Abstract
Small nucleolar RNAs (snoRNAs) are a large family of eukaryotic RNAs that function within the nucleolus in the biogenesis of ribosomes. One major class of snoRNAs is the box C/D snoRNAs named for their conserved box C and box D sequence elements. We have investigated the involvement of cis-acting sequences and intranuclear structures in the localization of box C/D snoRNAs to the nucleolus by assaying the intranuclear distribution of fluorescently labeled U3, U8, and U14 snoRNAs injected into Xenopus oocyte nuclei. Analysis of an extensive panel of U3 RNA variants showed that the box C/D motif, comprised of box C′, box D, and the 3′ terminal stem of U3, is necessary and sufficient for the nucleolar localization of U3 snoRNA. Disruption of the elements of the box C/D motif of U8 and U14 snoRNAs also prevented nucleolar localization, indicating that all box C/D snoRNAs use a common nucleolar-targeting mechanism. Finally, we found that wild-type box C/D snoRNAs transiently associate with coiled bodies before they localize to nucleoli and that variant RNAs that lack an intact box C/D motif are detained within coiled bodies. These results suggest that coiled bodies play a role in the biogenesis and/or intranuclear transport of box C/D snoRNAs.
INTRODUCTION
The generation of eukaryotic ribosomes takes place predominantly inside the nucleus within nucleoli. Nucleoli are composed of a complex mixture of macromolecules, and considerable intracellular trafficking of macromolecules is required to assemble functional nucleoli and to produce ribosomal subunits. Scores of ribosomal and nonribosomal proteins synthesized in the cytoplasm must move to nucleoli. Indeed, several nucleolar proteins have been demonstrated to shuttle continuously between the cytoplasm and nucleus (Borer et al., 1989; Meier and Blobel, 1992; Shaw and Jordan, 1995). In addition, >150 distinct small nucleolar RNAs (snoRNAs) are each targeted to nucleoli where they function in the modification and processing of rRNA (Bachellerie et al., 1995; Maxwell and Fournier, 1995; Smith and Steitz, 1997; Tollervey and Kiss, 1997). snoRNAs are produced in the nucleus and remain within the nucleus where they are matured at unidentified intranuclear sites (Terns and Dahlberg, 1994; Terns et al., 1995). Thus, production of functional snoRNAs involves transport from the nucleoplasm to nucleoli and may involve other nuclear structures.
The box C/D snoRNAs are one of two major classes of snoRNAs distinguished by the presence of conserved sequence elements and common secondary structures (Balakin et al., 1996). Box C/D snoRNAs each associate with common proteins including fibrillarin (Schimmang et al., 1989; Lapeyre et al., 1990; Baserga et al., 1991), Nop 56/58 (Wu et al., 1998; Lafontaine and Tollervey, 1999), and recently identified proteins (Caffarelli et al., 1998; Watkins et al., 1998). Only a few box C/D snoRNAs, namely, U3, U8, U14, and U22, are required for rRNA processing (Li et al., 1990; Savino and Gerbi, 1990; Hughes and Ares, 1991; Peculis and Steitz, 1993; Tycowski et al., 1994; Enright et al., 1996). The majority of box C/D snoRNAs function as guide RNAs to direct methylation of ribose 2′-hydroxyl groups at conserved positions in rRNA (Cavaille et al., 1996; Kiss-Laszlo et al., 1996; Tollervey, 1996; Tycowski et al., 1996).
The defining structural element of the box C/D snoRNAs is the box C/D motif. The box D element (core consensus sequence CUGA [Xia et al., 1997]) is generally found in a single-stranded region near the 3′ terminus of the box C/D RNAs. The box C element (core consensus sequence GANG [Xia et al., 1997]) exists in a single-stranded region opposite box D within the predicted secondary structure of most box C/D snoRNAs (Tycowski et al., 1993; Kiss-Laszlo et al., 1996; Watkins et al., 1996; Samarsky and Fournier, 1998). Thus the two common sequence elements, box C and box D, are generally distant from each other in the primary sequence of the snoRNAs but are brought into proximity in the folded RNAs as a result of the base pairing of complementary sequences flanking the box elements. The resultant structure, consisting of box C and box D and one or two adjacent helices, has been referred to as the stem-box structure (Qu et al., 1995), the terminal core motif (Xia et al., 1997), or, simply, the box C/D motif (Samarsky et al., 1998).
In this study, we have focused our analysis on the mechanisms governing the localization of the box C/D class of snoRNAs to the nucleolus. Extensive studies were performed using U3 snoRNA, which is the most abundant and well-characterized box C/D snoRNA. Comparison of U3 RNAs from diverse organisms reveals that U3 snoRNA contains a box D and two box C (referred to as boxes C and C′) sequence elements as well as U3-specific elements known as boxes A, A′, and B. A two-domain secondary structure is predicted for U3 RNA from all organisms examined. The 5′ domain contains sequences (boxes A and A′) that participate in base-paired interactions with 18S rRNA (Hughes, 1996; Mereau et al., 1997). The 3′ domain of U3 RNA is structurally diverse (Fournier et al., 1998) but invariably contains boxes B, C, C′, and D, which are protein-binding sites (Parker and Steitz, 1987; Jeppesen et al., 1988; Hartshorne and Agabian, 1994; Mereau et al., 1997), as well as a 3′ terminal stem. The “hinge,” a short, single-stranded sequence that links the 5′ and 3′ domains, recognizes complementary sequences in the 5′ external transcribed sequences of pre-rRNA (Beltrame and Tollervey, 1995; Mereau et al., 1997).
We have introduced substitution mutations throughout the U3 molecule including all conserved box elements, the 3′ terminal stem, and the hinge sequence, and have analyzed the intranuclear localization of these variant U3 RNAs after their injection into Xenopus oocyte nuclei. In addition, we have also examined the intranuclear localization of additional box C/D snoRNAs (U8 and U14) to test the generality of our observations. We have found that the targeting of box C/D snoRNAs to nucleoli depends on their common sequence elements (the box C/D motif) and is temperature dependent. Furthermore, we have characterized the association of the box C/D snoRNAs with an additional intranuclear organelle, the coiled body. Our results suggest that box C/D snoRNAs associate with coiled bodies transiently before localization to nucleoli. Important differences between the results obtained in this study and those of similar recent studies (Lange et al., 1998a–c; Samarsky et al., 1998) will be discussed.
MATERIALS AND METHODS
Generation of snoRNA Mutant Constructs
U3 Constructs.
A plasmid containing a genomic clone encoding Xenopus U3A snoRNA (Savino et al., 1992) was used as the source of wild-type U3 RNA-coding sequence. Templates used to transcribe wild-type or mutant U3 RNAs were DNA fragments generated by PCR amplification. DNA fragments encoding U3 RNAs with specific mutations were generated by PCR-based strategies described in detail below. In general, block substitution mutations were introduced in which each nucleotide of a conserved box element was replaced with a complementary nucleotide. The precise changes are noted in the specific description of the generation of each mutant below. The deoxyoligonucleotide primers used in PCR reactions to prepare wild-type and mutated Xenopus U3 templates are listed. SP6 promoter sequences are underlined, and sites of mutation are in bold. All PCR reactions were performed using Pfu DNA polymerase (Stratagene, La Jolla, CA) and an annealing temperature of 52°C.
5′ primers were as follows: 1) GATTTAGGTGACACTATAGAAGACTATACTTTCAGGGATCA; 2) GATTTAGGTGACACTATAGAAGACTAATGAATCAGGGATCA; 3) CAGTAAGACTATACT-TTCAGCCTAGTAAAGATTAGGTTGTACCTGGTGA; 4) GTGCT-CGAAAGTGTGTGACTTGAGTGTTACCACGAGGAAGAGC; 5) CTGAACTCACAAACCACCTCCTTCTGCGTCAGTGTTCTCTC ; 6) CGTCAGTGTTCTCTCCTCTCGCACTTGTGAGCTCACAGT-GCTG; 7) GGCTGCTGTTTGCTATACTACTTGCTTCTGCTCCC-CTTTA; 8) GATTTAGGTGACACTATAGACCACGAGGAAGA-GCG; and 9) AAAAAGAATTCCCAAATTCAGAAGTGACTGCG. 3′ primers were as follows: 10) GGGTGTCAGCCTGTGTTCTCTCCCTCC; 11) ACCACTCAGCCTGTGTTCTCTCCCTCC; 12) TCACCAGGTACAACCTAATCTTTACTAGGCTGAAAGTATAGTCT -TACTG; 13) GCTCTTCCTCGTGGTAACACTCAAGTCACACT-TTCGAGCACAT; 14) GAGAGAACACTGACGCAGAAGGAGGTGGTTTGTGAGTTCAG; 15) CAGCACTGTGAGCTCACAA-GTGCGAGAGGAGAGAACACTGACG; 16) TAAAGGGGAGCAGAAGCAAGTAGTATAGCAAACAGCAGC; 17) ACCACA-GTCGGTGTGTTC; 18) ACCACTCATCCTGTGTTCTCTCCC-TCC; 19) ACCACTCCGCCTGTGTTCTCTCCCTCC; 20) ACCA-CTGAGCCTGTGTTCTCTCCCTCC; 21) ACCACACAGCCTGTGTTCTCTCCCTCC; 22) ACCACTCAGCCTGTGTTCTCTCCCGA-AGG; and 23) AAAAAAAGCTTCAGCCCCACTTTTCCATTC.
Two different PCR strategies were used: one to introduce mutations near the termini of U3 and to generate subfragments of U3 and another to introduce mutations at internal positions within the U3-coding region.
Generation of Terminal U3 Mutations and U3 Subfragments.
Wild-type U3 transcription template DNA was generated by PCR amplification from wild-type U3 plasmid using oligonucleotides 1 + 11. Block substitutions of box A′ (nucleotides [nt] 8–12; UACUU to AUGAA), box D (nt 210–215; GGCUGA to CCGACU), and box D point mutants (see below) were generated by direct PCR amplification from wild-type U3 plasmid using the following primer pairs: box A′, 2 + 11; box D, 1 + 17; box D C212B, 1 + 18; box D U213G, 1 + 19; box D G214B, 1 + 20; and box D A215U, 1 + 21. The subfragment of U3 comprised of the 3′ domain (nucleotides 75–220) was generated using primers 8 + 11. The U3 subfragment containing box C′ and box D (nucleotides 75–104/GCUU tetraloop/198–220) was generated using primers 8 + 22 and the following oligonucleotide template: TAATACGACTCACTATAGGGAAGACTAC-CACGAGGAAGAGCGTCAGTGTTCTCTCCTTCGGGAGAGAA-CACAGGCTGAGTGGT. In all other cases, the wild-type U3 gene was used as the PCR template. The point mutation U213G in the subfragment containing box C′ and box D was produced using primers 8 + 19 and the unmutated subfragment as the template in a PCR reaction. All U3 mutant DNA fragments were subcloned into the SmaI site of pUC19 and confirmed by sequencing.
Generation of Internal U3 Mutations.
Mutations of the internal elements, box A (nt 17–28; GGAUCAUUUCUA to CCUAGUAAAGAU), hinge (nt 63–74; CUGAACUCACAA to GACUUGAGUGUU), box C′ (nt 80–87; GAGGAAGA to CUCCUUCU), box B (nt 106–114; GAGCGUGAA to CUCGCACUU), and box C (nt 157–165; UGAUGAACG to ACUACUUGC) of the U3 gene were produced by recombinant PCR (Higuchi, 1990). Block substitution of box A was accomplished by combining the products of two independent PCR reactions (from the wild-type U3 gene by the use of primer pairs 9 + 12 and 3 + 23) and PCR amplification using the outermost primers (9 + 23). The first series of PCR reactions to produce block substitutions in each of the following elements used the indicated primer pairs: the hinge region, primers 9 + 13 and 4 + 23; box C′, primers 9 + 14 and 5 + 23; box B, primers 9 + 15 and 6 + 23; and box C, primers 9 + 16 and 7 + 23. For each of these PCR reactions, the wild-type U3 plasmid was used as the template. Primers 9 + 23 were used to amplify from the combined products in the second step. All U3 mutant DNA fragments were digested with EcoRI and HindIII (sites incorporated into primers 9 + 23), subcloned into the EcoRI/HindIII site of pGEM3Zf−, and sequenced.
U14 Constructs.
pBT20, a plasmid containing the mouse U14.5 gene (Shanab and Maxwell, 1992), was used as a template to generate wild-type and variant U14 RNAs. Mutagenesis of the box C, box D, and 3′ terminal stem sequences of U14 genes was performed by PCR amplification using mutagenic primers. Base substitutions introduced were ACTACT (wild-type is TGATGA) in the position of box C and TCTAGA (wild-type is GTCTGA) for box D. To disrupt terminal stem formation in the U14 RNA, we substituted the 3′ terminal nucleotides of the U14 gene (GCGAAT) with CGCTTA.
In Vitro RNA Synthesis
PCR products (100 ng) or linearized plasmids (1 μg) were used as templates for in vitro transcription. Wild-type and mutant U3 and U14 RNAs were transcribed from PCR-derived DNA fragments. The following additional RNAs were prepared by in vitro transcription from plasmids as described previously: Xenopus U8 wild type and a box C mutant (Peculis and Steitz, 1994); Xenopus U8 box D mutant and Xenopus U3 terminal stem mutant (Terns et al., 1995); Xenopus U1, U1Sm−, and U6 (Terns et al., 1993); and DHFR mRNA, 5S rRNA, and tRNAiMet (Jarmolowski et al., 1994).
The reaction conditions used to generate capped, 32P-labeled RNAs by SP6 or T7 RNA polymerase were essentially as described previously (Terns et al., 1993). To generate fluorescein-labeled RNAs, we used an equal mixture (250 μM each) of UTP and fluorescein-12-UTP (Boehringer Mannheim, Indianapolis, IN). The RNAs were quantitated by measuring incorporation of trace amounts of [32P]GTP (ICN Pharmaceuticals, Costa Mesa, CA). RNA transcripts were either gel purified or purified by two successive isopropanol precipitations, suspended in distilled water, and stored at −20°C wrapped in aluminum foil to avoid exposure to light. The integrity and size of the RNA were assessed by electrophoresis on 8% denaturing polyacrylamide gels and ethidium bromide staining.
Injection of RNAs into Xenopus Oocytes
The method by which we microinject and micromanipulate oocytes has been described recently in detail (Terns and Goldfarb, 1998). In brief, stage V and VI Xenopus laevis oocytes were separated from each other and from the surrounding follicle cells by treatment with 2 mg/ml collagenase for 60–90 min. The collagenase-treated cells were washed thoroughly in MBSH buffer before microinjection. Injections were performed using the model PL1–100 picoinjector microinjector (Medical Systems Corporation, Greenvale, NY) and a glass needle with a tip of 10 μm inner diameter. The RNA sample was dried using a Savant (Farmingdale, NY) speed vacuum unit and resuspended in a solution of filter-sterilized blue dextran in water (20 mg/ml; 2 × 106 molecular weight; Sigma, St. Louis, MO). Oocyte nuclei were injected with 10 nl of solution containing 5 fmol of fluorescein-labeled RNA. In control experiments (e.g., see Figure 2B), 132 fmol of fluorescein-12-UTP was injected. The oocytes were maintained in MBSH buffer at 18°C. Nuclear injections were monitored by determining whether nuclei gained a blue appearance as the result of the injected blue dextrans (Jarmolowski et al., 1994).
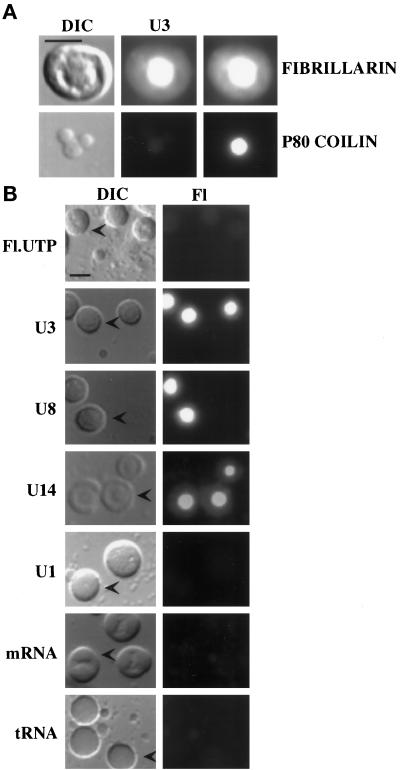
Figure 2.
Microinjected snoRNAs localize to the fibrillar regions of nucleoli. (A) U3 colocalizes with endogenous fibrillarin. Fluorescein-labeled U3 was injected into oocytes, and nuclear spreads were analyzed 1 h after injection. Top, a single, representative nucleolus is shown. Bottom, a single, coiled body with two snurposomes attached to its surface is shown. Left, differential interference contrast (DIC) images of the structures are shown. Right, indirect immunofluorescence using the anti-fibrillarin antibody 17C12 (Hultman et al., 1994) or antibody H1 (Tuma et al., 1993) directed against the coiled body marker protein p80 coilin confirm the identity of the structures as nucleoli and coiled bodies, respectively. Middle, the fluorescein signal from U3 RNA (U3) colocalized with fibrillarin in nucleoli but not p80 coilin in the coiled body. Bar, 10 μm. (B) Selective nucleolar localization of U3, U8, and U14 snoRNAs. RNAs (U3, U8, and U14 snoRNAs; U1 spliceosomal snRNA; DHFR mRNA; and tRNAiMet) were fluorescently labeled by in vitro transcription in the presence of fluorescein-12-UTP (see MATERIALS AND METHODS), and each RNA was injected into Xenopus oocytes. Fl.UTP indicates injection of fluorescein-12-UTP alone. Nuclear spreads were prepared 1 h after injection (exceptthat of tRNA that was analyzed after 15 min because this RNA is rapidly exported [Jarmolowski et al., 1994] and that of U14 that was analyzed after 4 h), and the RNAs were observed by fluorescence microscopy. Both DIC and fluorescence (Fl) images are shown for each sample. The arrowheads in the DIC images point to representative nucleoli. Bar, 10 μm.
For the analysis of RNA stability, nuclear retention, fibrillarin binding, and 5′ cap hypermethylation, oocytes were each injected with 10 nl of solution containing ∼1 fmol each of [32P]GTP-labeled U1, U8, and U6 RNAs and either [32P]GTP-labeled or fluorescein-12-UTP + [32P]GTP–labeled U3 RNA. After injection, the oocytes were incubated at 18°C for 8 h before they were manually dissected in J-buffer (Birkenmeier et al., 1978) into nuclear and cytoplasmic fractions. The nucleocytoplasmic distribution was determined by isolating the RNAs from two to four dissected nuclear and cytoplasmic fractions by proteinase K digestion, phenol extraction, and ethanol precipitation. The RNA equivalent of one nucleus or one cytoplasm was resolved on an 8% denaturing polyacrylamide gel and detected by autoradiography with an intensifying screen.
Immunoprecipitations
Antibodies used in these experiments included polyclonal antibodies directed against either the m7G (Munns et al., 1982) or m2,2,7G cap (Bringmann et al., 1983) and a monoclonal antibody (72B9) against fibrillarin (Reimer et al., 1987). Antibodies were coupled to preswollen protein A–Sepharose CL-4C beads (Sigma) by end-over-end rotation in 0.5 ml of Ipp-500 buffer (500 mM NaCl, 10 mM Tris-HCl, pH 8.0, 0.1% [vol/vol] NP-40, and 0.1% [wt/vol] sodium azide) at 4°C for 12–16 h. Immunoprecipitations with cap-specific antibodies were performed with purified nuclear RNAs (1 nuclear equivalent/reaction) in Net-2 buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, and 0.05% NP-40). Immunoprecipitations with fibrillarin antibodies were performed with nuclear extracts (5 nuclear equivalents/reaction) in Net-100 buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.5, and 0.05% NP-40). In both cases, immunoprecipitations were performed at 4°C for 12–16 h, supernatants were collected, and the beads were washed extensively. RNA from both supernatants and pellets were obtained by digestion with proteinase K, phenol extraction, and ethanol precipitation, and the purified RNAs were analyzed on 8% denaturing polyacrylamide gel by autoradiography.
Nuclear Spreads, Immunofluorescence, and Microscopy
After injection of fluorescently labeled RNAs into Xenopus oocytes, nuclear spreads were prepared from dissected nuclei as described (Gall et al., 1991; Wu et al., 1996). After centrifugation, the spreads were fixed in 2% paraformaldehyde in 1× PBS, pH 7.2, for 1 h. After washing with 1× PBS, mounts were made using 50% glycerol (in 1× PBS) containing 1 mg/ml phenylenediamine, pH 9, and were stored at −20°C.
Indirect immunofluorescence was performed on the nuclear spreads after fixation essentially as described (Wu and Gall, 1997). Briefly, slides were blocked using 10% horse serum (Hyclone, Logan, UT) in 1× PBS for 15 min at 37°C. After thorough washing with 1× PBS, the sample area was incubated with antibody (mAb 17C12) directed against fibrillarin at 1:1000 dilution in 1× PBS (Hultman et al., 1994) or with antibody (mAb H1) directed against the Xenopus p80 coilin homologue (Tuma et al., 1993) at 1:10 dilution in 1× PBS for 30 min at 37°C. The excess primary antibodies were washed away with 1× PBS, and the secondary antibody (Texas Red–conjugated anti-mouse antibody at 1:150 dilution in 1× PBS) was added and incubated at 37°C for 30 min. Excess secondary antibody was washed away with 1× PBS, and mounts were made as described above.
A Zeiss (Thornwood, NY) Axiovert S 100 inverted fluorescence microscope equipped with differential interference contrast optics was used for all observations. All images were acquired using a cooled charge-coupled device camera (Quantix-Photometrics) and IP Lab Spectrum software.
RESULTS
U3 RNA Synthesized with Fluorescein-12-UTP Retains Its Biological Properties
U3 RNA intranuclear transport was studied by examining the localization of fluorescein-labeled RNAs in nuclear spread preparations by fluorescence microscopy. As a first step, we compared the nuclear retention, 5′ cap hypermethylation, and fibrillarin binding of fluorescein-labeled U3 snoRNA with that of control RNA to verify that the fluorescein label did not affect known biological properties of the RNA.
U3 RNA injected into oocyte nuclei is stable and retained in the nucleus (Terns and Dahlberg, 1994; Terns et al., 1995). To ensure that incorporation of fluorescein-12-UTP into U3 RNA would not affect the stability or nuclear retention of the RNA, we injected RNA transcribed in vitro with fluorescein-12-UTP (see MATERIALS AND METHODS) into the nuclei of Xenopus oocytes. Eight hours after injection, RNAs present in the nuclear and cytoplasmic fractions were analyzed by PAGE. Coinjected control RNAs included a U1 small nuclear RNA (snRNA) mutant (U1sm−), U6 snRNA, and U8 snoRNA (Figure 1A). As expected, U1sm− that lacks the Sm protein-binding site was exported to the cytoplasm (and not reimported to the nucleus [Mattaj and De Robertis, 1985; Terns et al., 1993]). U6, an snRNA that is not exported to the cytoplasm (Hamm and Mattaj, 1989; Terns et al., 1993), and U8 snoRNA were retained within the nucleus (Figure 1A) (Terns et al., 1995). These controls ensure that, in each oocyte, RNAs were being exported (U1) and retained (U6 and U8) appropriately and that the nuclear injections and dissections were precise. We found that U3 snoRNA labeled with fluorescein-12-UTP (Figure 1A, lanes 2 and 3) is stable and retained within the nucleus similarly to nonfluorescein-labeled U3 RNA (lanes 5 and 6).
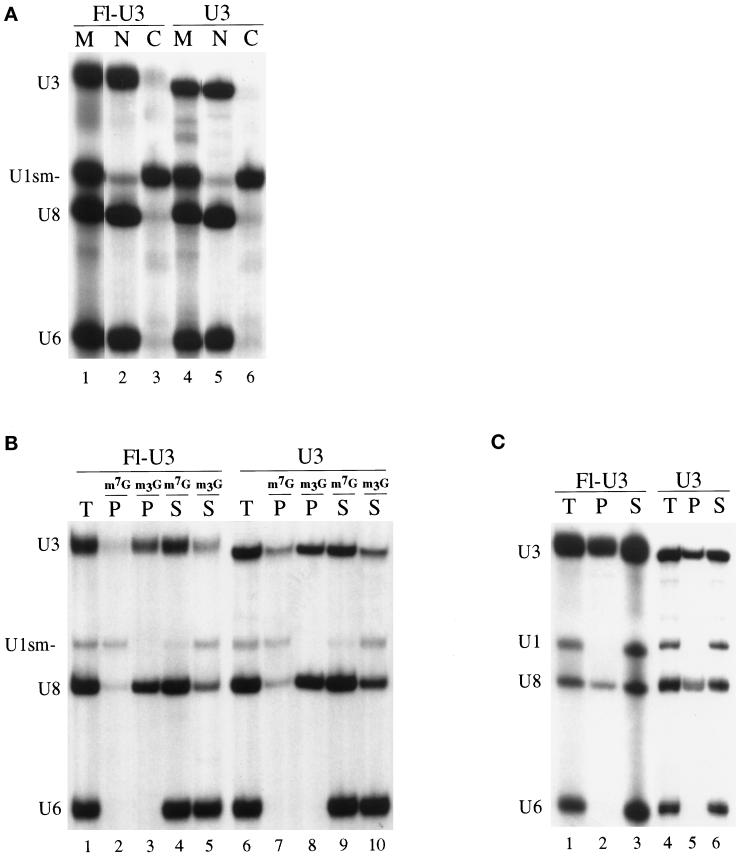
Figure 1.
Nuclear retention, cap hypermethylation, and fibrillarin binding of fluorescein-labeled U3 RNA in Xenopus oocytes. (A) Nucleocytoplasmic distribution of fluorescein-labeled U3 snoRNA (Fl-U3) or nonfluorescein-labeled U3 snoRNA (U3) and U1sm− snRNA, U8 snoRNA, and U6 snRNA after their injection into oocyte nuclei. 32P-labeled, m7G-capped Fl-U3, U3, U1sm−, U8, and U6 were synthesized in vitro, and mixtures of the RNAs were injected into nuclei of Xenopus oocytes. After 8 h of incubation at 18°C, the labeled RNAs present in the nuclear (N) and cytoplasmic (C) fractions of the oocytes were isolated and analyzed by electrophoresis in a denaturing polyacrylamide gel. Lanes 1 and 4 (M) show the RNAs before injection. Lanes 2 and 3 show the distribution of Fl-U3, and lanes 5 and 6 show that of nonfluorescein-labeled U3. (B) Identification of the 5′ cap structure (m7G or m2,2,7G) by immunoprecipitation. The RNAs present in the nucleus 4 h after injection were precipitated using anti-m7G (m7G) (Munns et al., 1982) or anti-m2,2,7G (m3G) (Bringmann et al., 1983) cap antibodies as indicated. The RNAs present in the total sample (T), precipitate (P), and supernatant (S) fractions were separated by gel electrophoresis as described in A. Lanes 1–5 show the precipitation profile of Fl-U3, and lanes 6–10 show that of nonfluorescein-labeled U3. (C) Determination of fibrillarin binding to fluorescein-labeled U3. Immunoprecipitations were performed on nuclear extracts prepared 4 h after injection of RNAs using anti-fibrillarin antibodies (72B9) (Reimer et al., 1987). RNAs present in the precipitate (P), 20% of the total sample (T), and 20% of the supernatant (S) were separated by gel electrophoresis as described in A. Lanes 1–3 show the precipitation profile of Fl-U3, and lanes 4–6 show that of nonfluorescein-labeled U3. Note that the gel electrophoretic mobility of the fluorescein-labeled U3 RNA is reduced relative to that of the unlabeled U3 in A–C.
We also tested for effects of fluorescein labeling on the ability of U3 RNA to undergo cap hypermethylation. U3 snoRNA is normally synthesized with a 5′ m7G cap that is hypermethylated to an m2,2,7G cap within the nucleus (Terns et al., 1995). Cap hypermethylation was assayed by immunoprecipitation of nuclear RNAs with antibodies that specifically recognize either the m7G (Munns et al., 1982) or m2,2,7G cap structures (Bringmann et al., 1983). The antibodies immunoprecipitated the fluorescein-12-UTP–labeled U3 and the nonfluorescein-labeled U3 to the same extent (Figure 1B, compare lanes 2 and 3 with lanes 7 and 8), indicating that fluorescein labeling does not interfere with cap hypermethylation.
Fibrillarin is a nucleolar protein that binds box C/D family snoRNAs including U3 (Baserga et al., 1991). We tested whether fluorescein-labeled U3 RNA associated with fibrillarin by assaying the ability of anti-fibrillarin antibodies to coimmunoprecipitate the RNA after injection of the RNA into oocyte nuclei. Monoclonal antibody 72B9 against fibrillarin (Reimer et al., 1987) immunoprecipitates U3 RNA labeled with fluorescein as well as nonfluorescein-labeled U3 (Figure 1C, compare lanes 2 and 3 with lanes 5 and 6). Thus, fluorescein-labeled U3 RNA interacts with fibrillarin.
Our results clearly show that labeling U3 RNA with fluorescein by transcription with fluorescein-12-UTP does not significantly alter any of the biological properties of the RNA that we examined, which included stability, nuclear retention, hypermethylation, and fibrillarin binding.
snoRNAs but Not Other Classes of RNA Localize to Nucleoli
U3 snoRNA has been shown to be involved in 18S rRNA processing within the nucleolus (Kass et al., 1990; Savino and Gerbi, 1990; Hughes and Ares, 1991; Beltrame and Tollervey, 1995). Amplified nucleoli and other nuclear structures including chromosomes, coiled bodies, and snurposomes can be observed and distinguished by morphology and molecular markers in oocyte nuclear spread preparations (Gall et al., 1991; Wu et al., 1993). For example, nucleoli can be identified by immunostaining using antibodies against nucleolar proteins such as fibrillarin (Shah et al., 1996). Fibrillarin is enriched in the dense fibrillar regions of nucleoli that can be distinguished as internal substructures by differential interference contrast microscopy (Figure 2A). Coiled bodies, which are highly conserved intranuclear organelles of unclear function (Gall et al., 1995; Matera, 1998), are distinguished by their size and morphology (Gall and Callan, 1989) and the presence of p80 coilin (Andrade et al., 1991). Fluorescein-labeled U3 RNA colocalized with endogenous fibrillarin within the dense fibrillar region of nucleoli and was not detectable in coiled bodies 1 h after injection into Xenopus oocytes (Figure 2A).
Other box C/D snoRNAs, namely, U8 and U14, also localized to the multiple amplified nucleoli after injection into oocyte nuclei (Figure 2B). As expected, a spliceosomal snRNA (U1 snRNA), an mRNA (dihydrofolate reductase mRNA), and a tRNA (tRNAiMet) did not localize to nucleoli after injection (Figure 2B). Furthermore, injection of fluorescein-12-UTP alone (at molar amounts that exceed those incorporated into the injected RNAs, see MATERIALS AND METHODS) did not result in significant fluorescence in nucleoli (Figure 2B). Thus, fluorescein-labeled box C/D snoRNAs (including U3, U8, and U14 RNA) specifically localize to the dense fibrillar region of nucleoli within the nucleus of Xenopus oocytes.
Nucleolar Localization of U3 snoRNA Is Temperature Dependent
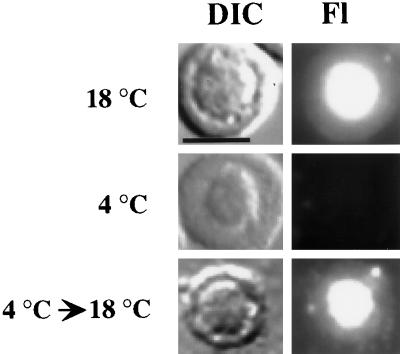
Energy-dependent, active transport processes in the cell are generally inhibited at low temperatures. To determine whether the nucleolar localization of U3 is a temperature-dependent process, we examined localization at reduced temperatures. Fluorescein-labeled U3 RNA was injected into the nuclei of oocytes that had been preincubated at 4°C for 1 h. The injections were performed at 4°C, and after injection the oocytes were maintained at 4°C for 1 h. U3 RNA was not localized to the nucleolus in oocytes that were maintained at 4°C (Figure 3). The inhibitory effect of temperature reduction on nucleolar localization was reversible because U3 RNA localized to the nucleolus in injected oocytes maintained at 4°C for 1 h and subsequently incubated at 18°C for 1 h (Figure 3) but not in oocytes maintained at 4°C for 2 h (our unpublished results). Thus, localization of U3 RNA to nucleoli is a temperature-dependent process.
Figure 3.
Effect of temperature reduction on the nucleolar localization of U3 snoRNA. Fluorescently labeled, in vitro–synthesized U3 RNA was injected into nuclei of oocytes that were maintained either at 18°C (top) or at 4°C (middle) for 1 h before nuclear spread preparations were analyzed. In the temperature-shift experiment (bottom), the oocytes were shifted to 18°C for 1 h after being maintained at 4°C for 1 h. Each DIC image shows a single nucleolus, and the RNA signal is shown in the corresponding Fl image. Bar, 10 μm.
Box C′ and Box D Are Required for Localization of U3 RNA to the Nucleolus
To identify cis-acting signals essential for nucleolar targeting of U3 snoRNA, we analyzed the localization of multiple sequence variants of U3. We began our analysis with block substitutions of the phylogenetically conserved sequence elements in U3: boxes A, A′, B, C, C′, and D (Figure 4A). A series of U3 mutants was generated in which each nucleotide of a given box element was replaced by its complement (see MATERIALS AND METHODS). In addition, similar substitutions were introduced into the single-stranded hinge region of the RNA. Although the primary sequence of the hinge is not conserved, there is evidence that it is a functionally important region of the RNA (Beltrame and Tollervey, 1995; Mereau et al., 1997). In addition, to determine whether the m7G cap (or its hypermethylated form) is required for nucleolar localization, we replaced the m7G cap of U3 RNA with a nonphysiological, nontrimethylatable ApppG cap.
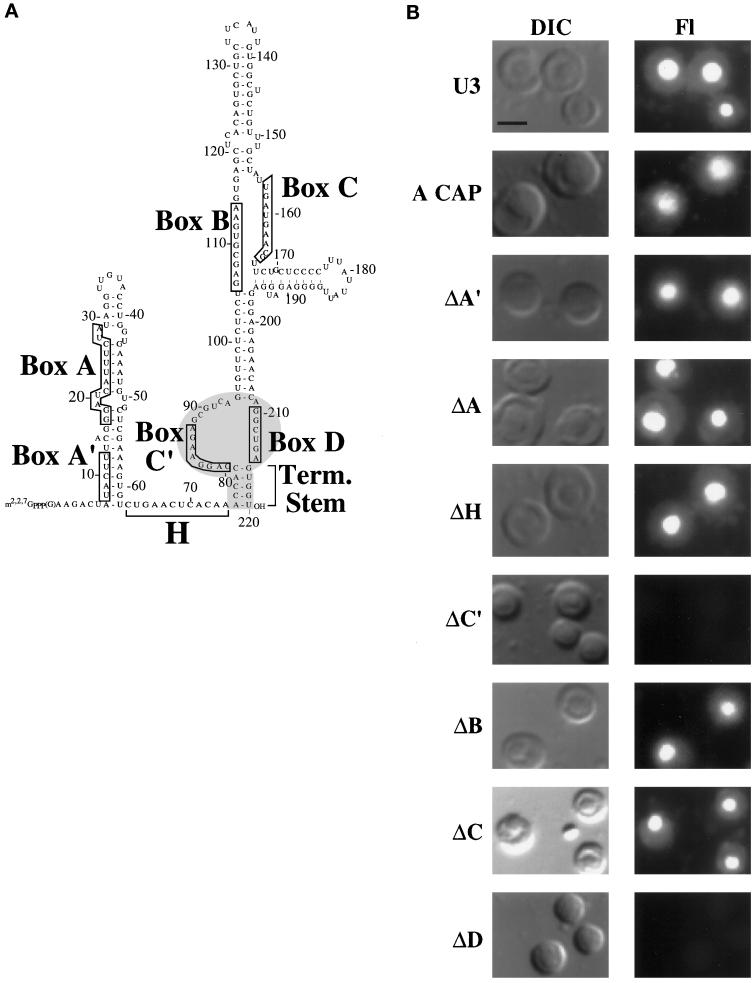
Figure 4.
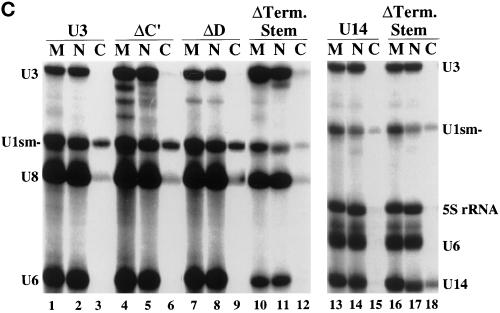
Identification of cis-acting sequences essential for the nucleolar localization of U3 snoRNA. (A) Sequence and secondary structure of Xenopus U3 RNA. The U3 box A, A′, B, C, C′, and D sequence elements are boxed, and the hinge (H) and 3′ terminal stem (Term. Stem) elements are marked by brackets. The box C/D motif of U3, comprised of box C′, box D, and the 3′ terminal stem, is shaded. (B) Mutation of box C′ or box D prevents nucleolar localization of U3 RNA. Fluorescently labeled, in vitro–transcribed wild-type and mutant RNAs were injected into late-stage oocyte nuclei, and nuclear spreads were made 1 h after injection and analyzed by fluorescence microscopy. “A cap” indicates wild-type U3 RNA synthesized with a 5′ ApppG cap instead of an m7G cap. A block substitution mutation in a given element is indicated by Δ (see MATERIALS AND METHODS for precise changes). The DIC images show two to four nucleoli, and the fluorescence (Fl) images show the RNA signals. Bar, 10 μm. (C) Nuclear retention and stability of U3 and U14 RNA variants that fail to localize to nucleoli. The nucleocytoplasmic distribution of wild-type U3 (lanes 2 and 3), of box C′ (lanes 5 and 6), box D (lanes 8 and 9), and the terminal stem (lanes 11 and 12) U3 mutants (see A), of wild-type U14 (lanes 14 and 15), and of the U14 terminal stem mutant (lanes 17 and 18) was determined 1 h after injection as described in Figure 1. Lanes 1, 4, 7, 10, 13, and 16 (M) show the RNAs before injection. When U14 (lanes 13–18) was analyzed, control U8 RNA was omitted (and 5S rRNA was substituted) because a U8 breakdown product migrated in the gel to a position similar to that of U14. The small amount of U14 terminal stem mutant RNA present in the cytoplasmic fraction (lane 18) is reproducible and reflects the requirement of an intact stem for nuclear retention of U14.
Among the eight variants analyzed, six were found in nucleoli in patterns similar to that in wild-type U3; substitution of boxes A, A′, B, and C and the hinge region did not prevent the nucleolar localization of U3 (Figure 4B). Likewise, ApppG-capped U3 RNA was also localized to nucleoli, indicating that the m7G or trimethylated cap is not essential for this process. On the other hand, disruption of box C′ and box D abolished the nucleolar localization of the RNA (Figure 4B) even at times up to 24 h after injection (our unpublished results). All experiments were repeated at least nine times, and nucleolar fluorescence intensity relative to that of wild-type was qualitatively consistent for a given RNA in various experiments. Importantly, the lack of nucleolar fluorescence does not simply reflect the lack of RNA in the nucleus because all of the variant U3 RNAs, including those that did not localize to nucleoli, were found to be stable and retained within the nucleus 1 h after injection (Figure 4C) (Terns et al., 1995). Our results clearly indicate that box C′ and box D are necessary for the nucleolar localization of U3 RNA.
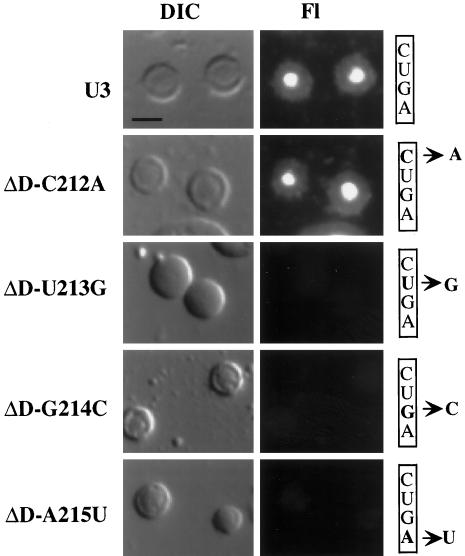
We performed a more detailed mutational analysis on box D, one of the elements found to be essential for nucleolar localization. The core box D element conserved among C/D family snoRNAs consists of CUGA (Xia et al., 1997). We tested the contribution that each of the four conserved nucleotides makes on nucleolar localization by analyzing U3 molecules with point mutations at each position. Substitution of C212 did not disrupt localization of U3 RNA to the nucleolus (Figure 5). However, mutations at U213, G214, and A215 each resulted in a loss of localization similar to that observed when the entire box D sequence was substituted (Figure 5). Thus, the conserved nucleotides UGA (213–215) of box D are each important for localization of U3 to the nucleolus.
Figure 5.
Effect of box D point mutations on the nucleolar localization of U3 snoRNA. Fluorescently labeled wild-type (U3) and variant U3 RNAs with single-nucleotide changes in the box D element (ΔD-C212B, -U213G, -G214B, and -A215U) were injected into oocytes, and nuclear spreads were made 1 h after injection. The particular nucleotides in box D (CUGA) that were altered are indicated on the right. The DIC images show two to three nucleoli, and the RNA signal is shown in the Fl images. Bar, 10 μm.
A Fragment of U3 Containing Box C′ and Box D Localizes to the Nucleolus
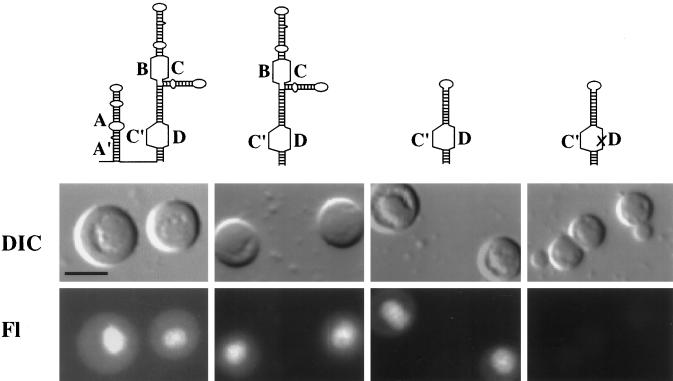
Box C′ and box D are the only conserved sequence elements that we found to be essential for nucleolar localization of U3 (Figure 4B). These sequence elements are present within the 3′ domain of the U3 molecule (nt 75–220; see Figure 4A and MATERIALS AND METHODS). We found that a fragment comprised of the 3′ domain of U3 that is stable and retained within the nucleus localized to the nucleolus (Figure 6 and our unpublished results). Stable subfragments of the 3′ domain were also analyzed. A subfragment comprised of box C′, box D, and flanking stems localized to the nucleolus (Figure 6). A single point mutation within the box D element of this subfragment abolished nucleolar localization (Figure 6). In addition, a subfragment containing box B, box C, and flanking stems does not localize to the nucleolus (our unpublished results). These results suggest that box C′ and box D are sufficient to direct nucleolar localization of U3 RNA.
Figure 6.
Analysis of the ability of stable subfragments of U3 snoRNA to localize to nucleoli. Fluorescently labeled, m7G-capped RNAs were injected into oocyte nuclei, and spreads were made 1 h after injection and examined by microscopy. Top, the diagrams represent the RNAs injected (for sequences, see MATERIALS AND METHODS). Left, wild-type U3 RNA is shown. Middle, a U3 fragment lacking the 5′ domain and hinge (second from left) localizes to nucleoli, as does a U3 subfragment containing box C′ and box D sequences and adjacent stems (second from right). Right, Nucleolar localization of this subfragment was abolished by a single point mutation (U213G; represented as an X) in the box D element of the RNA. Bar, 10 μm.
An Intact 3′ Terminal Stem Is Essential for Nucleolar Localization of U3 and U14
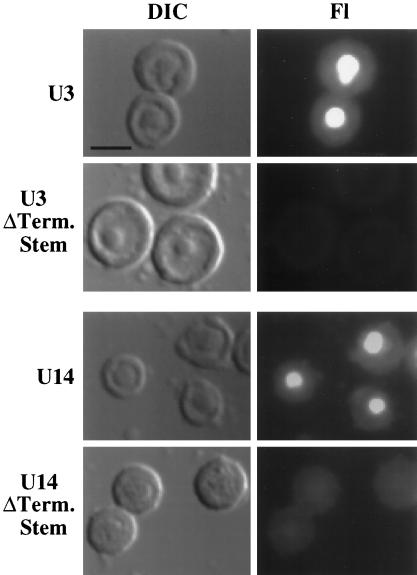
For many box C/D snoRNAs, the 3′ terminal sequences are predicted to be involved in the formation of a short-stem structure that results in positioning the box C and box D sequences adjacent to one another in the predicted secondary structures of the RNAs (Maxwell and Fournier, 1995; Tollervey and Kiss, 1997) (Figure 4A). Phylogenetically conserved 3′ terminal stems have been experimentally demonstrated for both U3 and U14 snoRNAs (Parker and Steitz, 1987; Jeppesen et al., 1988; Shanab and Maxwell, 1991; Baserga et al., 1992; Hartshorne and Agabian, 1994; Mereau et al., 1997). To determine whether the integrity of the 3′ terminal stem is important for nucleolar localization of box C/D snoRNAs, we examined the effect of disrupting the terminal stems of both U3 and U14 snoRNAs on nucleolar localization. Variant U3 and U14 snoRNAs predicted to be unable to form 3′ terminal stems were generated by substituting the nucleotides of one side of the stem with complementary sequences to abolish Watson–Crick base-pairing potential (see MATERIALS AND METHODS). In contrast to their wild-type U3 and U14 counterparts, both variant RNAs failed to localize to nucleoli (Figure 7). The lack of nucleolar signal did not simply reflect a lack of RNA in the nucleus at the time of analysis (Figure 4C). Thus, an intact 3′ terminal stem is essential for the nucleolar localization of both U3 and U14 snoRNAs.
Figure 7.
Disruption of the 3′ terminal stem structure prevents nucleolar localization of U3 and U14 RNA. Mutation of the 3′ terminal stem (ΔTerm. Stem) of U3 or U14 is predicted to disrupt stem formation completely (for sequences, see Figure 4A, MATERIALS AND METHODS, and Xia et al. [1997]). Fluorescently labeled, m7G-capped wild type (rows labeled U3 and U14) and stem mutants (rows labeled ΔTerm. Stem) were injected into oocyte nuclei, and spreads were made 1 h after injection and examined by microscopy. The DIC images show two to four nucleoli, and the RNA signal is shown in the Fl images. Bar, 10 μm.
Box C/D snoRNAs Transiently Localize to Coiled Bodies before Nucleoli
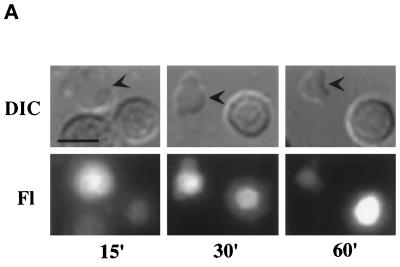
At very early time points (15 min after injection of RNA), before significant localization of U3 to nucleoli, the RNA was clearly observed within coiled bodies (Figure 8A). Over a time period of 1 h after injection, we observed an increase in U3 RNA present in nucleoli and a coincident decrease in the RNA observed in coiled bodies (Figure 8A). By 1 h after injection, U3 was observed predominantly in nucleoli (Figure 8A). Although U3 RNA remained in nucleoli for at least 24 h (our unpublished results), we only observed U3 in coiled bodies at early times after injection. Injection of fluorescein-12-UTP (as in Figure 2B) did not result in significant fluorescence in coiled bodies (our unpublished results). Our results suggest that box C/D snoRNAs transiently localize to coiled bodies before their localization to nucleoli.
Figure 8.
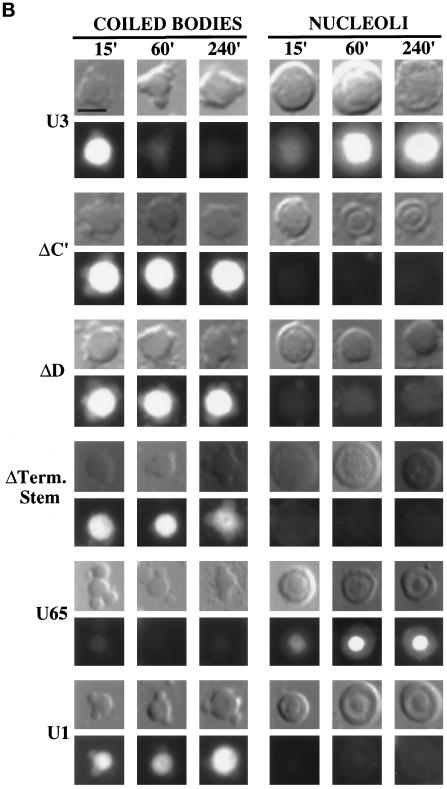
Dynamic association of U3 RNAs with coiled bodies. (A) The localization of U3 RNA to the coiled bodies is transient. Fluorescently labeled wild-type U3 RNA was injected into oocyte nuclei, and nuclear spreads were made at the times indicated in minutes. The arrowheads in the DIC images indicate coiled bodies. The other large structure(s) found in each image is a nucleolus. (B) U3 mutants that fail to localize to nucleoli (right) are retained for longer periods of time in coiled bodies (left). Wild-type U3, as well as box C′ (ΔC′), box D (ΔD), and the terminal stem (ΔTerm. Stem) mutants, was injected into oocyte nuclei, and nuclear spreads were made at the times indicated. The localization of U65 and U1 control RNAs revealed that U65 (a snoRNA of the box H/ACA class) localizes to nucleoli but not coiled bodies and that U1 (a spliceosomal snRNA) localizes to coiled bodies but not nucleoli over the time course analyzed. Bar, 10 μm.
Strikingly, U3 mutants that did not localize to nucleoli accumulated in coiled bodies (Figure 8B). U3 box C′, box D, and 3′ terminal stem mutants were not observed in nucleoli at any time tested. On the other hand, we observed these mutants within coiled bodies for at least 4 h after injection (Figure 8B), well beyond the 1 h time point when very little wild-type RNA remains in coiled bodies. The signal observed in coiled bodies 1 h after injection of other U3 mutants (boxes A, A′, B, and C and hinge) was similar to that of wild-type U3 RNA (our unpublished results). Furthermore, the subfragment of U3 RNA containing box B and box C, which does not localize to nucleoli, was found in coiled bodies for up to 4 h (our unpublished results). Thus, the snoRNA variants that lack intact box C/D motifs remained in coiled bodies longer than did wild-type RNAs. The box C/D motif (i.e., U3 box C′, box D, and the 3′ terminal stem; Figure 4A, shaded area) is essential for the localization of snoRNAs to nucleoli but is not required for localization to coiled bodies.
The specificity of the association of the RNAs with coiled bodies was supported by the localization observed with additional RNAs including a box H/ACA family snoRNA, U65 (Ganot et al., 1997), tRNA, and a spliceosomal snRNA, U1. Although U65 readily localized to nucleoli as expected, we did not detect U65 or tRNA in coiled bodies at any time point examined (Figure 8B and our unpublished results). The lack of association of U65 with coiled bodies is intriguing because U65 snoRNA-associated proteins including homologues of Cbf5p (Meier and Blobel, 1994) and GAR-1 (our unpublished results) have been observed in coiled bodies. In contrast, U1 RNA localized to coiled bodies for several hours but did not localize to nucleoli (weak localization of U1 to snurposomes [Wu et al., 1993] was observed at late time points [our unpublished results]). These controls clearly indicate that coiled bodies are not a simple default destination for injected RNAs.
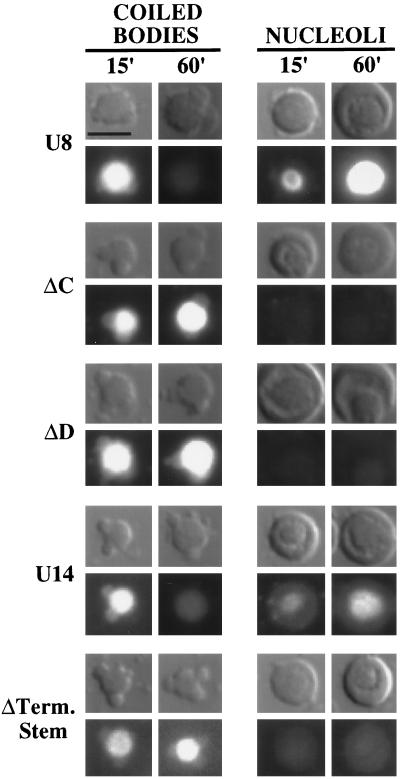
To examine the association of other box C/D snoRNAs with coiled bodies, we analyzed the localization of U8 and U14. As was the case for U3, a transient localization to coiled bodies was observed with U8 and U14 snoRNAs (Figure 9). Moreover, replacement of box C or box D sequences prevented localization of U8 to nucleoli, and the mutants were detained in coiled bodies longer than were wild-type U8 (Figure 9). The severe instability of box C and box D mutants of U14 RNA (even when artificially capped with m7GpppG or ApppG dinucleotides), coupled with the failure of these mutants to be retained in the nucleus, prevented meaningful analysis of these RNAs (our unpublished results). However, significant levels of the U14 terminal stem mutant are present in the nucleus 1 h after injection (Figure 4C), and although the RNA does not localize to nucleoli, it does localize to coiled bodies and remains in coiled bodies longer than does wild-type U14 RNA (Figure 9). Taken together, our results indicate that box C/D snoRNAs transiently localize to coiled bodies before nucleoli and that the box C/D motif is not necessary for the localization of RNAs to coiled bodies but is required for subsequent localization to nucleoli.
Figure 9.
Dynamic association of U8 and U14 RNAs with coiled bodies. Fluorescein-labeled, wild-type, or nucleolar localization-defective variant U8 and U14 RNAs (including U8 box C [ΔC] and box D [ΔD] mutants and the U14 terminal stem mutant [ΔTerm. Stem]) were injected into oocyte nuclei. Nuclear spreads were made at the times indicated in minutes, and the RNA signals were analyzed by fluorescence microscopy. Variant snoRNAs that fail to localize to nucleoli (right) are retained for longer periods of time in coiled bodies (left) than are wild-type RNAs. Bar, 10 μm.
DISCUSSION
In eukaryotic cells, mechanisms exist to ensure that RNAs are properly transported from the sites of their synthesis to the sites of their function. In this study, we have identified determinants governing the localization of members of the box C/D class of snoRNAs to the nucleolus. We have found that the nucleolar localization of box C/D snoRNAs requires cis-acting sequences and structural features that are common to this class of RNAs. Moreover, the RNAs apparently travel through coiled bodies en route to nucleoli.
The Box C/D Motif Targets Box C/D snoRNAs to the Nucleolus
The members of the box C/D snoRNA family contain two common sequence elements (box C and box D) that are brought together in the folded RNAs as a result of the base pairing of complementary sequences flanking the box elements. Our results indicate that the key elements of the box C/D motif (i.e., box C, box D, and a nearby stem; Figure 4A, shaded area) are each essential for localization of box C/D snoRNAs.
U3 snoRNA, the primary focus of this work, contains two conserved elements with homology to box C, termed box C and box C′, and a single box D. In U3 RNA, box C′ exists opposite box D in a conserved stem-loop motif (Figure 4A) (Parker and Steitz, 1987; Jeppesen et al., 1988; Tycowski et al., 1993; Hartshorne and Agabian, 1994; Mereau et al., 1997; Samarsky and Fournier, 1998). Interestingly, box C of U3 is found opposite box B in the predicted secondary structure (Mereau et al., 1997; Samarsky and Fournier, 1998), and this conserved box B/C motif is unique to U3 RNAs. In addition, box C′ (and not box C) of U3 functions in accumulation of the RNA in yeast similar to box C of other C/D snoRNAs (Samarsky and Fournier, 1998). Thus, the box C/D motif of U3 RNA is comprised of U3 box C′, box D, and the flanking stems including the 3′ terminal stem.
Our mutational analysis of U3 showed that substitution of sequences in box C′, box D, or the 3′ terminal stem but not of other elements (boxes A, A′, B, and C or hinge) disrupted localization of the RNA to the nucleolus (Figures 4B and 7). These variant RNAs were not observed in nucleoli even at times up to 24 h after injection, indicating that these mutations did not simply reduce the rate of localization to nucleoli but rather rendered the RNAs defective in their ability to localize to nucleoli (our unpublished results). A recent study of the localization of fluorescently labeled U3 RNA in Xenopus oocyte nuclear spreads concludes that box C (not box C′) and box D are the elements critical for nucleolar localization of U3 (Lange et al., 1998c). Furthermore, box C′ and the 3′ terminal stem were found not to be necessary for nucleolar localization (although it was noted that variable results were obtained with box C′ mutants in that study [Lange et al., 1998c]). It is difficult to reconcile the striking differences between our results and those of Lange et al. (1998c) given the similarity of the studies. In yeast, mutation of U3 box C′ results in the loss of RNA stability (Mereau et al., 1997; Samarsky and Fournier, 1998), and Lange et al. (1998c) postulated that the reduced localization that they occasionally observed with Xenopus box C′ mutants may reflect the lack of stability. However, we have consistently observed that essentially all of the Xenopus U3 box C′ mutant RNA injected into Xenopus oocytes is present at the time of analysis (Figure 4C) and that significant amounts of the RNA remain for several hours (with a half-life of ∼6 h) but that the RNA is not observed in nucleoli at times up to 24 h after injection (Figure 4B and our unpublished results). The lack of nucleolar localization of the variant U3 RNAs in our experiments clearly is not simply a reflection of degradation of the RNAs.
Importantly, our results indicate that box C (U3 box C′) and box D function together as parts of the box C/D motif to direct nucleolar localization of this class of RNAs. Neither element appears to be capable of supporting nucleolar localization in the absence of the other, because substitution of either element (block substitution of box C′ or block substitution or point mutation of box D) resulted in a loss of localization (Figures 4B and 5). Similarly we found that disruption of either box C or box D prevents localization of U8 to nucleoli (Figure 9). Furthermore, disruption of the 3′ terminal stem of U3 RNA, which likely maintains the proximity of box C′ and box D, abolished the nucleolar localization of the RNA (Figure 7). Our finding that an intact 3′ terminal stem was also essential for the nucleolar localization of U14 indicates that the presence of a stem that brings together the box C and box D elements in the folded RNA molecules is likely of general importance for the nucleoloar localization of box C/D snoRNAs. Lange and colleagues report the fundamentally different observation that neither box C′ nor the terminal stem of the box C/D motif of U3 (Lange et al., 1998c) nor the terminal stem of U14 (Lange et al., 1998b) are essential for nucleolar localization. In addition, although they also found that box C and box D are essential for nucleolar localization of U8 (Figure 9) (Lange et al., 1998b), Lange et al. (1998b,c) have suggested that the spatial relationship of U8 box C and box D sequence elements is not important. The secondary structure of U8 has not been experimentally determined, and it is possible and indeed might be predicted based on an alternate proposed structure model (Watkins et al., 1996) that the spatial relationship of box C and box D has not been significantly altered in the variants examined. The box C/D motif and specifically the terminal stem that is thought to maintain the spatial relationship of box C and box D have been demonstrated to be important for the stability of snoRNAs (Terns et al., 1995), snoRNA biogenesis (Baserga et al., 1992; Xia et al., 1997), small nucleolar ribonucleoprotein (snoRNP) assembly (Watkins et al., 1998), and the binding of box C/D–specific proteins (Caffarelli et al., 1998). Our results indicate that the box C/D motif is also essential for the transport of the box C/D family snoRNAs to the nucleolus.
Our findings further indicate that sequences common to box C/D snoRNAs, rather than sequences specific to particular snoRNAs, are responsible for localizing these RNAs to nucleoli. It is conceivable that mutations in sequences other than the box C/D motif have subtle, nonessential effects on the rate or extent of nucleolar localization that were not detected in our assay. However, none of the other variant RNAs tested exhibited obvious diminished nucleolar localization relative to that of wild-type U3 RNA. It is noteworthy that U3 mutations that are predicted to disrupt base pairing of U3 with pre-rRNA (Beltrame and Tollervey, 1995; Hughes, 1996; Mereau et al., 1997) did not prevent the nucleolar localization of the RNA (box A, box A′, and hinge mutants; Figure 4B). Furthermore, stable fragments of U3 RNA that lack all three of these known pre-rRNA–binding sequence elements were also targeted to nucleoli (Figure 6). These results indicate that base pairing with rRNA, which is essential for U3 function (Beltrame and Tollervey, 1995; Hughes, 1996; Mereau et al., 1997), is not essential for its localization to nucleoli. Likewise, sequences within two other box C/D snoRNAs (U14 and U8 snoRNA) that exhibit complementarity to pre-rRNA were shown recently to be dispensable for their nucleolar localization (Lange et al., 1998a–c; Samarsky et al., 1998). Finally, although box B and box C are important for U3 function in rRNA processing (Samarsky and Fournier, 1998), we have shown that box B and box C are not essential for localization to nucleoli.
In addition, our results indicate that the box C/D motif is sufficient to direct localization of RNA to the nucleolus (Figure 6). Recent work with a snoRNA in different experimental systems supports this conclusion (Samarsky et al., 1998). Samarsky et al. (1998) demonstrated that minimal RNAs derived from U14 that maintain the box C/D motif are localized to nucleoli in both yeast and mammalian cells. It was not possible for Samarsky et al. (1998) to assess the necessity of the box C/D motif for U14 localization in those systems because of the lack of stability of the relevant mutant RNAs. Together the results indicate that the highly conserved box C/D motif mediates the nucleolar targeting of snoRNAs in diverse cell types including Xenopus oocytes (U3, U8, and U14 [this study]), yeast, and mammalian somatic cells (U14 [Samarsky et al., 1998]).
5′ Cap Hypermethylation Is Not Essential for Nucleolar Localization of Box C/D snoRNAs
Certain box C/D snoRNAs, including U3 and U8 RNA, are synthesized with m7G caps that become hypermethylated in the nucleus (Tyc and Steitz, 1989; Terns and Dahlberg, 1994; Terns et al., 1995). There is evidence that cap hypermethylation of snoRNAs occurs in the nucleoplasm before their localization to nucleoli (Terns et al., 1995). We found that U3, U8, and U14 RNAs transcribed with either their natural m7GpppG cap structure or with a cap structure that cannot be trimethylated (ApppG) were comparably localized to nucleoli (Figure 4B and our unpublished results). Thus, our results and those of others (Lange et al., 1998a) directly demonstrate that 5′ cap hypermethylation is not essential for the nucleolar localization of the capped box C/D snoRNAs. This conclusion is perhaps not surprising given that the majority of vertebrate snoRNAs are processed out of introns and do not contain 5′ cap structures but are nevertheless targeted to nucleoli (Maxwell and Fournier, 1995; Tollervey and Kiss, 1997). Previously, it was suggested that cap hypermethylation is not essential for nucleolar localization of U8 RNA because ApppG-capped U8 RNA is functional in Xenopus oocytes (Peculis and Steitz, 1994). On the other hand, cap hypermethylation was reported to be essential for the localization of U3 and U8 RNAs to nucleoli of injected mammalian cells (Jacobson and Pederson, 1998). This latter study raises the possibility that the nucleolar localization of box C/D snoRNAs may depend on cap hypermethylation in some but not all cell types.
The Function of the Box C/D Nucleolar-Targeting Motif Is Likely Mediated by Specific RNA-Binding Proteins
It is clear from a number of studies that box C/D sequences play key roles in important aspects of snoRNA metabolism (Baserga et al., 1991; Huang et al., 1992; Peculis and Steitz, 1994; Caffarelli et al., 1996; Cavaille and Bachellerie, 1996; Kiss-Laszlo et al., 1996, 1998; Nicoloso et al., 1996; Watkins et al., 1996; Xia et al., 1997). In vivo and in vitro structure probing of U3 RNA from a diverse range of organisms demonstrates that both the box C′ and box D sequences of U3 RNA are phylogenetically conserved protein-binding sites (Parker and Steitz, 1987; Jeppesen et al., 1988; Hartshorne and Agabian, 1994; Mereau et al., 1997). Moreover, we have found via competition experiments that newly synthesized snoRNAs are actively retained in the nucleus by a mechanism that is both saturable and sequence specific (Terns et al., 1995), indicating that specific nuclear factors associate with snoRNAs to prevent them from exiting the nucleus. Recently, excellent candidates for protein mediators of box C/D function have been identified (Caffarelli et al., 1998; Watkins et al., 1998; Wu et al., 1998; Lafontaine and Tollervey, 1999). Further research will be required to understand how such RNA–protein interactions contribute to the maturation, transport, and function of box C/D snoRNAs.
Potential Role of Coiled Bodies in the Maturation and Transport of Box C/D snoRNAs
A major gap in our knowledge of RNA trafficking is how specific subcellular structures participate in RNA transport and localization. We have found that box C/D family snoRNAs associate with nucleoli and coiled bodies. snoRNAs have been observed both in nucleoli (Fischer et al., 1991; Puvion-Dutilleul et al., 1991; Matera et al., 1994; Samarsky et al., 1998) and in coiled bodies (Bauer et al., 1994; Jimenez-Garcia et al., 1994; Samarsky et al., 1998; Shaw et al., 1998) in diverse cell types by in situ hybridization and light and electron microscopy. However, because analysis of the steady-state distribution of endogenous RNAs yields little information about the mechanism of ongoing RNA transport, we have performed a temporal analysis of the intranuclear distribution of injected snoRNAs. We show that box C/D snoRNAs interact dynamically with coiled bodies (Figures 8 and 9). Our finding that the injected box C/D snoRNAs are readily detected first in coiled bodies and only later in nucleoli suggests that movement of the precursor U3, U8, and U14 snoRNAs through coiled bodies may be a step in the pathway toward nucleolar localization. Our observation that snoRNA mutants that fail to localize to nucleoli are detained in coiled bodies suggests that these mutant RNAs are stalled at an intermediate step in the nucleolar localization pathway.
In addition to box C/D snoRNAs, other nuclear RNAs may also transiently localize to coiled bodies. Indeed, we have found that the U1 spliceosomal snRNA is targeted to coiled bodies rapidly after injection into Xenopus oocytes before localization to snurposomes (Figure 8 and our unpublished results). Previous examination of the steady-state localization of U1 snRNA by in situ hybridization in Xenopus oocytes did not indicate significant amounts of the RNA in coiled bodies (Wu et al., 1991). However, U1 snRNA has been readily observed in coiled bodies of somatic cells where snRNAs are more actively synthesized (Carmo-Fonseca et al., 1992; Huang and Spector, 1992; Matera and Ward, 1993). In addition, studies with U7 snRNA indicate a dynamic localization with coiled bodies after injection into oocytes (Wu et al., 1996).
It is unclear why snoRNAs traverse coiled bodies. Numerous models for coiled body function have been proposed to account for the fact that these structures contain a multitude of factors including both snRNAs and snoRNAs and their associated proteins (Lamond and Carmo-Fonseca, 1993; Gall et al., 1995; Matera, 1998). Coiled bodies might be involved in the early metabolism of snoRNAs and may be sites of snoRNA modification or assembly into RNP complexes. Conceivably, the reason why mutant snoRNAs that do not localize to nucleoli are detained in coiled bodies is because they lack the ability (e.g., modifications or box C/D–binding factors) to exit these structures efficiently or to be transferred to nucleoli.
Coiled bodies could serve as intranuclear transport vehicles that shuttle components (including snoRNPs) to the sites of their function. We detected snoRNAs in coiled bodies at early but not late time points after injection, suggesting that the flow of snoRNAs between coiled bodies and nucleoli is unidirectional. The box C/D motif is not essential for localization of snoRNAs to coiled bodies, but our results suggest that the motif is required to move snoRNAs from coiled bodies to nucleoli. Coiled bodies have been detected in close association with snoRNA (and snRNA) gene loci (reviewed in Matera, 1998) and are often found physically associated with nucleoli (Raska et al., 1990; Lafarga et al., 1991; Malatesta et al., 1994; Ochs et al., 1994; Bohmann et al., 1995; Lyon et al., 1997). Thus, coiled bodies could conceivably be important in the synthesis, modification, and transport of snoRNAs and other nuclear RNAs.
ACKNOWLEDGMENTS
Special thanks to Joseph Gall for guidance in the preparation of oocyte nuclear spreads and for the generous loan of a specialized rotor. We also kindly thank Elisa Izzauralde (DHFR mRNA and tRNA), Brenda Peculis (U8), E. Stuart Maxwell (U14), and Paul Romaniuk (5S rRNA) for providing plasmids and Reinhard Lührmann (m2,2,7G cap), Elsebet Lund (m7G cap), Joseph Gall (p80 coilin mAb H1), and K. Michael Pollard and Eng Tan (fibrillarin mAbs 72B9 and 17C12) for providing antibodies used in this study. We are grateful to Claiborne Glover III, Pascale Legault, and Michael McEachern for critical reading of this manuscript and to James Griffith for technical assistance in generating many of the mutant templates used in this work. This work was supported in part by a University of Georgia Faculty Research Grant to R.T. and by a Basil O’Conner Starter Scholar Research Award from the March of Dimes Birth Defects Foundation, an American Cancer Society grant (NP-84149), and a National Science Foundation grant (RR61345F) to M.P.T.
REFERENCES
- Andrade L, Chan E, Raska I, Peebles C, Roos G, Tan E. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie JP, Michot B, Nicoloso M, Balakin A, Ni J, Fournier MJ. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- Balakin A, Smith L, Fournier M. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- Baserga SJ, Gilmore HM, Yang XW. Distinct molecular signals for nuclear import of the nucleolar snRNA, U3. Genes Dev. 1992;6:1120–1130. doi: 10.1101/gad.6.6.1120. [DOI] [PubMed] [Google Scholar]
- Baserga SJ, Yang XD, Steitz JA. An intact box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 1991;10:2645–2651. doi: 10.1002/j.1460-2075.1991.tb07807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Murphy C, Wu Z, Wu C, Gall J. In vitro assembly of coiled bodies in Xenopus egg extract. Mol Biol Cell. 1994;5:633–644. doi: 10.1091/mbc.5.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier EH, Brown DD, Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978;15:1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Bohmann K, Ferreira J, Lamond A. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer R, Lehner C, Eppenberger H, Nigg E. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- Bringmann P, Rinke J, Appel B, Reuter R, Luhrmann R. Purification of snRNPs U1, U2, U4, U5, and U6 with 2,2,7-trimethylguanosine-specific antibody and definition of their constituent proteins reacting with anti-Sm and anti-(U1) RNP antisera. EMBO J. 1983;2:1129–1135. doi: 10.1002/j.1460-2075.1983.tb01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoi I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E, Losito M, Giorgi C, Fatica A, Bozzoni I. In vivo identification of nuclear factors interacting with the conserved elements of box C/D small nucleolar RNAs. Mol Cell Biol. 1998;18:1023–1028. doi: 10.1128/mcb.18.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalno MT, Lamond A I. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J, Bachellerie J. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- Cavaille J, Nicoloso M, Bachellerie J. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- Enright C, Maxwell E, Sollner-Webb B. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA. 1996;2:1094–1099. [PMC free article] [PubMed] [Google Scholar]
- Fischer D, Weisenberger D, Scheer U. Assigning functions to nucleolar structures. Chromosoma. 1991;101:133–140. doi: 10.1007/BF00355363. [DOI] [PubMed] [Google Scholar]
- Fournier R, Brule F, Segault V, Mougin A, Branlant C. U3 snoRNA genes with and without intron in the Kluyveromyces genus: yeasts can accommodate great variations of the U3 snoRNA 3′-terminal domain. RNA. 1998;4:285–302. [PMC free article] [PubMed] [Google Scholar]
- Gall J, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- Gall JG, Callan HG. The sphere organelle contains small nuclear ribonucleoproteins. Proc Natl Acad Sci USA. 1989;86:6635–6639. doi: 10.1073/pnas.86.17.6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Murphy C, Callan HG, Wu Z. Lampbrush chromosomes. Methods Cell Biol. 1991;36:149–166. [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- Hamm J, Mattaj I. An abundant U6 snRNP found in germ cells and embryos of Xenopus laevis. EMBO J. 1989;8:4179–4187. doi: 10.1002/j.1460-2075.1989.tb08603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne T, Agabian N. A common core structure for U3 small nucleolar RNAs. Nucleic Acids Res. 1994;22:3354–3364. doi: 10.1093/nar/22.16.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R. PCR Protocols: A Guide to Methods and Applications, chap. 22. Academic Press; 1990. Recombinant PCR; pp. 177–183. [Google Scholar]
- Huang S, Spector DL. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc Natl Acad Sci USA. 1992;89:305–308. doi: 10.1073/pnas.89.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GM, Jarmolowski A, Struck JC, Fournier MJ. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′, 3′ terminal stem. Mol Cell Biol. 1992;12:4456–4463. doi: 10.1128/mcb.12.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- Hughes J, Ares MJ. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman P, Enestrom S, Turley SJ, Pollard KM. Selective induction of anti-fibrillarin autoantibodies by silver nitrate in mice. Clin Exp Immunol. 1994;96:285–291. doi: 10.1111/j.1365-2249.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M, Pederson T. A 7-methylguanosine cap commits U3 and U8 small nuclear RNAs to the nucleolar localization pathway. Nucleic Acids Res. 1998;26:756–760. doi: 10.1093/nar/26.3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A, Boelens W, Izaurralde E, Mattaj I. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen C, Stebbins-Boaz B, Gerbi S. Nucleotide sequence determination and secondary structure of Xenopus U3 snRNA. Nucleic Acids Res. 1988;16:2127–2148. doi: 10.1093/nar/16.5.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia LF, Segura-Valdez ML, Ochs R, Rothblum L, Hannan R, Spector D. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Bachellerie J, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga M, Andres M, Berciano M, Maquiera E. Organization of nucleoli and nuclear bodies in osmotically stimulated supraoptic neurons of the rat. J Comp Neurol. 1991;308:329–339. doi: 10.1002/cne.903080302. [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A, Carmo-Fonseca M. The coiled body. Trends Cell Biol. 1993;3:198–204. doi: 10.1016/0962-8924(93)90214-l. [DOI] [PubMed] [Google Scholar]
- Lange T, Borovjagin A, Gerbi S. Nucleolar localization elements in U8 snoRNA differ from sequences required for rRNA processing. RNA. 1998a;4:789–800. doi: 10.1017/s1355838298980438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Borovjagin A, Maxwell E, Gerbi S. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 1998b;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Ezrokhi M, Borovjagin A, Rivera-Leon R, North M, Gerbi S. Nucleolar localization elements of Xenopus laevis U3 small nucleolar RNA. Mol Biol Cell. 1998c;9:2973–2985. doi: 10.1091/mbc.9.10.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B, Mariottini P, Mathieu C, Ferrer P, Amaldi F, Amalric F, Caizergues-Ferrer M. Molecular cloning of Xenopus fibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol Cell Biol. 1990;10:430–434. doi: 10.1128/mcb.10.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HD, Zagorski J, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon C, Bohmann K, Sleeman J, Lamond A. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res. 1997;230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Zancanaro C, Martin T, Chan E, Amalric F, Luhrmann R, Vogel P, Fakan S. Is the coiled body involved in nucleolar functions? Exp Cell Res. 1994;211:415–419. doi: 10.1006/excr.1994.1106. [DOI] [PubMed] [Google Scholar]
- Matera A. Of coiled bodies, gems, and salmon. J Cell Biochem. 1998;69:1–13. [PubMed] [Google Scholar]
- Matera A, Tycowski K, Steitz J, Ward D. Organization of small nucleolar ribonucleoproteins (snoRNPs) by fluorescence in situ hybridization and immunocytochemistry. Mol Biol Cell. 1994;5:1289–1299. doi: 10.1091/mbc.5.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Ward DC. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol. 1993;121:715–727. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I, De Robertis E. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985;40:111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–933. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Meier U, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereau A, Fournier R, Gregoire A, Mougin A, Fabrizio P, Luhrmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and basepair interaction with the pre-ribosomal RNA. J Mol Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- Munns TW, Liszewski MK, Tellam JT, Sims HF, Rhoads RE. Antibody-nucleic acid complexes. Immunospecific retention of globin messenger ribonucleic acid with antibodies specific for 7-methylguanosine. Biochemistry. 1982;21:2922–2928. doi: 10.1021/bi00541a018. [DOI] [PubMed] [Google Scholar]
- Nicoloso M, Qu L, Michot B, Bachellerie J. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Stein TW, Jr, Tan EM. Coiled bodies in the nucleolus of breast cancer cells. J Cell Sci. 1994;107:385–399. doi: 10.1242/jcs.107.2.385. [DOI] [PubMed] [Google Scholar]
- Parker K, Steitz J. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987;7:2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes Dev. 1994;8:2241–2255. doi: 10.1101/gad.8.18.2241. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Mazan S, Nicoloso M, Christensen ME, Bachellerie JP. Localization of U3 RNA molecules in nucleoli of HeLa and mouse 3T3 cells by high resolution in situ hybridization. Eur J Cell Biol. 1991;56:178–186. [PubMed] [Google Scholar]
- Qu L, Henry Y, Nicoloso M, Michot B, Azum M, Renalier M, Caizergues-Ferrer M, Bachellerie J. U24, a novel intron-encoded small nucleolar RNA with two 12 nt long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res. 1995;23:2669–2676. doi: 10.1093/nar/23.14.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Ochs R, Andrade L, Chan E, Burlingame R, Peebles C, Gruol D, Tan E. Association between the nucleolus and the coiled body. J Struct Biol. 1990;104:120–127. doi: 10.1016/1047-8477(90)90066-l. [DOI] [PubMed] [Google Scholar]
- Reimer G, Pollard KM, Penning CA, Ochs RL, Lishwe MA, Busch H, Tan EM. Monoclonal autoantibody from a (New Zealand black X New Zealand white) F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 1987;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- Samarsky D, Fournier M. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:3431–3444. doi: 10.1128/mcb.18.6.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky D, Fournier M, Singer R, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R, Gerbi SA. In vivo disruption of Xenopus U3 snRNA affects rRNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R, Hitti Y, Gerbi SA. Genes for Xenopus laevis U3 small nuclear RNA. Nucleic Acids Res. 1992;20:5435–5442. doi: 10.1093/nar/20.20.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmang T, Tollervey D, Kern H, Frank R, Hurt E. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Terry C, Wells D, DiMario P. Structural changes in oocyte nucleoli of Xenopus laevis during oogenesis and meiotic maturation. Chromosoma. 1996;105:111–121. doi: 10.1007/BF02509521. [DOI] [PubMed] [Google Scholar]
- Shanab GM, Maxwell ES. Proposed secondary structure of eukaryotic U14 snRNA. Nucleic Acids Res. 1991;19:4891–4894. doi: 10.1093/nar/19.18.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanab GM, Maxwell ES. Determination of the nucleotide sequences in mouse U14 small nuclear RNA and 18S rRNA responsible for in vitro intermolecular base-pairing. Eur J Biochem. 1992;206:391–400. doi: 10.1111/j.1432-1033.1992.tb16939.x. [DOI] [PubMed] [Google Scholar]
- Shaw P, Beven A, Leader D, Brown J. Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J Cell Sci. 1998;111:2121–2128. doi: 10.1242/jcs.111.15.2121. [DOI] [PubMed] [Google Scholar]
- Shaw P, Jordan E. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Smith C, Steitz J. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Terns M, Goldfarb D. Nuclear transport of RNAs in microinjected Xenopus oocytes. Methods Cell Biol. 1998;53:559–589. doi: 10.1016/s0091-679x(08)60895-x. [DOI] [PubMed] [Google Scholar]
- Terns MP, Dahlberg JE. Retention and 5′ cap tri-methylation of U3 snRNA in the nucleus. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- Terns MP, Dahlberg JE, Lund E. Multiple cis-acting signals for export of pre-U1 snRNA from the nucleus. Genes Dev. 1993;7:1898–1908. doi: 10.1101/gad.7.10.1898. [DOI] [PubMed] [Google Scholar]
- Terns MP, Grimm C, Lund E, Dahlberg JE. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. Small nucleolar RNAs guide ribosomal RNA methylation. Science. 1996;273:1056–1057. doi: 10.1126/science.273.5278.1056. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Tuma R, Stolk J, Roth M. Identification and characterization of a sphere organelle protein. J Cell Biol. 1993;122:767–773. doi: 10.1083/jcb.122.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K, Steitz JA. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K, Shu M, Steitz J. Requirement for intron-encoded U22 small nucleolar RNA in 18S rRNA maturation. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- Tycowski K, Smith C, Shu M, Steitz J. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc Natl Acad Sci USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Steitz JA. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;7:1176–1190. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- Watkins N, Leverette R, Xia L, Andrews M, Maxwell E. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- Watkins N, Newman D, Kuhn J, Maxwell E. In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65-kDa box C/D-binding protein. RNA. 1998;4:582–593. doi: 10.1017/s1355838298980128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Murphy C, Gall J. The Sm binding site targets U7 snRNA to coiled bodies (spheres) of amphibian oocytes. RNA. 1996;2:811–823. [PMC free article] [PubMed] [Google Scholar]
- Wu P, Brockenbrough J, Metcalfe A, Chen S, Aris J. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Gall J. “Micronucleoli” in the Xenopus germinal vesicle. Chromosoma. 1997;105:438–443. doi: 10.1007/BF02510480. [DOI] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Wu CH, Tsvetkov A, Gall JG. Snurposomes and coiled bodies. Cold Spring Harb Symp Quant Biol. 1993;58:747–754. doi: 10.1101/sqb.1993.058.01.082. [DOI] [PubMed] [Google Scholar]
- Wu ZA, Murphy C, Callan HG, Gall JG. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J Cell Biol. 1991;113:465–483. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Watkins N, Maxwell E. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA. 1997;3:17–26. [PMC free article] [PubMed] [Google Scholar]