Abstract
JC virus (JCV)-specific cytotoxic T lymphocytes (CTL) in peripheral blood are associated with a favorable outcome in patients with progressive multifocal leukoencephalopathy (PML). However, the frequency of these cells in the peripheral blood mononuclear cells (PBMC) of PML patients is unknown. To develop a highly sensitive assay for detecting the cellular immune response against this virus, we performed a CTL epitope mapping study of JCV VP1 major capsid protein by using overlapping peptides. A novel HLA-A*0201-restricted epitope, the VP1p36 peptide SITEVECFL, was characterized. The cellular immune response against JCV was assessed in 32 study subjects. By combining the results of the 51Cr release assay on pooled peptides and staining with the HLA-A*0201/JCV VP1p36 tetramer, VP1-specific CTL were detected in 10 of 11 PML survivors (91%) versus only 1 of 11 PML progressors (9%, P = 0.0003). VP1-specific CTL were also detected in two of two patients recently diagnosed with PML and in four of four human immunodeficiency virus-positive patients with possible PML. The frequency of CTL specific for the novel VP1p36 and the previously described VP1p100 epitopes was determined. In two patients, the frequency of CTL specific for the VP1p36 or VP1p100 epitopes, as determined by fresh blood tetramer staining (FBTS), ranged from 1/6,000 to 1/24,000 PBMC. A CTL sorting technique combining tetramer staining and selection with immunomagnetic beads allowed the detection of epitope-specific CTL in two cases that were determined to be negative by FBTS. The phenotype of these CTL in vivo was consistent with activated memory cells. These data suggest that, although present in low numbers, JCV-specific CTL may be of central importance in the containment of JCV spread in immunosuppressed individuals.
JC virus (JCV) is the agent of progressive multifocal leukoencephalopathy (PML) (25), a deadly demyelinating disease of the central nervous system, for which there is no specific treatment. JCV infects more than 85% of the normal adult population (34), and its reactivation in the setting of AIDS, hemopathies, or in organ transplant recipients leads to a lytic infection of oligodendrocytes. Indeed, PML occurs in 5.1% of AIDS patients and in 0.07% of human immunodeficiency virus-negative (HIV−) people with hematologic malignancies (28). Before the availability of highly active antiretroviral therapy (HAART), only 10% of HIV+ PML patients survived more than 1 year (5). With HAART, this proportion has now increased to 50% (9). There is, thus far, no animal model of PML.
Immunosurveillance by cytotoxic T lymphocytes (CTL) has been demonstrated to be an important factor in preventing the onset of severe diseases in patients chronically infected by DNA viruses such as Epstein-Barr virus (EBV), cytomegalovirus (CMV), or herpes simplex virus (HSV) (7, 20, 27, 29, 32, 35). Indeed, a large proportion of the adult population is infected by these viruses, and yet they only cause severe diseases in a small subset of individuals, particularly in a setting of immunosuppression. In previous studies, we have demonstrated that JCV-specific CTL activity could be detected in JCV antigen-stimulated peripheral blood mononuclear cells (PBMC) of PML survivors but not in progressors (11, 18). Using a computer-assisted predictive analysis, we characterized the HLA-A*0201-restricted CTL epitope VP1p100. The HLA-A*0201/JCV VP1p100 tetramer was then used to detect VP1p100-specific CTL by flow cytometry (19). Since the presence of JCV-specific CTL was associated with a favorable clinical outcome in PML patients, we sought to define their frequency in vivo. To obtain the most sensitive assay, we first performed a CTL epitope mapping study of the whole VP1 protein. We chose to map this protein since, in our previous studies, the VP1 protein was more often recognized by CTL than was the T antigen (18). Moreover, seven HLA-A*0201-restricted candidate nonamer epitopes from three other proteins of JCV (T antigen, VP2, and VP3) failed to be recognized by CTL (19). We found a novel HLA-A*0201-restricted epitope, VP1p36. Then, by using fresh blood tetramer staining (FBTS) and an original CTL sorting (CTLS) technique combining tetramer staining and positive selection with immunomagnetic beads, we calculated the frequency of VP1p36- and VP1p100-specific CTL in the PBMC of an HIV+ PML survivor and two HIV+ patients with possible PML. We also characterized the phenotype of the JCV VP1p100-specific CTL of an HIV+ PML survivor in vivo.
MATERIALS AND METHODS
Selection of the study subjects.
To characterize in detail the immune response against VP1, the major capsid protein of JCV, a total of 32 subjects were enrolled in the present study, including 24 patients with proven PML (18 HIV+ and 6 HIV−), 4 HIV+ patients with possible PML, and 4 HIV+ individuals with other neurological diseases (OND) (i.e., HIV+ OND). The diagnosis of PML was ascertained by clinical and neuroradiological criteria and by brain biopsy or by positive JCV DNA PCR in the cerebrospinal fluid (CSF). Twenty-two of the study subjects were HLA-A*0201+, and ten were HLA-A*0201− (Table 1).
TABLE 1.
Clinical and laboratory data on 32 study subjects
| Diagnosisa | n | No. of subjects and HIV status | Mean age (yr) ± SD | Mean disease duration (mo)b ± SD | Mean HIV VL (cps/ml)c in plasma ± SD | Mean no. of CD4+T cell (count/μl) ± SD |
|---|---|---|---|---|---|---|
| PML S | 11 | 44.7 ± 8.1 | 80 ± 23.5 | 3,189 ± 9,766 | 428 ± 224 | |
| A*0201+ | 9 | 8 HIV+ 1 HIV− | ||||
| A*0201− | 2 | 2 HIV+ | ||||
| PML P | 11 | 40.2 ± 6.3 | 4 ± 2.6 | 50,150 ± 83,094 | 87 ± 112 | |
| A*0201+ | 6 | 3 HIV+ 3 HIV− | ||||
| A*0201− | 5 | 5 HIV+ | ||||
| PML E | 2 | 66.5 ± 19.1 | 9.7 ± 4.2 | NA | NA | |
| A*0201+ | 0 | |||||
| A*0201− | 2 | 2 HIV− | ||||
| Possible PML | 4 | 42.8 ± 4.3 | 26 ± 20.3 | 306 ± 436 | 481 ± 78 | |
| A*0201+ | 4 | 4 HIV+ | ||||
| A*0201− | 0 | |||||
| OND | 4 | 49.3 ± 7.1 | 40 ± 12.7 | 1,080 ± 2,008 | 549 ± 446 | |
| A*0201+ | 3 | 3 HIV+ | ||||
| A*0201− | 1 | 1 HIV+ | ||||
| Total | 32 |
S, survivors (alive and with inactive disease >12 months after diagnosis of PML); P, progressors; E, early.
That is, the interval between time of onset of PML and either death or the endpoint corresponding to 1 April 2003.
cps, copies; NA, not available.
Of the 18 HIV+ patients with PML, 10 were survivors whose disease had improved or remained stable 43 to 122 months after their initial diagnosis of PML. The eight HIV+ PML patients who had a progressive neurological disease had a fatal outcome in 1.4 to 9.8 months after their diagnosis. Of the 6 HIV− PML patients, one was a PML survivor. She had a history of non-Hodgkin's lymphoma and was neurologically stable 9.5 years after her initial diagnosis of PML. She had been treated with cytosine arabinoside (Ara-C). Two HIV− patients were recently diagnosed with PML. One had a quiescent non-Hodgkin's lymphoma. Detection of CTL was performed 5 months after the diagnosis of PML, when she was significantly improving from a neurological standpoint. Unfortunately, she died 6 months after disease onset from an unrelated medical condition. The second patient had dermatomyositis, and CTL detection was performed 4 months after the beginning of PML. She continued to improve 12 months after disease onset. The three other HIV− PML patients had progressive neurological disease and a fatal outcome 2.7 to 4.9 months after the diagnosis of PML. One had a history of autologous CD30+ stem cell transplant for multiple myeloma, and two were bone marrow transplant recipients for acute myeloid leukemia.
Four HIV+ patients presented with a leukoencephalopathy clinically and neuroradiologically undistinguishable from PML but with a negative JCV DNA PCR in the CSF. These patients may still have PML, but with a CSF JC viral load (VL) below the limit of detection of the PCR assay (17). Such patients have become more frequent since the availability of HAART in 1996 (3). They are referred to here as HIV+ possible-PML subjects (10).
Finally, four HIV+ OND patients were included as controls. They had histories of HIV encephalopathy, thoracic polyradiculitis, neurosyphilis, and CMV polyradiculopathy, respectively.
CTL epitope mapping of the JCV VP1 protein.
To cover the 354 amino acids (aa) of VP1 protein, 30 20-mer and 2 18-mer peptides overlapping by 8 aa were synthesized. Using these overlapping peptides, any potential nonamer CTL epitope of the VP1 protein could be identified. The amino acid sequence of the VP1 protein chosen for the present study was the one of the MAD-1 isolate (13), whose subtype corresponds to type 1 (4). However, the JCV subtype 2B, which has two amino acid changes (a threonine for a serine at amino acid position 117 and an alanine for a threonine at amino acid position 128 of the VP1 protein) compared to MAD-1, was found in 36% of the brains of PML patients versus only 5.1% of the samples of urine from healthy individuals (P < 0.001) (1). A third amino acid change compared to the MAD-1 sequence, an arginine instead of a lysine at amino acid position 345, was found in the brain of 11 of 11 PML patients (4). To examine whether these amino acid changes could account for a difference in the cellular immune response against the VP1 protein, two additional 20-mers (10′ and 11′) and one additional 18-mer (29′) bearing these amino acid changes were included in the present study. Peptides were divided into five pools: pool I, 20-mers 1 to 7; pool II, 20-mers 8 to 10, 10′, 11, and 11′; pool III, 20-mers 12 to 17; pool IV, 20-mers 18 to 24; and pool V, 20-mers 25 to 28 and 18-mers 29 and 29′. These pools were tested in a functional lysis assay. The peptides were stored at a concentration of 2 mg/ml in a solution of 10% dimethyl sulfoxide, 5% dithiothreitol, 2% fetal calf serum (FCS), and sterile H2O.
Functional lysis assay.
PBMC from a total of 20 study subjects, 10 HLA-A*0201+ and 10 HLA-A*0201−, were isolated by centrifugation over a Ficoll-diatrizoate gradient. Seven million cells were cultured in a well of a 12-well plate (Costar) with one of the five pools of overlapping 20-mer or 18-mer peptides at a final concentration of 10 μg/ml for each peptide. Therefore, the total peptide concentration in a well was 60 or 70 μg/ml, depending if the pool encompassed six or seven 20-mers. Cells were cultured in RPMI 1640-12% FCS at a density of 3.5 × 106 PBMC/ml. After 72 h, an equal volume of RPMI 1640-12% FCS containing 40 U of recombinant interleukin-2 (rIL-2)/ml was added to each culture well, and every 2 days thereafter half of the medium was replaced. After 12 to 14 days, the peptide-stimulated cells were analyzed in a 51Cr release assay (22). EBV-transformed autologous B-lymphoblastoid cells (B-LCL) were used as sources of target cells. Aliquots of 106 B-LCL were incubated overnight with peptide pools at a concentration of 60 to 70 μg/ml in a final volume of 1 ml. The peptide VP1p11B (ALSEGCTPYDIN) from the simian immunodeficiency virus was used as negative control. After a 16-h incubation period, target cells were labeled with 100 μCi of 51Cr for 90 min. These cells were washed, and aliquots of 104 cells were added to the effector cells as targets in 96-well U-bottom plates in a final volume of 200 μl/well. The assays were performed in duplicates. A specific lysis equal or superior to 10% at an effector/target (E:T) ratio of 20:1 was considered a positive result. When a positive result was obtained with a specific peptide pool, the assay was repeated with the individual 20-mer peptides of that pool.
Construction of the HLA-A*0201/JCV VP1p36 tetramer.
HLA-A*0201 protein expression and folding with human β2-microglobulin and peptide VP1p36, as well as tetramerization with phycoerythrin (PE)-labeled streptavidin (Prozyme), were performed as previously reported (2, 14, 19).
Staining and phenotypic analysis of JCV VP1p36-specific CD8+ T cells.
The monoclonal antibodies (MAbs) used for the present study were directly coupled to fluorescein isothiocyanate (FITC), allophycocyanin (APC), or PE-Texas red (ECD). The following MAbs were used: anti-CD3(SK7)-APC, anti-CD8α(SKI)-FITC (Becton Dickinson), and anti-CD8αβ(2ST8-5H7)-ECD (Beckman Coulter). The PE-coupled HLA-A*0201/JCV VP1p36 tetramer and the three MAbs noted above were used in four-color flow cytometric analyses. A total of 200 ng of the PE-coupled HLA-A*0201/JCV VP1p36 tetramer was used in conjunction with the directly labeled MAbs to stain 100 μl of fresh whole blood or 5 × 105 PBMC that were cultured in vitro with the VP1p36 peptide for 12 to 14 days. To remove red blood cells, fresh blood samples were lysed by using a TQ PREP Workstation (Beckman Coulter). The lysed samples were washed with phosphate-buffered saline (PBS) and centrifuged for 5 min at 300 × g.
Similarly, the stained cultured lymphocyte samples were washed in PBS and centrifuged for 5 min at 300 × g. The supernatants were decanted, and cells were resuspended in 0.5 ml of PBS containing 1.5% paraformaldehyde. Of the six HLA-A*0201+ PML progressors, five had passed away before the tetramer reagent was available. For these patients, frozen-thawed PBMC were used for tetramer staining. Frozen PBMC of a study subject known to harbor VP1p36-specific CTL were thawed and cultured in parallel as an internal positive control. VP1p36-specific CTL were consistently detected among the frozen-thawed PBMC of this positive control. The possibility of performing reliable tetramer staining on frozen-thawed PBMC has been previously reported (31). All tetramer staining assays were performed at room temperature. Samples were analyzed on a FACScalibur flow cytometry system (Becton Dickinson). The data presentation was prepared by using WinMDI software version 2.7 (Joseph Trotter) and Microsoft PowerPoint software version 2000 (Microsoft).
MHC class I typing.
The major histocompatibility complex (MHC) class I alleles expressed by the study subjects were determined by using standard serologic tissue typing procedures. Molecular analysis was performed on the PBMC of all HLA-A2+ subjects to determine their HLA-A*02 subtype.
Frequency of JCV VP1pept-specific CTL. (i) FBTS technique.
The number of tetramer-positive cells was directly calculated and is expressed as a percentage of CTL per CD8αβ+ T cells or PBMC in fresh blood.
(ii) CTLS technique.
Twenty to fifty million PBMC were isolated by using a Ficoll-diatrizoate gradient from HLA-A*0201+ individuals and then stained with 0.5 μg of PE-coupled HLA-A*0201/JCV VP1pept tetramer per 106 PBMC. After 10 min of incubation at 4°C, 2 μg of anti-PE microbeads (Miltenyi Biotec) were added per 106 PBMC, followed by incubation for 15 min at 4°C (23). After a wash with PBS, cells were sorted with an AUTOMACS cell sorter (Miltenyi Biotec), with two columns with a positive selection program specifically designed for rare events analysis. The tetramer-positive and -negative fractions were harvested. The positive fraction contained the tetramer-stained cells but also a substantial number of tetramer-negative cells because the latter were present at a much higher frequency than the tetramer-positive cells and were not all removed by the washing steps. In order to assess the precise number of HLA-A*0201/JCVpept tetramer-positive CD8αβ+ T cells, both the positive and the negative fractions were counted and then analyzed on a FACScalibur flow cytometry system. The tetramer-positive fraction was stained with anti-CD8α-FITC, anti-CD8αβ-ECD, and anti-CD3-APC MAbs but not the tetramer, whereas the negative fraction was stained with PE-conjugated tetramer and the three other MAbs. The frequency of peptide-specific CTL in fresh PBMC was calculated as follows: the frequency of JCV VP1pept-specific CTL = % tetramer-positive cells among the CD8αβ+ T cells in the positive fraction × (number of cells in the positive fraction/total number of cells), where the total number of cells is the number of cells in both the positive and negative fractions. For example, if 1% of the CD8αβ+ T cells in the positive fraction were tetramer positive and if there were 106 PBMC in the positive fraction and 49 × 106 PBMC in the negative fraction, then the frequency of epitope-specific CTL would be calculated as follows: (1% × 106)/[106 + (49 × 106)] = 0.02% = 1/5,000 PBMC. After a sorting step, PBMC of both fractions were cultured in RPMI 1640-12% FCS and stimulated on day 0 by VP1pept at 1 to 2.5 μg/ml, depending on the peptide used. Autologous PBMC irradiated with 3,000 rads were added as feeder cells on day 0 at a ratio of 10:1 in both sorted fractions. Then, 50 U of rIL-2/ml was added on day 3 in both sorted fractions, and the medium was changed every other day. The positive and negative fractions were assessed for the presence of VP1pept-specific cells by tetramer staining and by 51Cr release assay after an average of 14 days.
Statistical analysis.
The two-tailed Fisher exact test and the unpaired Student t test were used to compare the presence of JCV VP1-specific CTL and the CD4+-T-cell count and HIV VL values between PML survivors and progressors.
RESULTS
Cellular immune response against JCV VP1 protein.
To characterize every possible CTL epitope, we performed a mapping study of the JCV VP1 protein. To do this, we synthesized 30 20-mers and 2 18-mers overlapping by 8 aa spanning the entire 354-aa VP1 protein, as predicted by the nucleotides sequence from MAD-1. Using these overlapping peptides, any potential nonamer CTL epitope of the VP1 protein could be identified. We also added two 20-mers (10′ and 11′) and one 18-mer (29′) to account for three amino acid changes of the VP1 protein found at a higher frequency in the brain of PML patients (see Materials and Methods and references 1 and 4). This mapping study allowed us to identify any 9-mer epitope from the main reading frame but not hypothetical epitopes that would result from alternative reading frames.
PBMC from a total of 20 patients, including 10 HLA-A*0201+ and 10 HLA-A*0201− subjects, were stimulated with the five peptide pools and assessed as effector cells in a 51Cr release assay with peptide pools-pulsed autologous B-LCL as target cells. The clinical and laboratory data for the study subjects are presented in Table 1. Using this approach, CTL against VP1 protein were detected in 12 of 20 patients, including 6 of 9 PML survivors (67%), 1 of 6 PML progressors (17%), 2 of 2 HIV− patients who were recently diagnosed with PML and who were improving clinically, 1 of 1 HIV+ possible-PML subject, and 2 of 2 HIV+ OND subjects (Table 2). As expected, the CD4+-T-cell count was higher in HIV+ PML survivors (428 ± 24) than in HIV+ PML progressors (87 ± 12, P = 0.002). This indicates that the presence of CD4+ T cells is important for the proper functioning of virus-specific CTL (24). However, no statistically significant differences in HIV VL were detected in these two groups (3,189 ± 9,766 versus 50,150 ± 83,094; P = 0.092) (Table 1).
TABLE 2.
51Cr release CTL assay with pooled peptides of the JCV VP1 protein in PBMC of HLA-A*0201+ and HLA-A*0201− subjects
| Diagnosis (n) | HIV status and HLA typing | No. of subjects | No. positive/total no. tested from:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Pool I | Pool II | Pool III | Pool IV | Pool V | All pools | |||
| PML survivors (9) | HIV+ | 8 | ||||||
| A*0201+ | 6 | 3/6 | 1/6 | 2/6 | 1/6 | 0/6 | 3/6 | |
| A*0201− | 2 | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 2/2 | |
| HIV− | 1 | |||||||
| A*0201+ | 1 | 0/1 | 0/1 | 0/1 | 0/1 | 1/1 | 1/1 | |
| A*0201− | 0 | |||||||
| PML early (2) | HIV− | 2 | ||||||
| A*0201+ | 0 | |||||||
| A*0201− | 2 | 0/2 | 1/2 | 1/2 | 1/2 | 2/2 | 2/2 | |
| PML progressors (6) | HIV+ | 6 | ||||||
| A*0201+ | 1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | |
| A*0201− | 5 | 1/5 | 1/5 | 1/5 | 1/5 | 0/5 | 1/5 | |
| Possible PML (1) | HIV+ | 1 | ||||||
| A*0201+ | 1 | 1/1 | 1/1 | 0/1 | 0/1 | 0/1 | 1/1 | |
| A*0201− | 0 | |||||||
| OND (2) | HIV+ | 2 | ||||||
| A*0201+ | 1 | 1/1 | 0/1 | 0/1 | 0/1 | 0/1 | 1/1 | |
| A*0201− | 1 | 1/1 | 1/1 | 0/1 | 0/1 | 0/1 | 1/1 | |
| Total | HIV+ and HIV− | 20 | 8/20 | 6/20 | 5/20 | 4/20 | 4/20 | |
| A*0201+ | 10 | 5/10 | 2/10 | 2/10 | 1/10 | 1/10 | ||
| A*0201− | 10 | 3/10 | 4/10 | 3/10 | 3/10 | 3/10 | ||
In addition, JCV-specific CTL were also detected in one of one HIV+ possible-PML subject and two of two HIV+ OND subjects (Table 2). Among a total of 12 out of 20 subjects who exhibited a cellular immune response against VP1, 9 harbored CTL directed against more than one pool. Two HIV+ PML survivors had also CTL against two different epitopes contained in the same pool (pools I and II).
JCV sequences containing three amino acid changes in the VP1 protein compared to the reference MAD-1 have been found at a higher frequency in the brains of PML patients (1, 4). Therefore, we wanted to examine whether the apparent neurotropism of these JCV strains could be due to differences in the cellular immune response against their VP1 protein. The VP1pool II-stimulated PBMC of three patients who had a specific lysis against B-LCL pulsed with pool II, which encompasses the 20-mers 10, 10′, 11, and 11′, were tested against B-LCL pulsed with these different 20-mers separately. No specific lysis was detectable against any of these 20-mers. Moreover, VP1pool V-stimulated PBMC of three patients who had a specific lysis against B-LCL pulsed with pool V, which encompasses the 20-mers 29 and 29′, were assayed against B-LCL pulsed separately with the 20-mers 29 and 29′. There was no detectable specific lysis against these 20-mers either. These results suggest that these parts of the VP1 protein do not encompass significant immunogenic amino acid sequences.
Characterization of the HLA-A*0201-restricted epitope JCV VP1p36.
We decided to focus on HLA-A*0201-restricted epitopes, since this is the most frequent allele, present in ca. 40% of the North American population (8). To determine which epitope was restricted by the HLA-A*0201 allele, VP1pools-stimulated PBMC of HLA-A*0201+ subjects with detectable cytolytic activity against one or more peptide pools were assessed in a 51Cr release assay against HLA-A*0201 matched only or totally mismatched B-LCL. Pool I, which included seven 20-mers, elicited an HLA-A*0201-restricted CTL response. This pool was then broken into individual 20-mers which were assayed separately. The 20-mer “#3” elicited an HLA-A*0201-restricted response. This 20-mer contained an HLA-A*0201-restricted computer-predicted epitope that had not been synthesized in our previous study because it was not ranked among the 11 highest predicted epitopes (19). This 9-mer peptide, spanning aa 36 to 44 (SITEVECFL) of the VP1 protein (VP1p36), was synthesized and assessed for its capacity to be recognized by CTL of HLA-A*0201+ subjects with or without PML in a 51Cr release assay. Within #3, no other nonamer could fulfill the criteria for a potential A*0201-restricted CTL epitope (12, 16, 26, 30).
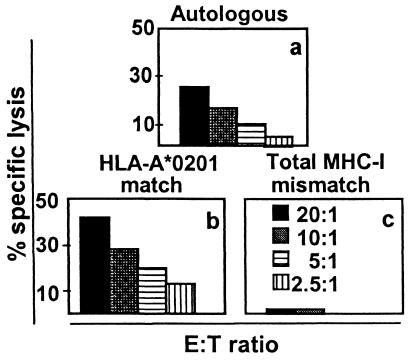
To confirm that effector cell recognition of the VP1p36 epitope was HLA-A*0201 restricted, B-LCL, which either (i) were autologous, (ii) shared only the A*0201 allele, or (iii) were fully MHC class I mismatched, were selected from a panel of previously characterized B-LCL, pulsed with VP1p36, and assessed as target cells in a standard 51Cr release assay. The autologous and allogeneic A*0201+ cells, but not the fully allogeneic target cells, were lysed by the A*0201+ effector cells. These experiments confirmed that the VP1p36-specific CTL were HLA-A*0201 restricted in their target cell recognition (Fig. 1).
FIG. 1.
CTL recognition of the JCV VP1p36 epitope SITEVECFL is HLA-A*0201 restricted. Autologous (a) and allogeneic HLA-A*0201-matched (b) target cells, but not fully MHC class I-mismatched target cells (c), pulsed with VP1p36 were lysed by JCV VP1p36-stimulated PBMC of an HIV+ PML survivor.
Pool II includes 20-mer #9 encompassing the previously described CTL epitope VP1p100. This pool elicited a strong cytolytic activity in the VP1pool II-stimulated PBMC of two HLA-A*0201+ patients, including one HIV+ PML survivor and one HIV+ possible-PML subject. Within pool II, no 20-mers other than #9 elicited any cytolytic activity. Within 20-mer #9, VP1p100 was the only nonamer that fulfilled the criteria for a potential A*0201-restricted CTL epitope (12, 16, 26, 30).
Detection of JCV VP1p36-specific CTL among VP1p36-stimulated PBMC of HLA-A*0201+ subjects.
To facilitate the detection and quantification of VP1p36-specific CTL in HLA-A*0201+ individuals, we constructed an HLA-A*0201/JCV VP1p36 tetramer. Ten fresh whole-blood samples and twenty-two VP1p36-stimulated PBMC specimens from HLA-A*0201+ study subjects were stained with the HLA-A*0201/JCV VP1p36 tetramer and then analyzed by flow cytometry, gating on the CD8+ CD3+ cells. Among these 22 subjects, 12 had their VP1p36-stimulated PBMC also tested for the presence of VP1p36-specific CTL by 51Cr release assay. VP1p36-stimulated PBMC of two additional subjects were tested by 51Cr release assay only (Table 3).
TABLE 3.
Detection of JCV VP1p36-specific effector cells in PBMC of HLA-A*0201+ subjects
| Diagnosis (n) | HIV status | No. of sub- jects | No. positive/total no. tested as determined witha:
|
||
|---|---|---|---|---|---|
| 51Cr | Fresh blood (TS) | Cultured cells (TS) | |||
| PML survivors (9) | HIV+ | 8 | 4/7 | 1/4 | 7/8 |
| HIV− | 1 | 0/1 | 0/1 | 1/1 | |
| PML progressors (6) | HIV+ | 3 | NA | NA | 0/3 |
| HIV− | 3 | NA | NA | 0/3 | |
| Possible PML (4) | HIV+ | 4 | 3/4 | 1/4 | 4/4 |
| OND (3) | HIV+ | 3 | 1/2 | 0/1 | 1/3 |
| Total | HIV+ and HIV− | 22 | 14 | 10 | 22 |
51Cr, 51Cr release CTL assay; TS, tetramer staining; NA, not applicable.
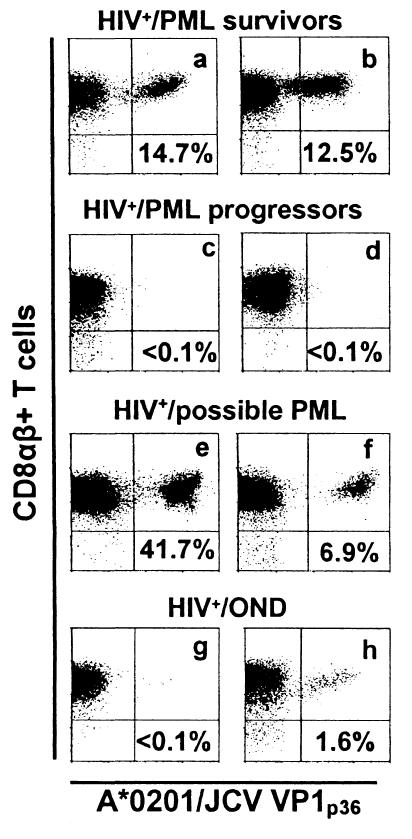
After in vitro VP1p36 stimulation, the lymphocytes of eight of nine PML survivors had between 0.1 and 18.5% (mean, 5.27%) VP1p36-specific CD8αβ+ T cells. However, tetramer staining could not be detected under the same conditions in the VP1p36-stimulated PBMC from six PML progressors (P = 0.0047). When the results of the 51Cr release assay on pooled peptides were combined with those of the tetramer staining assay, 10 of 11 PML survivors (91%) had evidence of JCV-specific CTL versus only 1 of 11 PML progressors (9%) (P = 0.0003). Tetramer binding was also detected in 1.8, 6.85, 7.43, and 41.7% of CD8αβ+ T cells from four of four HIV+ possible-PML individuals (mean, 14.4%). Finally, CTL were detectable in VP1p36-stimulated PBMC by tetramer staining in 1.6% of CD8αβ+ T cells from one of three HIV+ OND patients. This subject had also evidence of VP1p36-specific CTL as demonstrated by 51Cr release assay. A representative sample of the tetramer staining results is shown in Fig. 2.
FIG. 2.
Staining of JCV VP1p36-stimulated PBMC from HLA-A*0201+ individuals with the HLA-A*0201/JCV VP1p36 tetramer. The percentage of all CD8+ T cells that bind this tetramer is indicated in each panel. Tetramer-positive cells were detected in VP1p36-stimulated CD8+ T lymphocytes of HIV+ PML survivors (a and b), HIV+ possible-PML patients (e and f), and one HIV+ OND patient (h). However, negligible tetramer binding was seen in the CD8+ T lymphocytes of two HIV+ PML progressors (c and d) and an HIV+ OND patient (g). Cells were gated on CD3+ CD8+ lymphocytes. The results displayed represent staining with an anti-CD8αβ antibody and the HLA-A*0201/JCV VP1p36 tetramer.
To correlate the results of the 51Cr release and the tetramer staining assays, the percentage of specific lysis at an E:T ratio of 20:1 was compared to the percentage of tetramer staining CD8+ T cells in the same cell population in the VP1p36-stimulated PBMC of 12 HLA-A*0201+ study subjects. A linear correlation was observed (coefficient correlation, R = 0.72), suggesting that the tetramer staining and the functional lysis assays were measuring the same population of functionally active effector CTL (data not shown).
Frequency of JCV VP1pept-specific CTL in PBMC of one HIV+ PML survivor and two HIV+ possible-PML subjects.
To determine the frequency of JCV VP1pept-specific CTL in vivo, we used both FBTS and CTLS techniques. FBTS revealed few JCV VP1p36 and VP1p100 tetramer-positive cells in only one HIV+ PML survivor and JCV VP1p36 tetramer-positive cells in one HIV+ possible-PML subject (Table 4). Since the number of these tetramer-positive cells in vivo was at or even less than 0.1% of CD8αβ+ T cells, which is the lower limit of detection for this assay, we adapted a CTLS technique to confirm these results. A total of 2 × 107 to 5 × 107 fresh PBMC from the two patients mentioned above and an additional HIV+ possible-PML patient were stained directly with either the HLA-A*0201/JCV VP1p36 or VP1p100 PE-labeled tetramers and then sorted by using the AUTOMACS cell sorter and PE-labeled immunomagnetic beads (Table 4). These experiments showed 1 log of variability in the frequency of VP1p36-specific CTL between individuals, as measured by CTLS. The frequency of VP1p100-specific CTL was equivalent in two individuals and undetectable in the third. In addition, the frequency of epitope-specific CTL calculated by FBTS ranged from 0.1 to 1 log higher than when assessed by the CTLS technique. However, the CTLS technique allowed determination of the frequency of peptide-specific CTL in two HIV+ possible-PML subjects, even when FBTS was negative (Table 4). Therefore, the CTLS technique was more sensitive for detecting very rare events. Tetramer staining after in vitro stimulation with the respective epitope peptides is shown for these three patients in Fig. 2a, e, and f.
TABLE 4.
Determination of the frequency of JCV VP1p36- and VP1p100-specific CTL
| Method | Frequency (CTL/PBMC)a
|
|||||
|---|---|---|---|---|---|---|
| HIV+ PML survivor
|
HIV+ possible- PML 1
|
HIV+ possible-PML 2
|
||||
| p36 | p100 | p36 | p100 | p36 | p100 | |
| FBTS | 1/15,361 | 1/23,529 | 1/5,988 | - | NC | - |
| CTLS | 1/19,841 | 1/49,505 | 1/52,083 | 1/45,455 | 1/222,222 | - |
NC, not calculable because of nonhomgeneous tetramer population; -, negative.
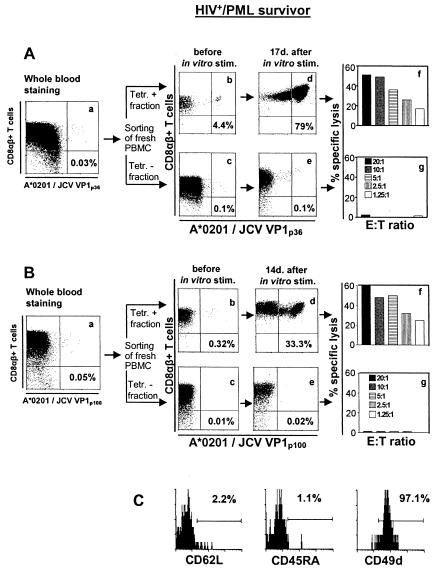
A detailed experiment comparing FBTS and CTLS techniques in an HIV+ PML survivor is shown in Fig. 3. FBTS performed in two separate experiments revealed that 0.03 and 0.05% of the CD8αβ+ T cells were JCV VP1p36 and JCV VP1p100 specific (Fig. 3Aa and Ba), corresponding to a frequency of 1/15,361 and 1/23,529 PBMC, respectively. After CTLS, 4.4% of the CD8αβ+ T cells were determined to be VP1p36+, whereas 0.32% of the CD8αβ+ T cells were VP1p100+ in the positive fraction (Fig. 3Ab and Bb) compared to negligible amounts in the negative fraction (Fig. 3Ac and Bc). To confirm that the tetramer-positive cells identified in vivo were functionally active CTL, we expanded the JCV VP1p36- and VP1p100-positive and -negative fractions. Sorted cells were stimulated with the VP1p36 or VP1p100 peptide at concentrations of 2.5 μg/ml for VP1p36 and 1 μg/ml for VP1p100. These concentrations were determined to yield the highest percentage of tetramer-staining cells for these two peptides (data not shown). Irradiated autologous feeder cells were added immediately after sorting, and human rIL-2 was added after 72 h. A massive expansion of VP1p36- or VP1p100-tetramer-staining cells was demonstrated after 2 weeks of stimulation in culture in the positive (Fig. 3Ad and Bd) but not in the negative (Fig. 3Ae and Be) sorted cell populations. The presence of functionally active effector cells in the positive (Fig. 3Af and Bf) but not the negative (Fig. 3Ag and Bg) sorted cell populations was further demonstrated in a 51Cr release assay.
FIG. 3.
Determination of the frequency and phenotype of JCV VP1p36- and -p100-specific CTL in an HIV+ PML survivor. Fresh blood was stained with either HLA-A*0201/JCV VP1p36 (Aa) or VP1p100 (Ba) PE-labeled tetramers. In addition, 5 × 107 fresh PBMC were stained with the same tetramers and sorted by using an AUTOMACS cell sorter with PE-labeled immunomagnetic beads. A positive (Ab and Bb) and a negative (Ac and Bc) fraction were collected and analyzed immediately after being sorted by flow cytometry. Sorted cells were stimulated in vitro in the presence of VP1p36 or VP1p100, feeder cells, and rIL-2 and then stained with their respective tetramers after 17 days (Ad and -e) or 14 days (Bd and -e). The percentage of all CD8+ T cells that bind the tetramers is indicated in each panel. These cells were then assessed for the presence of functionally active effector cells in a 51Cr release assay (Af and -g and Bf and -g). (C) Fresh PBMC were costained with the HLA-A*0201/JCV VP1p100 tetramer, sorted with immunomagnetic PE-labeled beads, and then stained with the activation markers CD62L, CD45RA, and CD49d. A total of 97% of the tetramer-positive cells were also CD49d positive compared to 2.2 and 1.1% that were positive for CD62L and CD45RA, respectively, indicating that these were activated memory cells. Tetr., tetramer.
Phenotype of JCV VP1p100-specific CTL in fresh blood.
To determine the phenotype of the VP1-specific CTL, 6 × 107 fresh PBMC from the HIV+ PML survivor described above were stained with the HLA-A*0201/JCV VP1p100 tetramer and sorted with PE-labeled immunomagnetic beads as described above. Cells were then divided into three groups and stained separately with the activation markers CD62L, CD45RA, and CD49d. CD49d was expressed on 97.1% of the tetramer-positive cells, whereas CD62L and CD45RA were present in only 2.2 and 1.1% of cells, respectively, indicating that the tetramer-positive cells were indeed activated memory cells (Fig. 3C).
DISCUSSION
One of our goals was to determine the frequency of JCV VP1-specific CTL in vivo. Therefore, we performed an epitope mapping study to design the most sensitive assay that could detect the presence of these CTL in the blood. Indeed, our previous approach by using a computerized predictive analysis proved to be of limited value since only 1 of 11 HLA-A*0201-restricted predicted epitopes, VP1p100, was recognized by the CTL (19).
Previous studies have shown that the eluted peptides from the HLA-A*0201 molecule were 9 aa long (12, 16). In another study, Parker et al. never found 8-mers but rarely found 10-mers. These 10-mers however had a lower affinity for the HLA-A*0201 molecule than the 9-mers (26). For these reasons, we focused our efforts on mapping 9-mer epitopes. The anchor positions of an HLA-A*0201-restricted nonamer epitope are at positions 2 and 9, which have a strong affinity for hydrophobic amino acids. The best consensus anchor residues for position 2 are leucine, isoleucine, or valine. Relaxed criteria include methionine and even the neutral amino acid threonine as alternate possibilities for this position. At position 9, the best anchor residues are leucine and valine, whereas isoleucine, alanine, and methionine may be alternate possibilities (12, 16, 26, 30). Based on these data, the 20-mers #3 and #9 could not contain any HLA-A*0201-restricted CTL epitopes other than VP1p36 SITEVECFL and VP1p100 ILMWEAVTL. For these reasons, it appears very unlikely that other HLA-A*0201-restricted epitopes are present in the VP1 protein.
The mapping study also allowed us to examine whether changes in the cellular immune response against VP1 protein could explain the differences in neurotropism attributed to some JCV subtypes. Indeed, JCV subtype 2B, which encompasses two amino acid changes in the VP1 protein compared to MAD-1 sequence, was found with a higher frequency in the brain of PML patients than in samples of urine of healthy individuals (P < 0.001) (1). Another amino acid change in the VP1 protein, compared to MAD-1, has been reported in the brains of 11 of 11 PML patients (4). Our results show that, in the 20-mers encompassing either the amino acid changes or the corresponding MAD-1 amino acid sequences, no epitopes were recognized by the CTL of any of the 17 PML patients who were included in the mapping study. Therefore, it seems unlikely that the apparent neurotropism of JCV strains containing these amino acid changes can be explained by differences in CTL responses against the VP1 protein.
Determination of the frequency of rare CTL in vivo is a challenging task. Tetramer staining in fresh blood would be a simple and rapid way to achieve this goal. However, the lower limit of detection of this technique is 0.1% of CD8αβ+ T cells. In a previous study, the amount of JCV VP1p100-specific CTL, as determined by FBTS, was always at or below the lower limit of detection of this assay, and the results were therefore considered negative (19). In the present study, some patients had rare JCV VP1p36- or VP1p100-specific CTL as determined by FBTS. To determine whether these were true effector cells, we adapted a CTLS technique with tetramer and immunomagnetic beads. The magnetic bead separation technique has been shown to be an efficient way to isolate tetramer-positive virus-specific CD8+ T cells that are readily detectable in nonstimulated PBMC (21). This CTLS technique confirmed that the cells detected by FBTS were indeed functionally active CTL specific for JCV VP1p36 and JCV VP1p100. Interestingly, CTLS appeared to be a more sensitive assay since it allowed us to determine the frequency of JCV VP1pept in two cases in which FBTS was negative. However, the frequency of JCV VP1pept as determined by CTLS was lower than that determined by FBTS (Table 4). This is probably explained by the fact that some tetramer-positive cells are retained in the columns of the AUTOMACS cell sorter. As additional evidence that these rare CTL could not have been generated de novo by mere in vitro stimulation with VP1pept, we characterized the phenotype of VP1p100-specific CTL in an HIV+ PML survivor in vivo and showed that these were indeed activated memory cells (Fig. 3C).
Combining the results of the 51Cr release and tetramer assays, CTL were detected in 10 of 11 PML survivors but in only 1 of 11 PML progressors (P = 0.0003). The demonstration that these JCV VP1-specific CTL are activated memory cells further underscores the importance of this cellular immune response in the prevention of disease progression (11, 15, 18, 19, 33). A strong CTL activity was detected in the VP1pools-stimulated PBMC of two of two HIV− patients who had been diagnosed recently with PML and who were improving clinically. In addition, four of four HIV+ possible-PML patients had JCV-specific CTL. Interestingly, these four patients were also tested sooner after the diagnosis of their leukoencephalopathy (16.5 months) than were HIV+ PML survivors (54.8 months, P = 0.0178). Moreover, the two HIV+ possible-PML subjects who had detectable CTL in fresh blood (Table 4) also had the shortest interval between the diagnosis and the assay (4.45 months). Together with the fact that a strong CTL response could be detected in two PML patients in the early stage of the disease, this suggests that the CTL response is an early and decisive event in the course of PML. This early CTL appearance seems to be associated with a favorable outcome and, possibly, with clearance of JCV from the CSF. Further studies that include larger numbers of such early-PML patients are warranted to confirm this observation.
The frequency of the CTL specific for the two HLA-A*0201-restricted epitopes that we identified is low compared to other DNA viruses such as EBV, CMV, and HSV. It has been demonstrated that CTL play an important role in the immune response against these viruses (7, 27, 29). In patients with genital herpes, the frequency of HSV-specific CTL ranged from 1/300 to 1/21,000 PBMC, as shown by limiting-dilution assay (LDA) (27). The frequency of CTL directed against epitopes restricted by different HLA alleles was compared by three different assays in healthy EBV carriers. The frequency of epitope-specific CTL calculated by LDA was between 1/100,000 and 1/1,379 PBMC. The enzyme-linked immunospot (ELISPOT) assay gave a frequency of virus-specific CTL 4.4 times higher than the LDA and, in turn, the tetramer-staining assay was 5.3 more sensitive than the ELISPOT assay for detecting these CTL. As measured by the tetramer-staining assay, the frequency of CTL specific for the same epitopes ranged from 1/5,600 to 1/88 PBMC. In only 1 of 13 cases were CTL not detectable by tetramer-staining assay and yet detectable by LDA (32). Tetramer staining done in a CMV-seropositive subject showed that the frequency of CTL specific for an immunodominant epitope of CMV was in the same range (20). The frequency of CMV-specific CTL was lower when estimated by LDA in two different studies, between 1/685 and 1/100,000 PBMC (6, 35). Compared to results obtained with the tetramer assay for EBV- and CMV-specific CTL, the frequency of JCV VP1-specific CTL was lower by ∼1 log. Yet, even in such low numbers, JCV-specific CTL seem to play a crucial role in limiting the progression of JCV-associated brain lesions in PML patients. JCV infects more than 85% of the healthy adult population. By analogy with other DNA viruses, such as EBV, CMV, and HSV, the presence of JCV-specific CTL might help protect individuals from developing PML, as is suggested by the detection of JCV-specific CTL in two of four HIV+ OND subjects. We are currently investigating whether such CTL are also present in healthy individuals.
Acknowledgments
This work was supported by Public Health Service grant RO1 NS/AI 41198, Dana-Farber Cancer Institute-Beth Israel Deaconess Medical Center-Children's Hospital Center for AIDS Research grant P30-AI28691, and a Milton Fund grant to I.J.K. R.A.D.P. is the recipient of a fellowship for advanced researcher from the Swiss National Science Foundation and a grant from the Eugenio Litta Foundation.
REFERENCES
- 1.Agostini, H. T., C. F. Ryschkewitsch, E. J. Singer, R. W. Baumhefner, and G. L. Stoner. 1998. JC virus type 2B is found more frequently in brain tissue of progressive multifocal leukoencephalopathy patients than in urine from controls. J. Hum. Virol. 1:200-206. [PubMed] [Google Scholar]
- 2.Altman, J. D., P. A. H. Moss, P. J. R. Goulder, D. H. Barouch, M. G. McHeyzer-Williams, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94-96. [DOI] [PubMed] [Google Scholar]
- 3.Ammassari, A., A. Cingolani, P. Pezzotti, D. A. De Luca, R. Murri, M. L. Giancola, L. M. Larocca, and A. Antinori. 2000. AIDS-related focal brain lesions in the era of highly active antiretroviral therapy. Neurology 55:1194-1200. [DOI] [PubMed] [Google Scholar]
- 4.Ault, G. S., and G. L. Stoner. 1992. Two major types of JC virus defined in progressive multifocal leukoencephalopathy brain by early and late coding region DNA sequences. J. Gen. Virol. 73:2669-2678. [DOI] [PubMed] [Google Scholar]
- 5.Berger, J. R., R. M. Levy, D. Flomenhoft, and M. Dobbs. 1998. Predictive factors for prolonged survival in acquired immunodeficiency syndrome-associated progressive multifocal leukoencephalopathy. Ann. Neurol. 44:341-349. [DOI] [PubMed] [Google Scholar]
- 6.Boppana, S. B., and W. J. Britt. 1996. Recognition of human cytomegalovirus gene products by HCMV-specific cytotoxic T cells. Virology 222:293-296. [DOI] [PubMed] [Google Scholar]
- 7.Borysiewicz, L. K., S. Morris, J. D. Page, and J. G. Sissons. 1983. Human cytomegalovirus-specific cytotoxic T lymphocytes: requirements for in vitro generation and specificity. Eur. J. Immunol. 13:804-809. [DOI] [PubMed] [Google Scholar]
- 8.Cao, K., J. Hollenbach, X. Shi, W. Shi, M. Chopek, and M. A. Fernandez-Vina. 2001. Analysis of the frequencies of HLA-A, -B, and -C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 62:1009-1030. [DOI] [PubMed] [Google Scholar]
- 9.Cinque, P., C. Pierotti, M. G. Vigano, A. Bestetti, C. Fausti, D. Bertelli, and A. Lazzarin. 2001. The good and evil of HAART in HIV-related progressive multifocal leukoencephalopathy. J. Neurovirol. 7:358-363. [DOI] [PubMed] [Google Scholar]
- 10.Cinque, P., I. J. Koralnik, and D. Clifford. 2003. The evolving face of human immunodeficiency virus-related progressive multifocal leukoencephalopathy: defining a consensus for terminology. J. Neurovirol. 9:88-92. [DOI] [PubMed] [Google Scholar]
- 11.Du Pasquier, R. A., K. W. Clark, P. S. Smith, J. T. Joseph, J. M. Mazullo, U. De Girolami, N. L. Letvin, and I. J. Koralnik. 2001. Favorable clinical outcome in HIV-infected individuals with progressive multifocal leukoencephalopathy correlates with JCV-specific cellular immune response. J. Neurovirol. 7:318-322. [DOI] [PubMed] [Google Scholar]
- 12.Falk, K., O. Rotzschke, S. Stevanovic, G. Jung, and H. G. Rammensee. 1991. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351:290-296. [DOI] [PubMed] [Google Scholar]
- 13.Frisque, R. J., G. L. Bream, and M. T. Cannella. 1984. Human polyomavirus JC virus genome. J. Virol. 51:458-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garboczi, D. N., D. T. Hung, and D. C. Wiley. 1992. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. USA 89:3429-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasnault, J., M. Kahraman, M. De Goer de Herve, J. Delfraissy, and Y. Taoufik. 2002. In HIV+ patients with progressive multifocal leukoencephalopathy, anti-JC virus CD4 T-cell responses are recovered following prolonged HAART. Fourth International Symposium on Neurovirology and 10th Conference on Neuroscience of HIV Infection, Dusseldorf, Germany.
- 16.Hunt, D. F., R. A. Henderson, J. Shabanowitz, K. Sakaguchi, H. Michel, N. Sevilir, A. L. Cox, E. Appella, and V. H. Engelhard. 1992. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255:1261-1263. [DOI] [PubMed] [Google Scholar]
- 17.Koralnik, I. J., D. Boden, V. X. Mai, C. I. Lord, and N. L. Letvin. 1999. JC virus DNA load in patients with or without progressive multifocal leukoencephalopathy. Neurology 52:253-260. [DOI] [PubMed] [Google Scholar]
- 18.Koralnik, I. J., R. A. Du Pasquier, and N. L. Letvin. 2001. JC virus-specific cytotoxic T lymphocytes in individuals with progressive multifocal leukoencephalopathy. J. Virol. 75:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koralnik, I. J., R. A. Du Pasquier, M. Kuroda, J. E. Schmitz, X. Dang, Y. Zheng, M. Lifton, and N. L. Letvin. 2002. Association of prolonged survival in HLA-A2+ progressive multifocal leukoencephalopathy patients with a cytotoxic T lymphocyte response specific for a dominant JC virus epitope. J. Immunol. 168:499-504. [DOI] [PubMed] [Google Scholar]
- 20.Kuzushima, K., N. Hayashi, H. Kimura, and T. Tsurumi. 2001. Efficient identification of HLA-A*2402-restricted cytomegalovirus-specific CD8+ T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood 98:1872-1881. [DOI] [PubMed] [Google Scholar]
- 21.McDermott, A. B., H. M. Spiegel, J. Irsch, G. S. Ogg, and D. F. Nixon. 2001. A simple and rapid magnetic bead separation technique for the isolation of tetramer-positive virus-specific CD8 T cells. AIDS 15:810-812. [DOI] [PubMed] [Google Scholar]
- 22.Miller, M. D., C. I. Lord, V. Stallard, G. P. Mazzara, and N. L. Letvin. 1990. The gag-specific cytotoxic T lymphocytes in rhesus monkeys infected with the simian immunodeficiency virus of macaques. J. Immunol. 144:122-128. [PubMed] [Google Scholar]
- 23.Miltenyi, S., W. Muller, W. Weichel, and A. Radbruch. 1990. High gradient magnetic cell separation with MACS. Cytometry 11:231-238. [DOI] [PubMed] [Google Scholar]
- 24.Oxenius, A., D. A. Price, P. J. Easterbrook, C. A. O'Callaghan, A. D. Kelleher, J. A. Whelan, G. Sontag, A. K. Sewell, and R. E. Phillips. 2000. Early highly active antiretroviral therapy for acute HIV-1 infection preserves immune function of CD8+ and CD4+ T lymphocytes. Proc. Natl. Acad. Sci. USA 97:3382-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padgett, B. L., D. L. Walker, G. M. ZuRhein, R. J. Eckroade, and B. H. Dessel. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet i:1257-1260. [DOI] [PubMed]
- 26.Parker, K. C., M. A. Bednarek, L. K. Hull, U. Utz, B. Cunningham, H. J. Zweerink, W. E. Biddison, and J. E. Coligan. 1992. Sequence motifs important for peptide binding to the human MHC class I molecule, HLA-A2. J. Immunol. 149:3580-3587. [PubMed] [Google Scholar]
- 27.Posavad, C. M., D. M. Koelle, and L. Corey. 1996. High frequency of CD8+ cytotoxic T-lymphocyte precursors specific for herpes simplex viruses in persons with genital herpes. J. Virol. 70:8165-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Power, C., J. G. Gladden, W. Halliday, M. R. Del Bigio, A. Nath, W. Ni, E. O. Major, J. Blanchard, and M. Mowat. 2000. AIDS- and non-AIDS-related PML association with distinct p53 polymorphism. Neurology 54:743-746. [DOI] [PubMed] [Google Scholar]
- 29.Rickinson, A. B., D. J. Moss, D. J. Allen, L. E. Wallace, M. Rowe, and M. A. Epstein. 1981. Reactivation of Epstein-Barr virus-specific cytotoxic T cells by in vitro stimulation with the autologous lymphoblastoid cell line. Int. J. Cancer 27:593-601. [DOI] [PubMed] [Google Scholar]
- 30.Ruppert, J., J. Sidney, E. Celis, R. T. Kubo, H. M. Grey, and A. Sette. 1993. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell 74:929-937. [DOI] [PubMed] [Google Scholar]
- 31.Seth, A., J. Markee, A. Hoering, A. Sevin, D. E. Sabath, J. E. Schmitz, M. J. Kuroda, M. A. Lifton, M. S. Hirsch, A. C. Collier, N. L. Letvin, and M. J. McElrath. 2001. Alterations in T-cell phenotype and human immunodeficiency virus type 1-specific cytotoxicity after potent antiretroviral therapy. J. Infect. Dis. 183:722-729. [DOI] [PubMed] [Google Scholar]
- 32.Tan, L. C., N. Gudgeon, N. E. Annels, P. Hansasuta, C. A. O'Callaghan, S. Rowland-Jones, A. J. McMichael, A. B. Rickinson, and M. F. Callan. 1999. A re-evaluation of the frequency of CD8+ T cells specific for EBV in healthy virus carriers. J. Immunol. 162:1827-1835. [PubMed] [Google Scholar]
- 33.Weber, F., C. Goldmann, M. Kramer, F. J. Kaup, M. Pickhardt, P. Young, H. Petry, T. Weber, and W. Luke. 2001. Cellular and humoral immune response in progressive multifocal leukoencephalopathy. Ann. Neurol. 49:636-642. [PubMed] [Google Scholar]
- 34.Weber, T., C. Trebst, S. Frye, P. Cinque, L. Vago, C. J. Sindic, W. J. Schulz-Schaeffer, H. A. Kretzschmar, W. Enzensberger, G. Hunsmann, and W. Luke. 1997. Analysis of the systemic and intrathecal humoral immune response in progressive multifocal leukoencephalopathy. J. Infect. Dis. 176:250-254. [DOI] [PubMed] [Google Scholar]
- 35.Wills, M. R., A. J. Carmichael, K. Mynard, X. Jin, M. P. Weekes, B. Plachter, and J. G. Sissons. 1996. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 70:7569-7579. [DOI] [PMC free article] [PubMed] [Google Scholar]