Abstract
Epstein-Barr virus OriP confers cell cycle-dependent DNA replication and stable maintenance on plasmids in EBNA1-positive cells. The dyad symmetry region of OriP contains four EBNA1 binding sites that are punctuated by 9-bp repeats referred to as nonamers. Previous work has shown that the nonamers bind to cellular factors associated with human telomeres and contribute to episomal maintenance of OriP. In this work, we show that substitution mutation of all three nonamer sites reduces both DNA replication and plasmid maintenance of OriP-containing plasmids by 2.5- to 5-fold. The nonamers were required for high-affinity binding of TRF1, TRF2, and hRap1 to the dyad symmetry element but were not essential for the binding of EBNA1 as determined by DNA affinity purification from nuclear extracts. Chromatin immunoprecipitation assays indicated that TRF1, TRF2, and hRap1 bound OriP in vivo. Cell cycle studies indicate that TRF2 binding to OriP peaks in G1/S while TRF1 binding peaks in G2/M. OriP replication was inhibited by transfection of full-length TRF1 but not by deletion mutants lacking the myb DNA binding domain. In contrast, OriP replication was not affected by transfection of full-length TRF2 or hRap1 but was potently inhibited by dominant-negative TRF2 or hRap1 amino-terminal truncation mutants. Knockdown experiments with short interfering RNAs (siRNAs) directed against TRF2 and hRap1 severely reduced OriP replication, while TRF1 siRNA had a modest stimulatory effect on OriP replication. These results indicate that TRF2 and hRap1 promote, while TRF1 antagonizes, OriP-dependent DNA replication and suggest that these telomeric factors contribute to the establishment of replication competence at OriP.
Epstein-Barr virus (EBV) is a lymphotropic gammaherpesvirus that can be cultured in latently infected B-cell lines as a multicopy extrachromosomal plasmid (reviewed in reference 27). The latent viral genome can be isolated from a variety of tumor tissues and is causally linked with Burkitt's lymphoma, nasopharyngeal carcinoma, and lymphoproliferative disorders in the immunosuppressed population (reviewed in reference 52). In most, if not all, latent infections the virus-encoded EBV nuclear antigen 1 (EBNA1) can be detected. Genetic and biochemical experiments have established that EBNA1 is essential for the maintenance of the viral genome during latency (34, 74). EBNA1 binds with high avidity to three regions of the viral genome, namely, the family of 30-bp repeats (FR), the dyad symmetry (DS) region, and the Q promoter (49). The FR and DS together comprise the viral origin of plasmid replication (OriP). The DS is required for DNA replication initiation, while FR stimulates DS replication and provides plasmid maintenance activity (3, 11, 29, 55, 73). The Q promoter has no known replication activity but is thought to autoregulate EBNA1 RNA transcription (62). EBNA1 binding to OriP is sufficient to confer plasmid replication and maintenance in most human and primate cell types but may be restricted in murine and hamster cells by unknown cellular factors (60, 74, 75). The precise mechanism by which EBNA1 stimulates replication and establishes plasmid maintenance has not been elucidated.
EBNA1 has structural similarity to the papillomavirus E2 protein, which is required for loading the E1 helicase on papillomavirus replication origins (6, 7). EBV does not encode a replicative helicase or ATPase dedicated to OriP-dependent latent cycle replication. However, cellular factors associated with replication origins also associate with OriP, including components of the origin recognition complex (ORC) and the minichromosome maintenance (MCM) replicative helicase complex (10, 13, 54). The C-terminal DNA binding domain of EBNA1 is necessary but not sufficient for replication or plasmid maintenance function (72). At least three other redundant regions in the N-terminal domain of EBNA1 provide an oligomerization function which correlates well with replication and plasmid maintenance activity (9, 37, 38). At least two cellular factors bind to these domains, SFp32 and EBP2, both of which have been implicated in other cellular processes including RNA processing (56, 63, 67). Overexpression of SFp32 can alter EBNA1 transcription activity in transient-transfection assays, and EBP2 can confer plasmid segregation properties on EBNA1 when coexpressed in Saccharomyces cerevisiae (26, 67, 69). In addition to these interacting proteins, EBNA1 also binds with high avidity to the nuclear transport factor importin α (or Rch1) through a region overlapping the EBNA1 nuclear localization signal (15, 28). EBNA1 binds to metaphase chromosomes, and genetic evidence indicates that this activity is required for plasmid maintenance (39). Substitution of the EBNA1 metaphase chromosome binding region with chromatin-associated protein histone H1 or HMG1 rescues EBNA1 replication and plasmid maintenance activity (24). These latter studies strongly implicate chromosome attachment as a primary mechanism for EBNA1-mediated plasmid maintenance.
OriP consists of two regions containing EBNA1 binding sites separated by an ∼1-kb spacer region. The FR consists of 20 30-bp imperfect repeats that bind EBNA1 with high affinity, while the DS consists of two pairs of low-affinity EBNA1 binding sites with a 21-bp spacing between the center of each paired site. Only one of the two pairs of the EBNA1 binding sites is essential for minimal replication activity, and small changes in the spacing of the paired EBNA1 binding sites can abolish DNA replication activity (3, 23, 31, 73). EBNA1 induces a strong bend in the DNA of the minimal replicator which correlates well with replication initiation activity (3). Three nonamer binding sites (TTAGGGTTA) straddle and separate the two pairs of EBNA1 binding sites in DS (12, 66). The nonamers resemble telomere repeats and can bind to TRF2 in vitro and in vivo (12). Deletion mutation of sequences overlapping two of the three nonamers reduces DNA replication, and site-directed mutations in all three nonamer sites reduced plasmid maintenance (12, 31, 66, 73). Plasmid maintenance function is strongly dependent upon FR, and at least seven EBNA1 binding sites are essential for this activity (71). EBNA1 bound to FR confers nuclear retention of plasmids, and this is also likely to increase the stability and transcription activity of transfected plasmid DNA (2, 29, 41, 61). The FR can also enhance replication of DS, as well as stimulate transcription from cis-linked heterologous promoters. EBV DNA is associated with the nuclear matrix, through a region that overlaps OriP, and it is possible that nuclear matrix attachment contributes to the plasmid maintenance function of the EBNA-OriP complex (25).
We have isolated several telomere repeat-associated factors that bind to DS in an EBNA1-dependent manner (12). We found that TRF2 can bind directly to the nonamer sites of DS and that overexpression of a dominant-negative form of TRF2 inhibits transient DNA replication. TRF2 was originally characterized as a telomere repeat binding factor that causes a dramatic dysregulation of telomere length and structure when inactivated by overexpression of a dominant-negative transgene (58, 65). Loss of TRF2 resulted in chromosome fusions and other chromosome aberrations associated with aberrant telomeres. TRF2 can stimulate T-loop formation in vitro and may facilitate strand invasion and base pairing of the G-rich single strand found at the ends of mammalian telomeres (19, 59). TRF2 is a member of the myb family of DNA binding domains and can homodimerize through a central region of the protein (9). TRF2 binds tightly to another myb domain protein, hRap1, which has additional sequence similarity to the budding yeast telomere binding protein, Rap1 (36). Yeast Rap1 binds to the telomere repeat sequence (TC1-3) found in budding yeast, but the human hRap1 appears to have lost its DNA binding activity and associates with telomeres primarily through its association with TRF2 (36, 42). In addition to TRF2, a second, less abundant telomere repeat binding factor has been identified, TRF1, which is also a member of the myb DNA binding family and binds with similar sequence specificity as does TRF2 (4, 5, 64). TRF1 promotes homologous parallel pairing of telomere repeats but is not sufficient for T-loop formation like TRF2 (18). TRF1 has several binding partners, including tankyrase, an ankyrin repeat protein with poly(ADP-ribose) polymerase activity (57), and the S-phase checkpoint protein NBS1, associated with Neijman breakage syndrome (68). It was previously shown that TRF2 and tankyrase bind to DS, but the role of hRap1 or the potential role of TRF1 was not characterized (12). In this study, we examined the role of the nonamer binding proteins and found that, in addition to TRF2, both TRF1 and hRap1 bind to DS in a nonamer-dependent manner. We also show that TRF2 and hRap1 promote DS replication, as well as plasmid maintenance, while TRF1 appears to inhibit OriP replication and opposes the activity of TRF2.
MATERIALS AND METHODS
Cells.
D98/HR1, HeLa, and 293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. Raji and ZKO-293 cells (kindly provided by H. Delecluse) were cultured in RPMI 1640 supplemented with 10% fetal bovine serum and antibiotics. For cell synchronization studies, exponentially growing Raji cells (2 × 105 to 3 × 105 cells/ml) were treated with thymidine (final concentration, 2 mM). After 12 to 14 h the thymidine medium was removed by centrifugation, and the cells were grown in thymidine-free medium for 10 h. Thymidine was again added (2 mM final concentration) and was removed after 14 to 16 h. After resuspension in thymidine-free medium, cells were harvested at 0.5 (G1/S), 2 (S), and 4 (G2/M) h after release. Synchronization was evaluated by staining with propidium iodide (Sigma) and fluorescence-activated cell sorting (FACS) analysis. For G0 arrest, Raji cells were cultured in serum-free RPMI for 24 h.
Plasmids.
OriPwt (N503) and OriPnm− (N506) were described previously (OriPnm− was referred to as OriPΔa,b,c) and are derivatives of pREP10 (Invitrogen) with enhanced green fluorescent protein (GFP) under the regulation of the respiratory syncytial virus promoter. The base substitutions in OriPnm− are indicated in Fig. 1C. OriPΔDS (N564) was generated by EcoRV and XbaI deletion of OriPwt (N503). Mammalian cell expression plasmids for TRF2 full-length (amino acids [aa] 1 to 500), TRF2ΔN/M (aa 45 to 454), TRF1 full-length (aa 1 to 440), TRF1ΔN (aa 66 to 440), TRF1ΔM (aa 1 to 350), TRF1ΔN/ΔM (aa 66 to 385), hRap1ΔB (aa 120 to 399), and hRap1ΔB/M (aa 240 to 399) were cloned into HindIII-BamHI sites of pFLAG-CMV-2 (Sigma). Full-length hRap1 (aa 1 to 399) was cloned into pBK-CMV (Stratagene) and was a gift of Titia de Lange (Rockefeller University). Short interfering RNA (siRNA) expression vectors were generated using the Hanon method (21, 47). Briefly, short hairpins of 27 to 29 nucleotides for TRF1, hRap1, and TRF2 were expressed by the U6 promoter in the pENTR/D-Topo vector (Invitrogen). pGEM1 plasmid containing the U6 promoter was used as the template for PCR with the Sp6 primer (CACCGATTTAGGTGACACTATAG) and TRF1 primer (AAAAAAGCTAGGTTCCATATTACAACTCAGATAAGCAAGCTTCCTTATCTAAGTTGCAACATGGAACCCAGCGGTGTTTCGTCCTTTCCACAA), TRF2 primer (AAAAAAACACCCAGCAATCGACTGCCTCTTCCAACCAAGCTTCGCTGGAAGAGGCAGTCAATCGCTGGGTGCGGTGTTTCGTCCTTTCCACAA), or hRap1 primer (AAAAAAAACCAACGCCTCCCTAGTATCCTCATCATCAAGCTTCATGATGAGGATACCAGAGAGGCATTGGTCGGTGTTTCGTCCTTTCCACAA).
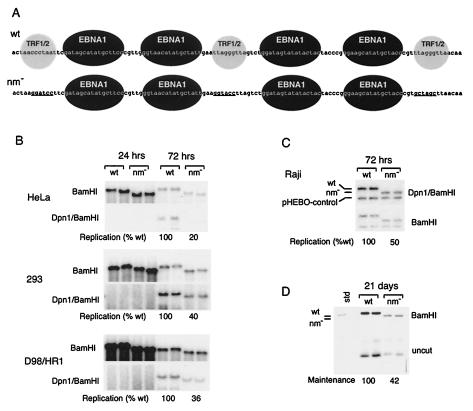
FIG. 1.
Nonamer repeats contribute to DNA replication and plasmid maintenance of OriP. (A) Sequences of OriPwt and OriPnm− with substitution mutations indicated by underlining. Black spheres indicate EBNA1 binding sites. Gray spheres indicate TRF2 and/or TRF1 binding sites. (B) HeLa, 293, and D98/HR1 cells were transfected with OriPwt or OriPnm− plasmid and assayed for DNA replication at 24 or 72 h posttransfection. BamHI digests are shown in the top panel and DpnI/BamHI double digests are indicated in the bottom panel for each cell type indicated to the left. (C) Raji cells were electroporated with OriPwt or OriPnm− and pHEBO control. Equal numbers of GFP-positive cells were selected by FACS and then assayed at 72 h posttransfection for DNA replication based on DpnI resistance. DpnI/BamHI digests (top panel) and BamHI digests (lower panel) were detected by Southern blotting and quantitated by PhosphorImager analysis. (D) Raji cells were electroporated and selected by FACS as described for panel A. After 21 days in culture without selection, DNA was extracted and analyzed by Southern blotting after BamHI digestion (top panel) or was uncut (bottom panel). Quantitation of Southern blots by PhosphorImager analysis is indicated below.
DNA affinity purification.
PCR fragments for pBKSII (BKS), DS, DSnm−, or ΔDS were amplified with one biotinylated primer and bound to streptavidin magnetic beads (Dynal) essentially as described previously (12). PCR fragments were amplified with the following primer pairs: for BKS, 5′-biotin-GTAAAACGACGGCCAGT and 5′-GGAAACAGCTATGACCATGATAC; for OriPwt, OriPnm−, and OriPΔDS, 5′-biotin-CGAAGGAGAATGAAGAAGCAGGCG and 5′-CGGGAGCAGACAAGCCCGTCAGGGCGC. Whole-cell extracts were generated by resuspending 5 × 109 Raji cells in 15 ml of HE1000 (20 mM HEPES [pH 7.9], 0.2 mM EDTA, 1 M NaCl, 0.05% Igepal, 5 mM β-mercaptoethanol, and protease inhibitor cocktail [Sigma]), followed by sonication at 4°C sufficient to reduce viscosity (5 ml of lysate was subjected to six bursts of 10-s pulses with a Branson sonicator set at maximum output for microtip). Whole-cell extracts were premixed with 400 μg of sonicated salmon sperm DNA for 15 min and then with beads bound to biotinylated DNA. Binding reaction mixtures were then diluted with HE0 (20 mM HEPES [pH 7.9], 0.2 mM EDTA, 0.05% Igepal [Sigma], 5 mM β-mercaptoethanol, and protease inhibitor cocktail [Sigma]) to a final concentration of 150 mM NaCl (5.6-fold) by 200-ml additions every 10 min during rotation at 25°C. Bound complexes were washed three times (low stringency) or five times (high stringency) with DN150 buffer (20 mM HEPES [pH 7.9], 0.2 mM EDTA, 20% glycerol, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 150 mM KCl, and 0.05% NP-40). Bound proteins were eluted with 2× sodium dodecyl sulfate (SDS) sample buffer (53) and analyzed by Western blotting.
Western blotting.
Antibodies to hemagglutinin (Roche Biosciences), Flag M2 (Sigma), EBNA1 (Advanced Biotechnologies, Inc.), PCNA (Santa Cruz Biotechnology), TRF2 (IMGENEX), and acetyl-histone H3 (Upstate Biotechnology) were purchased and used according to manufacturer specifications. Rabbit antibodies to hRap1 and TRF1 were generated against a full-length recombinant protein and affinity purified. Most antibodies were diluted 1:1,000 or 1:500 in TS buffer (20 mM Tris [pH 8.0], 200 mM NaCl) and 5% dried milk. Blots were washed once with TS, twice with TS containing 0.05% Igepal, and once with TS for 10 min each wash.
ChIP assay.
Chromatin immunoprecipitation (ChIP) was performed as described by the manufacturer (Upstate Biotechnology). Briefly, Raji cells were subjected to formaldehyde (1.0%) cross-linking for 15 min. Cross-linking was stopped by the addition of glycine to the final concentration of 0.125 M, and the cells were collected by centrifugation and washed twice with ice-cold phosphate-buffered saline containing 0.1 mM PMSF. About 106 cells were lysed in 100 μl of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.0], 1 mM PMSF) for 10 min on ice. The lysates were sheared by sonication to reduce the DNA length to between 400 and 800 bp. Cell debris was removed by centrifugation, and the supernatant was diluted 10-fold with immunoprecipitation dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8.0], 167 mM NaCl, 1 mM PMSF). The diluted extracts were precleared with a protein A-Sepharose (Amersham) slurry containing sonicated salmon sperm DNA for 30 min at 4°C with rotation. The beads were pelleted, and supernatant was immunoprecipitated with antibodies overnight at 4°C with rotation. Twenty microliters of the supernatant was kept as an input control. Immune complexes were collected with a salmon sperm DNA-protein A-Sepharose slurry for 1 h at 4°C with rotation. After several rounds of washing, immune complexes were eluted twice with 200 μl of elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature. Eluates were combined with 16 μl of 5 M NaCl and incubated at 65°C for at least 4 h to reverse cross-linking. Then, eluates were incubated with proteinase K for 1 h at 50°C. Immunoprecipitated DNA was purified by phenol-chloroform extraction and ethanol precipitation with 20 μg of glycogen. EBV sequences were amplified with primers spanning OriP (corresponding to EBV nucleotides 8586 to 9206) and the BZLF1 promoter (nucleotides 103254 to 103514).
To study in vivo association with nonamers of telomeric proteins, OriP or OriPnm− plasmid was transfected into D98/HR1 cells. Sixteen hours posttransfection, cells were trypsinized, washed with phosphate-buffered saline twice, and cultured in Dulbecco's modified Eagle's medium for another 2 days. Equal numbers of GFP-positive cells were isolated by FACS and subjected to the ChIP assay as described above. OriP-specific sequences were amplified with a DS-specific primer (CCCGTGACAGCTCATGGGGTGGGAGAT) and a Rep10-specific primer (CGGGAGCAGACAAGCCCGTCAGGGCGC). As a control, the plasmid Amp gene was amplified with primers oPL392 (TCCATAGGCTCCGCCCCCCTGACGAGCATC) and oPL391 (AGGTAACTGGCTTCAGCAGAGCGCAGATAC).
DNA replication assay.
For monolayers, 2 × 106 cells were transfected with 3 μg of DNA by using Lipofectamine 2000 (Invitrogen). For Raji cells, 107 cells were electroporated with 5 μg of OriP or OriPnm− plasmid together with control plasmid pHEBO. Twenty-four hours posttransfection GFP-positive cells were sorted by FACS and cultured in RPMI for another 2 days. Plasmid DNA from equal numbers of cells (2 × 106) was isolated by Hirt extraction. For a replication assay with siRNAs, OriP plasmid (3 μg) and a plasmid expressing siRNA (3 μg) were cotransfected into D98/HR1 cells with Lipofectamine 2000 (Invitrogen). Seventy-two hours posttransfection, GFP-positive cells were sorted by FACS. Equal amounts of cells were used to isolate plasmid DNA, which was assayed by Southern blotting. For replication assays with truncation mutants of TRF1, hRap1, and TRF2, D98 or ZKO-293 cells were transfected with 6 μg of DNA by the use of Lipofectamine 2000 (Invitrogen). The plasmids were extracted by a modified Hirt method (Invitrogen). The nucleic acid pellets were dissolved in 30 μl of Tris-EDTA. DNA was subjected to restriction digestion with DpnI and BamHI or with just BamHI. DNA samples were analyzed on 0.7% agarose gels in the absence of ethidium bromide and transferred to Zeta membranes (Bio-Rad). Radiographic images were quantified by PhosphorImager analysis. Plasmid maintenance assays were described previously (12). Briefly, equal numbers of transfected Raji cells were selected by FACS and then cultured for 21 days without selection. Plasmid DNA from equal numbers of cells (∼107) was isolated by Hirt extraction and then assayed directly as supercoiled DNA by Southern blotting or as BamHI digests. PhosphorImager analysis was used to quantify plasmid recovery.
RESULTS
Nonamers promote OriP-dependent DNA replication and plasmid maintenance.
It was previously found that the nonamers had a two- to threefold effect on plasmid maintenance in Akata Burkitt lymphoma cells (12). We further investigated the role of the nonamers in DNA replication of OriP in various cell types. Substitution mutations in all three nonamers were generated such that EBNA1 binding site spacing was not altered (Fig. 1A). Both OriPwt and OriPnm− plasmids also express EBNA1 to allow for replication in EBNA1-negative cell lines and GFP to allow for cell sorting of transfected cells. HeLa, 293, and EBV-positive D98/HR1 cells were transfected and assayed by Southern blotting of Hirt method-extracted DNA at 24 and 72 h posttransfection (Fig. 1B). Replication was assayed by resistance to the cytosine methylation-sensitive DpnI restriction enzyme. We found no detectable replication in all three cell lines at 24 h posttransfection. At 72 h posttransfection, we found that OriPwt replicated 5- to 2.5-fold better than OriPnm− did, with the greatest difference observed in HeLa cells. Interestingly, a similar difference in plasmid replication and recovery was observed when cells were assayed at 1 or 2 weeks posttransfection (data not shown). To examine this more carefully, EBV-positive Raji cells were transfected with OriPwt or OriPnm− plasmid and equal numbers of transfected cells were isolated by cell sorting for GFP expression. We found that the nonamer mutant (OriPnm−) had an approximately twofold reduction in replication efficiency when harvested 72 h posttransfection (Fig. 1C). When GFP-positive cells were cultured for 21 days posttransfection, we found that OriPwt was maintained approximately twofold better than the OriPnm− mutant was (Fig. 1D). These results suggest that nonamer repeats contribute to the establishment of a replication- and maintenance-competent plasmid, which is set at ∼72 h posttransfection, but then do not alter the stability or rate of plasmid loss in subsequent generations.
Nonamer repeats stabilize telomere repeat binding factor association with OriP.
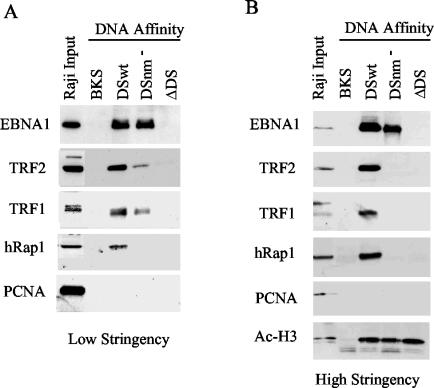
It was previously shown that mutations in the nonamer repeats disrupted the association of recombinant TRF2 with DS (12). However, it was not clear from these studies whether additional protein-protein associations found in vivo may allow telomere repeat factors to associate with OriP despite nonamer substitution mutations. To directly test this possibility, we measured protein association with DSwt and DSnm− by DNA affinity purification from cell extracts. We found that some telomere proteins remained with the insoluble chromatin fraction with typical nuclear extraction procedures. Consequently, we used whole-cell lysates derived by sonication in 1 M NaCl, which disrupts most chromatin-associated proteins. Biotinylated DNA was mixed with high-salt lysate and diluted stepwise, according to methods known to assemble chromatin on DNA templates. DNA-bound proteins were recovered on magnetic beads, washed three times (Fig. 2A) or five times (Fig. 2B), and analyzed by Western blotting with antibodies specific for telomere repeat-associated proteins (Fig. 2). We first confirmed that the binding of EBNA1 was specific for DS, but not for control DNA derived from BKS or from an OriP plasmid with a DS deletion (ΔDS). EBNA1 bound to DSwt and DSnm− similarly, although after high-stringency washing EBNA1 binding was stabilized modestly by the presence of the nonamers. In high-stringency-washed blots, TRF2, TRF1, and hRap1 bound exclusively to DSwt, with no detectable binding to BKS, DSnm−, or ΔDS (Fig. 2B). When complexes were washed under lower-stringency conditions, TRF2 and TRF1 also bound detectably to DSnm− but not to ΔDS or BKS control DNA (Fig. 2A). Control nuclear protein PCNA did not bind to any of the DNA templates under these conditions, while histone H3 bound equally well to DS, DSnm−, and ΔDS (Fig. 2B). Histone H3 did not bind to BKS, presumably because the template is too small or otherwise unfavorable for nucleosome assembly. These results indicate that TRF1, TRF2, and hRap1 have high affinity for nonamer sites but also reveal that TRF1 and TRF2 can bind to DSnm− with lower affinity, suggesting that unknown protein-protein interactions may stabilize these factors' association with DS in the absence of intact nonamers.
FIG. 2.
Telomere repeat binding factors interact with nonamer repeats in Raji cell nuclear extracts. Raji whole-cell extracts were subjected to DNA affinity purification with biotinylated DNA derived from pBKS, DSwt, DSnm−, or OriPΔDS. Affinity-purified proteins were washed under low (A)- or high (B)-stringency conditions and assayed by Western blotting with antibodies specific for EBNA1, TRF2, TRF1, hRap1, PCNA, and acetyl-histone H3.
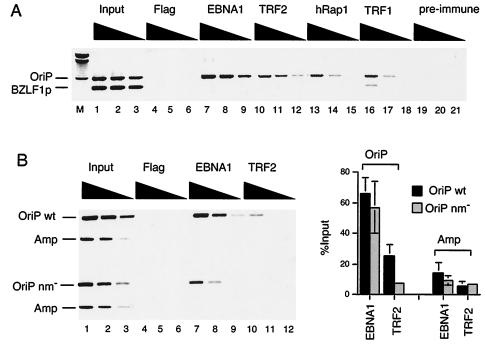
The telomere repeat factors TRF2, TRF1, and hRap1 were found to bind to DS efficiently by DNA affinity purification from Raji cell nuclear extracts. We next assayed the in vivo binding of these factors with OriP in the endogenous viral chromosome by the ChIP assay (Fig. 3A). Antibodies specific for telomere repeat proteins were tested for their ability to immunoprecipitate OriP-containing DNA but not DNA from another region of EBV overlapping the BZLF1 promoter (BZLF1p). As expected from previous studies, we found that EBNA1 and TRF2 antibodies specifically precipitated OriP-containing DNA but not BZLF1p DNA. We now show that antibodies to hRap1 and TRF1 specifically precipitate OriP DNA as well, indicating that these proteins associate with OriP in vivo in Raji cells. To test whether the nonamer sites were important for TRF2 binding in vivo, we used the ChIP assay on cells transfected with OriPwt- or OriPnm−-containing plasmids. We found that TRF2 bound efficiently to DSwt but was significantly reduced for binding to DSnm− (Fig. 3B, lane 10), similar to our findings with DNA affinity purification with nuclear extracts. Quantitation of several experiments indicates that TRF2 bound fivefold better to OriPwt than to OriPnm−, while there was only a minimum (5%) decrease in EBNA1 binding. This indicates that nonamer repeats are important for stable TRF2 binding in vivo as well as in vitro.
FIG. 3.
TRF1, TRF2, and hRap1 bind OriP in vivo. (A) Raji cells were formaldehyde cross-linked and subjected to ChIP assay with antibodies to EBNA1, TRF2, hRap1, TRF1, or control antisera to monoclonal Flag or from preimmune rabbit serum. Immunoprecipitated DNA was amplified with primers specific for OriP or the BZLF1 promoter region. (B) D98/HR1 cells were transfected with OriPwt or OriPnm− cells, sorted for GFP-positive cells, and assayed by ChIP 72 h posttransfection. Primers specific for plasmid-based OriP (OriPwt and OriPnm−) or the ampicillin gene (Amp) were used to amplify immunoprecipitated DNA. Quantification of several experiments is indicated in the bar graph to the right.
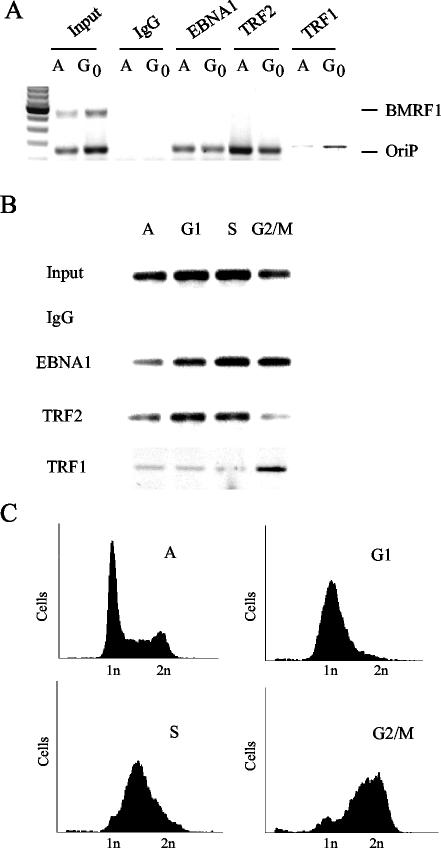
Cell cycle-dependent association of TRF1.
Since TRF1 and TRF2 have identical DNA binding specificities, we wanted to determine whether these proteins may interact with OriP at different stages of the cell cycle. We first compared exponentially growing asynchronous cells with cells arrested in G0 by serum starvation (Fig. 4A). The ChIP assay was used to compare the relative in vivo binding of EBNA1, TRF2, and TRF1 for either OriP or control BMRF1 promoter sequence. We found that EBNA1 binding to OriP was unchanged in asynchronous relative to G0-arrested cells. In contrast, TRF2 binding was slightly down regulated in G0 cells, while TRF1 binding to OriP was significantly elevated. Neither TRF1 nor TRF2 bound to the negative-control BMRF1 promoter region based on multiplex PCRs, indicating that these results are specific for OriP interactions. We next determined whether TRF1 or TRF2 binding to OriP was regulated across a normal cell cycle. Raji cells were synchronized by double thymidine block and then assayed by ChIP at various times in the cell cycle as determined by propidium iodide staining (Fig. 4C). Asynchronous cells, as well as G1 and S-phase enriched populations, had significantly more OriP bound to TRF2 than to TRF1. Interestingly, G2/M enriched cells were reversed, with more OriP bound to TRF1 than to TRF2. These results suggest that TRF2 and TRF1 binding to OriP is mutually exclusive and that a switch in nonamer binding factors at OriP may occur during the G2/M phase of the cell cycle.
FIG. 4.
Cell cycle-dependent binding of TRF1 to OriP. (A) Raji cells were arrested in G0 by serum starvation and assayed by ChIP for association of EBNA1, TRF2, or TRF1 with OriP or control BMRF1 promoter DNA. (B) Raji cells were synchronized by double thymidine block and isolated at various stages of the cell cycle. Asynchronous (A), G1, S, and G2/M phase-arrested cells were assayed by ChIP for EBNA1, TRF2, and TRF1 binding to OriP. (C) FACS profile of Raji cells synchronized in panel B as described above. Raji cells were stained with propidium iodide prior to analysis by FACS. IgG, immunoglobulin G.
Opposing activities of TRF1 and TRF2 at OriP.
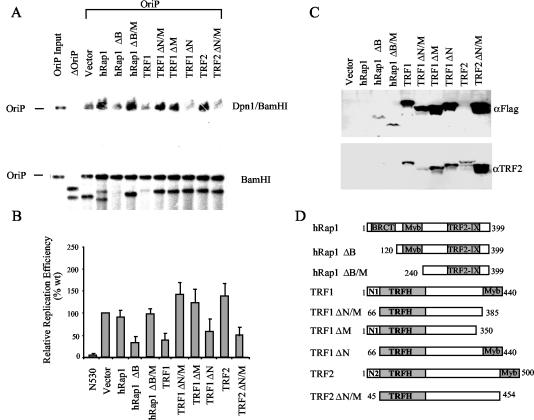
To determine what the effect of TRF1, TRF2, and hRap1 may be on OriP DNA replication activity, we assayed transient DNA replication in cells transfected with full-length versions and deletion mutants of these telomere-binding proteins (Fig. 5). We found that overexpression of full-length hRap1 and TRF2 had no effect on OriP DNA replication. Overexpression of an hRap1 amino-terminal BRCT domain (ΔB) truncation mutant resulted in a strong repression of OriP DNA replication. However, further truncation of hRap1, leading to the loss of the BRCT and myb domains (ΔB/M), relieved the inhibition of OriP replication. This suggests that the myb domain lacking the BRCT domain can function as a dominant-negative mutant of OriP DNA replication. Like that of full-length hRap1, full-length TRF2 expression had no effect on DNA replication. In contrast, deletion of the N-terminal and C-terminal myb domain (ΔN/M) resulted in a potent inhibition of OriP replication. This same deletion mutant has been shown to be a potent dominant-negative mutant of TRF2 at telomeres and leads to chromosome aberrations when expressed in some cell types (58).
FIG. 5.
Truncation mutants of telomere repeat factors inhibit OriP replication. (A) OriPwt or control plasmid ΔOriP was transfected into ZKO-293 cells with pCMV-Flag-2 vector or an expression plasmid for hRap1, TRF1, or TRF2 and their derivatives as indicated above each lane. Replicated DNA was measured by DpnI resistance in the top panel (DpnI/BamHI), and total extracted DNA is indicated in the bottom panel (BamHI digest). (B) PhosphorImager quantitation of at least three independent Southern blots is presented as a bar graph. (C) Transfected proteins were detected by Western blotting with anti-Flag antibody (full-length hRap1 is not Flag tagged), or the same blot was reprobed with anti-TRF2 (full-length TRF2 is not Flag tagged). (D) Schematic diagram of truncation mutants cloned into pCMV-Flag-2. The hRap1 BRCT (BRCA1 C-terminal homology), myb DNA binding, TRF2 interaction (TRF2-IX), and TRF homodimerization (TRFH) domains are indicated.
In contrast to full-length TRF2 and hRap1, full-length TRF1 had a strong inhibitory effect on OriP DNA replication (Fig. 5A). Deletion of the C-terminal myb domain (ΔM) completely eliminated this repression, indicating that the myb DNA binding domain is important for this inhibitory activity. Deletion of the TRF1 amino-terminal domain (ΔN), which has been shown to bind tankyrase 1 (57), had no effect on the ability of TRF1 to inhibit OriP replication. Combined deletion of N- and C-terminal myb domains (ΔN/M) eliminated repression activity, again suggesting that the myb DNA binding capacity of TRF1 is important for inhibiting OriP replication. All of the deletion mutants were expressed to detectable levels (Fig. 5B) and were not found to have significant toxicity in EBV-infected cells during the 72-h transient-replication assay. Together these results suggest that TRF2 and hRap1 promote OriP DNA replication, while TRF1 inhibits DNA replication, and that these proteins with identical DNA binding specificities have opposing activities at OriP.
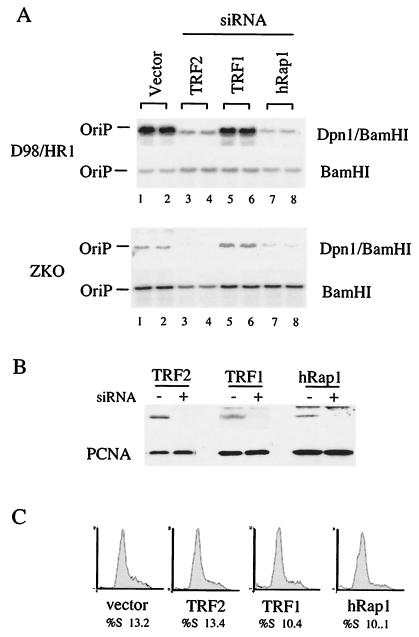
To further examine the role of telomere-binding proteins TRF1, TRF2, and hRap1 at OriP, we utilized RNA interference technology to knock down the expression of these proteins in EBV-positive D98/HR1 or ZKO-293 cells. Plasmids expressing small hairpin RNAs were designed to interfere with the expression of TRF1, TRF2, and hRap1 (Fig. 6). We found that reduction of TRF2 led to a profound decrease in OriP replication activity (Fig. 6A). A similar inhibition was found in cells knocked down for hRap1. In contrast, siRNA knockdown of TRF1 produced a slight increase in OriP replication. Western blots confirmed that TRF2, TRF1, and hRap1 protein levels were reduced in cells transfected with the specific RNA interference constructs (Fig. 6B). Reduction of these proteins did not have gross toxicity effects on these cells and did not alter the cell cycle profile significantly within the 72-h period of the replication assay, ruling out the possibility that these changes are reflecting pleiotropic effects on cellular metabolism and cycling (Fig. 6C). These results are consistent with our overexpression data suggesting that TRF2 and hRap1 promote OriP replication and further support our findings that TRF2 and TRF1 have opposing functions in OriP regulation.
FIG. 6.
siRNA knockdown mutants of TRF2 and hRap1 inhibit OriP replication. (A) D98/HR1 cells were cotransfected with OriPwt and siRNA expression plasmids for TRF2, TRF1, and hRap1. DpnI-resistant replicated DNA is indicated in the top panel (DpnI/BamHI), and total extracted DNA is indicated in the bottom panel (BamHI). (B) Western blots of cell extracts derived from siRNA-transfected cells (+) or vector-transfected cells (−) were probed with antibody specific to the siRNA target indicated above (top panel) or with antibody to PCNA (bottom panel). (C) FACS profile of propidium iodide-treated cells that were transfected with vector, TRF2, TRF1, or hRap1 siRNA plasmids, as indicated. The percentage of cells in S phase is indicated below.
DISCUSSION
The plasmid maintenance and DNA replication functions of OriP are thought to be important for genome stability during EBV latency in B lymphocytes. In this work we investigated the functional contribution of cellular telomeric factors in regulating OriP-dependent plasmid maintenance and DNA replication. We found that mutations in the telomere repeat sequences (also referred to as nonamers) reduced OriP replication and plasmid maintenance by two- to fivefold (Fig. 1). These sites were important for telomeric factor binding in vitro and in vivo (Fig. 2 and 3). Cell synchronization studies suggested that TRF2 and TRF1 bind OriP at different stages of the cell cycle (Fig. 4). A more direct role of telomere factors in OriP-dependent replication was revealed by exogenous expression of full-length and truncation mutants of telomeric factors, as well as by the use of siRNA knockdown experiments (Fig. 5 and 6). Our results suggest that telomere repeat binding factors play a direct role in the regulation of OriP replication and plasmid maintenance.
Mutations that affect OriP DNA replication efficiency are predicted to have a cumulative effect on plasmid maintenance over each generation. Contrary to this prediction, we found that mutations in the nonamers affected DNA replication within 72 h posttransfection but then did not accumulate further loss over subsequent generations, even after 3 weeks in culture (Fig. 1). This suggests that nonamer binding factors promote the establishment of a replicon at DS which, once established, maintains a stable replicating plasmid. In the intact EBV genome, OriP is thought to be used as a replication origin only 20% of the time, and viral genomes can be stably maintained even when DS is deleted (16, 45, 46). In contrast, genetic data indicate that DS is an EBNA1-dependent minimal replicator (73), and two-dimensional agarose gel electrophoresis experiments demonstrate that DNA replication initiates within or very close to DS in small plasmids carrying OriP (16). These observations suggest that DS has a high probability of becoming a replication origin but that other regions of the genome may function more actively on larger plasmids (32, 33). Chromatin replication initiates within loosely defined zones in higher eukaryotes, but the mechanism involved in selection of these zones is not completely understood (17). The establishment of an OriP replicon in plasmid-transfected cells has been characterized as a rare, stochastic event that is regulated by unknown epigenetic events (35). Epigenetic control of DNA replication and gene expression is typically transmitted through posttranslational modifications of chromatin-associated proteins which are thought to establish higher-ordered chromatin structures (51). One possible explanation to account for our findings is that telomere repeat binding factors influence the selection of OriP as a functional replicon and therefore are likely to influence the establishment of a chromatin structure or subnuclear localization favorable for replication and plasmid maintenance.
TRF1 and TRF2 bind to the identical telomere repeat consensus sequence found in the nonamers, and the two recombinant proteins can bind with similar affinities to OriP in vitro (12). TRF2 is the more abundant protein in asynchronous populations of most cell lines that we have examined and can be readily detected by ChIP assay bound to OriP in vivo. In contrast, TRF1 is less abundant and appears to associate with OriP under conditions where cells either have exited the cell cycle or are enriched in the G2/M stage of the cell cycle (Fig. 4). The TRF1 isoform Pin2 has been shown to be enriched prior to mitosis and plays a regulatory role in the entry of cells into mitosis (30, 77). We also found that ectopic expression of full-length TRF1 inhibits OriP replication. TRF1 may block OriP replication by displacing TRF2 or may have a more active role in preventing replication initiation. TRF1 has also been shown to promote microtubule assembly and is thought to play a role in telomere segregation during mitosis (43, 44). A similar role of TRF1 may also exist at OriP, where TRF1 may help to segregate plasmids by tethering them to microtubules during mitosis.
Our data indicate that TRF2 and hRap1 promote OriP replication and plasmid maintenance. Dominant-negative mutants of TRF2 and siRNA knockdown of TRF2 lead to a significant loss of OriP replication function (Fig. 6). We also note that TRF2 dominant-negative and siRNA knockdown mutants have significantly greater effects on OriP replication than does mutation of the nonamer sites. Inhibition of TRF2 is likely to influence numerous cellular processes, and therefore some of the inhibition of OriP replication may be an indirect consequence of cell cycle checkpoint control. While dominant-negative TRF2 can cause cell cycle arrest in some cell types, we did not observe a gross loss of viability or cell cycle progression in EBV-positive Raji and D98/HR1 cells used in these replication assays (Fig. 6C). We consider it more likely that TRF2 has a direct role in establishing OriP replication competence primarily through its DNA binding to the nonamers, but it may also have important protein-protein interactions at OriP that are nonamer independent. Evidence from DNA affinity purification indicates that TRF1 and TRF2 can associate with DSnm− under low-stringency wash conditions (Fig. 2A), and previous work demonstrated that recombinant TRF2 binds cooperatively with EBNA1 at DS, suggesting that these two proteins may make direct contact (12). One possible function of TRF2 may be to stabilize EBNA1 binding to DS in vivo, and this could account for the higher efficiency of plasmid replication. However, TRF1 can also bind cooperatively with EBNA1, and its overexpression inhibits replication. Alternatively, TRF2 may function at OriP by stabilizing DNA replication intermediates (as is proposed for the MRE11 complex [48]), perhaps facilitating strand invasion (as is proposed for TRF2 at T loops of telomeres [59]), or helping to recruit the ORC to establish a stable replication origin at or near DS. Future experiments will help to distinguish between these possibilities.
We also found that hRap1 was important for OriP replication. DNA affinity purification and in vivo ChIP assay revealed that hRap1 binds specifically to OriP, with a similar affinity as that of TRF2. TRF2 and hRap1 form a stable protein complex, and it is not surprising that these proteins copurify with OriP. Overexpression of hRap1 truncation mutants lacking the N-terminal BRCT domain potently inhibits OriP replication, much like the dominant-negative mutant of TRF2 does. Overexpression of full-length hRap1 or truncation of the myb domain had no significant effect on OriP replication. A positively active role of hRap1 at OriP was best demonstrated by siRNA knockdown experiments, which led to a decrease in OriP replication similar to that seen for TRF2, suggesting that these two proteins function as partners. Interestingly, the myb domain of hRap1 does not mediate DNA binding to telomeric repeats and is unlikely to bind DNA directly (22). The BRCT domain has sequence similarity to numerous other BRCT-containing proteins, including BRCA1, but has no known function other than to mediate protein-protein interactions (76). hRap1 has some homology to yeast Rap1, but it is not clear that these proteins share any functional properties beyond localization at telomeres. Interestingly, yeast Rap1 binds yeast telomeric repeats and can function as an accessory protein at several replication origins and silencing elements (14, 42). The hRap1 myb domain may function more like the related SANT domains found in numerous chromatin-associated proteins, like Spt20, ADA2, and NCoR (1). The SANT domain of NCoR has been shown to bind to histone tails (20), and it will be interesting to determine if the hRap1 myb domain has a similar chromatin-specific function.
OriP requires only one viral protein, EBNA1, to function as a replication origin and plasmid maintenance element. EBNA1 resembles the papillomavirus E2 protein known to load the E1 helicase protein onto the replication origin of papillomaviruses (6, 7). For EBV, the replicative helicase is thought to be the cellular MCM complex, which can also associate with OriP in vivo based on ChIP studies (10, 13, 54). It is not clear how these better-characterized replication proteins interact with telomere repeat proteins or whether telomere repeat proteins may regulate the association of these replication proteins with OriP. It has been shown that MCM and ORC proteins are located at telomeric and subtelomeric regions of yeast chromosomes (70) and that telomere replication must also be integrated with telomerase activity and telomere-binding protein function (40, 50). In this respect, OriP may be a valuable model system to investigate questions regarding DNA replication at telomeres. In both cases, it is likely that telomere-binding factors are protecting the distorted DNA structures from DNA repair enzyme modification. Our data are consistent with a role of the telomere repeat factors stabilizing the DNA replication intermediates found at OriP and required for faithful propagation of the genome during replication, condensation, and segregation of DNA in proliferating cells.
Acknowledgments
This work was supported by a grant from the NIH (CA93606) to P.M.L. Z.D. is a fellow of the Leukemia/Lymphoma Society of America.
We thank the National Cell Culture Center for providing Raji cells and the Wistar Institute Core Facilities for cell sorting and FACS analysis.
REFERENCES
- 1.Aasland, R., A. F. Stewart, and T. Gibson. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87-88. [PubMed] [Google Scholar]
- 2.Aiyar, A., C. Tyree, and B. Sugden. 1998. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 17:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashaw, J. M., and J. L. Yates. 2001. Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J. Virol. 75:10603-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi, A., S. Smith, L. Chong, P. Elias, and T. de Lange. 1997. TRF1 is a dimer and bends telomeric DNA. EMBO J. 16:1785-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchi, A., R. M. Stansel, L. Fairall, J. D. Griffith, D. Rhodes, and T. de Lange. 1999. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 18:5735-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. [DOI] [PubMed] [Google Scholar]
- 7.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, W. J. Furey, A. M. Edwards, and L. Frappier. 1995. Crystal structure of the DNA binding domain of the Epstein-Barr virus origin binding protein EBNA-1. Cell 83:39-46. [DOI] [PubMed] [Google Scholar]
- 8.Broccoli, D., A. Smogorzewska, L. Chong, and T. de Lange. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17:231-235. [DOI] [PubMed] [Google Scholar]
- 9.Ceccarelli, D. F., and L. Frappier. 2000. Functional analyses of the EBNA1 origin DNA binding protein of Epstein-Barr virus. J. Virol. 74:4939-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chittenden, T., S. Lupton, and A. J. Levine. 1989. Functional limits of oriP, the Epstein-Barr virus plasmid origin of replication. J. Virol. 63:3016-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, Z., L. Lezina, C. J. Chen, S. Shtivelband, W. So, and P. M. Lieberman. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9:493-503. [DOI] [PubMed] [Google Scholar]
- 13.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 14.Diffley, J. F., and B. Stillman. 1989. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science 246:1034-1038. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, N., E. Kremmer, G. Lautshcam, N. Mueller-Lantzsch, and F. A. Grasser. 1997. Epstein-Barr virus nuclear antigen 1 forms a complex with the nuclear transporter karyopherin α2. J. Biol. Chem. 272:3999-4005. [DOI] [PubMed] [Google Scholar]
- 16.Gahn, T. A., and C. L. Schildkraut. 1989. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell 58:527-535. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffith, J., A. Bianchi, and T. de Lange. 1998. TRF1 promotes parallel pairing of telomeric tracts in vitro. J. Mol. Biol. 278:79-88. [DOI] [PubMed] [Google Scholar]
- 19.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 20.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond, S. M., A. A. Caudy, and G. J. Hannon. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2:110-119. [DOI] [PubMed] [Google Scholar]
- 22.Hanaoka, S., A. Nagadoi, S. Yoshimura, S. Aimoto, B. Li, T. de Lange, and Y. Nishimura. 2001. NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J. Mol. Biol. 312:167-175. [DOI] [PubMed] [Google Scholar]
- 23.Harrison, S., K. Fisenne, and J. Hearing. 1994. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J. Virol. 68:1913-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jankelevich, S., J. L. Kolman, J. W. Bodnar, and G. Miller. 1992. A nuclear matrix attachment region organizes the Epstein-Barr viral plasmid in Raji cells into a single DNA domain. EMBO J. 11:1165-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 15:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 28.Kim, A. L., M. Maher, J. B. Hayman, J. Ozer, D. Zerby, J. L. Yates, and P. M. Lieberman. 1997. An imperfect correlation between the DNA replication activity of Epstein-Barr virus nuclear antigen 1 (EBNA1) and the binding to the nuclear import receptor, Rch1/importin alpha. Virology 239:340-351. [DOI] [PubMed] [Google Scholar]
- 29.Kirchmaier, A. L., and B. Sugden. 1995. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J. Virol. 69:1280-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishi, S., G. Wulf, M. Nakamura, and K. P. Lu. 2001. Telomeric protein Pin2/TRF1 induces mitotic entry and apoptosis in cells with short telomeres and is down-regulated in human breast tumors. Oncogene 20:1497-1508. [DOI] [PubMed] [Google Scholar]
- 31.Koons, M. D., S. V. Scoy, and J. Hearing. 2001. The replicator of the Epstein-Barr virus latent cycle origin of DNA replication, oriP, is composed of multiple functional elements. J. Virol. 75:10582-10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krysan, P. J., and M. P. Calos. 1991. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol. Cell. Biol. 11:1464-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krysan, P. J., S. B. Haase, and M. P. Calos. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol. 9:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, M. A., M. E. Diamond, and J. L. Yates. 1999. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J. Virol. 73:2974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leight, E. R., and B. Sugden. 2001. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol. Cell. Biol. 21:4149-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, B., S. Oestreich, and T. de Lange. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101:471-483. [DOI] [PubMed] [Google Scholar]
- 37.Mackey, D., T. Middleton, and B. Sugden. 1995. Multiple regions within EBNA1 can link DNAs. J. Virol. 69:6199-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J.-C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin, A. A., I. Dionne, R. J. Wellinger, and C. Holm. 2000. The function of DNA polymerase α at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Middleton, T., and B. Sugden. 1994. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J. Virol. 68:4067-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morse, R. 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 16:51-53. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura, M., X. Z. Zhou, S. Kishi, I. Kosugi, Y. Tsutsui, and K. P. Lu. 2001. A specific interaction between the telomeric protein Pin2/TRF1 and the mitotic spindle. Curr. Biol. 11:1512-1516. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura, M., X. Z. Zhou, S. Kishi, and K. P. Lu. 2002. Involvement of the telomeric protein Pin2/TRF1 in the regulation of the mitotic spindle. FEBS Lett. 514:193-198. [DOI] [PubMed] [Google Scholar]
- 45.Norio, P., and C. L. Schildkraut. 2001. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science 294:2361-2364. [DOI] [PubMed] [Google Scholar]
- 46.Norio, P., C. L. Schildkraut, and J. L. Yates. 2000. Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein-Barr virus chromosome after infection of established cell lines. J. Virol. 74:8563-8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrini, J. H. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293-296. [DOI] [PubMed] [Google Scholar]
- 49.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42:859-868. [DOI] [PubMed] [Google Scholar]
- 50.Ray, S., Z. Karamysheva, L. Wang, D. E. Shippen, and C. M. Price. 2002. Interactions between telomerase and primase physically link the telomere and chromosome replication machinery. Mol. Cell. Biol. 22:5859-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richards, E. J., and S. C. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108:489-500. [DOI] [PubMed] [Google Scholar]
- 52.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shirakata, M., and K. Hirai. 1998. Identification of minimal oriP of Epstein-Barr virus required for DNA replication. J. Biochem. (Tokyo) 123:175-181. [DOI] [PubMed] [Google Scholar]
- 56.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, S., I. Giriat, A. Schmitt, and T. de Lange. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282:1484-1487. [DOI] [PubMed] [Google Scholar]
- 58.Smogorzewska, A., B. van Steensel, A. Bianchi, S. Oelmann, M. R. Schaefer, G. Schnapp, and T. de Lange. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stansel, R. M., R. de Lange, and J. D. Griffith. 2001. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20:5532-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugden, B., K. Marsh, and J. Yates. 1985. A vector that replicates as a plasmid and can be efficiently selected in B lymphocytes transformed by Epstein-Barr virus. Mol. Cell. Biol. 5:410-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugden, B., and N. Warren. 1989. A promoter of Esptein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J. Virol. 63:2644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsai, C. N., S. T. Liu, and Y. S. Chang. 1995. Identification of a novel promoter located within the BamHI Q region of the Epstein-Barr virus genome for the EBNA 1 gene. DNA Cell Biol. 14:767-776. [DOI] [PubMed] [Google Scholar]
- 63.Van Scoy, S., I. Watakabe, A. R. Krainer, and J. Hearing. 2000. Human p32: a coactivator for Epstein-Barr virus nuclear antigen-1-mediated transcriptional activation and possible role in viral latent cycle DNA replication. Virology 275:145-157. [DOI] [PubMed] [Google Scholar]
- 64.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 65.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 66.Vogel, M., K. Wittmann, E. Endl, G. Glaser, R. Knuchel, H. Wolf, and H. H. Niller. 1998. Plasmid maintenance assay based on green fluorescent protein and FACS of mammalian cells. BioTechniques 24:540-544. [PubMed] [Google Scholar]
- 67.Wang, Y., J. E. Finan, J. M. Middledorp, and S. D. Hayward. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236:18-29. [DOI] [PubMed] [Google Scholar]
- 68.Wu, G., W. H. Lee, and P. L. Chen. 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 275:30618-30622. [DOI] [PubMed] [Google Scholar]
- 69.Wu, H., D. F. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wyrick, J. J., J. G. Aparicio, T. Chen, J. D. Barnett, E. G. Jennings, R. A. Young, S. P. Bell, and O. M. Aparicio. 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294:2357-2360. [DOI] [PubMed] [Google Scholar]
- 71.Wysokenski, D. A., and J. L. Yates. 1989. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J. Virol. 63:2657-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yates, J., and S. M. Camiolo. 1988. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. Cancer Cells 6:197-205. [Google Scholar]
- 73.Yates, J. L., S. M. Camiolo, and J. M. Bashaw. 2000. The minimal replicator of Epstein-Barr virus oriP. J. Virol. 74:4512-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yates, J. L., N. Warren, P. Reisman, and B. Sugden. 1984. A cis-acting element from Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, X., S. Morera, P. A. Bates, P. C. Whitehead, A. I. Coffer, K. Hainbucher, R. A. Nash, M. J. E. Sternberg, T. Lindahl, and P. S. Freemont. 1998. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. EMBO J. 17:6404-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou, X. Z., and K. P. Lu. 2001. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107:347-359. [DOI] [PubMed] [Google Scholar]