal promoter activity of the truncated promoters seen in other cells in vivo was not seen in TG neurons.
FIG. 7.
Whole-ganglion histochemical and immunohistochemical detection of β-Gal and ICP0 protein. Mice were infected with 2 ×105 PFU of the mutant or wild-type isolates, and at 4 days p.i. TG were processed for the detection of β-Gal activity or ICP0 protein as detailed in Materials and Methods. (Left three panels) Whole mounts of TG processed for the detection of ICP0 protein. A brown precipitate indicates the presence of ICP0 protein. Numerous neurons and support cells gave positive results in all samples. (Right four panels) Whole mounts of ganglia processed for detection of β-Gal (indicated by a blue precipitate). The top panels demonstrate that the 562-bp promoter was expressed in numerous cells regardless of whether the reporter gene was inserted into the gC or gJ locus. The inset shows a high-power-magnification field of a positive neuron. The lower panels depict whole mounts of ganglia infected with the truncated promoter mutants. Cells giving positive results (arrows) were very rare. The insets depict high-power-magnification fields of neurons giving positive results.
The lack of expression from the truncated promoters was not due to a lack of expression of the leaky late class of proteins.
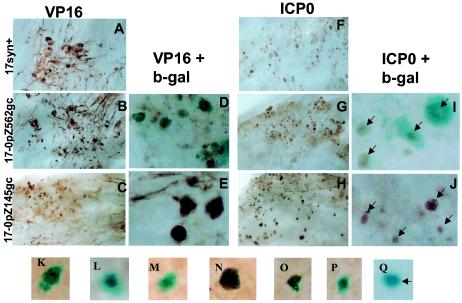
In tissue culture, the truncated ICP0 promoters expressed with leaky late kinetics (see above). It was formally possible that these mutants had some unknown mutation that precluded leaky late or late gene expression in TG. In this case, IE and early proteins would be expressed and thus detected by the anti-HSV antibody but the expression of leaky late or late promoters (including the truncated ICP0 promoters) would be impaired. To test this possibility, immunohistochemical staining of whole ganglia with an antibody directed against VP16 (a leaky late protein) was performed. Widespread staining for VP16 was observed in ganglia infected with the wild-type strain (Fig. 8A) or the promoter/reporter mutants (panels B and C), confirming that all the viruses expressed leaky late genes in TG. In ganglia infected with strain 17-0pZ562gJ, dual staining for β-Gal and VP16 revealed that most cells positive for VP16 were also positive for β-Gal. This was revealed by the presence of a brown precipitate (marking VP16 positive cells) that colocalized with the blue precipitate produced by cleavage of X-Gal (panel D). VP16-positive cells were numerous in ganglia infected with 17-0pZ145gC, but these cells were not positive for β-Gal (panel E).
FIG. 8.
Colocalization of ICP0 or VP16 protein and β-Gal activity. Mice were infected with 2 × 105 PFU of wild-type or promoter/reporter strains and processed for the detection of specific viral proteins and β-Gal. (A, B, and C) Whole mounts of ganglia stained immunohistochemically for VP16 revealed widespread positive signal in numerous neurons and support cells (brown precipitate). (D and E) Micrographs of ganglia processed for the simultaneous expression of VP16 and β-Gal. Virtually all neurons positive for VP16 (brown precipitate) in the ganglia infected with 17-0pZ562gC (D) were also positive for β-Gal (blue precipitate). (E) No dually stained cells were evident in the ganglion infected with 17-0pZ145gC. (F, G, and H) A similar analysis performed with antibody specific for ICP0. Positive signal for ICP0 (brown precipitate) was widespread throughout all of the ganglia, regardless of the identity of the infecting strain. (I) β-Gal expression (blue precipitate) colocalized with ICP0 in the great majority of cells giving positive results in ganglia infected with the 562-bp promoter/reporter virus (arrows). No β-Gal signal was evident in ICP0-positive cells (brown precipitate) (arrows in panel J) in ganglia infected with the truncated promoter mutant (arrows). (K to Q) Mice were similarly infected with a LAT null mutant carrying the 562-bp promoter/reporter, maintained for 40 days p.i., and subjected to the HS reactivation stress, and TG were stained for viral protein and β-Gal at 22 h post-HS. Viral protein and β-Gal colocalized in these reactivating neurons (K to P), and only one neuron expressed β-Gal in the absence of detectable viral proteins (O).
Colocalization of ICP0 protein and β-Gal in TG.
A similar strategy was employed to examine the extent to which β-Gal and ICP0 protein were colocalized within the ganglia. Additional TG were processed first for the histochemical detection of β-Gal and subsequently for the immunohistochemical detection of ICP0. Lower-power micrographs showing that the ganglia contained many cells positive for ICP0 regardless of the infecting mutant are presented in Fig. 8F to H. In TG infected with strain 17-0pZ562gJ or 17-0pZ562gC, >95% of cells positive for ICP0 protein also contained detectable β-Gal activity. The converse was also true in that those cells that expressed β-Gal also contained detectable levels of ICP0. However, dual staining of cells was quite rare in ganglia infected with 17-0pZ145gC. In those ganglia, the great majority (>95%) of the cells positive for ICP0 did not contain detectable β-Gal activity (panel J).
Similar studies were performed at days 6 and 12 p.i. with the mutants containing the 562-bp ICP0 promoter/reporter constructs. The number of cells positive for β-Gal expression was decreased at day 6, and this was coincident with a decrease in the number of cells positive for viral proteins. By 12 days p.i., positive cells were extremely rare. At both of these later time points β-Gal activity and ICP0 protein expression occurred within the same cells, demonstrating that the kinetics of shutoff of the promoter gene reflected the shutoff of the native gene.
Expression of the ICP0 promoter during reactivation in vivo.
Additional mice infected with strain 17syn+, 17-0pZ562gC, or 17-0pZ562gJ as described above were maintained for a minimum of 40 days p.i. and subjected to HS to reactivate the virus in vivo (55). At 22 h post-HS, TG were removed and either processed to detect reactivation of HSV or stained to detect the expression of β-Gal and viral proteins. Both 17-0pZ562gC and 17-0pZ562gJ reactivated efficiently, with virus detected in 70 to 80% of the animals tested; this result was similar to that seen with the 17syn+ control group (80%). As measured by this assay, therefore, disruption of either the gC or gJ locus did not negatively impact the ability of the mutants to reactivate in vivo.
The whole-ganglion staining procedures demonstrated a close correlation between the number of ganglia in which at least one or more reactivating neurons (viral protein positive) were detected and the frequency with which reactivation was detected by standard virus assays. Our findings are tabulated in Table 1. Of 29 ganglia latently infected with strain 17-0pZ562.gJ, 24 contained at least one neuron expressing lytic viral proteins at 22 h post-HS (83%). These represented ganglia from 12 of 15 (80%) different mice. In the latently infected mice that were not subjected to the HS induction procedure, no viral protein-positive cells (0/26 ganglia) were detected in this experiment. During the course of these studies many additional ganglia from similarly infected mice were examined (data not shown), and two contained a single neuron positive for β-Gal activity. These neurons did not contain detectable viral protein. A similar percentage (85% [22/26]) of ganglia containing reactivating neurons was found in the second experiment with mice latently infected with 17-0pZ562gJ at 22 h post-HS.
TABLE 1.
Detection of ICP0 protein and ICP0 promoter activity during HSV reactivation in vivoa
| Strain | No. (%) of reactivating neurons containing indicated precipitate(s)/ total no. of neurons detected
|
No. (%) of ganglia containing one or more neurons positive for either viral protein or β-Gal/total no. of ganglia examined | ||
|---|---|---|---|---|
| Blue | Blue + brown | Brown | ||
| 17-0pZ562gJb | 45/113 (40) | 68/113 (60) | 0/113 (0) | 24/29 (83) |
| 17-0pZ562gJc | 26/77 (34) | 51/77 (66) | 0/77 (0) | 22/26 (85) |
| 17AH-0pZ562gJc | 1/14 (7) | 13/14 (93) | 0/14 (0) | 7/24 (29) |
Mice were infected with strain 17-0pZ562gJ and the corresponding LAT null mutant 17AH-0pZ562gJ, maintained for at least 40 days p.i., and subjected to the HS in vivo reactivation procedure. At 23 h poststress, ganglia were removed and processed to detect viral protein (brown precipitate) and promoter reporter activity (β-Gal; blue precipitate).
Experiment 1.
Experiment 2.
The presence of β-Gal activity driven by the full-length promoter did not correlate with the presence of ICP0 in reactivating neurons.
In contrast to the close correlation between protein and β-Gal expression observed during acute infection, during reactivation β-Gal and viral protein expression correlated less well. In a total of 29 ganglia examined in experiment 1, 113 neurons expressing either β-Gal or β-Gal plus viral proteins were detected. Neurons expressing viral proteins in the absence of β-Gal were not detected. Both viral protein expression and β-Gal expression were detected in 68 neurons, and the remaining 45 (40%) neurons giving positive results expressed only β-Gal. Similar findings were obtained in a second experiment, in which 66% of the neurons giving positive results expressed both β-Gal and viral proteins and 34% of neurons giving positive results were positive only for β-Gal (see Table 1).
These findings suggested two possibilities. Either the ICP0 promoter was active in some neurons that did not progress to the production of any detectable viral proteins, or, unlike the results seen at the acute stage of infection (Fig. 6), the detection of β-Gal in reactivating neurons was more sensitive than the detection of ICP0. This latter possibility might result from the masking of the brown precipitate by a heavy blue precipitate, by differential stabilities of β-Gal and viral protein, or by incomplete penetration of the antibodies into the tissues. Fortuitously, the availability of a recently derived mutant in which this same 562-bp ICP0 promoter/β-Gal reporter cassette is expressed from the gJ locus on a latency-associated-transcript (LAT) null mutant background allowed us to distinguish between these possibilities.
The mutant 17AH-0pZ562gJ strain was generated using the previously described LAT null mutant 17AH (65) as the parental strain. In vivo reactivation was induced in mice latently infected with this mutant as described above, and ganglia were examined at 22 h poststress for the presence of viral proteins and β-Gal. As shown in Table 1, 29% of the ganglia showed evidence of reactivating neurons. The lower frequency of reactivation induced by the LAT null mutant was not unexpected, as LAT null mutants establish far fewer latent infections in mice (55, 65). Of the reactivating neurons, 93% contained both the blue and brown precipitates, demonstrating a high degree of concordance between the reporter gene and the presence of viral protein. Only one reactivating neuron contained only the blue precipitate indicative of reporter gene expression in the absence of detectable viral protein. Representative reactivating neurons are shown in Fig. 8, panels K through Q. Panels K through P show neurons that contain both blue and brown precipitate, whereas panel Q illustrates the single neuron found to contain only blue precipitate. Thus, the antibodies penetrated the tissue efficiently; there was no evidence of differential protein stability, and the blue precipitate did not prevent detection of the brown precipitate.
DISCUSSION
Considerable phenotypic evidence supports the hypothesis that ICP0 plays some direct role in latency. However, it is still not clear whether ICP0 initiates reactivation from latency, is required solely for the efficient establishment of latency, or is needed for progression to virus production once reactivation has been initiated. Attempts to employ ICP0 null mutants to study its role in vivo have been complicated by the loss of the many important functions this protein performs during the infection cycle (7, 10, 27, 33, 37). In this report, the pattern of gene expression from the ICP0 promoter during the acute stage of infection (and during reactivation from latency in vivo) was examined by the utilization of recombinant promoter/reporter viral mutants. If the expression of ICP0 is a precipitating event in the initiation of reactivation, as has been hypothesized (reviewed in reference 19), analysis of its promoter should allow the cis-acting sequences that respond to reactivation triggers to be deduced.
The use of a promoter/reporter strategy eliminates the problems that would be caused by altered regulation of the native ICP0 gene, but there are no published studies that demonstrate that such a strategy can accurately reflect expression from the native ICP0 locus in TG neurons in vivo. The recent development of a whole-ganglion immunohistochemistry procedure made practical the direct comparison of expression of the native protein to that of the reporter β-Gal gene at the single-cell level in entire ganglia. Our findings permit us to draw the following three conclusions. First, a DNA fragment from bp −562 through the methionine start codon of the ICP0 gene contains all sequences required for proper ICP0 expression in diverse cell types in vitro and in vivo, including sensory neurons of the TG. Second, sequences upstream of bp −145 in the ICP0 promoter are required for basal expression in neurons during acute infection but not for basal expression in certain other cell types in vivo. Third, in direct contrast to the coexpression of the native protein and the β-Gal reporter seen during the acute phase of infection, nearly twice as many neurons expressed the ICP0 promoter/reporter during reactivation in vivo as expressed detectable ICP0 protein.
Expression of a 562-bp ICP0 promoter is identical to that of the native ICP0 locus in vitro and in vivo.
Several groups have employed promoter/reporter viruses to investigate promoter function in cell cultures and in vivo (5, 14, 17, 25, 30, 31, 36). A major drawback to such studies has been the uncertainty that the expression from promoter/reporter construct accurately reflects expression of the native gene. Indeed, it has been reported that the genomic context in which a promoter/reporter construct resides can exert a great influence on the expression of the reporter gene (23). Synchronous infection is possible in cell cultures, and so direct comparisons of transcription from the native and reporter promoters are also possible. We found that as would be expected from the presence of TAATGARAT sequences that confer IE gene kinetics (6, 35, 44), in infected-tissue cell cultures the 562-bp ICP0 promoter was expressed with IE kinetics that were indistinguishable from those of expression of the native ICP0 mRNA. There was no difference in the behavior of this promoter in the context of either the gC or the gJ locus, reducing concerns about the influence of promoter context on promoter function. The expression of promoters truncated at bp −129 or −145 was delayed, did not reach maximal levels until after 6 h p.i., and was reduced in the presence of inhibitors of DNA replication. Therefore, the truncated ICP0 promoters were expressed with leaky-late kinetics in cultured RSCs and in SK-N-SH neuroblastoma cells.
The expression of β-Gal in peripheral tissues in vivo was widespread and indistinguishable from the expression of viral proteins. This was true regardless of whether the 562-bp or the truncated ICP0 promoters were employed to drive β-Gal expression. It is likely that the expression from the truncated promoters in this tissue was similar to the leaky late expression of β-Gal seen in cell cultures.
In mouse TG, a very different pattern of expression was observed. The full-length ICP0 promoter drove the expression of β-Gal in virtually all TG neurons in which viral proteins could be detected. In contrast, expression from the truncated promoters was extremely limited in TG. Less than 1% of the neurons that contained detectable viral proteins also contained detectable β-Gal. The lack of expression was not caused by an abortive infection of neurons that did not progress to leaky late gene expression, because the expression of VP16, a leaky late gene, was readily detectable in numerous cells within the ganglia. This result was unexpected, as previous work by O'Rourke and O'Hare demonstrated that a ∼90-bp promoter that retained an inverted CCAAT box at bp −71 yielded strong basal expression of the ICP0 promoter equivalent to that of the simian virus 40 early-region enhancer/promoter in cell cultures (46).
The structure of the truncated promoters resembles that of early or leaky late promoters, at least superficially, in that they contain functional CCAAT boxes, TATA boxes, several potential binding sites for transcription factors (e.g., two known SP1 binding sites), and the native ICP0 5′ untranslated region and mRNA start site. However, they must lack sequences that are very important for expression in neurons but not in certain other cell types. Likely candidates include the initiator-like sequences near the mRNA start sites of leaky late promoters previously described (38, 39) as well as downstream elements that have been shown to be important for the full expression of this class of HSV genes (3, 4, 72). Alternately, these promoters may contain neural tissue-specific down-regulatory signals that can be overcome by upstream sequences.
During latency, no expression of ICP0 protein was detected and only two neurons positive for β-Gal expressed from the full-length ICP0 promoter/reporter were detected. Schimeld et al. also did not detect β-Gal-positive neurons with a virtually identical reporter mutant in strain SC16 during latency (58), but Thackray and Field did detect many β-Gal-positive neurons with that same SC16 mutant during latency (62). It may be that the different strains of mice or different routes of inoculation employed by Thackray and Field resulted in this observation. Loiacono et al. employed transgenic mice expressing β-Gal from the ICP0 and other promoters to examine promoter function in neurons in vivo in the absence of viral gene products. Expression of β-Gal from the ICP0 promoter was detected in some sensory neurons in several transgenic lines (41). Very rare neurons express viral proteins during latency (21, 52, 59), and the differences noted between these studies may reflect differences in the probability of detecting such rare events in the various systems.
Evidence for a posttranscriptional constraint on ICP0 protein production during reactivation.
Following HS of mice latently infected with the full-length promoter/reporter viruses, both β-Gal and ICP0 were detectable in very rare neurons within TG. About 80 to 85% of the mice examined had one or more HSV protein-positive neurons in their TG at 23 h post-HS, and this was similar to the percentage of the same group of mice in which virus was detected in TG at 23 h post-HS. Of interest, nearly twice as many neurons contained detectable β-Gal than contained detectable viral proteins including ICP0. One trivial explanation for this observation is that the detection of β-Gal was more sensitive than the detection of viral protein with whole-ganglion procedures. This seems an unlikely explanation, as the concordance between viral protein and β-Gal in TG neurons during the acute stage was virtually 100%. It is also not likely that these neurons represent the very early stages of viral reactivation (e.g., prior to detectable viral protein production), as the progression to antigen production was quite rapid (∼4 h) following HS and the intensity of the β-Gal staining in many neurons giving positive results was quite intense, suggesting the promoter had been active for a significant amount of time. Sequences that may contribute to the proper regulation of ICP0 may be contained within the introns of this gene, but these sequences appear to promote rather than inhibit gene expression (24, 48). Importantly, we found concordant expression of β-Gal and ICP0 protein during reactivation in 93% of reactivating neurons with the 562-bp promoter/reporter on the genetic background of a LAT null viral mutant, which argues strongly against a trivial explanation for this difference.
One intriguing possibility is that this phenomenon, which was observed only during reactivation from latency, represents a heretofore-unrecognized posttranscriptional restraint on the expression of ICP0 following a reactivation trigger. Such a restraint might serve to limit the number of neurons in which reactivation occurs during any given reactivation event, thereby preserving the latent pool. Further experimentation will determine whether this posttranscriptional constraint on ICP0 protein expression is mediated by LAT.
That there are differences in the functions of viral promoters in various cell types and/or under various physiologic conditions is not surprising. That sequences upstream of bp −145 in the ICP0 promoter are required specifically for expression in neuronal tissues, but not in other cell types, of mice was unexpected. Likewise, that significantly more neurons were found to express the ICP0 promoter/reporter in neurons in vivo following a reactivation stimulus than expressed ICP0 protein was also not predicted. These results emphasize the importance of studying viral promoters in the context of the viral genome during all phases of infection, including acute replication at the body surface, lytic infection of TG cells, maintenance of latency, and reactivation in vivo.
Acknowledgments
We thank Elisabeth Burger and Rebecca Haas for excellent technical assistance.
This work was funded by Public Health Service grant RO1 EY 13168 and also by RO1 AI 32121.
REFERENCES
- 1.apRhys, C. M., D. M. Ciufo, E. A. O'Neill, T. J. Kelly, and G. S. Hayward. 1989. Overlapping octamer and TAATGARAT motifs in the VF65-response elements in herpes simplex virus immediate-early promoters represent independent binding sites for cellular nuclear factor III. J. Virol. 63:2798-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balan, P., N. Davis-Poynter, S. Bell, H. Atkinson, H. Browne, and T. Minson. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 75:1245-1258. [DOI] [PubMed] [Google Scholar]
- 3.Blair, E. D., C. C. Blair, and E. K. Wagner. 1987. Herpes simplex virus virion stimulatory protein mRNA leader contains sequence elements which increase both virus-induced transcription and mRNA stability. J. Virol. 61:2499-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blair, E. D., and E. K. Wagner. 1986. A single regulatory region modulates both cis activation and trans activation of the herpes simplex virus VP5 promoter in transient-expression assays in vivo. J. Virol. 60:460-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom, D. C., N. T. Maidment, A. Tan, V. B. Dissette, L. T. Feldman, and J. G. Stevens. 1995. Long-term expression of a reporter gene from latent herpes simplex virus in the rat hippocampus. Brain Res. Mol. Brain Res. 31:48-60. [DOI] [PubMed] [Google Scholar]
- 6.Bzik, D. J., and C. M. Preston. 1986. Analysis of DNA sequences which regulate the transcription of herpes simplex virus immediate early gene 3: DNA sequences required for enhancer-like activity and response to trans-activation by a virion polypeptide. Nucleic Acids Res. 14:929-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, W., T. L. Astor, L. M. Liptak, C. Cho, D. M. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, W. Z., and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clements, G. B., and N. D. Stow. 1989. A herpes simplex virus type 1 mutant containing a deletion within immediate early gene 1 is latency-competent in mice. J. Gen. Virol. 70:2501-2506. [DOI] [PubMed] [Google Scholar]
- 11.Davido, D. J., and D. A. Leib. 1996. Role of cis-acting sequences of the ICPO promoter of herpes simplex virus type 1 in viral pathogenesis, latency and reactivation. J. Gen. Virol. 77:1853-1863. [DOI] [PubMed] [Google Scholar]
- 12.Desai, P., R. Ramakrishnan, Z. W. Lin, B. Osak, J. C. Glorioso, and M. Levine. 1993. The RR1 gene of herpes simplex virus type 1 is uniquely trans activated by ICP0 during infection. J. Virol. 67:6125-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devireddy, L. R., and C. J. Jones. 2000. Olf-1, a neuron-specific transcription factor, can activate the herpes simplex virus type 1-infected cell protein 0 promoter. J. Biol. Chem. 275:77-81. [DOI] [PubMed] [Google Scholar]
- 14.Dobson, A. T., T. P. Margolis, W. A. Gomes, and L. T. Feldman. 1995. In vivo deletion analysis of the herpes simplex virus type 1 latency-associated transcript promoter. J. Virol. 69:2264-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douville, P., M. Hagmann, O. Georgiev, and W. Schaffner. 1995. Positive and negative regulation at the herpes simplex virus ICP4 and ICP0 TAATGARAT motifs. Virology 207:107-116. [DOI] [PubMed] [Google Scholar]
- 16.Dudov, K. P., and R. P. Perry. 1984. The gene family encoding the mouse ribosomal protein L32 contains a uniquely expressed intron-containing gene and an unmutated processed gene. Cell 37:457-468. [DOI] [PubMed] [Google Scholar]
- 17.Ecob-Prince, M. S., K. Hassan, M. T. Denheen, and C. M. Preston. 1995. Expression of beta-galactosidase in neurons of dorsal root ganglia which are latently infected with herpes simplex virus type 1. J. Gen. Virol. 76:1527-1532. [DOI] [PubMed] [Google Scholar]
- 18.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 20.Everett, R. D., W. C. Earnshaw, A. F. Pluta, T. Sternsdorf, A. M. Ainsztein, M. Carmena, S. Ruchaud, W. L. Hsu, and A. Orr. 1999. A dynamic connection between centromeres and ND10 proteins. J. Cell Sci. 112:3443-3454. [DOI] [PubMed] [Google Scholar]
- 21.Feldman, L. T., A. R. Ellison, C. C. Voytek, L. Yang, P. Krause, and T. P. Margolis. 2002. Spontaneous molecular reactivation of herpes simplex virus type 1 latency in mice. Proc. Natl. Acad. Sci. USA 99:978-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelman, I. H., and S. Silverstein. 1986. Co-ordinate regulation of herpes simplex virus gene expression is mediated by the functional interaction of two immediate early gene products. J. Mol. Biol. 191:395-409. [DOI] [PubMed] [Google Scholar]
- 23.Goodart, S. A., J. F. Guzowski, M. K. Rice, and E. K. Wagner. 1992. Effect of genomic location on expression of beta-galactosidase mRNA controlled by the herpes simplex virus type 1 UL38 promoter. J. Virol. 66:2973-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu, W., Q. Huang, and G. S. Hayward. 1995. Multiple tandemly repeated binding sites for the YY1 repressor and transcription factors AP-1 and SP-1 are clustered within intron-1 of the gene encoding the IE110 transactivator of herpes simplex virus type 1. J. Biomed. Sci. 2:203-226. [DOI] [PubMed] [Google Scholar]
- 25.Guzowski, J. F., and E. K. Wagner. 1993. Mutational analysis of the herpes simplex virus type 1 strict late UL38 promoter/leader reveals two regions critical in transcriptional regulation. J. Virol. 67:5098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halford, W. P., C. D. Kemp, J. A. Isler, D. J. Davido, and P. A. Schaffer. 2001. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J. Virol. 75:6143-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halford, W. P., and P. A. Schaffer. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75:3240-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris, R. A., R. D. Everett, X. Zhu, S. Silverstein, and C. M. Preston. 1989. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J. Virol. 63:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang, C. B., B. Horsburgh, E. Pelosi, S. Roberts, P. Digard, and D. M. Coen. 1994. A net +1 frameshift permits synthesis of thymidine kinase from a drug-resistant herpes simplex virus mutant. Proc. Natl. Acad. Sci. USA 91:5461-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarman, R. G., E. K. Wagner, and D. C. Bloom. 1999. LAT expression during an acute HSV infection in the mouse. Virology 262:384-397. [DOI] [PubMed] [Google Scholar]
- 32.Jones, K. A., and R. Tjian. 1985. Sp1 binds to promoter sequences and activates herpes simplex virus “immediate-early” gene transcription in vitro. Nature 317:179-182. [DOI] [PubMed] [Google Scholar]
- 33.Katz, J. P., E. T. Bodin, and D. M. Coen. 1990. Quantitative polymerase chain reaction analysis of herpes simplex virus DNA in ganglia of mice infected with replication-incompetent mutants. J. Virol. 64:4288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kemp, L. M., C. L. Dent, and D. S. Latchman. 1990. Octamer motif mediates transcriptional repression of HSV immediate-early genes and octamer-containing cellular promoters in neuronal cells. Neuron 4:215-222. [DOI] [PubMed] [Google Scholar]
- 35.Kristie, T. M., and B. Roizman. 1987. Host cell proteins bind to the cis-acting site required for virion-mediated induction of herpes simplex virus 1 alpha genes. Proc. Natl. Acad. Sci. USA 84:71-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lachmann, R. H., M. Sadarangani, H. R. Atkinson, and S. Efstathiou. 1999. An analysis of herpes simplex virus gene expression during latency establishment and reactivation. J. Gen. Virol. 80:1271-1282. [DOI] [PubMed] [Google Scholar]
- 37.Leib, D. A., D. M. Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieu, P. T., N. T. Pande, M. K. Rice, and E. K. Wagner. 2000. The exchange of cognate TATA boxes results in a corresponding change in the strength of two HSV-1 early promoters. Virus Genes 20:5-10. [DOI] [PubMed] [Google Scholar]
- 39.Lieu, P. T., and E. K. Wagner. 2000. Two leaky-late HSV-1 promoters differ significantly in structural architecture. Virology 272:191-203. [DOI] [PubMed] [Google Scholar]
- 40.Lillycrop, K. A., J. K. Estridge, and D. S. Latchman. 1993. The octamer binding protein Oct-2 inhibits transactivation of the herpes simplex virus immediate-early genes by the virion protein Vmw65. Virology 196:888-891. [DOI] [PubMed] [Google Scholar]
- 41.Loiacono, C. M., R. Myers, and W. J. Mitchell. 2002. Neurons differentially activate the herpes simplex virus type 1 immediate-early gene ICP0 and ICP27 promoters in transgenic mice. J. Virol. 76:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luque, J. M., W. B. Adams, and J. G. Nicholls. 1998. Procedures for whole-mount immunohistochemistry and in situ hybridization of immature mammalian CNS. Brain Res. Protoc. 2:165-173. [DOI] [PubMed] [Google Scholar]
- 43.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 44.McKnight, J. L., T. M. Kristie, and B. Roizman. 1987. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc. Natl. Acad. Sci. USA 84:7061-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Rourke, D., and P. O'Hare. 1993. Mutually exclusive binding of two cellular factors within a critical promoter region of the gene for the IE110k protein of herpes simplex virus. J. Virol. 67:7201-7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry, L. J., and D. J. McGeoch. 1988. The DNA sequences of the long repeat region and adjoining parts of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:2831-2846. [DOI] [PubMed] [Google Scholar]
- 48.Poon, A. P., S. J. Silverstein, and B. Roizman. 2002. An early regulatory function required in a cell type-dependent manner is expressed by the genomic but not the cDNA copy of the herpes simplex virus 1 gene encoding infected cell protein 0. J. Virol. 76:9744-9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roizman, B. 1999. HSV gene functions: what have we learned that could be generally applicable to its near and distant cousins? Acta Virol. 43:75-80. [PubMed] [Google Scholar]
- 50.Sawtell, N. M. 1997. Comprehensive quantification of herpes simplex virus latency at the single-cell level. J. Virol. 71:5423-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawtell, N. M. 1998. The probability of in vivo reactivation of herpes simplex virus type 1 increases with the number of latently infected neurons in the ganglia. J. Virol. 72:6888-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawtell, N. M. 2003. Quantitative analysis of herpes simplex virus reactivation in vivo demonstrates that reactivation in the nervous system is not inhibited at early times postinoculation. J. Virol. 77:4127-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sawtell, N. M., D. I. Bernstein, and L. R. Stanberry. 1999. A temporal analysis of acyclovir inhibition of induced herpes simplex virus type 1 In vivo reactivation in the mouse trigeminal ganglia. J. Infect. Dis. 180:821-823. [DOI] [PubMed] [Google Scholar]
- 54.Sawtell, N. M., D. K. Poon, C. S. Tansky, and R. L. Thompson. 1998. The latent herpes simplex virus type 1 genome copy number in individual neurons is virus strain specific and correlates with reactivation. J. Virol. 72:5343-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawtell, N. M., and R. L. Thompson. 1992. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J. Virol. 66:2157-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawtell, N. M., and R. L. Thompson. 1992. Rapid in vivo reactivation of herpes simplex virus in latently infected murine ganglionic neurons after transient hyperthermia. J. Virol. 66:2150-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawtell, N. M., R. L. Thompson, L. R. Stanberry, and D. I. Bernstein. 2001. Early intervention with high-dose acyclovir treatment during primary herpes simplex virus infection reduces latency and subsequent reactivation in the nervous system in vivo. J. Infect. Dis. 184:964-971. [DOI] [PubMed] [Google Scholar]
- 58.Shimeld, C., S. Efstathiou, and T. Hill. 2001. Tracking the spread of a lacZ-tagged herpes simplex virus type 1 between the eye and the nervous system of the mouse: comparison of primary and recurrent infection. J. Virol. 75:5252-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimeld, C., J. L. Whiteland, N. A. Williams, D. L. Easty, and T. J. Hill. 1996. Reactivation of herpes simplex virus type 1 in the mouse trigeminal ganglion: an in vivo study of virus antigen and immune cell infiltration. J. Gen. Virol. 77:2583-2590. [DOI] [PubMed] [Google Scholar]
- 59a.Simmons, A., B. Slobedman, P. Speck, J. Arthur, and S. Efstathiou. 1992. Two patterns of persistence of herpes simplex virus DNA sequences in the nervous systems of latently infected mice. J. Gen. Virol. 73:1287-1291. [DOI] [PubMed]
- 60.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 61.Sunstrum, J. C., C. E. Chrisp, M. Levine, and J. C. Glorioso. 1988. Pathogenicity of glycoprotein C negative mutants of herpes simplex virus type 1 for the mouse central nervous system. Virus Res. 11:17-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thackray, A. M., and H. J. Field. 2000. The effects of antiviral therapy on the distribution of herpes simplex virus type 1 to ganglionic neurons and its consequences during, immediately following and several months after treatment. J. Gen. Virol. 81:2385-2396. [DOI] [PubMed] [Google Scholar]
- 63.Thompson, R. L., G. V. Devi-Rao, J. G. Stevens, and E. K. Wagner. 1985. Rescue of a herpes simplex virus type 1 neurovirulence function with a cloned DNA fragment. J. Virol. 55:504-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson, R. L., S. K. Rogers, and M. A. Zerhusen. 1989. Herpes simplex virus neurovirulence and productive infection of neural cells is associated with a function which maps between 0.82 and 0.832 map units on the HSV genome. Virology 172:435-450. [DOI] [PubMed] [Google Scholar]
- 65.Thompson, R. L., and N. M. Sawtell. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75:6660-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson, R. L., and N. M. Sawtell. 1997. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J. Virol. 71:5432-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson, R. L., and N. M. Sawtell. 2000. Replication of herpes simplex virus type 1 within trigeminal ganglia is required for high frequency but not high viral genome copy number latency. J. Virol. 74:965-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson, R. L., and J. G. Stevens. 1983. Biological characterization of a herpes simplex virus intertypic recombinant which is completely and specifically non-neurovirulent. Virology 131:171-179. [DOI] [PubMed] [Google Scholar]
- 69.Thompson, R. L., and J. G. Stevens. 1983. Replication at body temperature selects a neurovirulent herpes simplex virus type 2. Infect. Immun. 41:855-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson, R. L., E. K. Wagner, and J. G. Stevens. 1983. Physical location of a herpes simplex virus type-1 gene function(s) specifically associated with a 10 million-fold increase in HSV neurovirulence. Virology 131:180-192. [DOI] [PubMed] [Google Scholar]
- 71.Wagner, E. K., and D. C. Bloom. 1997. Experimental investigation of herpes simplex virus latency. Clin. Microbiol. Rev. 10:419-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wagner, E. K., J. F. Guzowski, and J. Singh. 1995. Transcription of the herpes simplex virus genome during productive and latent infection. Prog. Nucleic Acid Res. Mol. Biol. 51:123-165. [DOI] [PubMed] [Google Scholar]
- 73.Weir, J. P. 2001. Regulation of herpes simplex virus gene expression. Gene 271:117-130. [DOI] [PubMed] [Google Scholar]
- 74.Wilcox, C. L., R. L. Smith, R. D. Everett, and D. Mysofski. 1997. The herpes simplex virus type 1 immediate-early protein ICP0 is necessary for the efficient establishment of latent infection. J. Virol. 71:6777-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu, X., J. Chen, C. S. H. Young, and S. Silverstein. 1990. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J. Virol. 64:4489-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]