Abstract
Canine parvovirus (CPV) and feline panleukopenia virus (FPV) differ in their ability to infect dogs and dog cells. Canine cell infection is a specific property of CPV and depends on the ability of the virus to bind the canine transferrin receptor (TfR), as well as other unidentified factors. Three regions in the capsid structure, located around VP2 residues 93, 300, and 323, can all influence canine TfR binding and canine cell infection. These regions were compared in the CPV and FPV capsid structures that have been determined, as well as in two new structures of CPV capsids that contain substitutions of the VP2 Asn-93 to Asp and Arg, respectively. The new structures, determined by X-ray crystallography to 3.2 and 3.3 Å resolutions, respectively, clearly showed differences in the interactions of residue 93 with an adjacent loop on the capsid surface. Each of the three regions show small differences in structure, but each appears to be structurally independent of the others, and the changes likely act together to affect the ability of the capsid to bind the canine TfR and to infect canine cells. This emphasizes the complex nature of capsid alterations that change the virus-cell interaction to allow infection of cells from different hosts.
The Parvoviridae are spherical, nonenveloped, T=1 icosahedral viruses that infect a wide range of natural hosts, including humans, monkeys, pigs, dogs, cats, mink, and mice. These viruses cause a variety of serious diseases, especially in the young of the species that they infect. Parvoviral capsids are ∼260 Å in diameter and encapsidate a single-stranded DNA genome of ∼5,000 bases. The capsid is assembled from a total of 60 copies of two overlapping structural proteins, VP1 and VP2, where VP1 contains an additional N-terminal domain of 143 amino acid residues in canine parvovirus (CPV) and 227 residues in the human parvovirus B19.
The high-resolution X-ray structures of DNA-containing (full) and empty capsids of several members of the Parvoviridae, as well as those of mutant capsids, have been determined (1-3, 21, 32, 33, 39, 43; Y. Tao, M. Agbandje-McKenna, C. R. Parrish, and M. G. Rossmann, unpublished data). The VP2 common region is made up of a core eight-stranded β-barrel domain, with two-thirds of the polypeptide sequence being loop insertions between the strands of the β-barrel. The loop insertions form elaborate decorations on the surface of the capsids and result in large spike protrusions at or surrounding the icosahedral threefold axes (1, 3, 33, 39, 43). Other prominent features of the parvoviral capsid include a canyon-like depression that encircles the icosahedral fivefold axes and a dimple-like depression at the twofold axes. Although the conserved β-barrel core forms the contiguous parvoviral capsid, surface variations control many biological differences, including tissue tropism, pathogenicity, and antigenicity, both among members and between strains of the same virus (1-3, 5, 33, 43).
CPV is of particular interest for host range studies since it emerged in the 1970s as a host range variant of feline panleukopenia virus (FPV) (31). FPV naturally infects cats and some carnivores but not dogs, whereas the original 1978 strain of CPV (CPV type 2) infects dogs and feline cells in vitro but does not infect cats (14, 38). Within 3 years of first emerging, the CPV type 2 strain was replaced in nature by an antigenically variant strain designated CPV type 2a, which had gained the ability to infect cats (30). Comparison of the CPV and FPV amino acid sequences showed that they contain several amino acid differences within the overlapping VP1 and VP2 sequences, a number of which determine their tissue tropic phenotype. The amino acid type(s) at VP2 residues 93 and 323 have been shown to be the most important in controlling CPV host range and a CPV-specific antigenic site on the capsids (9, 15). Residues 80, 564, and 568 are important for efficient viral replication in cats (37, 38). In addition to these residues that differ between FPV and CPV, surface residues in the “shoulder” region of the threefold spikes adjacent to VP2 residue 300 (the “300 region”), which are the same in the two viruses, also play a role in CPV host range determination (28, 37). The 300 region is structurally proximal to residues 80, 564, and 568. The high-resolution structures of CPV and two site-directed mutants, CPV-A300D and CPV-N93K (21, 39; Tao et al., unpublished), and that of FPV (1) shows that small surface differences at or near these capsid amino acids are associated with the observed in vitro tissue tropism and in vivo pathogenic properties of the viruses.
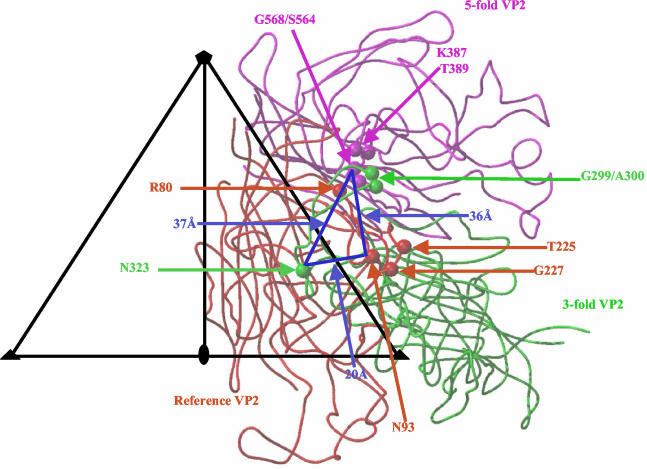
CPV and FPV bind to the feline transferrin receptor (TfR) and use that receptor to infect feline cells (27). The canine TfR controls the host range for canine cells by binding specifically to CPV but not to FPV capsids (15). To analyze the effect of the host range controlling amino acids and adjacent regions on receptor binding and canine cell infection, CPV and FPV capsid mutants were prepared with changes at residues 93 and 323, a surface loop adjacent to residue 93 (referred to as loop 2) and in the 300 region (15, 16). Binding studies with these mutants showed that, in the FPV background, changing both residues 93 and 323 together to Asn (the CPV amino acid type) allowed the virus to bind the canine TfR and infect dog cells but that individual changes of either residue 93 or 323 did not allow either binding or canine cell infection (16). Canine TfR binding and canine cell infection was also affected by changes in the 300 region, as exemplified by a change of VP2 residue 299 from Gly to Glu and of VP2 residue 300 from Ala to Asp (15, 28). Interestingly, the CPV type 2a strain contained several amino acid substitutions within and close to the 300 region, including changes of residues 87 (Met to Leu), 300 (Ala to Gly), and 305 (Asp to Tyr) (29, 30, 37). Thus, in CPV capsids the three different regions that control host range also control canine TfR binding (15, 16, 28). The capsid regions containing these residues are separated from each other by ca. 20 to 37 Å (Fig. 1) and suggest contact with a large area on the receptor molecule. This possibility is consistent with mutational analysis of the canine and feline TfRs that show that three sites within the apical domains can affect capsid binding and cell infection by CPV and FPV (26).
FIG. 1.
Receptor attachment requirements displayed on the CPV capsid surface. The viral icosahedral asymmetric unit (triangle bounded by two threefold axes [filled triangles] and a fivefold axis [filled pentagon] divided by a line drawn to the twofold axis [filled oval]) showing the reference (in red), and threefold (in green) and fivefold (in magenta) related VP2s. Residues 80, 93, 225, and 227 are highlighted in the reference VP2. Residues 299, 300, and 323 are highlighted in the threefold VP2. Residues 387, 389, 564, and 568 are highlighted in the fivefold VP2. The blue triangle connects residues (93, 323, and 568) in the three regions involved in receptor binding; distance measurements between them are indicated.
Residue 93 also forms a CPV-specific antigenic epitope (9). In all of the available structures of FPV and in the CPV-N93K mutant structure, the K93 forms two hydrogen bonds with main chain residues in loop 2 (containing residues 222 to 230), and these bonds cannot be formed by the Asn normally found in that position of CPV (1, 32; Tao et al., unpublished). The K93 residue in CPV-N93K or in the wild-type (wt) FPV changes the topology of this capsid region compared to CPV, pushing loop 2 away from the main chain of residue 93. It has been postulated that the host range effects of residue 93 and its role in a CPV-specific antigenic epitope is due to specific surface properties conferred by the presence of the asparagine residue (1, 16). We determined here the structures of two additional mutants of CPV—CPV-N93D and CPV-N93R—in order to further define the role(s) of amino acids with different interactive capabilities at position 93 on the functioning of the capsids. The phenotypes of these CPV mutants with respect to host cell binding, infectivity, and antigenicity have been reported elsewhere (16) and were utilized in our discussions. The new mutant structure were also compared to those of several previous structures of wt CPV and FPV capsids, as well as the structures of CPV mutants or of capsids determined under different pH conditions or in the absence of calcium ions (1, 21, 32; Tao et al., unpublished). The results show that small changes in the capsid structure at three separate sites on a raised region likely act together to control interactions with the host receptor and that two of those sites are also recognized by neutralizing monoclonal antibodies (MAbs) (34), showing a coincidence between receptor and antibody binding sites in this viral system.
MATERIALS AND METHODS
Cells and viruses.
NLFK cells were cultured in a 50% mixture of McCoy's A5 and Leibovitz L15 media with 5% fetal bovine serum. The CPV mutants, CPV-N93D and CPV-N93R, were prepared by site-directed mutagenesis of M13 single-stranded DNA (16). A fragment from PstI (nucleotide 3059) to EcoRV (nucleotide 40011) was cloned back into the infectious plasmid clones of CPV (29). Plasmids were sequenced to confirm the changes made and then transfected into NLFK cells to prepare the viruses. Infected cells were harvested, and the virus samples were purified by using polyethylene glycol (PEG) precipitation and sucrose gradient banding as previously described (1) and dialyzed against 20 mM Tris-HCl for crystallization.
Crystallization, X-ray data collection, and processing.
Crystals of empty particles of CPV-N93D and full DNA containing particles of CPV-N93R were grown at 20°C with 1% PEG 8000 and 8 mM CaCl2 in 20 mM Tris-HCl (pH 7.5) by using the sitting-drop vapor diffusion technique. The crystals were cryoprotected with 30% (vol/vol) ethylene glycol and 5% PEG 8000 in 20 mM Tris-HCl (pH 7.5) containing 8 mM CaCl2 and then flash frozen in liquid nitrogen vapor prior to data collection. X-ray diffraction data sets for both mutants were collected at the F1 station of the Cornell High Energy Synchrotron Source. For CPV-N93D, a total of 492 images were collected, from 15 crystals, by using a 0.1-mm-diameter collimator, at a wavelength of λ = 0.938 Å, on an ADSC Quantum 4 charge-coupled device detector. The crystal-to-detector distance was 300 mm, the oscillation angle was 0.25°, and the exposure time for each image was 60 s. A total of 498 images were collected for CPV-N93R on an ADSC Quantum 210 charge-coupled device detector, with a crystal-to-detector distance of 250 mm, by using a 0.1 mm collimator, with an oscillation angle of 0.25° for each image. The wavelength was 0.950 Å. The diffraction images for both mutants were initially indexed by using the program DENZO, and the cell parameters were further refined by using the program SCALEPACK (25). Interestingly, CPV-N93D and CPV-N93R each crystallized in different crystal systems, neither of which was isomorphous with those of previous CPV or FPV crystals (1, 32, 40). CPV-N93D crystallized in a monoclinic C2 space group, with the unit cell parameters a = 440.448, b = 246.814, c = 443.877 Å and β = 93.54°, whereas CPV-N93R was in an orthorhombic P212121 space group, with the unit cell parameters a = 372.417, b = 373.017, c = 377.084 Å (see Table 1 for data statistics). The calculated Matthews coefficients (22) were 3.0 Å3/Da for CPV-N93D and 3.3 Å3/Da for CPV-N93R, corresponding to solvent contents of 57.5 and 61%, respectively. Both mutant crystal forms had one particle present in their asymmetric unit. The observed intensities for each mutant were integrated, scaled, and reduced by using the HKL suite programs DENZO and SCALEPACK (25) and converted into structure factor amplitudes by using the CCP4 program TRUNCATE (8). Both data sets were scaled to 3.2 Å (CPV-N93D) and 3.3 Å (CPV-N93R) resolution with Rsym of 14.3% (61.2% completeness) and 13.1% (70.5% completeness), respectively. The data collection and processing statistics are presented in Table 1.
TABLE 1.
Diffraction data processing statistics
| CPV-N93D diffraction data
|
CPV-N93R diffraction data
|
|||||
|---|---|---|---|---|---|---|
| Resolution range (Å) | Rsyma | Completeness (%) | Resolution range (Å) | Rsyma | Completeness (%) | |
| 25.00-6.85 | 0.092 | 68.0 | 10.00-6.48 | 0.096 | 69.0 | |
| 6.85-5.46 | 0.119 | 71.3 | 6.48-5.40 | 0.107 | 75.4 | |
| 5.46-4.77 | 0.134 | 69.8 | 5.40-4.80 | 0.114 | 74.3 | |
| 4.77-4.34 | 0.142 | 64.6 | 4.80-4.40 | 0.120 | 70.8 | |
| 4.34-4.03 | 0.155 | 60.3 | 4.40-4.11 | 0.133 | 71.2 | |
| 4.03-3.79 | 0.173 | 56.7 | 4.11-3.88 | 0.149 | 72.5 | |
| 3.79-3.60 | 0.190 | 55.7 | 3.88-3.70 | 0.164 | 72.4 | |
| 3.60-3.45 | 0.217 | 55.4 | 3.70-3.54 | 0.179 | 72.1 | |
| 3.45-3.31 | 0.261 | 55.1 | 3.54-3.41 | 0.191 | 66.0 | |
| 3.31-3.20 | 0.297 | 54.8 | 3.41-3.30 | 0.198 | 61.2 | |
| Overall | 0.143 | 61.2 | Overall | 0.131 | 70.5 | |
Rsym = Σhkl|Ihkl − <Ihkl>|/ Σhkl<Ihkl>, where Ihkl is the measured intensity for reflection hkl and <Ihkl> is the mean of the intensities of all observations of reflection hkl.
Structure solution and refinement.
The structures of CPV-N93D and CPV-N93R were determined by the molecular replacement method with the CCP4 program AMoRe (24) by using the coordinates of the FPV VP2 (PDB: 1C8E) (32) as a search model. A self-rotation function was calculated for CPV-N93D and CPV-N93R by using the GLRF program (35) to verify that the five-, three-, and twofold axes were consistent with icosahedral symmetry. The FPV VP2 model was expanded to 60 subunits in a standard orientation by using icosahedral symmetry operators and used to calculate structure factors in the 10- to 5-Å resolution range for cross-rotation and translation function calculations in AMoRe. The highest peak obtained for the cross-rotation function was used in a translation function calculation. The translation search was carried out by using the structure factor correlation-coefficient function as a key parameter in the AMoRe program, which gave a clear highest peak with a correlation coefficient (CC) of 56.4% and an R-factor (Σhkl| |Fo|−|Fc| |/Σhkl|Fo|) of 36.8% for CPV-N93D and a CC of 59.0% and an R-factor of 35.2% for CPV-N93R. A further rigid-body refinement was carried out with the FITING function in AMoRe, which improved the CC to 67.1% and the R-factor to 32.0% for CPV-N93D and the CC to 69.6% and the R-factor to 30.6% for CPV-N93R. The FPV particle was then rotated and translated into the unit cells of the mutants according to the final solutions obtained, with an orientation, in Eulerian angles, of 353.94, 50.05, and 308.08° and a fractional coordinate position of 0.2353, 0.000, and 0.2488 for CPV-N93D and an orientation, in Eulerian angles, of 155.35, 72.65, and 205.59° and a fractional coordinate position of 0.4967, 0.0025, and 0.4949 for CPV-N93R.
The models were improved by iterative cycles of refinement and rebuilding, constrained with 60-fold noncrystallographic symmetry (NCS) operators generated by the CNS program (7). The correctly oriented and positioned models of CPV-N93D and CPV-N93R were subjected to rigid-body refinement with the CNS program (7) for all reflections up to a 4-Å resolution, resulting in R-factors of 0.29 and 0.26, respectively. Further crystallographic refinement, with all data to 3.2 and 3.3 Å resolutions for CPV-N93D and CPV-N93R, respectively, included simulated annealing (at 2,000 K) with torsion angle molecular dynamics, restrained individual B-factor refinement, conjugate gradient minimization, and bulk solvent correction against the maximum-likelihood target function. The model was examined at each cycle of the refinement by visual inspection of the 2Fo-Fc and Fo-Fc electron density maps by using the program O, and the refinement was interspersed by manual rebuilding of the model, also by using O (17, 18). Several cycles of refinement (using all reflections) and model rebuilding were performed until the R-factor for the model could not be improved any further. During the final stages of refinement solvent water molecules were incorporated into the model only if they were at hydrogen bond-forming distances from the appropriate protein atoms. 2Fo-Fc maps were also used to check the consistency in the peaks. To improve the quality of the density maps for model rebuilding, molecular averaging was carried out by using the CNS program, with 60 NCS operators. The initial NCS-averaging mask covering one VP2 monomer and the solvent-flattening mask, also covering one VP2 monomer, was generated with the CNS program. The CPV-N93D and CPV-N93R structure models were refined to final R-factors of 0.252 (Rfree = 0.253) (6) and 0.200 (Rfree = 0.202), respectively (Table 2).
TABLE 2.
Refinement statistics for host-range mutant models
| Statistic | Host range mutant
|
|
|---|---|---|
| CPV-N93D | CPV-N93R | |
| Resolution (Å) | 25-3.2 | 10-3.3 |
| Space group/crystal system | C2/monoclinic | P212121/orthorhombic |
| Rfactor (%)a | 0.252 | 0.200 |
| Rfree (%)b | 0.253 | 0.202 |
| No. of reflections | 8,508,415 | 5,882,971 |
| No. of independent reflections | 474,357 | 448,898 |
| No. of protein atoms | 4,329 | 4,322 |
| No. of solvent molecules | 65 | 84 |
| RMSD, bond length (Å) | 0.009 | 0.008 |
| RMSD, bond angle (°) | 1.52 | 1.42 |
| Avg B factor, main chain (Å2) | 37.40 | 31.67 |
| Avg B factor, side chain (Å2) | 38.08 | 31.58 |
| Avg B factor for protein/water (Å2) | 37.74/36.96 | 31.63/32.36 |
| Residues in the most/additional allowed regions (%) | 83/17 | 84/16 |
Rfactor = Σhkl| |Fo|−|Fc| |/∑hkl|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
Rfree is the same as Rfactor but is calculated with a 5% randomly selected fraction of the reflection data not included in the refinement (6).
The stereochemistry of the refined structures was analyzed by using PROCHECK (19), which showed no residues in disallowed regions of the Ramachandran plot. Final refinement statistics are given in Table 2. The atomic coordinates of the capsid mutants have been deposited (1P5w and 1P5y) in the Protein Data Bank (http://www.rcsb.org). Figures were generated with several combined uses of BOBSCRIPT (11) and Raster3D (23).
RESULTS AND DISCUSSION
Here we report the structures of mutants of the CPV type 2 strain containing substitutions of VP2 residue 93 from Asn to Asp (CPV-N93D) and Arg (CPV-N93R), determined to 3.2- and 3.3-Å resolutions, respectively, by using molecular replacement. Although these capsids each differed at only one amino acid position in their primary sequence, they crystallized in different space groups under the same crystallization condition, a feature of the parvoviruses that suggests that many different crystallization contacts can be made between the capsids (1, 32).
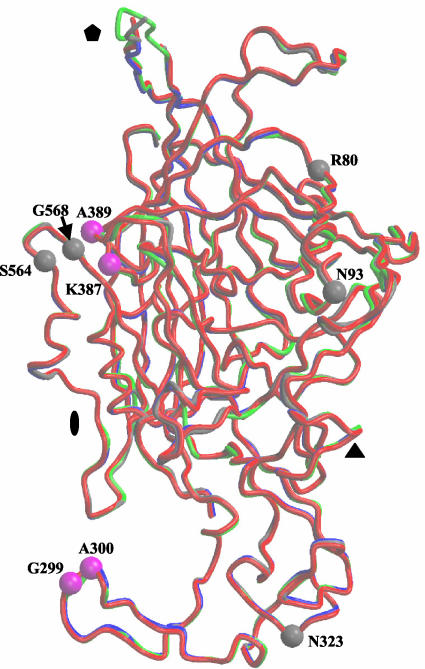
The Cα backbones of wt CPV, wt FPV, and the two mutant CPVs could be superimposed onto each other with a root-means-square deviation (RMSD) of <0.5 Å (Fig. 2). However, some of the surface loop regions connecting the strands that form the core capsid showed differences of ≥1.0 Å (Fig. 2). The most disparate regions were on the wall of the depression spanning the icosahedral twofold axes of the capsids that showed variation of up to 5 Å between CPV and FPV and also within the flexible top of the β-ribbon forming the cylindrical channels at the icosahedral fivefold axes (Fig. 2). Both of these regions represent highly flexible structures, with high B-factors during refinement (7), and residues with values >50 Å2 were omitted. The differences at the twofold axes have been discussed elsewhere (1, 32). The fivefold differences appear to be related to whether the capsids contain DNA (full) or are empty since the two structures containing DNA, CPV, and CPV-N93R showed the smallest difference (∼1.5 Å), whereas variation of that structure in CPV and the empty capsid structures of FPV and CPV-N93D were up to 4.0 and 3.5 Å, respectively. The reasons for this difference in the VP2 topology at the fivefold axis is not completely clear but may be associated with the exposure of N-terminal peptides of VP2 and of the 5′ end of the viral DNA in full capsids, which are predicted to pass through the channel at the fivefold axis (3, 39). Thus, it is possible that full capsids are “open” at their fivefold axes because they have undergone a conformational change to enable genomic DNA entry and exposure of the VP2 N termini, whereas empty capsids remain “closed.”
FIG. 2.
Superimposition of the backbone atoms of CPV (gray), FPV (blue), CPV-N93D (red), and CPV-N93R (green). These color designations for the four capsid structures are used, respectively, in Fig. 4, 5, and 6. The icosahedral twofold (filled oval), threefold (filled triangle), and fivefold (filled pentagon) axes are shown. Positions of amino acids involved in host range determination between CPV and FPV that differ (80, 93, 323, 564, and 568) or are the same type (299, 300, 387, and 389) are shown for a VP2 monomer of CPV in gray and magenta, respectively.
The new CPV mutant structures were compared to those of the wt CPV type 2 and FPV capsids previously determined at neutral or acidic pH or with Ca2+ ions removed (32), as well as to the structures of a CPV-A300D mutant (21) and a CPV-N93K mutant (Tao et al., unpublished). The structures are interpreted in light of the functional differences between the highly homologous CPV and FPV, and with the mutants, including changes in canine TfR binding, canine cell infectivity, and the reactivity with anti-capsid antibodies (9, 16, 26, 34).
Variation in the capsid structure due to differences of residue 93.
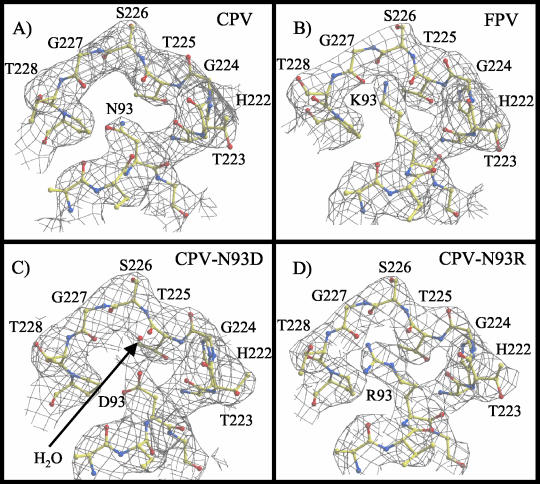
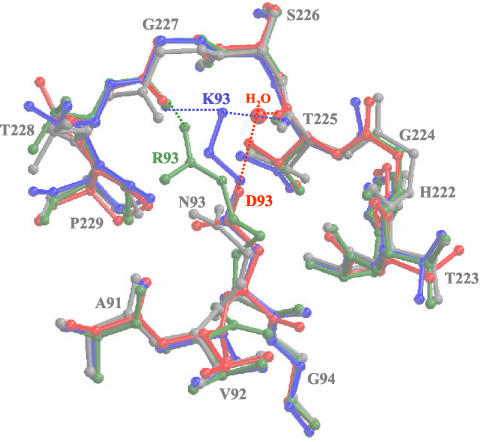
Residue 93 is a critical determinant that affects canine TfR binding, host range for canine cells, and reactivity with CPV-specific antibodies (16, 34). The structural differences between the surfaces of the capsids of CPV and FPV and the two CPV mutants in the vicinity of residue 93 were relatively small. There were differences in the electron densities of the side chains for residue 93 (Fig. 3 and 4). The capsid structure of each virus showed a distinct location for the side chain of residue 93 (Fig. 4) and its interactions with neighboring residues, and also a small displacement (∼1 Å) in the surrounding loop 2 in FPV and in the CPV mutants compared to wt CPV. The amino group of the K93 side chain in FPV forms two hydrogen bonds with the backbone carbonyl oxygen atoms of residues T225 and G227 in the adjacent loop 2 of the same VP2 monomer, whereas the shorter N93 side chain in CPV did not form equivalent bonds (Fig. 4) (1). In the CPV-N93R capsid, one of the terminal amino groups of the longer Arg side chain is able to form a single hydrogen bond with the backbone carbonyl oxygen of residue G227 (Fig. 4). In the CPV-N93D capsid, the Asp side chain that is the same length as the Asn in the wt virus is clearly associated with electron density (assigned as a water molecule) that was also coordinated by the main chain carbonyl of residue T225 (Fig. 3 and 4). This density has been modeled as a water molecule as that was the most consistent with B-factor refinement parameters, although it might be a Ca2+ ion that was present at 8 mM in the crystallization solution.
FIG. 3.
Electron density at VP2 residue 93 in CPV and FPV, as well as in the CPV-N93D and CPV-N93R mutant capsids. (A) Electron density of CPV at VP2 amino acid N93 and the surrounding capsid surface regions, including the tops of loop 1 and loop 2. (B) Electron density of the FPV at K93, with surrounding regions equivalent to those shown in panel A. (C and D) Electron density for the CPV-N93D and CPV-N93R mutants with the same views as in panels A and B. The water molecule modeled in CPV-N93D is indicated with a red ball and labeled.
FIG. 4.
Comparison of the models for wt CPV, wt FPV, CPV-N93D, and CPV-N93R. The models for the four viruses at VP2 residue 93 and its surrounding surface loops are superimposed. The colors are as defined in Fig. 2. The water molecule in the CPV-N93D structure is shown as a red ball. Regions of H-bonding interactions are indicated by the dotted lines in the same colors as those used for the respective structures.
The small displacement (∼1 Å) in the surface region compared to the wt CPV capsid (Fig. 4) was also seen in both the CPV-N93K mutant structure (Tao et al., unpublished) and in the structure of FPV that was determined at low pH and without Ca2+ ions (32). In both cases this was due to the same lysine interaction with loop 2 as was seen for the wt FPV (32; Tao et al., unpublished). The Cα positions of various FPV structures (FPV at pH 6.2; FPV crystallized without Ca2+ at pH 7.5, and FPV crystallized with Ca2+ at pH 7.5) (32) differ only slightly from the wt FPV, with a shift of ∼0.5 Å at the Cα position of residue K93 and ∼0.7 Å at the top of loop 2. The CPV-A300D and low-pH CPV structures are similar to wt CPV in this capsid region (21, 32), with Ca positions for N93 within 0.37 Å. Thus, substitutions in CPV with Lys, Arg, and Asp at position 93 all resulted in similar topologies in the capsid structures that differ from that of the wt CPV (Fig. 4). In these CPV structures there is a small depression or gap on the surface of the capsid at the tip of the threefold spike between loops 1 and loop 2 due to the lack of interaction between the N93 side chain and the neighboring loop 2 (Fig. 3 and 4). No depression is present in FPV or in the CPV-N93D, CPV-N93R, or CPV-N93K (Tao et al., unpublished) mutant structures, and the structural differences in the two new mutant structures appear due to the changes in the interacting properties of the substituted residues at position 93.
The N93D and N93R changes had two major effects on capsid binding properties. They reduced binding to the canine TfR and also to CPV-specific MAbs (15, 16), likely by changing the interactions of those molecules with the exposed surface of the capsid. This suggests that the canine TfR and antibody both make a contact with the Asn side chain on the wt CPV capsid that is partially or completely inhibited by the FPV K93 or by the CPV mutant substitutions at position 93. The effects of the CPV N93D and N93R mutations on binding may be due to the need for the receptor or the MAb to insert into the small depression that is present in the capsid between N93 and loop 2 in wt CPV (Fig. 3). However, CPV mutants N93A and N93V, with side chains that would occupy less space than the N93 in wt CPV, create a larger gap in the surface, and these mutants also showed reduced binding to the canine TfR and CPV specific MAbs (16). Another possibility is that the small change in the position of loop 2 is responsible for that decreased receptor and antibody binding. However, when mutants CPV-N93A or CPV-N93V mutants (16) with small hydrophobic side chains were examined, these mutants also showed changes in antibody binding. These side chains would be unable to engage in the type of hydrogen bonding interactions observed in FPV and CPV-N93D and CPV-N93R mutants and thus would not be expected to affect the location of loop 2. The role(s) of residue 93 and the loop 2 region (Fig. 4) in receptor and antibody binding was also examined in parallel studies by mutational analysis of CPV residues in the loop 2 region, with the changes G224E, G224R, and P229A (16). Although these changes did not dramatically affect canine cell infection, they did prevent binding of antibodies that normally recognize this antigenic site (designated site “A”) (16, 34). Those loop 2 mutants also showed that the “A” antigenic site could be divided into two regions, with one epitope recognized by the CPV-specific antibodies that are strongly affected by the structure around residue 93 and closely adjacent mutations (G224E and G224R). A second epitope recognized by certain CPV-FPV common antibodies was only affected by a CPV-H222Y substitution, which did not affect the binding of the CPV-specific antibodies.
Combined effects of mutations on receptor binding and canine cell infection.
We have shown in previous studies that substitutions in three capsid regions, defined by VP2 residue 93, the region around residue 300, and residue 323 can cause differences in both canine TfR binding and canine cell infection by the viruses (15, 16). In an FPV background changes of both residue Lys-93 to Asn and Asp-323 to Asn are required be introduced together to give that virus the ability to bind the canine TfR, and neither of these changes alone affects canine TfR binding or cell infection (16). However, these residues are separated by ∼20 Å in the structure (Fig. 1). There are a number of possible explanations for the need for the coordinated changes; the two changes may each independently affect binding of the virus to the same ligand, or it may be that the change of residue 323 is required to compensate for a long-range structural effect induced by the substitution of residue 93.
The possibility of long-range effects have been suggested in studies of the parvovirus minute virus of mice (MVM), where mutations in the host range and pathogenicity determining residues could be complemented by forward selected changes (3) of residues that were distant in primary sequence but were relatively proximal in the structure (3). In that case the compensatory mutations could act together to affect the interaction of the capsid with a host cell factor that determined tissue tropism (3).
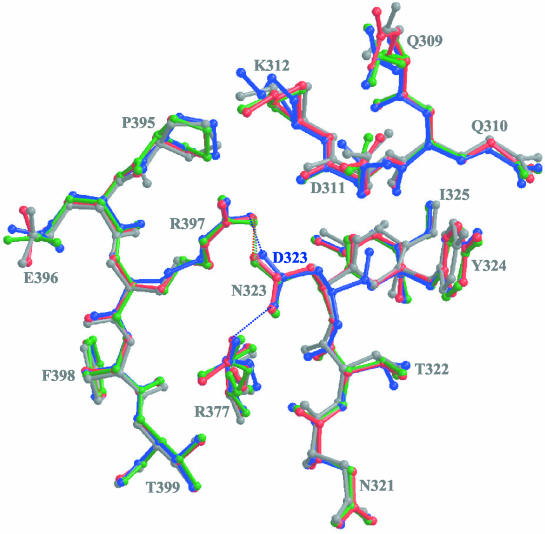
When we examined the structures of CPV, FPV, CPV-N93D, and CPV-N93R around amino acid position 93, there were no significant effects seen beyond those of the immediately adjacent loop 2 (Fig. 4). Residue 323 is situated ∼20 Å away from residue 93 in the capsid structures, and the Cα of the Asn or Asp at position 323 in wt CPV and its mutants or in FPV, respectively, were superimposable in all of the structures (Fig. 5). However, both terminal oxygens of the D323 in FPV are able to form hydrogen bonding interactions with terminal amino groups in residues R377 and R397 in a threefold symmetry-related VP2 contact region (Fig. 5). The terminal oxygens of the N323 residue in CPV, CPV-N93D, and CPV-N93R are also able to hydrogen bond with a terminal amino group of R397 but do not interact with residue R377. The effect of pH and Ca2+ binding on the CPV and FPV structures of the capsid region containing residue 323 have been extensively discussed elsewhere (32). Residues R377 and R397 do not differ between FPV and CPV, and both residues control the ability of the viruses to interact with sialic acid residues in erythrocytes during hemagglutination of red blood cells (36). Thus, the intramolecular interactions between these residues and 323 most likely directly affect the contact between the capsids of CPV and FPV and the canine TfR, rather than acting to compensate for the changes of residue 93.
FIG. 5.
Comparison of the models for wt CPV, wt FPV, CPV-N93D, and CPV-N93R. The models for the four viruses at VP2 residue 323 (threefold monomer) and its surrounding surface loops are superimposed. Residues 309 to 312 and 321 to 325 (threefold monomer) and residues 377 and 395 to 399 (reference monomer) are shown. Residues 377 and 397 are involved in the determination of hemaglutination characteristics of CPV and FPV. The view is similar to that used in Fig. 1. Colors are as defined in Fig. 2. Regions of H-bonding interactions are indicated by the dotted lines in the same colors as those used for the respective structures.
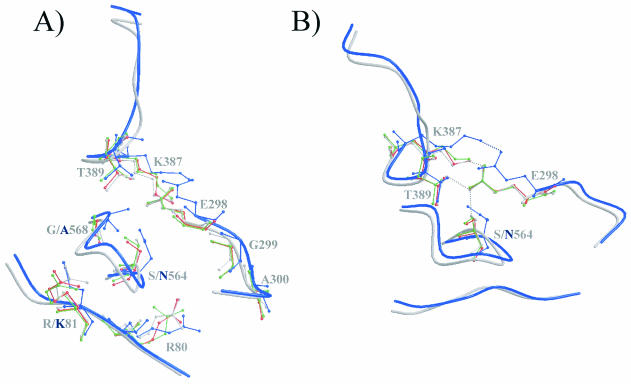
Mutational analysis and binding experiments identified a third region in the capsid structure that affects canine TfR binding (15, 16), and this was seen in studies which showed that the change in CPV of VP2 residue 299 from Gly to Glu resulted in the virus losing the ability to bind the canine TfR (15). CPV and FPV mutants with changes in residues 93 and 323 (16) also revealed the involvement of a third region determining canine TfR binding. This was seen in FPV, where both residues 93 and 323 had to be changed to the CPV sequences to achieve canine TfR binding, whereas the reciprocal double mutant of CPV (CPV-N93K/N323D) did not completely lose the ability to bind the canine TfR. Candidates for residues causing these secondary effects on TfR binding are in the 300 region and, in particular, the CPV to FPV differences observed at residues 80 (Arg to Lys), 564 (Ser to Asn), and 568 (Gly to Ala), which have previously also been implicated in feline tropism (37, 38). These residues, although distant from one another in the primary sequence, are clustered together in icosahedrally symmetry-related molecules within the shoulder region of the threefold spike of the capsid (Fig. 1 and 6), close to the VP2 residues such as Gly-299, which had a strong effect on canine TfR binding and canine cell infection when changed to a Glu (15). CPV and FPV show small structural differences in both the main chains and the side chains of amino acids in these intertwining loops (Fig. 6) and also show differences of up to 1.4 Å between CPV and FPV in a neighboring surface loop from residues 387 to 394, which contains no amino acid sequence variations between the viruses (Fig. 6). Previous mutational analyses of two residues in this loop, K387 and T389, in which both residues were changed to an Ala, resulted in a nonviable CPV phenotype (28). The side chains of these residues appear to form hydrogen-bonding interactions with E298 in the wt CPV structure, CPV-N93D and CPV-N93R (Fig. 6). In the FPV capsid structure, E298 is only able to form a hydrogen bond with K387 because of the altered conformation of this loop region (Fig. 6B). Another difference between CPV and FPV is seen at residue 564, where the terminal oxygen of the Ser side chain is able to form a hydrogen bond with E298 in CPV and the Asn in FPV is not (Fig. 6B). In all of the available CPV and FPV structures, residue 568, situated approximately in the center of this cluster of 300 region residues, is ∼36 and ∼37 Å distant from residues 93 and 323, respectively (Fig. 1). The single change of Gly-299 to Glu in CPV inhibits canine TfR binding by itself (15), and no obvious structural interaction can be detected between the various capsids at this residue (Fig. 6A) at the resolution of the solved structures. This indicates that there are direct interactions between the 300 region of the capsid (Fig. 6A) and the canine TfR that are independent of both residues 93 and 323. Thus, it appears that several small changes of the viral surface occurring as result of multiple side chain and main chain differences of CPV mutant and FPV capsid structures all act together to determine virus binding to the canine TfR and control viral host range.
FIG. 6.
Comparison of the models for wt CPV, wt FPV, CPV-N93D, and CPV-N93R. (A) Backbone trace of CPV and FPV at VP2 regions containing residues 80 and 81 (reference monomer), 298 to 300 (threefold related), and 387 to 392 and 564 to 568 (fivefold related). The surface view is similar to that used in Fig. 1. Colors are as defined in Fig. 2. Residues that differ between CPV and FPV or that might be engaged in interactions between adjacent VP2 monomers are shown in ball-and-stick format. (B) Some of the residues in panel A have been removed, and the figure was rotated to show the possible interaction(s) between residues E298, K387, and T389 and between residues E298 and S564. Regions of H-bonding interactions are indicated by the dotted lines in the same colors as used for the respective structures.
Control of viral host range and antibody binding.
How do these distinct structural regions of the capsid affect the host cell receptor binding and canine host range? All of the mutant capsids generated thus far still infect feline cells and appear to bind the feline TfR, showing that the specific interaction with the canine TfR involves residues that are not important for the feline TfR binding. Comparison of the amino acid sequences of the canine and feline TfRs and the mapping of residues onto the available human TfR structure shows that the receptors differ in ∼10% of their amino acid sequence and that capsid binding is determined by the apical domain of the receptor (20, 26). Recombination and mutational mapping of the specific sequences of the canine and feline TfRs shows that three separate sequences within the apical domain can all affect virus binding (26). Since the apical domain of the human TfR is large enough to interact simultaneously with all three different regions of the capsid that affect canine TfR binding (4, 20), it is possible that the three positions in the capsid discussed here are all involved in separate receptor-capsid interactions. These observations indicate that the emergence from FPV of CPV as a successful canine pathogen required a series of changes in the feline virus to accommodate the differences between the canine and feline TfR structures. Subsequently, the emergence and rapid selection of the CPV type 2a virus included changes of several other positions near the shoulder of the threefold spike (i.e., VP2 residues 87, 300, and 305), and these changes increased the efficiency of infection of cells expressing the canine TfR (15).
Previous analyses of parvoviral capsid determinants of tissue tropism and host range have suggested that restriction between highly homologous strains such as CPV and FPV and between MVMi and MVMp occur after cell binding but before viral genome transcription (3, 13). Our further examination of the cell and receptor interactions of CPV and FPV indicates that their host range differences are due to direct contact with host cell receptor molecules. Thus, the identification of the canine and feline TFRs as the infectious cell receptors for CPV and FPV continues to provide a means for relating the surface property differences for these viral capsids to receptor interactions that determine tropism. To reconcile a block in cell infection after entry but before genome replication with the model where the receptor contacts these regions of the capsid that differ between CPV and FPV, the receptor-virus interaction on the cell surface must initiate specific conformational changes required for permissive interaction. In infection with mutant capsids or of nonpermissive cells, the required structural rearrangements are disabled, although the exact point of the blockage is still unknown.
Two of the regions affecting receptor binding are also major binding sites for neutralizing antibodies, since the antigenic sites A and B (34) coincide with the regions around residue 93 and 300. The coincidence of receptor and antibody binding sites has been observed for other viruses, including foot-and-mouth-disease virus, equine rhinitis A virus, and rhinovirus (see, for example, references 10, 12, 41, and 42), in which antigenic variation could occur while the virus retained the ability to bind the host receptor. This is also the case for CPV and FPV, for which antigenic variation in both the A and the B antigenic sites has been seen in nature or after selection of MAb neutralization escape mutants, whereas the virus retained the ability to bind the feline or canine TfR and infect cells (15, 16).
The natural adaptation of a virus to a new host is a very rare event, suggesting that there are high barriers that prevent viruses from gaining the ability to infect and spread naturally in hosts to which they are not adapted. The emergence of CPV as a new pathogen of dogs is one of only a few contemporary examples of this type of viral emergence. Our analysis, reported here and elsewhere (15, 16), indicates that the adaptation involved a series of small but specific changes of the capsid surface that enabled it to bind the canine TfR and to use it to efficiently infect dog cells. We are now seeking to understand the mechanisms by which the capsid-receptor interaction leads to cell infection and the role of the different structures in controlling the process.
Acknowledgments
MacCHESS staff at the Cornell High Energy Synchrotron Source (CHESS) provided help during data collection, and Lana Walsh helped in obtaining beam time. David Duda and Robert McKenna assisted in data collection at CHESS, and Robert McKenna, Alan Simpson, and Vijay Reddy provided helpful discussions related to Molecular Replacement procedures.
This study was supported by grant AI33468 from the National Institutes of Health to, C.R.P. and M.A.-M.
REFERENCES
- 1.Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155-171. [DOI] [PubMed] [Google Scholar]
- 2.Agbandje, M., C. R. Parrish, and M. G. Rossmann. 1995. The structure of parvoviruses. Semin. Virol. 6:299-309. [Google Scholar]
- 3.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, M. J., J. A. Lebron, and P. J. Bjorkman. 2000. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature 403:46-53. [DOI] [PubMed] [Google Scholar]
- 5.Bergeron, J., B. Hebert, and P. Tijssen. 1996. Genome organization of the Kresse strain of porcine parvovirus: identification of the allotropic determinant and comparison with those of NADL-2 and field isolates. J. Virol. 70:2508-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunger, A. T. 1992. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-474. [DOI] [PubMed] [Google Scholar]
- 7.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 8.CCP4. 1994. The CCP4 site: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 9.Chang, S. F., J. Y. Sgro, and C. R. Parrish. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 66:6858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Che, Z., N. H. Olson, D. Leippe, W. M. Lee, A. G. Mosser, R. R. Rueckert, T. S. Baker, and T. J. Smith. 1998. Antibody-mediated neutralization of human rhinovirus 14 explored by means of cryoelectron microscopy and X-ray crystallography of virus-Fab complexes. J. Virol. 72:4610-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esnouf, R. 1999. BOBSCRIPT. Further additions to MOLSCRIPT version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D 55:938-940. [DOI] [PubMed] [Google Scholar]
- 12.Hewat, E. A., N. Verdaguer, I. Fita, W. Blakemore, S. Brookes, A. King, J. Newman, E. Domingo, M. G. Mateu, and D. I. Stuart. 1997. Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: positioning of a highly mobile antigenic loop. EMBO J. 16:1492-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horiuchi, M., N. Ishiguro, H. Goto, and M. Shinagawa. 1992. Characterization of the stage(s) in the virus replication cycle at which the host-cell specificity of the feline parvovirus sub-group is regulated in canine cells. Virology 189:600-608. [DOI] [PubMed] [Google Scholar]
- 14.Horiuchi, H. M., Goto, N. Ishiguro, and M. Shinagawa. 1994. Mapping of determinants of the host range for canine cells in the genome of canine parvovirus using canine parvovirus/mink enteritis virus chimeric viruses. J. Gen. Virol. 75:1319-1328. [DOI] [PubMed] [Google Scholar]
- 15.Hueffer, K., J. S. Parker, W. S. Weichert, R. E. Geisel, J. Y. Sgro, and C. R. Parrish. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hueffer, K., L. Govindasamy, M. Agbandje-McKenna, and C. Colin. Parrish. 2003. Combinations of two capsid regions control host range and specific transferrin receptor binding by canine parvovirus. J. Virol. 77:10099-10105. [DOI] [PMC free article] [PubMed]
- 17.Jones, T., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 97:110-119. [DOI] [PubMed] [Google Scholar]
- 18.Kleywegt, G. J., and T. A. Jones. 1997. Model building and refinement practice. Methods Enzymol. 277:208-230. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 20.Lawrence, C. M., S. Ray, M. Babyonyshev, R. Galluser, D. W. Borhani, and S. C. Harrison. 1999. Crystal structure of the ectodomain of human transferrin receptor. Science 286:779-782. [DOI] [PubMed] [Google Scholar]
- 21.Llamas-Saiz, A. L., M. Agbandje-McKenna, J. S. L. Parker, A. T. M. Wahid, C. R. Parrish, and M. G. Rossmann. 1996. Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology 225:65-71. [DOI] [PubMed] [Google Scholar]
- 22.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33:491-497. [DOI] [PubMed] [Google Scholar]
- 23.Merritt, E. A., and D. J. Bacon. 1997. Raster3D-photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 24.Navaza, J., and P. Saludjian. 1997. AMoRe: an automated molecular replacement package. Methods Enzymol. 276:581-594. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski, Z., and W. Minor. 1997. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 26.Palermo, L., K. Hueffer, and C. R. Parrish. 2003. Residues in the apical domain of the feline and canine transferrin receptors control host-specific binding and cell infection of canine and feline parvoviruses. J. Virol. 77:8915-8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker, J. S. L., W. J. Murphy, D. Wang, S. J. O'Brien, and C. R. Parrish. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker, J. S. L., and C. R. Parrish. 1997. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 71:9214-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parrish, C. R. 1991. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology 183:195-205. [DOI] [PubMed] [Google Scholar]
- 30.Parrish, C. R., C. Aquadro, M. L. Strassheim, J. F. Evermann, J.-Y. Sgro, and H. Mohammed. 1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 65:6544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parrish, C. R., C. F. Aquadro, and L. E. Carmichael. 1988. Canine host range and a specific epitope map along with variant sequences in the capsid protein gene of canine parvovirus and related feline, mink and raccoon parvoviruses. Virology 166:293-307. [DOI] [PubMed] [Google Scholar]
- 32.Simpson, A. A., V. Chandrasekar, B. Hebert, G. M. Sullivan, M. G. Rossmann, and C. R. Parrish. 2000. Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses. J. Mol. Biol. 300:597-610. [DOI] [PubMed] [Google Scholar]
- 33.Simpson, A. A., B. Hebert, G. M. Sullivan, C. R. Parrish, Z. Zadori, P. Tijssen, and M. G. Rossmann. 2002. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 315:1189-1198. [DOI] [PubMed] [Google Scholar]
- 34.Strassheim, L. S., A. Gruenberg, P. Veijalainen, J.-Y. Sgro, and C. R. Parrish. 1994. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198:175-184. [DOI] [PubMed] [Google Scholar]
- 35.Tong, L., and M. G. Rossmann. 1990. The locked rotation function. Acta Crystallogr. A 46:783-792. [DOI] [PubMed] [Google Scholar]
- 36.Tresnan, D. B., L. Southard, W. Weichert, J. Y. Sgro, and C. R. Parrish. 1995. Analysis of the cell and erythrocyte binding activities of the dimple and canyon regions of the canine parvovirus capsid. Virology 211:123-132. [DOI] [PubMed] [Google Scholar]
- 37.Truyen, U., M. Agbandje, and C. R. Parrish. 1994. Characterization of the feline host range and a specific epitope of feline panleukopenia virus. Virology 200:494-503. [DOI] [PubMed] [Google Scholar]
- 38.Truyen, U., J. F. Evermann, E. Vieler, and C. R. Parrish. 1996. Evolution of canine parvovirus involved loss and gain of feline host range. Virology 215:186-189. [DOI] [PubMed] [Google Scholar]
- 39.Tsao, J., M. S. Chapman, M. Agbandje, W. Keller, K. Smith, H. Wu, M. Luo, T. J. Smith, M. G. Rossmann, R. W. Compans, and C. R. Parrish. 1991. The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456-1464. [DOI] [PubMed] [Google Scholar]
- 40.Tsao, J., M. S. Chapman, H. Wu, M. Agbandje, W. Keller, and M. G. Rossmann. 1992. Structure determination of monoclinic canine parvovirus. Acta Crystallogr. B 48:75-88. [DOI] [PubMed] [Google Scholar]
- 41.Verdaguer, N., N. Sevilla, M. L. Valero, D. Stuart, E. Brocchi, D. Andreu, E. Giralt, E. Domingo, M. G. Mateu, and I. Fita. 1998. A similar pattern of interaction for different antibodies with a major antigenic site of foot-and-mouth disease virus: implications for intratypic antigenic variation. J. Virol. 72:739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warner, S., C. A. Hartley, R. A. Stevenson, N. Ficorilli, A. Varrasso, M. J. Studdert, and B. S. Crabb. 2001. Evidence that equine rhinitis A virus VP1 is a target of neutralizing antibodies and participates directly in receptor binding. J. Virol. 75:9274-9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie, W. Q., Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]