Abstract
Lentiviruses exist in vivo as a population of related, nonidentical genotypes, commonly referred to as quasispecies. The quasispecies structure is characteristic of complex adaptive systems and contributes to the high rate of evolution in lentiviruses that confounds efforts to develop effective vaccines and antiviral therapies. Here, we describe analyses of genetic data from longitudinal studies of genetic variation in a lentivirus regulatory protein, Rev, over the course of disease in ponies experimentally infected with equine infectious anemia virus. As observed with other lentivirus data, the Rev variants exhibited a quasispecies character. Phylogenetic and partition analyses suggested that the Rev quasispecies comprised two distinct subpopulations that coexisted during infection. One subpopulation appeared to accumulate changes in a linear, time-dependent manner, while the other evolved radially from a common variant. Over time, the two subpopulations cycled in predominance coincident with changes in the disease state, suggesting that the two groups differed in selective advantage. Transient expression assays indicated the two populations differed significantly in Rev nuclear export activity. Chimeric proviral clones containing Rev genotypes representative of each population differed in rate and overall level of virus replication in vitro. The coexistence of genetically distinct viral subpopulations that differ in phenotype provides great adaptability to environmental changes within the infected host. A quasispecies model with multiple subpopulations may provide additional insight into the nature of lentivirus reservoirs and the evolution of antigenic and drug-resistant variants.
Lentiviruses exhibit high mutation rates and exist in vivo as a population of closely related viral genotypes that are commonly referred to as a quasispecies (11, 13, 19). The population structure of a quasispecies consists of a master sequence, which is the dominant genotype, and the mutant spectrum, which includes reservoirs of genotypic and phenotypic variants with the potential to become dominant in the face of environmental change. Studies have demonstrated that the evolution of viral quasispecies adheres to basic principles of population genetics; however, quasispecies theory diverges from population genetics in that the entire deme of related sequences, rather than the individual replicon, is the main target of selection (14). The quasispecies occupies a region on a fitness landscape where adaptive mutations move the quasispecies toward a fitness peak, thereby increasing the mean fitness of the quasispecies. Understanding the principles that shape the evolution of viral quasispecies is becoming increasingly important as molecular information is used to model disease progression and to predict the emergence of antigenic and drug-resistant variants.
Experimental infection of ponies with equine infectious anemia virus (EIAV) provides an excellent system for longitudinal studies of lentivirus evolution during disease progression. EIAV is a lentivirus that is genetically (21), antigenically (31), and morphologically (15) related to human immunodeficiency virus type 1 (HIV-1). In ponies, EIAV induces a reproducible clinical disease course with progression of clearly demarcated stages of disease (17, 20, 22, 29). Upon infection, animals may suffer an early episode of acute illness, including fever and thrombocytopenia associated with high levels of virus replication. Resolution of acute illness occurs concurrently with the appearance of a virus-specific cytotoxic T-lymphocyte (CTL) response and a decrease in the level of plasma viremia (16, 30). In the chronic stage, recurrent episodes of high-titer viremia and associated disease often occur within the first year after initial infection but generally abate in frequency and severity with time. Thereafter, most horses enter an inapparent carrier state with no apparent compromises in their long-term health and activity. Nonetheless, these individuals do not clear the virus and remain persistently infected throughout their life (18, 23, 26, 32, 38).
We and others have observed genetic variation in the region of the EIAV genome where the second exon of Rev overlaps the cytoplasmic portion of the transmembrane protein (2, 5, 27). Rev is a regulatory protein that controls nuclear export of unspliced and singly spliced viral mRNAs, which are essential for virus replication. Therefore, factors that modulate Rev activity and, consequently, alter levels of viral gene expression may be important in regulating virus replication in vivo. To examine this possibility, we recently undertook a comprehensive longitudinal analysis of genetic and biological variation in Rev throughout a clinically dynamic disease course in one pony experimentally infected with the virulent EIAVWyo2078 (4). The pony experienced multiple febrile cycles during the first 4 months of infection. The onset of the inapparent period coincided with the appearance of detectable neutralizing antibody, consistent with previous evidence that clinical quiescence is associated with the development of broadly reacting virus neutralizing antibody and maturation of the immune response (16). Changes in the genotype and phenotype of Rev quasispecies correlated with changes in clinical stages of EIAV infection: Rev variants predominant during chronic febrile periods had significantly higher Rev-mediated nuclear export activity than the variants predominant during the inapparent stage of disease. Therefore, the onset of clinical latency was associated with the maturation of the host immune response and a significant decrease in Rev activity.
In the present study, we used phylogenetic and biological analyses to characterize the evolution of EIAV Rev variants in vivo. Results identified two distinct subpopulations of rev that evolved in different patterns, coexisted during disease, and differed in phenotype. A quasispecies comprised of multiple subpopulations that occupy different regions on a fitness landscape could allow a virus to adapt rapidly to changes in the host environment by expanding minor, less fit populations that may be near the new fitness peaks. Such a model may provide new insight into the factors that shape the evolution of viral quasispecies during progression of disease.
MATERIALS AND METHODS
Experimental infections and identification of Rev variants.
The virulent Wyoming strain of EIAV was used to infect pony 524, and sequential serum samples were collected from different stages of clinical disease, as previously described (4). Pony 625 was inoculated intravenously with 2 ml of heparinized plasma collected from pony 524 at 878 days postinfection. These inocula are heterogeneous populations of virus, similar to a natural infection.
Viral RNA isolated from the inoculum and from sera collected during variable stages of disease was reverse transcribed and amplified with primers to conserved regions of EIAV (Fig. 1). PCR amplification conditions consisted of 37 cycles of denaturation at 94°C for 2 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min. The initial and final cycles contained a prolonged extension at 72°C for 5 min. Then 2 μl of PCR product was cloned into pGEM-T vectors as recommended by the manufacturer (Promega, Madison, Wis.) and transformed into Escherichia coli DH5α. Clones were sequenced bidirectionally with primers to vector sequences flanking the insert. Sequencing was performed by the Iowa State University DNA Synthesis and Sequencing Facility with an automated DNA sequencer. Sequences were aligned by MacVector and AssemblyLIGN software (Oxford Molecular, Beaverton, Oreg.).
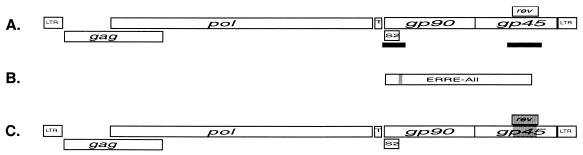
FIG. 1.
(A) Organization of EIAV open reading frames, with solid black lines indicating regions amplified, cloned, and sequenced. The minimal EIAV Rev-responsive element (RRE; see below) is contained within the S2/gp90 amplified region, which spans nucleotides 5233 to 5589. The rev/gp45 amplified region spans nucleotides 7066 to 7680. (B) The EIAV Rev reporter plasmid pERRE-All contains nucleotides 5280 to 7534. The small shaded area indicates the 57-nucleotide region (nucleotides 5485 to 5540) within ERRE-All that was found to comprise a minimal RRE (6). (C) Representation of the Rev chimeric proviral clones. The shaded area in the overlapping rev/tm region indicates the portion containing Rev variant sequences. Numbering is based on that of Kawakami et al. (21).
For pony 524, we analyzed 61 rev clones from the inoculum and 23 to 25 clones from each of 11 sequential serum samples taken at 12, 35, 67, 89, 118, 201, 289, 385, 437, 754, and 800 days postinfection. In addition, 20 clones spanning the EIAV Rev-responsive element (RRE) were analyzed from the inoculum and serum samples collected at 12, 118, 385, and 800 days postinfection. Sixteen clones were analyzed from the pony 625 inoculum and 13 to 21 clones from each of five sequential serum samples collected at 14, 25, 46, 67, and 77 days postinfection. Rev amino acid variants were named in the order they were identified, with identical variants given the same name. The names for pony 625 Rev variants were continued from the numbering of pony 524 variants.
Analysis of viral quasispecies.
Phylogenetic analyses were used to characterize the genetic relationship among Rev amino acid variants in each horse. The neighbor-joining distance method based on p-distance was used to construct unrooted phylogenetic trees from the Rev amino acid sequences with Mega versions 1.01 and 2.1 (24, 25). Other methods for estimating amino acid substitutions between clones, i.e., number of amino acid differences, Poisson-corrected distance, and gamma distance, were also used and resulted in trees nearly identical to those presented in this paper. Tree-Puzzle (36) was used to assess the support for particular clades in the phylogenetic tree with likelihood mapping (37). Briefly, the relative support, based on a maximum-likelihood analysis, is inferred for each of the three possible tree topologies for random quartets of sequences. The data are reported as the percentage of random quartets supporting the hypothesized clades; ambiguous results were not counted as supportive of any clade (37). Tree-Puzzle can use any of the standard maximum-likelihood models of protein evolution. We used the JTT model with empirical amino acid frequencies estimated from the data and no rate variation among sites.
The program PAQ, for partition analysis of quasispecies (3), was used to group the rev genotypes that were most similar at each time point. The program uses the Hamming distance as a measure of distance between rev variants and a nonhierarchical clustering method to identify discrete and cohesive partitions of closely related sequences, similar to other clustering algorithms (28). However, PAQ does not select a predetermined number of partitions, and overlap between partitions is allowed. The optimal output maximizes the number of variants contained within the partition while minimizing the partition radius and any overlap between partitions. The central genotype of each group is the sequence most representative of all variants within the group. A standard comparison of two population means was used to determine whether distances within each group were statistically different from distances between two groups. A z-statistic, assuming a normal distribution, was calculated with the mean and variance of the two populations and tested under the criterion of P < 0.01.
Construction of Rev expression plasmids and CAT assays.
Selected Rev variants were subcloned into a Rev expression vector and assayed for nuclear export activity as previously described (5). pERevWT, which was previously described as pcH21 (4), contains Rev cDNA in the pCR3 background (Invitrogen, Carlsbad, Calif.). The rev exon 2 was removed by digestion with the restriction endonuclease ApaI. The digested plasmid was gel purified, dephosphorylated, and ligated with the exon 2 sequence of rev variants as described (4, 5). All clones were confirmed by sequencing.
Rev-dependent nuclear export activity was determined in transient transfection assays with the pERRE-All reporter plasmid (5), a derivative of pDM138 with an intron containing the chloramphenicol acetyltransferase (CAT) gene and EIAV nucleotides 5280 to 7534 (Fig. 1b). 293T cells were transfected with 1 μg of either pcDNA3 or Rev variant plasmid, 0.2 μg of pERRE-All reporter plasmid, 0.2 μg of pCH110 (Amersham Pharmacia Biotech, Piscataway, N.J.), and 0.6 μg of pUC19 by calcium phosphate coprecipitation. Each experiment also included a sham group that contained no reporter plasmid but an additional 0.2 μg of pUC19. Two days posttransfection, cells were harvested, resuspended in 0.3 ml of 0.25 M Tris (pH 7.5), lysed by freeze/thawing, and assayed for β-galactosidase activity to normalize CAT assays for transfection efficiency. Normalized lysates were assayed for CAT activity in a 0.1-ml volume with 3 μl of [14C]chloramphenicol and 1 mM acetyl-coenzyme A. Acetylated products were separated by thin-layer chromatography, and the percentage of acetylation was quantified by phosphorimager. In some experiments, the amount of CAT enzyme was quantified with a commercial enzyme-linked immunosorbent assay (ELISA) kit (Roche, Indianapolis, Ind.). Experiments were performed in triplicate, and results summarize a minimum of nine transfections. Statistical significance was determined by analysis of variance.
Construction and analysis of chimeric virus.
Chimeric proviruses were constructed in the backbone of pSPEIAV19, an infectious molecular clone of EIAV (34). To facilitate cloning, pSPEIAV19 was digested with SphI and EcoRI and the 3′ half of the EIAV provirus was subcloned into pLG338/30 (34), generating a shuttle plasmid that included the rev/tm overlapping region. The shuttle plasmid was digested with ApaI and BstBI to remove the rev/tm region. This region was replaced with the similar 403-bp insert isolated from Rev variant cDNAs by ApaI and BstBI digestion and gel purification. The SphI-EcoRI fragment containing rev/tm variants was excised from the shuttle plasmid, isolated, and reinserted into the pSPEIAV19 backbone, generating full-length proviral clones that differed only in the rev/tm overlapping region (Fig. 1c). Chimeric proviral plasmid DNAs were purified and confirmed by sequence analysis.
To generate chimeric virus stocks, 1.9 μg of proviral DNA and 0.1 μg of pSV5neo were cotransfected into canine fetal thymus cells (Cf2th; ATCC CRL-1430) with TransIT-LT1 (Mirus Corporation, Madison, Wis.). At 2 days posttransfection, cells were passed into 100-mm tissue culture dishes and grown in Dulbecco's modified Eagle's medium supplemented with 2-mg/ml G418. Surviving cell colonies were tested for supernatant reverse transcriptase (RT) activity (8), and RT-positive clones were used for production of chimeric virus stocks. The identity of the rev/tm insert in the virus stock was confirmed by sequence analysis. To assess the effect of variation on virus replication, chimeric virus stocks were normalized by RT activity and 104 cpm was inoculated onto equine dermal cells (ED; ATCC CCL57) seeded the previous day at 105 cells/well in 24-well tissue culture plates in Dulbecco's modified Eagle's medium supplemented with 20% fetal bovine serum and penicillin/streptomycin. Supernatant was collected every 2 days, and virus replication was quantified by supernatant RT activity (8).
Nucleotide sequence accession numbers.
GenBank accession numbers are AF314257 to AF314404 and AF512608 to AF512669.
RESULTS
Subpopulations identified in Rev quasispecies of EIAV-infected horses.
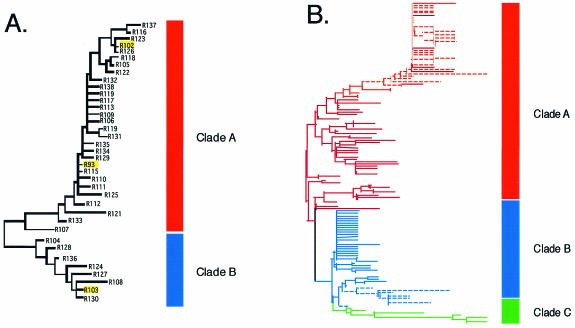
Following experimental infection, pony 524 experienced a variable clinical disease course characterized by recurring fever cycles interspersed with afebrile periods ranging from days to months (Fig. 2). The onset of the inapparent stage was associated with the maturation of the host immune response and a significant decrease in Rev activity (4). Here, we examined the evolution of EIAV Rev quasispecies during infection of pony 524. A neighbor-joining tree was constructed to elucidate the genetic relatedness of the entire data set of pony 524 Rev amino acid variants. The phylogenetic tree displayed a distinct branching pattern of Rev variants into two major clades, designated clade A and clade B, and a minor clade, designated clade C (Fig. 3). However, the combination of low genetic diversity and short but numerous sequences resulted in low bootstrap support for the branch point separating clades A and B (10%).
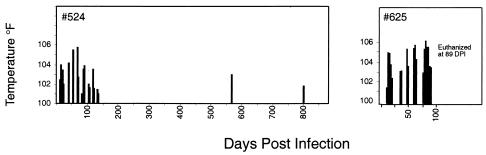
FIG. 2.
Febrile response of ponies 524 and 625 following experimental infection. The temperature of the ponies was measured daily during clinical episodes and intermittently during chronic clinical disease. Febrile episodes (defined as 100°F or higher) are shown for each pony.
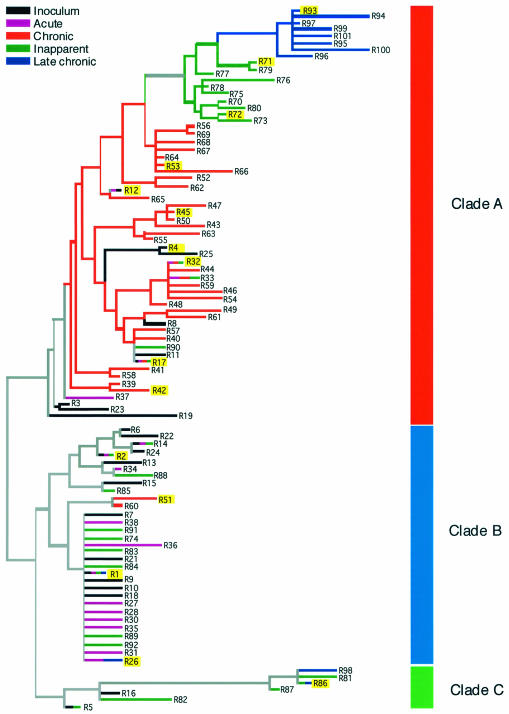
FIG. 3.
Neighbor-joining tree of the pony 524 Rev amino acid variants based on p distance. The tree is divided into three clades, designated clades A, B, and C. The stage of disease from which the Rev variants were isolated is indicated by the color of its branch. Multicolored branches indicate variants isolated at multiple stages of disease. Highlighted variants were assayed for Rev activity.
To test whether clade A and clade B were indeed monophyletic, likelihood mapping (37) with Tree Puzzle (36) was employed. Of the 1,000 random quartets sampled, 89.3% supported the distinct clades A and B. In addition, when separate phylogenetic trees were inferred with neighbor-joining for each time point sampled, the bootstrap support for clades A and B steadily increased from ≈50% at early time points to 99% and 100% for the last two time points, respectively (data not shown). Therefore, despite the low bootstrap support, the phylogenetic tree (Fig. 3) captures the basic evolutionary patterns of the Rev variants isolated from pony 524.
Both clade A and clade B contained variants from the inoculum and from each stage of disease; however, there were marked differences in topology between the clades that suggested they had evolved under different selective mechanisms. Clade A was characterized by transience and divergence of Rev variants over the course of infection such that late time points appear to nest as monophyletic groups within earlier time points. Genetic variation within this clade was relatively large, with 8 amino acid differences between R3 and R94. In contrast, all of the sequences in clade B were genetically very similar, with a maximum of three amino acid differences between any two variants. Clade B variants came from all stages of disease and appeared to evolve radially away from R1, the major founder variant in the inoculum (4).
To confirm this, we again applied likelihood mapping, this time independently within clade A and clade B. The results supported increased diversity and linear evolution in clade A, contrasting with low diversity and radial evolution in clade B (not shown). The minor clade, clade C, contained only seven variants. The genetic variation within clade C was the same as clade A (eight amino acid differences), but unlike clade A, there was no incremental progression of variation. It is possible that additional clade C variants were present throughout all stages of infection, but existed at levels too low to be detected by our sampling procedures. Therefore, the origin and biological significance of this minor clade are uncertain at this time.
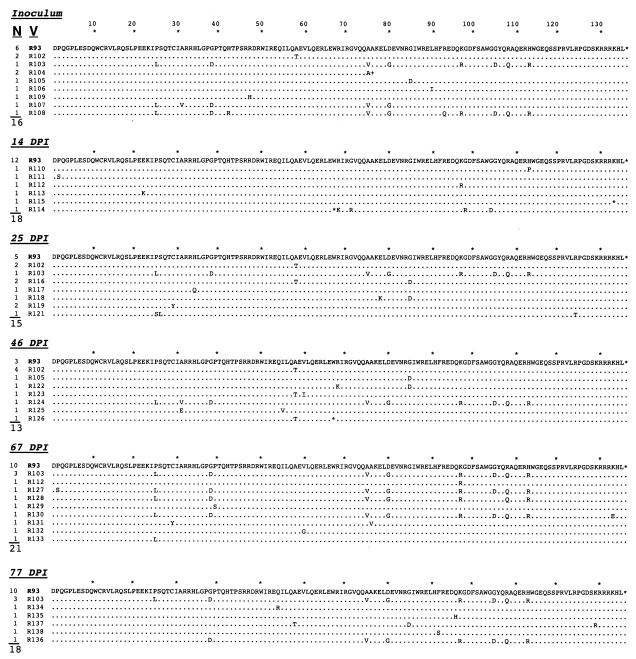
The population structure of rev quasispecies was examined in a second horse, pony 625, which was inoculated with serum from pony 524 collected at 878 days postinfection. Pony 625 underwent several febrile cycles over the acute and chronic stages of disease, became moribund, and was euthanized at 89 days postinfection (Fig. 2). Viral RNA was isolated from the inoculum and from serum samples collected at sequential times postinfection. Sequence analysis identified a heterogeneous population of Rev variants in the inoculum and throughout infection (Fig. 4). The predominant sequence at every time point examined was R93, a variant that was predominant in pony 524 late in infection (4). A minor group of related sequences, represented by R103, was also present in the inoculum and most time points examined (except 14 days postinfection). R103 differed from R93 at eight amino acid positions, but only differed by two amino acids from R1 (which was not observed in pony 625).
FIG. 4.
Frequency and alignment of Rev amino acid (a.a.) sequences obtained from the pony 625 inoculum and sequential serum samples. The frequency (N) and variant identity (V) are given for each time point, and the total number of clones sequenced is indicated below the N column. Sequences are compared to R93, which is the dominant variant in the pony 625 inoculum. For individual sequences, a dot indicates an amino acid identical to R93 at that position, an asterisk indicates a premature stop codon in the rev open reading frame, and a plus sign indicates a frameshift. DPI, days postinfection.
Phylogenetic analysis of pony 625 variants identified two clades, designated clade A and clade B (Fig. 5a); clade A included R93 and related variants, and clade B included R103 and related variants. The variation between these two clades was more distinct and was supported by higher bootstrap values (82%). When the entire data sets of variants from each pony were analyzed together, pony 625 clade A variants grouped with pony 524 clade A variants and pony 625 clade B variants grouped with pony 524 clade B variants (Fig. 5b). Thus, the two subpopulations of Rev showed greater similarity between ponies than within ponies.
FIG. 5.
Phylogenetic analysis of Rev variants. (A) Neighbor-joining tree of the pony 625 Rev amino acid variants based on p distance. The tree is divided into 2 clades, designated clades A and clade B. The Rev variants highlighted by yellow boxes were biologically assayed for Rev activity. (B) Neighbor-joining tree of the combined pony 524 and pony 625 Rev amino acid variants based on p distance. Solid lines indicate pony 524 variants, and dashed lines indicate pony 625 variants.
Subpopulations of Rev quasispecies coexist in vivo.
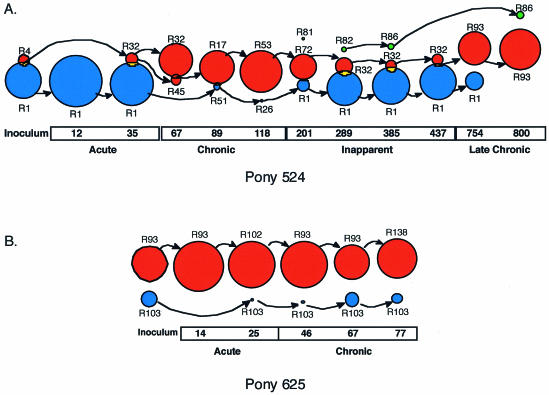
In each horse, most time points sampled contained variants representative of both subpopulations of Rev quasispecies. To more specifically characterize the temporal relationship between the two subpopulations, we developed a computer program, designated partition analysis of quasispecies (PAQ), to detect natural partitions, or groups, of nucleotide variants at each time point (3). The program uses the Hamming distance as a measure of distance between rev/tm genotypes and employs a nonhierarchical clustering method to define grouping of the variants, represented graphically as circles. Each group contains a central genotype, which is the genotype most representative of all other variants within the group. Application of PAQ to the rev/tm data sets indicated that for each pony, most time points could be partitioned into two or three groups, each with a radius of three nucleotide differences or less (Fig. 6). Statistical analysis confirmed that the mean distance among variants within a group was significantly smaller than the mean distance among variants between groups (P < 0.01). In both ponies, there was 90 to 100% correlation between the partition grouping and the phylogenetic grouping of the variants. Therefore, the PAQ analyses supported the phylogenetic results, providing further evidence that multiple subpopulations of EIAV Rev variants coexisted in vivo.
FIG. 6.
Partition analysis identifies two coexisting subpopulations of rev variants. The groups present at each time point were found with the program PAQ (3). The relative size of each group represents the proportion of the population contained within the group at that time point. The central Rev variant for each group is shown, and the arrows show from which group the central variant likely evolved. Groups that overlap indicate that both groups share at least one variant. For each pony, variants within the same group were generally found within the same clade of the phylogenetic trees (shown in Fig. 3 and 5) and the color of the group corresponds to the clade from which most variants were found. The different groups were designated group 1 (red), group 2 (blue), and group 3 (green). The day postinfection and clinical stage of infection are indicated. (A) Partition analysis of pony 524 rev nucleotide variants at sequential times following infection. (B) Partition analysis of pony 625 rev nucleotide variants at sequential times following infection.
PAQ analysis of the pony 524 rev/tm data set identified three partition groups, designated groups 1, 2, and 3, which corresponded to clades A, B, and C, respectively, in the Rev phylogenetic tree. All groups were defined with a similar radius value; however, the proportion of sequences found within each group was highly variable over the course of infection (Fig. 6a). The greatest proportion of variants in the inoculum and in samples isolated during the acute stage partitioned into group 2, with R1 as the predominant Rev variant and central genotype at each of those time points. The chronic stage of disease was characterized by an expansion of group 1 and a concomitant decrease in the proportion of variants in group 2. Throughout infection, the proportion of variants in group 1 was minor when group 2 variants were predominant and predominant when group 2 variants were minor. The fact that changes in predominance coincided with changes in the clinical stage of disease suggested that groups 1 and 2 represent two distinct subpopulations that differ in selective advantage. Group 1/clade A variants appeared to have had a selective advantage in the in vivo environment present during the chronic and late chronic stages of disease, whereas group 2/clade B variants were better suited to the environment present during the acute and inapparent stages.
Analysis of the pony 625 rev/tm data set with PAQ identified two groups of viral variants, designated groups 1 and 2 (Fig. 6b). As with pony 524, there was concordance between the phylogenetic grouping and the partitioning. Clade A/group 1 variants were predominant in the inoculum and at each time point sampled, while clade B/group 2 variants persisted as a minor population. The two ponies differed with respect to the subpopulation that was predominant in the inoculum and during the acute stage of disease. Clade B/group 2 was predominant in the pony 524 inoculum and during the acute stage of disease, whereas clade A/group 1 was predominant in the pony 625 inoculum and acute stage of disease. From these data, it appears that predominance of the Rev subpopulations during the acute stage of infection was not due to selective pressures exerted during transmission, but rather to a founder effect from the inoculum. In contrast to pony 524, dynamic changes in predominance between the two groups were not observed in pony 625. This may be due, in part, to the fact that the clinical disease course in pony 625 did not progress beyond the chronic stage of disease. Despite these differences, the PAQ analysis of pony 625 variants clearly demonstrated that minor subpopulations of rev/tm quasispecies can persist in EIAV-infected horses.
Subpopulations of Rev differ in phenotype.
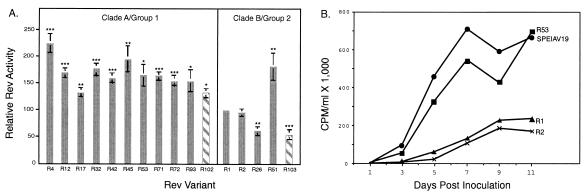
The phylogenetic and partition analyses identified two subpopulations of Rev variants that appeared to coexist and differ in selective advantage. To determine whether there were phenotypic differences between the two Rev subpopulations, we used a chloramphenicol acetyltransferase (CAT) reporter system to quantitate the nuclear export activity of the central genotype from each of the partitions as well as representative variants from the phylogenetic trees. A total of 17 variants from ponies 524 and 625 were tested for Rev phenotype. These variants represented 67% of the rev quasispecies sampled in pony 524 and 54% of the population sampled in pony 625. For each variant, CAT activity was normalized to the activity level of R1, the founder variant in pony 524 (Fig. 7a), and results are reported as Rev activity relative to R1. For 16 of the 17 variants tested, Rev activity segregated according to the genetic groups identified by phylogenetic and partition analyses. Furthermore, Rev activity of all variants from clade A was significantly higher than the activity of all variants from clade B (P < 0.0001), and the nuclear export activity of Rev variants within group 1 was significantly higher than the activity of variants within group 2 (P < 0.0001).
FIG. 7.
Subpopulations of Rev differ in phenotype. (A) Nuclear export activity of pony 524 variants (solid bars) and pony 625 variants (striped bars) are reported relative to the activity of R1. Results represent the mean activity of at least 9 independent transfections ± standard error of the mean. Variants that differed significantly from the activity of R1 are indicated; *, P < 0.05; **, P < 0.005; and ***, P < 0.00005. (B) In vitro replication of Rev chimeric virus. Chimeric proviruses representative of clade A/group 1 variants (R53) and clade B/group 2 variants (R1 and R2) were constructed in the backbone of pSPEIAV19. Virus stocks were produced from stably transfected cell lines, normalized by RT activity, and inoculated in duplicate onto equine dermal cells. Supernatant was collected every other day and assayed for virion production by RT activity (8). RT activity was quantified with a phosphorimager, and the results of a representative experiment are shown.
To assess if differences observed in transient assays would be biologically significant in the context of replicating virus, we constructed chimeric viruses in which rev/tm variants were inserted in the backbone of the infectious molecular clone of EIAV, pSPEIAV19 (34). Chimeric viruses generated from Rev variants representative of either clade A (R53) or clade B (R1, R2) were assayed for replication phenotype in equine dermal (ED) cells. R1 and R2 chimeric proviruses replicated more slowly and to lower levels than parental SPEIAV19 virus or R53 chimeric provirus in ED cells (Fig. 6b). Hence, the differences in Rev phenotype detected in transient transfection assays translated to differences in levels of virus replication in vitro. These differences in phenotype support our interpretation of PAQ that changes in predominance of groups 1 and 2 over the course of infection were due to differences in selective advantage.
Variation in Rev is not associated with covariation in the RRE.
Rev is a nucleocytoplasmic shuttling protein that binds to a specific sequence on the viral pre-mRNA, designated the Rev-responsive element (RRE). We recently mapped the EIAV RRE to a 57-nucleotide purine-rich sequence in the 5′ portion of env, in a region that also encodes the first exon of Rev (6). It is possible that the genotypic variation we observed in Rev exon 2 was accompanied by compensatory covariation in the RRE. If so, our biological assessment of Rev activity may not accurately reflect Rev activity in vivo.
To detect covariation in Rev and the RRE, a 230-nucleotide region encompassing the RRE was amplified by RT-PCR and cloned, and individual clones were sequenced. RRE variation in pony 524 was analyzed with five sequential plasma samples, representing the inoculum and each clinical stage of disease. Four clones were sequenced from each time point, and there was 100% nucleotide sequence identity among the 20 clones (not shown). Furthermore, the RRE sequence found in the 20 clones was identical to the RRE in the Rev reporter assay used for biological characterization of Rev variations. The finding that the RRE does not covary with Rev indicates that observed differences in Rev phenotype are due to the observed genetic changes in Rev. The high degree of genetic conservation in the RRE is in marked contrast to that found in Rev exon 2 and provides additional evidence that genetic and phenotypic variation in Rev may contribute to EIAV selection in vivo.
DISCUSSION
These studies provide experimental evidence that a viral quasispecies may comprise multiple subpopulations that evolve independently and possess distinct selective advantages. Phylogenetic analysis of the Rev quasispecies in two EIAV-infected ponies identified two subpopulations of variants that differed in their genetic divergence and pattern of evolution. Multiple subpopulations of Rev variants were also detected with partition analyses, which showed that the subpopulations coexisted in vivo and fluctuated in predominance over the course of disease. Transient expression assays demonstrated that the two subpopulations differed in Rev-dependent nuclear export activity. Chimeric proviral clones containing rev/tm sequences representative of each subpopulation demonstrated that the differences in nuclear export activity also translated to differences in the rate and overall level of replication in vitro. Taken together, these results indicate that the rev/tm quasispecies formed two subpopulations that coexisted over the course of infection and differed in Rev phenotype. Subpopulations that differ in phenotype would be predicted to occupy different regions on the fitness landscape and rapidly respond to changes in the landscape topology. Such a model may provide additional insight as to how viruses adapt so quickly to changes in the host environment.
Several studies have reported the presence of genetically distinct populations of HIV-1 in different infected organs and even in different regions within the same organ (1, 35, 39-41). Anatomical sequestration of virus populations may provide a biological niche that could lead to the subdivision of the quasispecies into smaller, semi-isolated groups that differ in phenotype and/or selective advantage. For example, the differential expression of HIV-1 coreceptors on CD4+ subsets was associated with the coexistence and independent evolution of HIV-1 viruses that differed in syncytium-inducing phenotype (39). Our analyses of the evolution of Rev quasispecies during progression of EIA disease included over 400 rev/tm sequences. Although they were derived from only two experimentally infected ponies and the genetic variation within the entire data set of rev/tm variants was relatively low, each of the genotypic and phenotypic assays pointed to the same conclusion: two coexisting subpopulations within the Rev quasispecies that differed not only genetically but also phenotypically. Furthermore, the changes in population size coincided with changes in disease state. Together, these data strongly suggest the presence of subpopulations of Rev that occupied distinct biological niches, which differed in selective advantage during progression of disease.
Genotypes representative of the two subpopulations of Rev quasispecies exhibited significant differences in biological activity, as measured by transient transfection assays and chimeric provirus replication experiments. The chronic stage of disease was predominated by variants that had higher Rev activity, while the inapparent stage of disease was characterized by a predominance of variants with lower Rev activity. These findings agree with the statistical analyses of quasispecies Rev activity in pony 524, which showed that the clinical stages of EIAV infection were correlated with Rev activity (4). The fact that similar conclusions were reached via different computational and genetic approaches supports the hypothesis that Rev phenotype contributes to variant selection in vivo. At this time, however, the in vivo selection pressures on these two subpopulations of Rev variants are unknown.
Recent studies by Bobbitt et al. (7) demonstrated that variation in HIV-1 Rev activity can alter sensitivity to CTL killing by downmodulating viral late gene expression. A single amino acid substitution in HIV-1 Rev resulted in a two- to threefold reduction in Gag expression and a significant decrease in CTL killing. Furthermore, primary HIV-1 isolates from asymptomatic patients showed reduced Rev activity, reduced levels of Gag expression, and increased resistance to CTL killing. Similar to our results, these findings indicate that limited amino acid changes in Rev can attenuate activity and may confer a selective advantage in vivo. While changes in the cytoplasmic region of TM and/or changes in cytotoxic T-cell epitopes may also contribute to variant selection, it is reasonable to hypothesize that attenuation of Rev activity may be advantageous for viral persistence in the presence of a significant host immune response. Ongoing studies in additional EIAV-infected horses will increase our understanding of the role of Rev variation in immune evasion and viral persistence.
The coexistence of multiple subpopulations within a viral quasispecies implies the persistence of minor subpopulations of viral variants that have reduced fitness relative to variants in the predominant population. This impacts our understanding of how virus populations evolve and how their evolution may be modeled. Multiple coexisting subpopulations that differ in selective advantage would occupy different regions on a fitness landscape and would allow the virus to adapt rapidly to changes in the host environment by expanding minor populations that may be near new fitness peaks. This may be especially relevant in modeling reservoirs of persistent replicating virus and the emergence of virus variants. For example, failure of antiviral drug therapy can arise from de novo generation of resistant variants due to incomplete suppression of replicating virus or from selection of a minor pool of existing resistant variants.
In a quasispecies model where there is only one population of variants, existing resistant variants are rare, and drug resistance would result from the accumulation and fixation of mutations that reduce susceptibility to therapy. In a model with multiple subpopulations, an existing minor pool of less fit genotypes may include resistant variants or variants that require fewer point mutations to confer drug resistance. In this model, the changes in fitness landscape following the onset of antiviral therapy would induce rapid expansion of the minor subpopulation of resistant variants and persistence of drug-sensitive variants as the minor population. The latter model may help explain the rapid rebound of wild-type virus observed in some HIV-1 patients undergoing structured treatment interruptions (9, 10, 12, 33). Further examination of the genetic and phenotypic complexity of virus populations in vivo will aid our understanding of quasispecies structure and lentivirus evolution.
Acknowledgments
We thank Susan Payne for the generous gift of pSPEIAV19. Gavin Naylor and Curtis Eckerman are thanked for assistance with the phylogenetic analyses and for critical reviews of the manuscript.
This work was partially supported by NSF grant DGE9972653 and by USDA grant 96-358204-3847.
REFERENCES
- 1.Ait-Khaled, M., J. E. McLaughlin, M. A. Johnson, and V. C. Emery. 1995. Distinct HIV-1 long terminal repeat quasispecies present in nervous tissues compared to that in lung, blood and lymphoid tissues of an AIDS patient. AIDS 9:675-683. [DOI] [PubMed] [Google Scholar]
- 2.Alexandersen, S., and S. Carpenter. 1991. Characterization of variable regions in the envelope and S3 open reading frame of equine infectious anemia virus. J. Virol. 65:4255-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baccam, P., R. J. Thompson, O. Fedrigo, S. Carpenter, and J. L. Cornette. 2001. PAQ: Partition analysis of quasispecies. Bioinformatics 17:16-22. [DOI] [PubMed] [Google Scholar]
- 4.Belshan, M., P. Baccam, J. L. Oaks, B. A. Sponseller, S. C. Murphy, J. Cornette, and S. Carpenter. 2001. Genetic and biological variation in equine infectious anemia virus Rev correlates with variable stages of clinical disease in an experimentally infected pony. Virology 279:185-200. [DOI] [PubMed] [Google Scholar]
- 5.Belshan, M., M. E. Harris, A. E. Shoemaker, T. J. Hope, and S. Carpenter. 1998. Biological characterization of Rev variation in equine infectious anemia virus. J. Virol. 72:4421-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belshan, M., G. S. Park, P. Bilodeau, C. M. Stoltzfus, and S. Carpenter. 2000. Binding of equine infectious anemia virus Rev to an exon splicing enhancer mediates alternative splicing and nuclear export of viral mRNAs. Mol. Cell. Biol. 20:3550-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobbitt, K. R., M. M. Addo, M. Altfeld, T. Filzen, A. A. Onafuwa, B. D. Walker, and K. L. Collins. 2003. Rev activity determines sensitivity of HIV-1-infected primary T cells to CTL killing. Immunity 18:289-299. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter, S., and B. Chesebro. 1989. Change in host cell tropism associated with in vitro replication of equine infectious anemia virus. J. Virol. 63:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun, T. W., R. T. Davey, M. Ostrowski, J. S. Justement, D. Engel, J. I. Mullins, and A. S. Fauci. 2000. Relationship between pre-existing viral reservoirs and the reemergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 6:757-761. [DOI] [PubMed] [Google Scholar]
- 10.Davey, T., N. Bhat, C. Yoder, T.-W. Chun, J. A. Metcalf, R. Dewar, V. Natarajan, R. A. Lempicki, J. W. Adelsberger, K. D. Miller, J. A. Kovacs, M. A. Polis, R. E. Walker, J. Falloon, H. Masur, D. Gee, M. Baseler, D. S. Dimitrov, A. S. Fauci, and H. C. Lane. 1999. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc. Natl. Acad. Sci. USA 96:15109-15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domingo, E., C. Escarmis, N. Sevilla, A. Moya, S. F. Elena, J. Quer, I. S. Novella, and J. J. Holland. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859-864. [DOI] [PubMed] [Google Scholar]
- 12.Dybul, M., M. Dacher, M. A. Jensen, C. W. Hallahan, T.-W. Chun, M. Belson, B. Hidalgo, D. C. Nickle, C. Yoder, J. A. Metcalf, R. T. Davey, L. Ehler, D. Kress-Rock, E. Nies-Kraske, S. Liu, J. I. Mullins, and S. Fauci. 2003. Genetic characterization of rebounding human immunodeficiency virus type 1 in plasma during multiple interruptions of highly active antiretroviral therapy. J. Virol. 77:3229-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eigen, M. 1993. Viral quasispecies. Sci. Am. 269:42-49. [DOI] [PubMed] [Google Scholar]
- 14.Eigen, M., and P. Schuster. 1977. The hypercycle. A principle of natural self-organization. Part A: emergence of the hypercycle. Naturwissenschaften 64:541-565. [DOI] [PubMed] [Google Scholar]
- 15.Gonda, M. A., H. P. Charman, J. L. Walker, and L. Coggins. 1978. Scannning and transmission electron microscopic study of equine infectious anemia virus. Am. J. Vet. Res. 39:731-740. [PubMed] [Google Scholar]
- 16.Hammond, S. A., S. J. Cook, D. L. Lichtenstein, C. J. Issel, and R. C. Montelaro. 1997. Maturation of the cellular and humoral immune responses to persistant infection in horses by equine infectious anemia virus is a complex and lengthy process. J. Virol. 71:3840-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond, S. A., F. Li, B. M. McKeon, S. J. Cook, C. J. Issel, and R. C. Montelaro. 2000. Immune responses and viral replication in long-term inapparent carrier ponies inoculated with equine infectious anemia virus. J. Virol. 74:5968-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrold, S. M., S. J. Cook, R. F. Cook, K. E. Rushlow, C. J. Issel, and R. C. Montelaro. 2000. Tissue sites of persistent infection and active replication of equine infectious anemia virus during acute disease and asymptomatic infection in experimentally infected equids. J. Virol. 74:3112-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holland, J. J., J. C. De La Torre, and D. A. Steinhauer. 1992. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 176:1-20. [DOI] [PubMed] [Google Scholar]
- 20.Issel, C. J., and L. Coggins. 1979. Equine infectious anemia: current knowledge. J. Am. Vet. Med. Assoc. 174:727-733. [PubMed] [Google Scholar]
- 21.Kawakami, T., L. Sherman, J. Dahlbert, A. Gazit, A. Yaniv, S. R. Tronick, and S. A. Aaronson. 1987. Nucleotide sequence analysis of equine infectious anemia proviral DNA. Virology 158:300-312. [DOI] [PubMed] [Google Scholar]
- 22.Kono, Y. 1969. Viremia and immunological responses in horses infected with equine infectius anemia virus. Natl. Inst. Anim. Health Q. 9:1-9. [PubMed] [Google Scholar]
- 23.Kono, Y., K. Hirasawa, Y. Fukunaga, and T. Taniguchi. 1976. Recrudescence of equine infectious anemia by treatment with immunosuppressive drugs. Natl. Inst. Anim. Health Q. 16:8-15. [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 26.Leroux, C., J. K. Craigo, C. J. Issel, and R. C. Montelaro. 2001. Equine infectious anemia virus genomic evolution in progressor and nonprogressor ponies. J. Virol. 75:4570-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leroux, C., C. J. Issel, and R. C. Montelaro. 1997. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J. Virol. 71:9627-9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacQueen, J. 1965. Some methods for classification and analysis of multivariate observations. Proc. 5th Berkeley Symp. Math. Stat. Probab. 1:281-297. [Google Scholar]
- 29.McGuire, T. C., K. O'Rourke, and W. P. Cheevers. 1987. A review of antigenic variation by equine infectious anemia virus. Contrib. Microbiol. Immunol. 8:77-89. [PubMed] [Google Scholar]
- 30.McGuire, T. C., D. B. Tumas, K. M. Byrne, M. T. Hines, S. R. Leib, A. L. Brassfield, K. I. O'Rourke, and L. E. Perryman. 1994. Major histocompatability complex-restricted CD8+ cytotoxic T lymphocytes from horses with equine infectious anemia virus recognize Env and Gag/PR proteins. J. Virol. 68:1459-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montelaro, R. C., W. G. Robey, M. D. West, C. J. Issel, and P. J. Fischinger. 1988. Characterization of the serological cross-reactivity between glycoproteins of the human immunodeficiency virus and equine infectious anemia virus. J. Gen. Virol. 69:1711-1717. [DOI] [PubMed] [Google Scholar]
- 32.Oaks, J. L., T. C. McGuire, C. Ulibarri, and T. B. Crawford. 1998. Equine infectious anemia virus is found in tissue macrophages during subclinical infection. J. Virol. 72:7263-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortiz, G. M., M. Ellons, J. Brancato, H. T. T. Vo, R. L. Zinn, D. E. Clarkson, K. Van Loon, S. Bonhoeffer, G. D. Miralles, D. Montefiori, J. A. Bartlett, and D. F. Nixon. 2001. Structured antiretroviral treatment interruptions in chronically HIV-1-infected subjects. Proc. Natl. Acad. Sci. USA 98:13288-13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne, S. L., J. Rausch, K. Rushlow, R. C. Montelaro, C. Issel, M. Flaherty, S. Perry, D. Sellon, and F. Fuller. 1994. Characterization of infectious molecular clones of equine infectious anaemia virus. J. Gen. Virol. 75:425-429. [DOI] [PubMed] [Google Scholar]
- 35.Shapshak, P., D. M. Segal, K. A. Crandall, R. K. Fujimura, B. Zhang, K. Xin, K. Okuda, C. K. Petito, C. Eisdorfer, and K. Goodkin. 1999. Independent evolution of HIV type 1 in different brain regions. AIDS Res. Hum. Retrovir. 15:811-820. [DOI] [PubMed] [Google Scholar]
- 36.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 37.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tumas, D. B., M. T. Hines, L. E. Perryman, W. C. Davis, and T. C. McGuire. 1994. Corticosteroid immunosuppression and monoclonal antibody-mediated CD5+ T lymphocyte depletion in normal and equine infectious anaemia virus-carrier horses. J. Gen. Virol. 75:959-968. [DOI] [PubMed] [Google Scholar]
- 39.van Rij, R. P., H. Blaak, J. A. Visser, M. Brouwer, R. Rientsma, S. Broersen, A.-M. de Roda Husman, and H. Schuitemaker. 2000. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J. Clin. Investig. 106:1039-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van't Wout, A. B., L. J. Ran, C. L. Kuiken, N. A. Kootstra, S. T. Pals, and H. Schuitemaker. 1998. Analysis of the temporal relationship betweeen human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J. Virol. 72:488-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong, J. K., C. C. Ignacio, F. Torriani, D. Havlir, N. J. Fitch, and D. D. Richman. 1997. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J. Virol. 71:2059-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]