Abstract
The spread of vaccinia virus in cell cultures is mediated by virions that adhere to the tips of specialized actin-containing microvilli and also by virions that are released into the medium. The use of a small plaque-forming A36R gene deletion mutant to select spontaneous second-site mutants exhibiting enhanced virus release was described previously. Two types of mutations were found: C-terminal truncations of the A33R envelope protein and a single amino acid substitution of the B5R envelope protein. In the present study, we transferred each type of mutation into a wild-type virus background in order to study their effects in vitro and in vivo. The two new mutants conserved the enhanced virus release properties of the original isolates; the A33R mutant produced considerably more extracellular virus than the B5R mutant. The extracellular virus particles contained the truncated A33R protein in one case and the mutated B5R protein in the other. Remarkably, both mutants failed to form actin tails and specialized microvilli, despite the presence of an intact A36R gene. The synthesis of the A36R protein as well as its physical association with the mutated or wild-type A33R protein was demonstrated. Moreover, the A36R protein was tyrosine phosphorylated, a step mediated by a membrane-associated Src kinase that regulates the nucleation of actin polymerization. The presence of large numbers of adherent virions on the cell surface argued against rapid dissociation as having a key role in preventing actin tail formation. Thus, the A33R and B5R proteins may be more directly involved in the formation or stabilization of actin tails than had been previously thought. When mice were inoculated intranasally, the A33R mutant was highly attenuated and the B5R mutant was mildly attenuated compared to wild-type virus. Enhanced virus release, therefore, did not compensate for the loss of actin tails and specialized microvilli.
Vaccinia virus replicates in cytoplasmic factories where infectious particles called intracellular mature virions (IMV) are assembled (15). IMV are wrapped by trans-Golgi apparatus or endosomal cisternae to form intracellular enveloped virions (IEV) (7, 25, 27), which are transported along microtubules to the cell periphery (9, 18, 30, 31). The outer of the two acquired IEV membranes fuses with the plasma membrane to form cell-associated enveloped virions (CEV), which adhere to the cell surface, and extracellular enveloped virions (EEV), which are released into the medium. In some cells, extracellular virions also may form by budding of IMV at the plasma membrane (28). CEV are primarily responsible for cell-to-cell spread (1), a process that is greatly enhanced by their attachment to the tips of specialized microvilli, which appear as motile actin tails when viewed by fluorescence microscopy (3, 8, 26).
Vaccinia virus mutants that exhibit altered plaque phenotypes have been isolated. Mutations in the A33R, A34R, and A36R genes that interfere with the formation of actin-containing microvilli result in a small-plaque phenotype and reduced virulence (19, 21, 23, 35, 37). Cells infected with some vaccinia virus strains, notably IHD, release large numbers of EEV that provide long-range spread and form elongated comet-shaped plaques in cell monolayers covered by liquid medium (17). The IHD phenotype is caused in large part by a point mutation in the A34R envelope protein (2). Mutations in envelope proteins encoded by the A33R and B5R open reading frames (ORFs) also can increase the amounts of EEV in the medium (12, 19).
In a previous study, Katz et al. described the use of a small plaque-forming A36R deletion mutant to isolate spontaneous second-site mutants exhibiting enhanced virus spread (11). The second-site mutations, however, did not correct the defect in actin tail formation but instead caused the release of large numbers of EEV. Of five such viruses isolated, four had mutations that truncated the C terminus of the A33R envelope protein, and one had a point mutation in the B5R envelope protein. Analysis of the effects of these mutations on virus trafficking, however, was compromised by the absence of the A36R gene. For the present study, we substituted the mutated A33R or B5R gene for the normal one in the genome of vaccinia virus containing an intact A36R gene. The resulting mutant viruses formed large numbers of CEV and EEV and consequently produced comet-shaped plaques. Despite the synthesis and tyrosine phosphorylation of the A36R protein, neither actin tails nor specialized microvilli were detected. Thus, tyrosine phosphorylation of the A36R protein regulates the nucleation of actin polymerization, but cooperation of the A33R and B5R proteins is required for actin tail formation.
MATERIALS AND METHODS
Cells and viruses.
BS-C-1 and RK13 cells were grown in Earle's minimum essential medium (E-MEM; Quality Biologicals, Gaithersburg, Md.), and HeLa cells were grown in Dulbecco's modified Eagle's medium (D-MEM; Quality Biologicals). Both media were supplemented with 10% fetal bovine serum (HyClone, Logan, Utah). Vaccinia virus strain WR was used, as were WR mutants with deletions of the A33R (19), A36R (23), or B5R (34) genes, referred to as vΔΑ33R, vΔΑ36R, and vΔΒ5R, respectively. The double mutants vΔΑ36R.c1 and vΔΑ36R.c3 were previously described (11).
Plaque assays.
Following adsorption of the virus to monolayers of BS-C-1 cells for 1 h at 37°C, E-MEM containing 2% fetal bovine serum was added. Two days later, the medium was removed, and crystal violet (0.1% in 20% ethanol) was added. After 30 min, the cells were washed and dried. When a semisolid overlay was used, 0.6% SeaKem ME agarose (BioWhittaker Molecular Applications, Rockland, Maine) and 5% fetal bovine serum were included in the medium. After two days, the overlaid cells were fixed with formaldehyde (20% in saline), the semisolid overlay was removed, and the cells were stained with crystal violet.
One-step growth.
BS-C-1 cells were infected at a multiplicity of 5 PFU per cell for 1 h and then washed twice with fresh medium to remove nonadsorbed virus. At various times thereafter, the medium was collected and clarified by low-speed centrifugation to sediment the detached cells and debris. The adherent cells were scraped into 1 ml of fresh medium and combined with the pellet obtained by centrifugation of the medium. The cells were frozen and thawed three times and sonicated for 30 s. Virus titers in cell lysates and media were determined by plaque assays with an agar overlay.
Transfection.
Approximately 2 μg of PCR product in 100 μl of Opti-MEM (Invitrogen, Carlsbad, Calif.) was mixed with 8 μl of Lipofectamine 2000 (Invitrogen, Gaithersburg, Md.) in 100 μl of Opti-MEM and kept at room temperature for 15 min. Then, 800 μl of Opti-MEM was added. BS-C-1 cells were infected at a multiplicity of 1 PFU per 20 cells. After 2 h, the cells were washed with Opti-MEM, and the DNA-Lipofectamine mixture was added to the cell monolayer. Three hours later, E-MEM containing 2% fetal bovine serum was added. The cultures were harvested after 2 days at 37°C, and the virus was plaque purified.
Sequencing of viral DNA.
Virus-infected cells were treated with protease, and DNA was purified by using a QIA DNA blood minikit (Qiagen, Hilden, Germany) as suggested by the manufacturer. Primers with the 5′-to-3′ sequences GGTCGTTAGTAGGGAGGAGAACAAAG and GCGCAAGCACTAGGCATCAGTTC were used to amplify the A33R ORF, and those with the sequences ATTGATGTTTTTAACGCTACAATC and GTACATCTCATTGTCATTTACAAC were used to amplify the B5R ORF. Pfu Turbo DNA polymerase (Stratagene, La Jolla, Calif.) was added to the PCR mixture; the extension reaction times for the A33R and B5R ORFs were 45 and 80 s, respectively. The PCR products were purified by using Wizard PCR Preps DNA purification resin (Promega, Madison, Wis.), sequenced by using a BigDye terminator cycle sequencing v2 Ready Reaction kit (Applied Biosystems, Foster City, Calif.), and analyzed by using a model 3100 genetic analyzer (Applied Biosystems).
Immunoprecipitation.
Confluent BS-C-1 cells (in 35-mm-diameter dishes) were infected at a multiplicity of 5 PFU per cell. At 5 h after infection, the medium (1 ml) was replaced either with E-MEM that had been supplemented with 2.5% fetal bovine serum or, for radiolabeling, with methionine- and cysteine-free D-MEM (Sigma, St. Louis, Mo.) that had been supplemented with 2% dialyzed fetal bovine serum (Invitrogen) and 70 μCi of 35S-protein labeling mix (Easy Tag; Perkin-Elmer, Life Sciences, Boston, Mass.). The cells were harvested 20 h after infection, washed with phosphate-buffered saline (PBS), and suspended in 200 μl of lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% NP-40) containing protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). After 30 min at 4°C, the extract was centrifuged at 20,000 × g for 30 min, and 200 μl of lysis buffer and 50 μl of protein A-agarose (Roche Diagnostics) were added to the supernatant. The mixture was rotated for 2 h at 4°C and centrifuged at 1,000 × g for 1 min. The supernatant was incubated for 20 h at 4°C with 2 μl of serum from a rabbit that had been immunized with a synthetic peptide comprised of amino acids 84 to100 of the A33R protein. Protein A-agarose was then added, and incubation was continued for 2 h at 4°C. The complex was washed several times with PBS and dissolved in 50 μl of SDS-protein gel loading buffer (Quality Biologicals). Proteins (labeled or unlabeled) were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). For radiolabeled samples, polyacrylamide gels were dried and exposed to Kodak BioMax MR film. Unlabeled samples were transferred to NitroPure nitrocellulose membranes (Osmonics, Westborough, Mass.). The membranes were incubated with anti-A36R antibody that had been conjugated to horseradish peroxidase by using an EZ-Link activated peroxidase antibody-labeling kit (Pierce). Bound antibodies were detected with chemiluminescence reagents (Pierce).
Western blot analysis.
BS-C-1 cells were infected at a multiplicity of 5 PFU per cell. After 22 h, the cells were harvested in 2× Tris-glycine SDS sample buffer (Invitrogen). Following several passages through a syringe to shear the DNA, lysates were diluted 1:2 in water or in 4% mercaptoethanol and then heated for 5 min at 100°C. Following SDS-PAGE in Tris-glycine 4 to 20% polyacrylamide gels (Novex; Invitrogen), the polypeptides were transferred to NitroPure nitrocellulose membranes. The membranes were incubated with rabbit antiserum as dexcribed above, followed by horseradish peroxidase-conjugated donkey anti-rabbit antibody (Amersham, Life Sciences). Bound antibodies were detected with chemiluminescence reagents. The molecular masses of viral proteins were estimated by comparison with Full-Range-Rainbow protein markers (Amersham, Life Sciences).
CsCl gradient analysis of virus.
Monolayers of RK13 cells were infected at a multiplicity of 5 PFU per cell. After 5 h, the medium (10 ml) was replaced with methionine- and cysteine-free D-MEM (Invitrogen) that had been supplemented with 2% dialyzed fetal bovine serum and 900 μCi of 35S-protein labeling mix (Easy Tag). At 40 h after infection, the medium was harvested, and cells and large debris were removed by low-speed centrifugation. The cells were scraped, collected by low-speed centrifugation, resuspended in swelling buffer (10 mM Tris [pH 9.0]), and disrupted by Dounce homogenization. After removal of the nuclei by low-speed centrifugation, the cytoplasmic fraction was obtained. Virus from the cytoplasm or the culture medium was centrifuged through a 36% sucrose cushion in an SW41 rotor at 32,000 rpm for 12 min at 4°C. The virus pellets were resuspended in swelling buffer and layered over CsCl step gradients as previously described (5). After centrifugation at 25,000 rpm for 95 min in an SW41 rotor at 20°C, fractions were collected from the bottom of the tube, and the amount of radioactivity was determined by scintillation counting.
Confocal microscopy.
HeLa cells were grown on coverslips and infected at a multiplicity of 5 PFU per cell. After 20 h, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.1%Triton X-100 in PBS, and stained with rat anti-B5R monoclonal antibody (MAb) 19C2 (25) for 1 h at 37°C. The cells were washed with PBS and reacted with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-rat immunoglobulins (Dako, Glostrup, Denmark). Actin filaments were stained with rhodamine-conjugated phalloidin (Molecular Probes), and coverslips were mounted in mowoil containing 1 μg of 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI) (Molecular Probes) per ml to visualize DNA in the nucleus and viral factories. Alternatively, the Triton X-100 step was omitted, and the nonpermeabilized cells were stained with rat anti-B5R MAb 19C2 and then treated with 0.05% NP-40 prior to rhodamine-conjugated phalloidin staining. Images were collected on a Leica TSC-NT/SP inverted laser scanning confocal microscope system with an attached argon ion laser (Coherent Inc.). Images were processed by using Adobe Photoshop version 6.0.
Fluorescence microscopy of live cells.
Time-lapse confocal microscopy was carried out essentially as described previously (31). HeLa cells were plated at ∼80% confluence on ΔTC3 dishes (Bioptechs, Inc.). On successive days, the cells were transfected with pEGFP-actin (Clontech) by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions, infected with 0.2 PFU of virus per cell, incubated overnight, stained with 6 μg of Hoechst 33258 (Pierce) per ml of D-MEM for 20 min, washed three times for 5 min each time with D-MEM, and visualized by using a Bio-Rad MicroRadiance confocal scanning system attached to a Zeiss Axiovert 135 microscope. During microscopy, cells were maintained on a heated ΔTC3 stage (Bioptechs) with the temperature set at 37°C and perfused at a rate of 0.1 ml/min with D-MEM supplemented with 2.5% fetal calf serum and 25 mM HEPES.
Scanning electron microscopy.
BS-C-1 cells grown on coverslips were infected at a multiplicity of 5 PFU per cell for 20 h before fixation with 2% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.4). The cells were then washed with cacodylate buffer, postfixed with 1% osmium tetroxide, and dehydrated in ethanol. The samples were critical point dried, mounted on stubs, coated with gold-palladium alloy, and viewed on a Hitachi S-4700 field emission scanning electron microscope at an accelerating voltage of 3 kV.
Virulence of mutant viruses for mice.
BS-C-1 cells were harvested 2 days after infection by low-speed centrifugation. The cells were suspended in swelling buffer and Dounce homogenized. The cytoplasm was separated from the nuclei by low-speed centrifugation and layered on 36% sucrose in swelling buffer. After centrifugation at 13,500 rpm in an SW28.1 rotor for 80 min at 4°C, the virus was suspended in a small volume of buffer, and its infectivity was determined by plaque assays. The virulence of vaccinia virus for mice was then determined essentially as described previously (13, 29, 33). Groups (n = 4) of 5-week-old female BALB/c mice were anesthetized and inoculated intranasally with various concentrations of purified virus in 20 μl of swelling buffer. Mice were weighed daily, and those that had lost more than 30% of their initial body weight were sacrificed. Statistical significance was determined by Fisher's least-significant-difference test.
RESULTS
Isolation of viruses with mutated A33R or B5R genes.
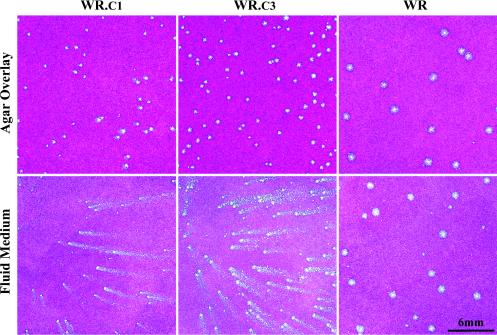
Katz et al. previously selected viruses that produced large numbers of EEV and comet-forming plaques by passage of a virus that had a deletion of the A36R gene and that made small, non-comet-forming plaques (11). Sequence analysis indicated that several of the comet-forming viruses had C-terminal truncations in A33R and that one had a point mutation in B5R. To further study the effects of these mutations on virus spread, we transferred these mutations into the genome of vaccinia virus with an otherwise normal genetic background. This was accomplished by transfecting a PCR copy of the A33R ORF from A33R truncation mutant ΔA36R.c1 into cells infected with wild-type vaccinia virus strain WR. Approximately 18% of the resulting plaques formed comets on cell monolayers covered with a liquid overlay, compared to only 0.2% when a PCR product containing an intact A33R gene was transfected. In agar overlay cultures, the plaques of the recombinant viruses were slightly smaller than those of the wild-type virus, facilitating their plaque purification. The A33R ORFs of five such isolates were found to be identical in sequence to that of vΔA36R.c1. The plaque morphologies under liquid and agar overlays of one of these isolates, WR.c1, were compared to those of WR in Fig. 1. The other WR.c1 clones had plaque morphologies that were identical to the one shown.
FIG. 1.
Plaque morphologies of virus clones. BS-C-1 cell monolayers were infected with WR.c1, WR.c3, or vaccinia virus strain WR. Following adsorption, fluid- or agar-containing overlays were added. After 2 days of incubation at 37°C, the cultures were fixed and stained with crystal violet.
A similar procedure was used to transfer the mutated B5R ORF from vΔA36R.c3 to wild-type vaccinia virus strain WR. In this case, 11% of the plaques had a comet-forming phenotype, whereas none had this phenotype when the transfected PCR product contained a wild-type B5R ORF. Again, the plaques of the recombinant viruses in agar overlay cultures were smaller than those of the wild-type virus, assisting in their purification. The B5R ORFs of five such isolates were sequenced and found to be identical to that of vΔA36R.c3. The plaque morphologies under liquid and agar overlays of one of these isolates, WR.c3, were compared to those of WR and WR.c1 in Fig. 1. The other WR.c3 clones had plaque morphologies that were identical to the one shown.
Analysis of mutated A33R and B5R proteins expressed by WR.c1 and WR.c3, respectively.
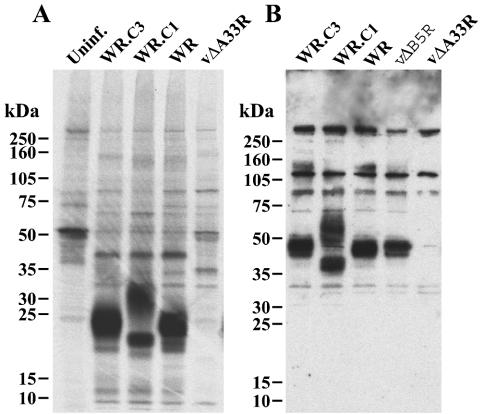
The A33R ORF encodes a type II integral membrane glycoprotein that migrates diffusely with mobilities of 23 to 28 kDa and approximately 50 kDa under reducing and nonreducing conditions, respectively (20). We used rabbit antiserum prepared against a synthetic peptide derived from amino acids 84 to 100 of A33R to characterize the A33R protein produced by the mutant viruses. Unexpectedly, the peptide antiserum as well as a mouse MAb reacted well only with the nonreduced A33R protein. Therefore, metabolically labeled proteins from cells infected with WR, WR.c1, or WR.c3 were first immunoprecipitated with the rabbit antiserum and then treated with SDS and mercaptoethanol. On SDS-PAGE, prominent bands of 23 to 28 kDa, corresponding to the mass expected for the reduced A33R glycoprotein, were detected in extracts from cells infected with WR and WR.c3 but not in those from uninfected cells or cells infected with an A33R deletion mutant (Fig. 2A). A prominent band of about 21 kDa, corresponding to the decrease in mass caused by the truncation of 40 amino acids from the C terminus of the A33R protein, was detected on analysis of extracts from cells infected with WR.c1. An additional diffuse band, with an estimated mass of greater than 30 kDa, was also detected but varied in intensity in different experiments. The nonreduced forms of the A33R protein were analyzed by SDS-PAGE and by Western blotting of lysates with A33R peptide antiserum. Under nonreducing conditions, the rabbit antiserum reacted with the A33R dimer of approximately 50 kDa from cells infected with WR, WR.c3, or a B5R deletion mutant but not with that from cells infected with an A33R deletion mutant (Fig. 2B) or uninfected cells (data not shown). The A33R protein of WRc.1 migrated at about 40 kDa, corresponding to the truncated dimer (Fig. 2B). In addition, there was a band of about 60 kDa, corresponding to a dimer containing the 30-kDa band seen under reducing conditions. At present, we are uncertain of the origin of the extra band, which was also detected when lysates of cells infected with ΔA36R.c1 were examined (11).
FIG. 2.
SDS-PAGE analysis of the A33R proteins made by the mutant viruses. (A) BS-C-1 cells were left uninfected (Uninf.) or were infected with wild-type virus (WR) or mutants (WR.c3, WR.c1, or vΔA33R) and metabolically labeled with 35S-amino acids. Lysates were prepared, and proteins were precipitated with antibodies against a synthetic peptide comprised of amino acids 84 to 100 of A33R. The bound proteins were analyzed by SDS-PAGE under reducing conditions and exposed to X-ray film. (B) Lysates from cells infected with the above viruses as well as vΔB5R were dissociated with SDS under nonreducing conditions, and the polypeptides were separated by SDS-PAGE. The polypeptides were transferred to nitrocellulose membranes, which were then incubated with the above antibodies. Peroxidase-conjugated anti-rabbit immunoglobulins were added, and bound antibodies were detected with chemiluminescence reagents. The positions and masses of marker proteins are indicated in both panels.
The B5R ORF encodes a type I membrane glycoprotein that migrates with an estimated mass of about 42 kDa (4, 10). As the B5R mutation in WR.c3 is a substitution of a serine for a proline, Western blot analysis did not reveal any change in mobility for the B5R protein expressed in cells infected with WR.c1, WR.c3, WR, or vΔA33R (data not shown). In contrast, this protein was not detected in cells infected with a B5R deletion mutant.
Formation of intracellular and extracellular viruses.
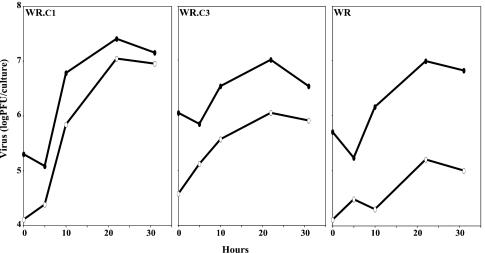
The replication of WR, WR.c1, and WR.c3 under one-step growth conditions in BS-C-1 cells was determined. The titers of viruses in the cell lysates and clarified media were compared, although the former values might have been underestimated because of the failure to release and disaggregate all viruses from membranes and other particulate materials by freezing-thawing and sonication. In repeated experiments, the yields of cell-associated viruses were similar for WR, WR.c1, and WR.c3, although the yield was about 0.5 log unit higher for WR.c1 than for the others in the experiment depicted in Fig. 3. In contrast, there was a consistent difference in the relative amounts of viruses in the clarified media. The percentages of released viruses, averaged from two experiments, were 49, 15, and 2% for WRc.1, WRc.3, and WR, respectively. The relative production of released viruses, WR.c1 > WR.c3 > WR, is the same as had been found for the corresponding mutants with deletions of A36R genes (11). The greater release of extracellular viruses by WR.c1 and WR.c3 than by WR correlated with comet formation by the two mutants (Fig. 1).
FIG. 3.
Formation of intra- and extracellular viruses. BS-C-1 cells were infected with WR.c1, WR.c3, or vaccinia virus strain WR at a multiplicity of 5 PFU per cell. At the indicated times, titers of viruses in the fluid medium (open ovals) and in the cells (filled ovals) were determined by plaque assays.
The experiments were repeated with RK13 cells, which are known to release larger amounts of EEV than other cell lines (16). RK13 cells were infected with WR, WR.c1, or WR.c3 and labeled with a mixture of [35S]methionine and [35S]cysteine from 5 to 35 h after infection. The percentages of total virus infectivity in the media were 82.7, 42.6, and 3.8% for WR.c1, WR.c3, and WR, respectively. Again, we believe that the amounts of cell-associated viruses were underestimated so that the relative amounts in the media were too high. In each case, however, these numbers were approximately twice those obtained in BS-C-1 cells, so that the relative abilities of the wild-type and mutant strains to release extracellular viruses were the same.
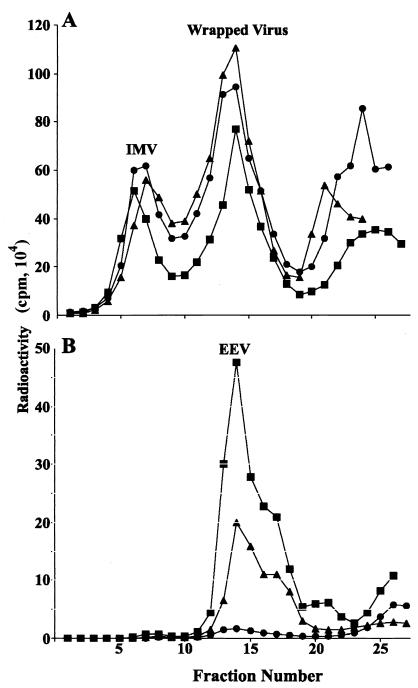
We next centrifuged the intra- and extracellular fractions through a sucrose cushion and applied the concentrated viruses to CsCl gradients. The percentages of total recovered radioactive intracellular viruses in the lower buoyant density peak (refractive index, 1.357), representing membrane-associated virions, were similar (67 to 72%) for the three viruses (Fig. 4A). However, there is no evidence that these viruses represent predominantly IEV, as they could include large numbers of IMV peripherally associated with membranes as well as CEV dislodged from the cell surface. Most of the viruses in the media were in the EEV peak (refractive index, 1.357), indicating that little cell lysis had occurred (Fig. 4B). EEV represented 25% of total WR.c1 compared to 9% of WR.c3 and less than 1% of WR. Although the absolute numbers of EEV estimated from the total infectivity and from radioactivity measurements of peak fractions differed, the trend for WR.c1 > WR.c3 > WR was the same. The presence of the B5R protein and full-length or truncated A33R proteins on EEV was confirmed by Western blotting (data not shown).
FIG. 4.
Analysis of cell-associated and released virions by CsCl centrifugation. RK13 cells were infected with WR.c1 (▪), WR.c3 (▴), or vaccinia virus strain WR (•) and labeled with a mixture of [35S]methionine and [35S]cysteine. Virus particles associated with the cells (A) and in the culture medium (B) were concentrated by sedimentation through a sucrose cushion, applied to CsCl gradients, and centrifuged. Fractions were collected from the bottom of the tube, and radioactivity was determined by scintillation counting.
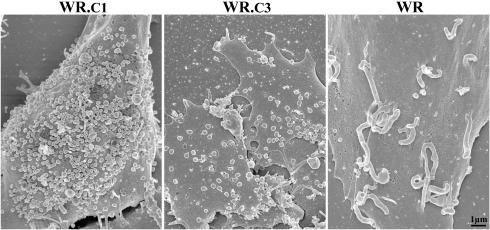
The above experiments did not ascertain the relative amounts of CEV associated with cells infected with the three viruses, as they might have been discarded during clarification of the lysates and would not have been separated from IEV in any case. To visualize CEV, scanning electron microscopy of intact cells was performed, and representative images are shown in Fig. 5. Most strikingly, whereas many of the CEV on cells infected with WR were located at the tips of microvilli, they were not found on cells infected with either of the mutants (Fig. 5). Although there was some cell-to-cell variation, WR.c1 produced the largest number of cell surface virions, followed by WR.c3 and then WR. Thus, the two mutant viruses appeared to produce the largest numbers of CEV as well as EEV.
FIG. 5.
Scanning electron microscopy of infected cells. BS-C-1 cells were infected with WR.c1, WR.c3, or vaccinia virus strain WR at a multiplicity of 5 PFU per cell. After incubation for 20 h at 37°C, the cells were fixed, coated with gold-palladium alloy, and viewed with a Hitachi S-4700 field emission scanning electron microscope at an accelerating voltage of 3 kV.
Actin tail formation.
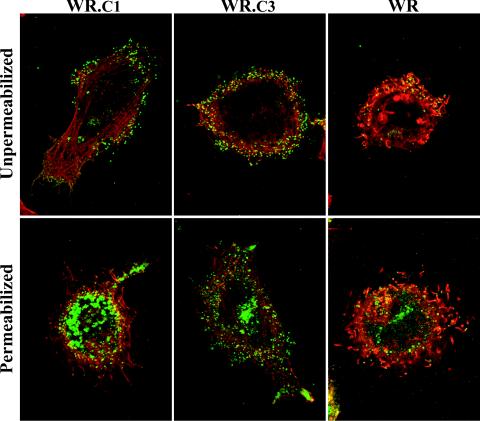
The absence of specialized microvilli suggested a corresponding defect in actin tail formation. To assess this possibility, infected HeLa cells were fixed with paraformaldehyde and then incubated with a rat MAb to the vaccinia virus B5R protein followed by FITC-conjugated anti-rat immunoglobulin, either before or after permeabilization. The cells were also stained with rhodamine-conjugated phalloidin to visualize actin tails and with DAPI to discern nuclei and DNA-containing viral factories. The cells infected with WR displayed numerous actin tails that were associated with virus particles at the cell surface and that stained with or without cell permeabilization (Fig. 6). Numerous CEV were also detected on the surface of cells infected with WR.c1 or WR.c3, but the characteristic thick elongated actin tails were not found (Fig. 6), although some actin fibers seen may have represented short atypical tails.
FIG. 6.
Visualization of actin tails by confocal microscopy. HeLa cells were infected with WR.c1, WR.c3, or vaccinia virus strain WR. One set of cultures was permeabilized with Triton X-100, and one set was not. Enveloped virus particles were stained with a rat monoclonal antibody to B5R followed by FITC-conjugated anti-rat antibodies (green). The cells were also stained with rhodamine-conjugated phalloidin (red) and DAPI (blue) to visualize F-actin and DNA, respectively. The DAPI stain is not shown.
The inability of the mutant viruses to form actin tails was also demonstrated by confocal microscopy of living cells. Cells transfected with a plasmid encoding actin-GFP were infected with WR, WR.c1, or WR.c3. Infected cells were identified with Hoechst dye, which stained the viral factories, and the formation of actin tails was assessed by capturing one image per 6 s for 10 min. At least one motile actin tail was visualized in each WR-infected cell that expressed actin-GFP. In contrast, we were unable to detect motile actin tails in any of the cells infected with either WR.c1 or WR.c3. Individual time-lapse series were further analyzed by creating maximum-intensity projections comprising images from all time points. With this technique, actin tails appeared as long, thick fluorescent structures only in cells infected with WR (data not shown).
Synthesis and tyrosine phosphorylation of the A36R protein.
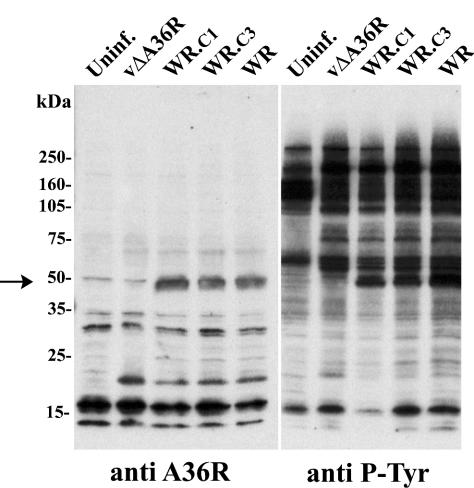
In a series of elegant experiments on vaccinia virus actin tail formation, Frischknecht and coworkers (6) demonstrated that the nucleation of actin polymerization is regulated by the phosphorylation of Tyr112 and Tyr132 located in the cytoplasmic domain of the A36R protein. We wanted to find out whether the absence of actin tails in WR.c1- and WR.c3-infected cells was due to a failure to stably express or tyrosine phosphorylate the A36R protein. Cell lysates were prepared from uninfected or infected BS-C-1 cells, and the proteins were resolved by SDS-PAGE and blotted onto nitrocellulose. Half of the blot was probed with antibody against the A36R protein, revealing a band of approximately 50 kDa from cells infected with WR, WR.c1, or WR.c3 but not from uninfected cells or cells infected with vΔA36R (Fig. 7). Thus, there appeared to be no defect in the synthesis or stability of the A36R protein in cells infected with WR.c1 or WR.c3. The other half of the blot was probed with a phosphotyrosine (P-Tyr)-specific antibody. A specific 50-kDa band, corresponding to phosphorylated A36R, was detected among the proteins from cells infected with WR, WR.c1, or WR.c3 but not from uninfected cells or cells infected with vΔA36R. These results indicated that the failure to form actin tails in cells infected with mutants WR.c1 and WR.c3 was not due to a block in the synthesis or tyrosine phosphorylation of the A36R protein.
FIG. 7.
Synthesis and tyrosine phosphorylation of the A36R protein. Infected or mock-infected (Uninf.) HeLa cells were incubated for 20 h at 37°C, harvested, and analyzed by SDS-PAGE. The polypeptides were transferred to nitrocellulose membranes. One half of each membrane was incubated with a P-Tyr-specific mouse MAb (anti P-Tyr), and the other half was incubated with rabbit antibodies against A36R (anti A36R). The arrow indicates the A36R band. The positions and masses of marker proteins are indicated on the left.
Coimmunoprecipitation of A33R and A36R proteins.
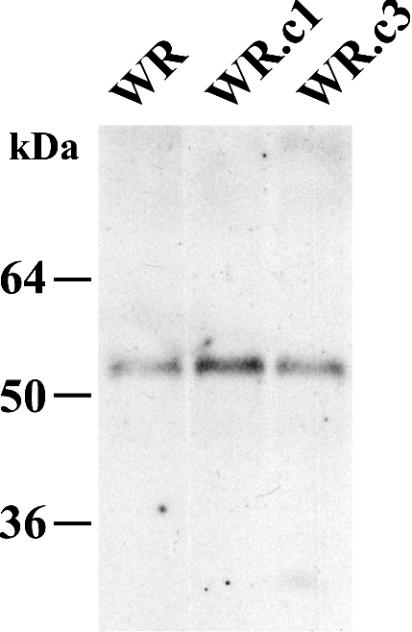
Previous studies demonstrated that the A33R and A36R proteins interact through their cytoplasmic tails and that this interaction is important for the incorporation of A36R into the viral envelope (21, 32, 36). We wanted to determine whether the A33R and A36R proteins produced by WR.c1 and WR.c3 were still capable of interacting with each other. Lysates from cells infected with WR, WR.c1, or WR.c3 were incubated with the anti-A33R MAb followed by protein A-Sepharose. Bound proteins were separated by SDS-PAGE, blotted onto nitrocellulose membranes, and probed with a horseradish peroxidase-conjugated anti-A36R antibody. For all three viruses, the A36R protein was coimmunoprecipitated by the anti-A33R MAb (Fig. 8), indicating that the A33R and A36R proteins produced by WR.c1 and WR.c3 formed a complex.
FIG. 8.
Coimmunoprecipitation of the A33R and A36R proteins. Lysates from HeLa cells that were infected with the indicated recombinant viruses were immunoprecipitated with an anti-A33R MAb. Immune complexes were analyzed by SDS-PAGE followed by Western blotting with a horseradish peroxidase-conjugated anti-A36R antibody. The masses and migation positions of marker proteins are indicated on the left.
Virulence for mice.
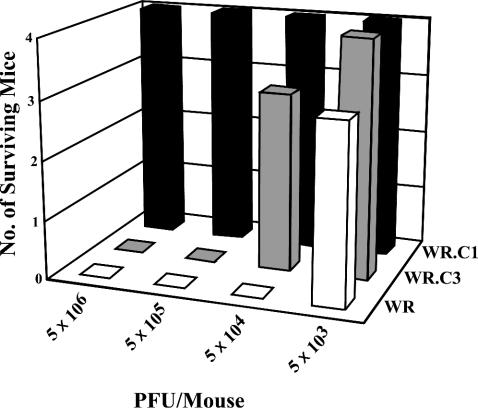
We next compared the virulence of the mutant and wild-type viruses by using a well-characterized murine intranasal infection model (13, 29, 33) in order to determine whether increased virus release compensated for the absence of actin tails and specialized microvilli. Mice were inoculated by the intranasal route with 5 × 103, 5 × 104, 5 × 105, or 5 × 106 PFU of purified virus and weighed daily. The wild-type WR virus was the most virulent, causing the death of one mouse inoculated with 5 × 103 PFU and the deaths of all mice inoculated with 5 × 104 PFU or higher (Fig. 9). WR.c3 was mildly attenuated, with three surviving mice at 5 × 104 and no survivors at 5 × 105 or higher (Fig. 9). WR.c1, however, was highly attenuated, with no deaths even at 5 × 106 PFU (Fig. 9).
FIG. 9.
Attenuation of mutant viruses, as determined by survival of mice. Mice were inoculated intranasally with purified virus preparations containing 5 × 106, 5 × 105, 5 × 104, or 5 × 103 PFU of wild-type vaccinia virus (WR) or mutant WR.c1 or WR.c3. Mice were weighed daily, and those that had lost more than 30% of their initial weight were sacrificed. The survival of mice was determined for 14 days after inoculation.
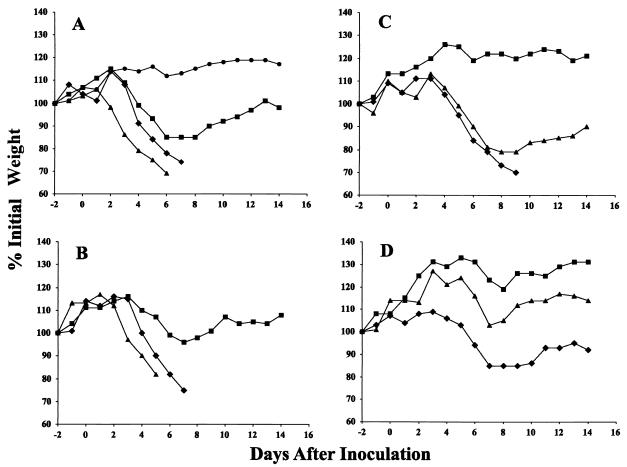
The weight losses of the surviving mice are shown in Fig. 10. The lowest weights were found between days 6 and 9, while mice surviving 9 days did recover. The weight differences between mice infected with WR.c1 and those infected with WR were highly significant at all doses except the highest. At 5 × 103, 5 × 104, and 5 × 105 PFU, the P values were equal or less than 0.001 for the critical weight loss days. The weight differences between the mice infected with WR.c3 and those infected with WR, however, reached significance only with the dose of 5 × 103 PFU; the P values were equal to or less than 0.027 for days 6 through 10.
FIG. 10.
Virulence of mutant viruses for mice, as measured by weight loss. Purified virus preparations containing 5 × 106 PFU (A), 5 × 105 PFU (B), 5 × 104 PFU (C), or 5 × 103 PFU (D) of WR.c1 (▪), WR.c3 (▴), or vaccinia virus strain WR (⧫) were inoculated intranasally into mice. The weights of the four mice in each group were determined daily, and mice that had lost more than 30% of their initial weight were sacrificed. Percent weight relative to the initial average weight of surviving mice is plotted against days before and after infection. The weight of a group of uninfected control mice (•) was also monitored and is shown in panel A.
DISCUSSION
Vaccinia virus spreads efficiently in tissue cultures by two methods: direct cell-to-cell transfer mediated by CEV at the tips of actin-containing microvilli and long-range spread accomplished by the release of EEV into the medium. Vaccinia virus strain WR, used as the parent virus for all mutants in the present study, relies predominantly on CEV-mediated spread (1, 2). In a previous study (11), Katz et al. used a vaccinia virus mutant which was unable to form actin tails because of a deletion of the A36R gene to select viruses with second-site mutations that increased their ability to spread in tissue cultures. The enhanced spread was due to the greater release of EEV caused by C-terminal truncations of the A33R protein or a serine-to-proline substitution in the B5R protein. As the mutants lacked the A36R gene, we were unable to determine the effects of these mutations on actin tail formation. To overcome this limitation, we transferred two of the mutations into wild-type vaccinia virus. The resulting viruses, WR.c1 and WR.c3, with mutations in the A33R ORF and the B5R ORF, respectively, exhibited enhanced release phenotypes, as shown by the formation of comets in cell monolayers covered with a liquid overlay and by the production of increased amounts of infectious extracellular viruses. The extracellular viruses sedimented as EEV in CsCl gradients and contained the mutated or wild-type A33R and B5R proteins, as determined by Western blotting. Katz et al. had speculated that the A33R and B5R mutations weakened the adherence of CEV to the cell membrane (11). However, in the present study, more CEV were found on the surface of cells infected with WR.c1 and WR.c3 than on that of cells infected with WR. The mutated proteins might somehow increase the efficiency of wrapping, the transport of IEV to the periphery, or the externalization of virions. Discrimination among these possibilities is difficult with presently available methods.
Interestingly, actin tails were not detected by confocal microscopy of fixed or living cells infected with either mutant, and specialized microvilli were not observed by scanning electron microscopy. The mechanism of vaccinia virus actin tail formation has been intensively investigated. Although the A33R, A34R, and A36R membrane proteins are all required for actin tail formation, only A36R has been shown to have a direct role in the process (19, 21, 23, 35, 37). The phosphorylation of A36R Tyr112 and Tyr132 by a Src family kinase that probably resides in the plasma membrane results in the recruitment of the adaptor protein Nck, WIP, N-WASP, and Grb2, which leads to activation of the Arp2/3 complex and nucleation of actin polymerization (6, 14, 24). Conservative mutations of Tyr112 and Tyr132 resulted in a specific block in actin tail formation by recombinant viruses and a reduction in plaque size (18, 30).
Several possibilities could account for the absence of actin tails in cells infected with mutant WR.c1 or WR.c3. It has been shown that the A36R protein interacts with the A33R protein (21) and that this interaction is required for the incorporation of the former into IEV membranes (36). A major interaction site was mapped to the cytoplasmic domain of the A33R protein (32), which was unaltered by the C-terminal truncation of the A33R protein and should not have been affected at all by the B5R mutation. Indeed, our coimmunoprecipitation experiments indicated the presence of a complex containing the A33R and A36R proteins in cells infected with the mutant viruses. A second possibility was that the A36R protein was not tyrosine phosphorylated. However, this idea was ruled out by Western blotting with a P-Tyr-specific antibody. Moreover, the latter result also implied that the A36R protein was associated with the plasma membrane during fusion of the IEV outer membrane. Another possibility was that CEV was dissociated from the plasma membrane too rapidly to allow actin tail formation. Scanning electron microscopy, however, revealed numerous CEV still attached to the plasma membrane. Indeed, WR.c1 especially appeared to produce more CEV than did wild-type virus. We are left, therefore, with the hypothesis that the A33R and B5R proteins participate in either the nucleation or the stability of actin tails.
In spite of the enhanced spread of the WR.c1 and WR.c3 mutants in cell cultures, their virulence for mice inoculated intranasally was significantly reduced. This finding suggests that direct cell-to-cell spread of virus mediated by actin-containing microvilli is important in vivo as well as in vitro. Another possibility is that antibody more effectively prevents the spread of released virus than of cell-associated virus. Further studies are needed to evaluate these mechanisms as well as to compare the titers of the mutant and wild-type viruses in the different organs of infected mice.
Acknowledgments
We thank members of the Laboratory of Viral Diseases, especially Norman Cooper for tissue culture cells, Patricia Szajner for advice on immunoprecipitation procedures, and Christiana Fogg for statistical analysis. Tim Maugel of the Laboratory for Biological Ultrastructure at the University of Maryland helped with the scanning electron microscopy. Some of the work was performed in the NIAID Imaging Facility with the guidance of Owen Schwartz.
REFERENCES
- 1.Blasco, R., and B. Moss. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 66:4170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasco, R., J. R. Sisler, and B. Moss. 1993. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J. Virol. 67:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378:636-638. [DOI] [PubMed] [Google Scholar]
- 4.Engelstad, M., S. T. Howard, and G. L. Smith. 1992. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology 188:801-810. [DOI] [PubMed] [Google Scholar]
- 5.Engelstad, M., and G. L. Smith. 1993. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology 194:627-637. [DOI] [PubMed] [Google Scholar]
- 6.Frischknecht, F., V. Moreau, S. Rottger, S. Gonfloni, I. Reckmann, G. Superti-Furga, and M. Way. 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401:926-929. [DOI] [PubMed] [Google Scholar]
- 7.Hiller, G., and K. Weber. 1985. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J. Virol. 55:651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiller, G., K. Weber, L. Schneider, C. Parajsz, and C. Jungwirth. 1979. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology 98:142-153. [DOI] [PubMed] [Google Scholar]
- 9.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaacs, S. N., E. J. Wolffe, L. G. Payne, and B. Moss. 1992. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 66:7217-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz, E., E. Wolffe, and B. Moss. 2002. Identification of second-site mutations that enhance release and spread of vaccinia virus. J. Virol. 76:11637-11644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathew, E. C., C. M. Sanderson, R. Hollinshead, and G. L. Smith. 2001. A mutational analysis of the vaccinia virus B5R protein. J. Gen. Virol. 82:1199-1213. [DOI] [PubMed] [Google Scholar]
- 13.Moore, J. B., and G. L. Smith. 1992. Steroid hormone synthesis by a vaccinia enzyme—a new type of virus virulence factor. EMBO J. 11:1973-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreau, V., F. Frischknecht, I. Reckmann, R. Vincentelli, G. Rabut, D. Stewart, and M. Way. 2000. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat. Cell Biol. 2:441-448. [DOI] [PubMed] [Google Scholar]
- 15.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 16.Payne, L. G. 1979. Identification of the vaccinia virus hemagglutinin polypeptide from a cell system yielding large amounts of extracellular enveloped virus. J. Virol. 31:147-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne, L. G. 1980. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J. Gen. Virol. 50:89-100. [DOI] [PubMed] [Google Scholar]
- 18.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmström, F. Frischknecht, M. Zettl, T. Zimmerman, and M. Way. 2001. Kinesin dependent movement on microtubules precedes actin based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 19.Roper, R., E. J. Wolffe, A. Weisberg, and B. Moss. 1998. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 72:4192-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roper, R. L., L. G. Payne, and B. Moss. 1996. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol. 70:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rottger, S., F. Frischknecht, I. Reckmann, G. L. Smith, and M. Way. 1999. Interactions between vaccinia virus intracellular enveloped virion (IEV) membrane proteins and their roles in IEV assembly and actin tail formation. J. Virol. 73:2863-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Sanderson, C. M., F. Frischknecht, M. Way, M. Hollinshead, and G. L. Smith. 1998. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J. Gen. Virol. 79:1415-1425. [DOI] [PubMed] [Google Scholar]
- 24.Scaplehorn, N., A. Holmstrom, V. Moreau, F. Frischknecht, I. Reckmann, and M. Way. 2002. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr. Biol. 12:740-745. [DOI] [PubMed] [Google Scholar]
- 25.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans-Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stokes, G. V. 1976. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J. Virol. 18:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 28.Tsutsui, K. 1983. Release of vaccinia virus from FL cells infected with the IHD-W strain. J. Electron Microsc. 32:125-140. [PubMed] [Google Scholar]
- 29.Turner, G. S. 1967. Respiratory infection of mice with vaccinia virus. J. Gen. Virol. 1:399-402. [DOI] [PubMed] [Google Scholar]
- 30.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward, B. M., A. S. Weisberg, and B. Moss. 2003. Mapping and functional analysis of interaction sites within the cytoplasmic domains of the vaccinia virus A33R and A36R envelope proteins. J. Virol. 77:4113-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williamson, J. D., R. W. Reith, L. J. Jeffrey, J. R. Arrand, and M. Mackett. 1990. Biological characterization of recombinant vaccinia viruses in mice infected by the respiratory route. J. Gen. Virol. 71:2761-2767. [DOI] [PubMed] [Google Scholar]
- 34.Wolffe, E. J., S. N. Isaacs, and B. Moss. 1993. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J. Virol. 67:4732-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolffe, E. J., E. Katz, A. Weisberg, and B. Moss. 1997. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J. Virol. 71:3904-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolffe, E. J., A. Weisberg, and B. Moss. 2001. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J. Virol. 75:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolffe, E. J., A. S. Weisberg, and B. Moss. 1998. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology 244:20-26. [DOI] [PubMed] [Google Scholar]