Abstract
Molecular events and the interdependence of the two rotavirus nonstructural proteins, NSP5 and NSP2, in producing viroplasm-like structures (VLS) were previously evaluated by using transient cellular coexpression of the genes for the two proteins, and VLS domains as well as the NSP2-binding region of NSP5 were mapped in the context of NSP2. Review of the previous studies led us to postulate that NSP2 binding of NSP5 may block the N terminus of NSP5 or render it inaccessible and that any similar N-terminal blockage may render NSP5 alone capable of producing VLS independent of NSP2. This possibility was addressed in this report by using two forms of NSP5-green fluorescent protein (GFP) chimeras wherein GFP is fused at either the N or the C terminus of NSP5 (GFP-NSP5 and NSP5-GFP) and evaluating their VLS-forming capability (by light and electron microscopy) and phosphorylation and multimerization potential independent of NSP2. Our results demonstrate that NSP5 alone can form VLS when the N terminus is blocked by fusion with a nonrotavirus protein (GFP-NSP5) but the C terminus is unmodified. Only GFP-NSP5 was able to undergo hyperphosphorylation and multimerization with the native form of NSP5, emphasizing the importance of an unmodified C terminus for these events. Deletion analysis of NSP5 mapped the essential signals for VLS formation to the C terminus and clearly suggested that hyperphosphorylation of NSP5 is not required for VLS formation. The present study emphasizes in general that when fusion proteins are used for functional studies, constructs that represent fusions at both the N and the C termini of the protein should be evaluated.
One of the members of the family Reoviridae, rotavirus, is known to cause severe gastroenteritis in children and animals (14). The virus contains 11 double-stranded RNA segments that code for six structural proteins, namely, VP1 to VP4, VP6, and VP7, and six nonstructural proteins, NSP1 to NSP6 (9, 14). During infection, viral mRNAs direct the synthesis of viral proteins and serve as templates for genomic double-stranded RNA. Since rotavirus is a cytoplasmic virus, the processes of viral RNA packaging, assortment, replication, and assembly of the double-layered particle are thought to occur in large discrete cytoplasmic electrodense particulate structures called viroplasms (11, 23). Similar structures called viroplasm-like structures (VLS) have been observed by cotransfection of NSP5 and NSP2 expression plasmids, wherein the expressed NSP2 protein binds to the N-terminal region of NSP5, an established requirement in facilitating VLS formation (11, 20). Several rotavirus proteins localize to viroplasms during infection (3, 12, 19, 23). During infection, the NSP5 protein is synthesized as a 26- to 28-kDa protein and, after undergoing O-linked glycosylation and phosphorylation at specific serines or threonines, reaches a size of 32 to 34 kDa (1, 2, 4-6, 11, 13, 20). Studies so far have shown that NSP2 binding of NSP5 leads to phosphorylation of NSP5 (1, 2, 11, 20). The C-terminal region of NSP5 is responsible for hyperphosphorylation, multimerization, and interaction with NSP6 (21). However, according to a recent study, NSP5 phosphorylation per se is not essential for viroplasm formation in rotavirus infection (8).
Previous gene cotransfection studies with rotavirus NSP5 were performed by expressing either the native form of NSP5 (both termini unmodified) or a modified form of NSP5 in which the C terminus is fused to green fluorescent protein (GFP) (NSP5-GFP) by using expression plasmid pEGFP-N1 (8, 11, 12, 21). We noticed in retrospect that in the previous studies, another type of chimeric NSP5 construct, i.e., NSP5 with the N terminus fused to GFP (GFP-NSP5) but with the C terminus unmodified, was lacking. This construct could resemble NSP2-bound NSP5, which facilitates VLS formation, as in both cases the N terminus of NSP5 would be somewhat inaccessible but the C terminus would be unmodified. Hence, in this report, we used both forms of chimeras of NSP5 and GFP (i.e., GFP-NSP5 and NSP5-GFP) in the absence of NSP2 and compared their potentials to form VLS and undergo phosphorylation and multimerization in a transient expression assay. Our results demonstrated for the first time that NSP5 alone is capable of forming VLS when (i) its N terminus is blocked, in this case by fusion with either GFP or a hemagglutinin (HA) tag, and (ii) its C terminus is unmodified. The GFP-NSP5 construct physiologically mimicked NSP2-NSP5 interdependence by forming VLS and was able to undergo hyperphosphorylation as well as multimerization with the native form of NSP5. Furthermore, this type of chimera, wherein the N terminus is fused or blocked but the C terminus is unmodified, provided an opportunity to identify a minimal domain required to form VLS in rotavirus NSP5 in the absence of NSP2. The results described in this report also emphasize that NSP5 phosphorylation is not required for VLS formation, as reported by others recently (8).
MATERIALS AND METHODS
Cells and viruses.
MA104 cells (American Type Culture Collection, Manassas, Va.) and COS-7L cells (Gibco-BRL, Gaithersburg, Md.) were routinely cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 2 mM l-glutamine, and 50 mg of gentamicin/ml. Simian rotavirus strain SA11 (American Type Culture Collection) was propagated in MA104 cells as described previously (16).
Construction of plasmids.
We used rotavirus strain SA11 to amplify and clone various NSP5 and NSP2 gene constructs described in this report. Using a pair of NSP5 open reading frame (ORF) sequence-specific reverse transcription-PCR primers (17) (Table 1), we amplified the gene from rotavirus-infected cellular RNA. The amplified product was unidirectionally cloned into EcoRI- and PstI-digested vector pEGFP-C2 for N-terminal GFP fusion and EcoRI- and BamHI-digested vector pEGFP-N3 for C-terminal GFP fusion (Clontech, Palo Alto, Calif.). Several deletions in the ORF were generated by PCR with pEGFP-NSP5 as a template and with appropriate primers, followed by unidirectional cloning into the respective vectors as described above (Table 1). To express NSP5 in its native form, we cloned the ORF into vector pCMV-Script (Stratagene, La Jolla, Calif.) at EcoRI and XhoI sites to produce pCMV-NSP5. HA-NSP5 fusion chimeras were constructed with vectors pCMV and pEGFP-N3 by cloning a PCR fragment amplified with a 5′ primer that contained a start codon (underlined) and the sequence for the 11-amino-acid epitope for HA fused to the 5′ of NSP5 ORF sequence (5′-GCCGAATTCGATGTACCCATACGATGTTCCAGATTACGCCTCGTTGTCTCTCAGTATTGACGTGACGAGTCTTCC-3′) and with two different 3′ primers, one for pCMV (3′-5255; Table 1) and another for pEGFP-N3 (3′-5872; Table 1), such that the expressed NSP5 protein has either the N terminus alone fused (in pCMV) or both the N and the C termini fused (HA-NSP5-GFP) to a foreign protein sequence. The NSP2 gene was amplified by gene-specific reverse transcription-PCR from RNA isolated from virus-infected cells as described previously (15) and was cloned in pDS1Red-1 (Clontech) such that the C terminus of NSP2 is fused to the red fluorescent protein (NSP2-Red), leaving the N terminus free for NSP5 interaction. Primers complementary to the 5′ end (5′-GTTCGAATTCTGATGGCTGAGCTAGCTTGC-3′) and the 3′ end (5′-CGGTGGATCCCGAACGCCAACTTGAGAAAC-3′) of the ORF were used in the amplification. Amplified products were digested and cloned into the EcoRI and BamHI sites of pDS1Red-1. The recombinant plasmids were selected on Luria broth plates containing 50 μg of kanamycin/ml.
TABLE 1.
Primers used in this study
| Vector | Fusion | Gene amplified | Primer
|
|
|---|---|---|---|---|
| Designation | Sequence | |||
| pEGFP-C2 (EcoRI-PstI cloning) | GFP-NSP5 | ORF | 5′-5254 | 5′-CGCGAATTCATGTCTCTCAGTATTGACGTG |
| 3′-5255 | 5′-GGCCTGCAGTTACAAATCTTCAATCAA | |||
| NΔ65 | 5′-5647 | 5′-CGCGAATTCGCTTCAAACAATCCA | ||
| 3′-5255 | 5′-GGCCTGCAGTTACAAATCTTCAATCAA | |||
| NΔ130 | 5′-6498 | 5′-CGCGAATTCGATACTAAAAAGGAG | ||
| 3′-5255 | 5′-GGCCTGCAGTTACAAATCTTCAATCAA | |||
| NCΔ65 | 5′-5647 | 5′-CGCGAATTCGCTTCAAACAATCCA | ||
| 3′-6499 | 5′-GGCCTGCAGTTAATCCGTAGATATTGA | |||
| CΔ66 | 5′-5254 | 5′-CGCGAATTCATGTCTCTCAGTATTGACGTG | ||
| 3′-6499 | 5′-GGCCTGCAGTTAATCCGTAGATATTGA | |||
| CΔ130 | 5′-5254 | 5′-CGCGAATTCATGTCTCTCAGTATTGACGTG | ||
| 3′-8923 | 5′-GGCCTGCAGAGAATCAGATGGTCC | |||
| pEGFP-N3 (EcoRI-BamHI cloning) | NSP5-GFP | ORF | 5′-5871 | 5′-CGCTCGAATTCTATGTCTCTCAGCATTGAC |
| 3′-5872 | 5′-GATGGATCCCAAATCTTCTATCAATTG | |||
| NΔ65 | 5′-8250 | 5′-CGCTCGAATTCTGCTTCAAACGAT | ||
| 3′-5872 | 5′-GATGGATCCCAAATCTTCTATCAATTG | |||
| NΔ130 | 5′-8251 | 5′-CGCTCGAATTCTGATACTAAAAAG | ||
| 3′-5872 | 5′-GATGGATCCCAAATCTTCTATCAATTG | |||
| CΔ66 | 5′-5871 | 5′-CGCTCGAATTCTATGTCTCTCAGCATTGAC | ||
| 3′-8252 | 5′-GATGGATCCATCCGTAGATATTGA | |||
Transfection and immunofluorescence assay.
Transient transfection was performed with MA104 and COS-7L cells maintained for 24 to 36 h in six-well culture plates and chamber slides (Labtek). Cells were transfected with 2 to 5 μg of plasmid DNA by use of Lipofectamine Plus reagent in accordance with the manufacturer's instructions (Invitrogen). Slides were fixed with ice-cold acetone and mounted with Vectashield (Vector Laboratories, Burlingame, Calif.). To compare the morphologies of VLS in transfection and rotavirus infection, cells transfected with GFP-NSP5 plasmid DNA were infected 18 h posttransfection with rotavirus strain SA11 at a multiplicity of infection (MOI) of 1 (18). Cells were fixed 12 h postinfection with ice-cold ethanol, rinsed with phosphate-buffered saline (PBS), permeabilized with 0.3% Triton X-100-PBS for 5 min at room temperature, and washed twice with PBS. Cells were incubated with a guinea pig anti-NSP5 antibody at 37°C for 1 h and washed three times with PBS. Cells were incubated with Texas red-labeled anti-guinea pig secondary antibodies (Vector Laboratories) for 1 h at 37°C and washed three times with PBS. Visualization, analysis, and photography were all performed with a Carl Zeiss laser scanning confocal microscope (model LSM5 PASCAL) equipped with a microprocessor. Images were transferred to Adobe Photoshop version 5.0 (for the PC) for labeling and printing.
Transmission electron microscopy (TEM).
To compare the morphological similarities between the viroplasms formed during infection and the VLS formed during GFP-NSP5, GFP-NΔ65 (NΔ65 is NSP5 with a deletion of the N-terminal 65 amino acids), and NSP5-NSP2 cotransfections, infected and transfected cells were pelleted and fixed for 2 to 3 h with electron microscopy fixative (2% glutaraldehyde-2% paraformaldehyde in 0.1 M sodium cacodylate buffer [pH 7.3]). Samples were transferred to and stored in PBS at 4°C until processed. Cells were subsequently postfixed with 2% osmium tetroxide, dehydrated with a graded series of alcohol, and embedded in epoxy resin. Thin sections were stained with uranyl acetate and lead citrate and were examined for virus particles with a Zeiss EM 912 Omega electron microscope.
Immunoblotting and immunoprecipitation analysis.
NSP5-transfected MA104 and COS-7L cells were washed with PBS, centrifuged at 3,000 × g in a refrigerated centrifuge (Sorvall), and extracted with radioimmunoprecipitation (RIPA) buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1.0% NP-40, 0.5% sodium deoxycholate). To confirm that the higher-molecular-weight forms of the protein observed in NSP5 transfections were hyperphosphorylated products related to NSP5, mock- and NSP5-transfected cytolysates were immunoprecipitated with 2 μl of an antiphosphoserine mouse monoclonal antibody (Abcam) by overnight incubation at 4°C. To analyze the potential of both GFP-NSP5 and NSP5-GFP for multimerization with the native form of NSP5, each GFP chimeric construct was cotransfected along with pCMV-NSP5, and the lysates were incubated overnight at 4°C with 2 μl of an anti-GFP mouse monoclonal antibody (Clontech). The immune complexes were collected with the addition of Sepharose beads (Pharmacia) by centrifugation at 3,000 × g for 5 min. Sample buffer (40 μl) was added to the Sepharose beads, and the samples were denatured for 5 min in a boiling water bath, subjected to electrophoresis, and analyzed by immunoblotting with an anti-NSP5 antibody as a probe. Briefly, the samples were resolved by sodium dodecyl sulfate-4 to 20% polyacrylamide gel electrophoresis (Invitrogen) and transferred to a polyvinylidene difluoride membrane (Millipore). The NSP5 phosphorylated forms and multimerization were detected by incubation of the individual blot with 1:1,000-diluted guinea pig anti-NSP5 antibody overnight at 4°C (13). Secondary incubation with 1:10,000-diluted peroxidase-conjugated anti-guinea pig secondary antibodies (Chemicon) was carried out at room temperature for 1 h. Signals were detected by enhanced chemiluminescence (Pierce).
The effect of NSP2 or a phosphatase inhibitor (okadaic acid) on a mobility shift indicative of the level of phosphorylation of NSP5-GFP, GFP-NSP5, and pCMV-NSP5 was assessed essentially as follows. Okadaic acid (Sigma) was added at 0.5 μM to MA104 cells and at 0.1 μM to COS-7L cells 2 h prior to and during the entire 24-h transfection phase. The NSP2-Red plasmid was cotransfected with NSP5 plasmids to observe any mobility shift (phosphorylation status) of NSP5 protein and consequently an effect on its ability to form VLS.
RESULTS
A blocked N terminus and an unmodified C terminus are sufficient for NSP5 to form VLS in transfected cells.
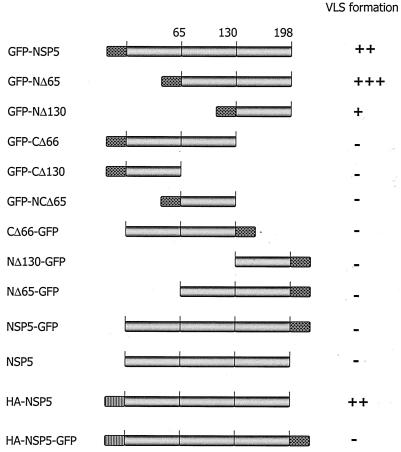
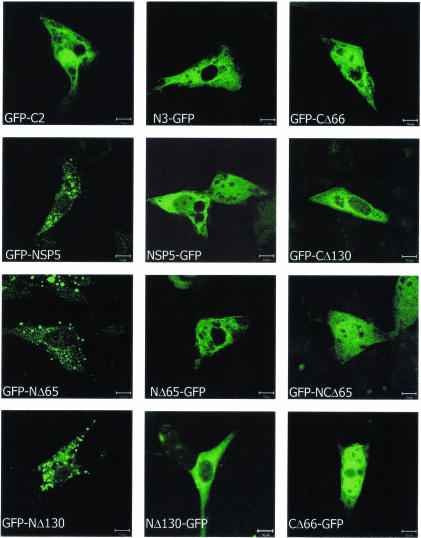
Rotavirus NSP2 binds to NSP5 in the N-terminal region and facilitates the formation of VLS in NSP5-NSP2-cotransfected cells, suggesting that NSP5 with the N-terminal region blocked, as achieved by NSP2 binding, but with the C-terminal region unmodified, is crucial. If this is true, then VLS formation perhaps could be achieved, even in the absence of NSP2, if the N terminus of NSP5 were blocked by fusion with a nonrotavirus marker. Therefore, to elucidate the role of unmodified N and C termini of NSP5 in VLS formation, we generated chimeras of NSP5 by fusing GFP to either the N or the C terminus (Fig. 1). We transfected MA104 cells (a simian kidney cell line) with either GFP-NSP5 or NSP5-GFP constructs. Cells were analyzed 24 h posttransfection for chimeric NSP5 intracellular expression and localization by confocal microscopy. Surprisingly, we observed that recombinant GFP-NSP5 (NSP5 with its N terminus blocked and an unmodified C terminus) formed discrete cytoplasmic vesicular structures, which we termed VLS (Fig. 2). In contrast, the NSP5-GFP construct (NSP5 with a free N terminus but with its C terminus blocked) showed a diffuse cytoplasmic pattern (Fig. 2). These results clearly support the idea that a blocked N terminus and an unmodified C terminus can impart VLS activity to NSP5 without the help of NSP2.
FIG. 1.
Schematic representation of various NSP5 constructs used in this study. Rotavirus NSP5 was fused with GFP (checkered box) at either the N terminus (GFP-NSP5) or the C terminus (NSP5-GFP). N-terminal fusion of an HA tag is indicated by a hatched box. The VLS-forming abilities of the various NSP5 constructs are shown on the right (−, negative with diffuse localization; +, positive, few and small; ++, positive, moderate in size and number; +++, positive, intense and large VLS).
FIG. 2.
Patterns of localization of transiently expressed full-length NSP5 and deletion constructs. Plasmid-transfected MA104 cells were fixed after 18 h and analyzed by confocal microscopy. GFP-C2 and N3-GFP represent GFP expression alone from the parent empty vectors that were used in the GFP-NSP5 and NSP5-GFP constructs and in the related deletion constructs (as labeled on the images) to serve as GFP controls. Note that only the N-terminal fusion constructs containing the C terminus resulted in VLS formation. Bars, 10 μm.
Analysis of NSP5 domains essential for VLS formation.
To identify which region of NSP5 alone is responsible for the formation of discrete structures, we generated a series of eight deletion chimeras of NSP5 by fusing GFP to either the N or the C terminus (Fig. 1). Upon individual transfection of these plasmid chimeras, protein expression, localization, and appearance were analyzed by confocal microscopy. The results are shown in Fig. 2. GFP-NSP5, GFP-NΔ65, and GFP-NΔ130 produced VLS. The two C-terminal deletion constructs, GFP-CΔ66 and GFP-CΔ130, and a double-deletion construct in which both N- and C-terminal regions were lacking (GFP-NCΔ65) developed a diffuse cytoplasmic expression pattern. NSP5-GFP and related deletion constructs (NΔ65-GFP, NΔ130-GFP, and CΔ66-GFP) all failed to form VLS as well. GFP-C2 and N3-GFP represent the two respective “empty” plasmids that served as controls for transfection and that were used to construct GFP-NSP5, NSP5-GFP, and related deletion constructs (Fig. 2).
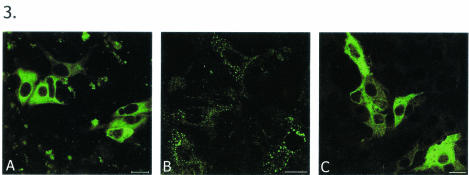
To confirm the above observations that NSP5 alone can form VLS and that this activity is not an artifact of GFP, we constructed another set of NSP5 expression plasmids. One expressed the native form of NSP5 (pCMV-NSP5), which by itself does not form VLS (Fig. 3A), and another, a chimeric form of NSP5 in which an HA tag is fused to the N terminus of NSP5 (HA-NSP5) but in which the C terminus is unmodified. Transient expression of the latter construct alone also resulted in the formation of VLS (Fig. 3B) similar to that seen with GFP-NSP5. Furthermore, to check whether the unmodified C terminus of NSP5 also plays a role in the formation of VLS, nonrotavirus protein sequences were fused at both termini of NSP5 (HA-NSP5-GFP, as shown in Fig. 1). Interestingly, this type of chimeric NSP5 protein showed a diffuse cytoplasmic pattern, suggesting the indispensability of an unmodified C terminus in the formation of VLS (Fig. 3C).
FIG. 3.
Analysis of a fused N terminus and an unmodified C terminus of NSP5 for VLS formation. MA104 cells were transfected with NSP5-containing plasmids, fixed, and immunostained with anti-NSP5 antibody. (A) Pattern of localization of native form of NSP5. (B) Pattern of expression of HA-NSP5. Note that this construct, in which the N terminus of NSP5 is fused, is able to form VLS. (C) Lack of VLS-forming ability of HA-NSP5-GFP, in which both the N and the C termini are fused to a nonrotavirus protein. Bars, 10 μm.
VLS formed by GFP-NSP5 expression alone and infection-associated viroplasms have similar morphologies and locations.
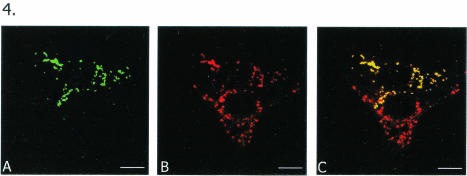
To find out whether VLS do resemble infection-associated viroplasms in morphology and location, cells initially transfected with the full-length construct GFP-NSP5 for 18 h were infected with rotavirus SA11, and the samples were processed for analysis at the light microscope level. Striking similarities in the morphologies and cytoplasmic locations of VLS and viroplasms were seen in two adjacent cells, one transfected (top) and the other infected (bottom), shown in Fig. 4. The top cell expresses green GFP-NSP5 (Fig. 4A), and the bottom cell has infection-associated viroplasms immunostained in red by the anti-NSP5 antibody (Fig. 4B). The anti-NSP5 antibody also stained GFP-NSP5 to appear red in the top cell (Fig. 4B); this cell may or may not have been infected with the virus—most likely not, given the low MOI (1) coupled with a low efficiency of transfection, making a cotransfected-infected cell a rare occurrence. Thus, in the overlay (Fig. 4C), GFP-NSP5-associated VLS turned yellow due to the dual staining (green and red) of GFP-NSP5 in the top cell, whereas the infection-associated viroplasms remained red. Figure 4C thus provided an opportunity to examine the morphological similarities between VLS and viroplasms and their similar cytoplasmic locations in adjacent cells.
FIG. 4.
Morphologic similarities and cytoplasmic localization of VLS formed by GFP-NSP5 and infection-associated viroplasms in two adjacent cells at the light microscopic level. MA104 cells were first infected with rotavirus SA11 and then transfected with pGFP-NSP5. (A) Top cell expresses green GFP-NSP5. (B) Bottom cell demonstrates infection-associated viroplasms immunostained in red by anti-NSP5 antibody. Anti-NSP5 antibody also stained GFP-NSP5 in red in the top cell; this cell may or may not have been infected with the virus—most likely not, given the low MOI (1) coupled with a low efficiency of transfection, making a cotransfected-infected cell a rare occurrence. (C) Overlay shows GFP-NSP5-associated VLS in yellow due to the dual staining (green and red) of the chimeric protein in the top cell, whereas the infection-associated viroplasms remain red. Note morphologic similarities between VLS and viroplasms as well as their similar locations in adjacent cells. Bars, 10 μm.
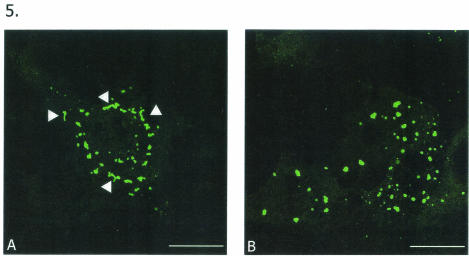
Since GFP-NΔ65 expression resulted in much more intense and profuse VLS formation, we also compared the morphologies of these structures and those formed by GFP-NSP5. Interestingly, GFP-NSP5, in addition to forming punctate VLS, also formed slender, tubular filamentous structures (Fig. 5A). In contrast, GFP-NΔ65 showed intense formation of spherical and punctate VLS similar to those seen during infection (Fig. 5B).
FIG. 5.
Comparison of VLS formed by full-length and NΔ65 NSP5 constructs. MA104 cells were transfected with GFP-NSP5 (A) and GFP-NΔ65 (B) and observed for localization patterns. Both constructs resulted in VLS formation, with GFP-NΔ65 VLS much more closely resembling viroplasms. However, note that GFP-NSP5 also produced slender, tubular structures (arrowheads). Bars, 10 μm.
TEM.
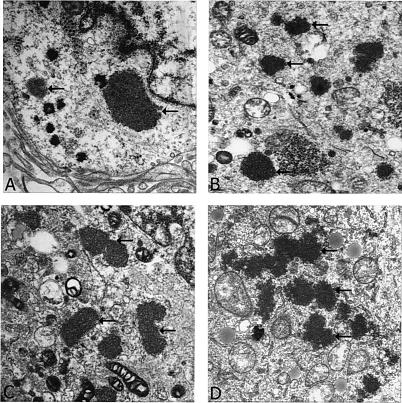
The results so far reported here suggest that specific modifications of the NSP5 protein are critical for the formation of VLS and that such structures resemble infection-associated viroplasms in morphology and location. However, such observations were based solely on confocal light microscopy analysis. Since the observations reported here are novel and certainly add to the understanding of how VLS form, we extended these studies to TEM as well to corroborate the light microscopy results. Our TEM observations clearly suggest that rotavirus infection (Fig. 6A) as well as coexpression of NSP5-NSP2 (Fig. 6B) or individual expression of GFP-NSP5 (Fig. 6C) or GFP-NΔ65 (Fig. 6D) produced electron-dense structures that are similar in morphology and location to the electron-dense viroplasmic structures previously reported for rotavirus-infected cells (10). Thus, the TEM studies described here not only agree with previous observations but also corroborate our light microscopy observations reported above.
FIG. 6.
Analysis of viroplasms and VLS by transmission electron microscopy. (A) Virus-infected cells. (B) Cells cotransfected with NSP5-GFP and NSP2 expression plasmids. (C) Capability of GFP-NSP5 alone to form VLS in tranfected cells. (D) GFP-NΔ65-transfected cells, in which we previously observed (by confocal imaging) intense particulate VLS. Arrows indicate electron-dense viroplasms and VLS.
Phosphorylation status of full-length and truncated NSP5 chimeric proteins.
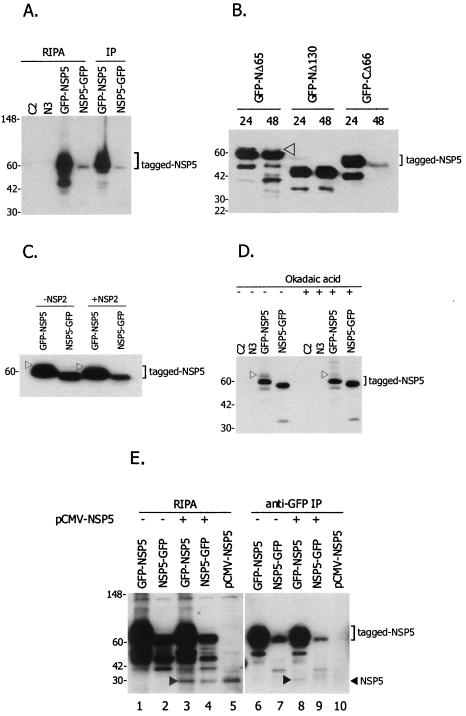
Analysis of the levels of phosphorylation of the GFP-fused full-length NSP5 construct and the various deletion clones was carried out to identify the domain responsible for the hyperphosphorylation of NSP5 following transfection of COS-7L cells with the plasmids. Immunoprecipitation of transfected cell lysates with an antiphosphoserine antibody followed by immunoblot detection of the samples with an anti-NSP5 antibody clearly indicated that the higher-molecular-weight band above the major GFP-NSP5 band was the hyperphosphorylated form of NSP5 and that such a high-molecular-weight band was lacking in NSP5-GFP (Fig. 7A, IP lanes). The lack of such band in NSP5-GFP could be a true reflection of the lack of hyperphosphorylation of this form of NSP5 or could be due to a low level of expression of NSP5-GFP in this experiment. GFP-C2 and N3-GFP empty vectors were used as negative controls for tranfection (Fig. 7A).
FIG. 7.
Phosphorylation and multimerization analysis of NSP5 fusion proteins. (A) Effects of N- and C-terminal fusions of NSP5 on phosphorylation. COS-7L cells were transfected with GFP-NSP5 and NSP5-GFP plasmids; one portion of the lysates was prepared in RIPA buffer (RIPA samples), and the other was immunoprecipitated (IP) with antiphosphoserine antibody (IP samples). Both RIPA and IP samples were separated on 4 to 20% acrylamide gels, and NSP5 was detected by using a guinea pig anti-NSP5 antibody. Only the GFP-NSP5 fusion construct revealed higher-molecular-weight forms of NSP5, suggesting hyperphosphorylation. Lanes C2 and N3 contained cytolysates from GFP-C2 and N3-GFP parent empty vector transfections, which served as negative controls. Protein molecular mass markers in kilodaltons are shown on the left. (B) NSP5 deletion constructs and hyperphosphorylation. COS-7L cells were transfected with GFP-NSP5 constructs, and lysates were subjected to immunoblot analysis with anti-NSP5 antibody. Only GFP-NΔ65 resulted in hyphosphorylated forms of NSP5; no detectable phosphorylation was observed with GFP-NΔ130 and GFP-CΔ66. The arrowhead indicates the higher-molecular-weight form, which represents the hyperphosphorylated band. Protein molecular mass markers in kilodaltons are shown on the left. (C) Immunoblot analysis demonstrating the effect of NSP2 on phosphorylation levels. To demonstrate whether the levels of phosphorylation of GFP-NSP5 and NSP5-GFP could be affected by NSP2, cotransfections with NSP2-Red were performed, and immunoblot analysis was done with anti-NSP5 antibody. An arrowhead indicates the higher-molecular-weight form, which represents the hyperphosphorylated band. No significant difference was observed in the levels of phosphorylation of NSP5, even with the addition of NSP2. A protein molecular mass marker in kilodaltons is shown on the left. (D) Immunoblot analysis demonstrating the effect of okadaic acid on NSP5 phosphorylation levels. COS-7L cells, not treated or treated with 0.1 μM okadaic acid, were transfected with GFP-NSP5 and NSP5-GFP, and lysates were analyzed with anti-NSP5 antibody. An arrowhead indicates the hyperphosphorylated form. No significant difference was observed in the levels of phosphorylation of NSP5 with or without okadaic acid, suggesting that threshold levels of phosphorylation were attained for GFP-NSP5. Lanes C2 and N3 contained lysates from GFP-C2 and N3-GFP parent empty vector transfections, which served as negative controls. Protein molecular mass markers in kilodaltons are shown on the left. (E) Multimerization of GFP-NSP5 with the native form of NSP5. COS-7L cells were transfected with GFP-NSP5 and NSP5-GFP plasmid DNAs, with or without pCMV-NSP5 (expressing the native form of NSP5), and immunoprecipitated with anti-GFP antibody. RIPA samples (lanes 1 to 5) and IP samples (lanes 6 to 10) were separated on 4 to 20% acrylamide gels, and NSP5 was detected with a guinea pig anti-NSP5 antibody. Lanes 3 to 5 demonstrate the presence of the native form of NSP5 in the lysates, indicating the expression of the protein (arrowhead). Only GFP-NSP5 was able to bind to the native form of NSP5, as indicated by the arrowhead in lane 8. Protein molecular mass markers in kilodaltons are shown on the left.
Once we established that the higher-molecular-weight form truly represented the phosphorylated form of NSP5, in the following NSP5-specific immunoblot experiments, where appropriate, such forms are designated phosphorylated forms (without antiphosphoserine immunoprecipitation; Fig. 7B to D, arrowheads). Among the deletion constructs of NSP5, transfected lysates of only the GFP-NΔ65 construct demonstrated hyperphosphorylated forms (Fig. 7B). The GFP-NΔ130, GFP-CΔ66, and NSP5-GFP constructs demonstrated a single form of the expressed protein, indicating either no phosphorylation or undetectable levels of phosphorylation (Fig. 7B). Furthermore, the role of NSP2 in the hyperphosphorylation of NSP5 was assessed by cotransfecting NSP2 with free (pCMV-NSP5) and GFP-fused (GFP-NSP5 and NSP5-GFP) NSP5 constructs. No significant difference was observed in the levels of phosphorylation of GFP-NSP5 and NSP5-GFP when cotransfected with NSP2 (Fig. 7C). The addition of okadaic acid did not enhance the phosphorylation profile for tagged NSP5. The slowly moving phosphorylated form of GFP-NSP5 is shown in Fig. 7D (arrowhead). When GFP-NSP5- and NSP5-GFP-containing lysates were subjected to a longer run time in the gels to separate the higher-molecular-weight phosphorylated forms from the major protein band (as in Fig. 7C and D), we noticed that the NSP5-GFP fusion construct migrated faster than the GFP-NSP5 fusion construct, even though the sizes and sequences of the cloned NSP5 gene are identical in the GFP-NSP5 and NSP5-GFP constructs. It was previously reported that the C-terminal amino acids of NSP5 are critical for phosphorylation of the protein (21). Since the C terminus of NSP5 is fused to GFP in the NSP5-GFP construct, we believe that the differential migration between the two forms likely is due to the difference in their basal phosphorylation levels.
GFP-NSP5 associates with the native form of NSP5.
To analyze whether there is any difference in the multimerization potential of NSP5 depending on whether the C or the N terminus is unmodified, immunoprecipitation and subsequent immunoblot analysis were performed following plasmid DNA transfections. Briefly, NSP5-GFP and GFP-NSP5 were transfected alone or each was cotransfected with pCMV-NSP5, and cell lysates were prepared in RIPA buffer. These samples were analyzed by immunoblotting with an anti-NSP5 antibody to confirm the expression of NSP5-related proteins in the transfected cell lysates (Fig. 7E, lanes 1 to 5). Once this finding was established, aliquots of the same cell lysates were immunoprecipitated with an anti-GFP antibody and subjected to immunoblotting with an anti-NSP5 antibody to assess whether the native form of NSP5 can be pulled down along with the GFP-fused forms, a result which would be indicative of multimerization of tagged NSP5 with the native form (Fig. 7E, lanes 6 to 10). From this experiment, it is clear that even though the individual levels of expression varied among GFP-NSP5, NSP5-GFP, and NSP5 (Fig. 7E, lanes 1, 2 and 5), the relative ratios of GFP-NSP5 to NSP5 and of NSP5-GFP to NSP5 appeared somewhat similar based on the band intensities (lanes 3 and 4). Based on these immunoprecipitation results, the fact that only in the presence of both GFP-NSP5 and NSP5, in which both forms of NSP5 have an unmodified C terminus, was the native form of NSP5 pulled down (Fig. 7E, lane 8, arrowhead) suggests that the free C terminus promotes NSP5 multimerization.
DISCUSSION
This study indicates that in a transient transfection assay, rotavirus NSP5 by itself can form VLS without the help of NSP2, provided its N terminus is blocked by fusion with any nonrotavirus polypeptide (GFP or HA) but its C terminus remains unmodified. Analysis with full-length and deletion versions of NSP5 with a GFP tag at locations distinctly different from those used in previous work unraveled new features of rotavirus NSP5 and its true potential in generating VLS without the help of NSP2.
In light of this finding, it is reasonable to rationalize that the NSP5 N-terminal modification (fusion) is sufficient to replace the NSP2 cotransfection requirement in the transient transfection assay with regard to VLS formation. However, due to the lack of a reverse genetic approach for rotaviruses, this hypothesis could not be tested. However, the success of rotavirus VP4 gene silencing in a recently described small interfering RNA knockout approach (7) might offer possibilities for using NSP2-specific small interfering RNA to knock out gene expression during rotavirus infection and study the role of NSP2 in NSP5-dependent viroplasm formation. It will be interesting to determine whether viroplasms are formed in such a scenario or whether viroplasm formation is blocked due to the lack of NSP2, although one could speculate that the lack of expression of NSP2 certainly would lead to a diffuse NSP5 pattern. Nonetheless, we caution readers that our report of the capability of a specific form of chimeric NSP5 in forming VLS in no way undermines the importance of crucial functions of NSP2 in rotavirus biology as well as its requirement in viroplasm formation during infection. Our analysis rather provides new insights into the intrinsic properties of NSP5 with respect to VLS formation.
The GFP-NSP5 construct used in this study produced a vesicular pattern morphologically similar to that reported previously in NSP5-NSP2 cotransfection studies as well as to that of infection-associated viroplasms, whereas NSP5-GFP alone produced a diffuse cytoplasmic distribution, as was observed with this type of construct in a previous study (8). The ability of the GFP-NSP5 construct alone to form VLS emphasized the importance of the context or nature of both the N and the C termini of NSP5, which normally is regulated during infection by NSP2. The C-terminal 68-amino-acid construct (GFP-NΔ130) of NSP5 was able to form VLS, suggesting that the VLS-imparting signals of NSP5 are embedded in its C terminus, as was suggested recently by another group as well (8). Thus, by using the GFP-NSP5 chimera, we were able to map the critical domains of NSP5 responsible for VLS formation and hyperphosphorylation independent of NSP2.
The GFP-NΔ130 construct did not undergo hyperphosphorylation but still was able to form VLS, suggesting a lack of association between phosphorylation and formation of VLS. Furthermore, a substantial reduction in phosphorylation of GFP-NΔ65 compared to GFP-NSP5 was evident in our phosphorylation analysis. This result could be attributed to the facts that almost 50% of the serine residues (20 of 43) of NSP5 are present in the N-terminal 65 amino acids and that this is the region that has been deleted in GFP-NΔ65. However, it is this construct that shows the most intense and profuse VLS formation. In addition, our experiments with okadaic acid did not reveal enhanced phosphorylation of the tagged NSP5 proteins and did not affect the ability of NSP5 to form VLS, a possibility that was proposed by others recently (8).
Hyperphosphorylation of NSP5 so far has been thought to be NSP2 dependent (11, 21, 22). In the present analysis, the full-length NSP5-GFP chimera expression studies revealed that when the N terminus of NSP5 was fused to GFP and the C terminus was unmodified, hyperphosphorylation of the protein was observed, whereas no phosphorylation was observed when the N terminus was unmodified and the C terminus was fused to GFP. Furthermore, the levels of phosphorylation of GFP-NΔ65 which, in spite of a lack of the NSP2-binding domain, had undergone hyperphosphorylation and the levels of phosphorylation of NSP5 (GFP-NSP5 and NSP5-GFP) were not affected even after cotransfection with NSP2, indicating that threshold phosphorylation levels were achieved for GFP-NSP5. These results clearly suggest that for NSP5 phosphorylation events, it appears that NSP5 N-terminal fusion replaces the NSP2 requirement. Based on these findings, the precise role of NSP2 in the phosphorylation of NSP5 may be to render the N terminus of NSP5 occupied to provide an optimal structural conformation for NSP5, perhaps for cellular kinases to act upon. On the other hand, it appears that the multimerization of NSP5 is an important prerequisite for VLS formation in the noninfection scenario, based on the different multimerization capabilities found for GFP-NSP5 and NSP5-GFP in the present study.
In conclusion, our findings suggest that the N terminus of NSP5 should be somehow inaccessible, i.e., kept “occupied,” and that the C terminus should be unmodified, presumably for multimerization, in order to form VLS. Moreover, based on the results discussed in this report, NSP5 (hyper)phosphorylation appears not to be an important event for VLS formation, an observation that has been gaining acceptance in the field recently (8). All of the observations reported here together with those from recent studies on NSP5 (8) suggest that with regard to VLS formation, rotavirus recruits its own protein, NSP2, to ensure effective N-terminal blocking of NSP5; that properties such as VLS formation, phosphorylation, and multimerization are all intrinsic to NSP5 alone; and that NSP2 has other crucial functions in viral infection (22). The present study also sheds light on another important aspect: that, in general, when functions and patterns of expression of a given protein are elucidated by recombinant methods, such as fusion with a foreign protein of interest, constructs that represent fusions at both the N and the C termini should be evaluated for a better understanding of the associated molecular events.
Acknowledgments
We thank Oscar Burrone, ICGEB, Trieste, Italy, for the generous gift of rotavirus NSP5 antisera, I. Som for constructing some of the NSP5-GFP fusion plasmids used in this report, and Marilyn Lundquist for electron microscopy sample preparation. Critical reviews of the manuscript by Tahir Malik and Zhiping Ye are duly acknowledged.
The Center Director's Targeted Research Award, CBER, FDA, to C.D.A. and an ORISE postdoctoral fellowship to K.V.K.M. supported this work.
REFERENCES
- 1.Afrikanova, I., M. C. Miozzo, S. Giambiagi, and O. R. Burrone. 1996. Phosphorylation generates different forms of rotavirus NSP5. J. Gen. Virol. 77:2059-2065. [DOI] [PubMed] [Google Scholar]
- 2.Afrikanova, I., E. Fabbretti, M. C. Miozzo, and O. R. Burrone. 1998. Rotavirus NSP5 phosphorylation is upregulated by interaction with NSP2. J. Gen. Virol. 79:2679-2686. [DOI] [PubMed] [Google Scholar]
- 3.Berois, M., C. Sapin, I. Erk, D. Poncet, and J. Cohen. 2003. Rotavirus nonstructural protein NSP5 interacts with major core protein VP2. J. Virol. 77:1757-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackhall, J., A. Fuentes, K. Hansen, and G. Magnusson. 1997. Serine protein kinase activity associated with rotavirus phosphoprotein NSP5. J. Virol. 71:138-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackhall, J., M. Munoz, A. Fuentes, and G. Magnusson. 1998. Analysis of rotavirus nonstructural protein NSP5 phosphorylation. J. Virol. 72:6398-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chnaiderman, J., M. Barro, and E. Spencer. 2002. NSP5 phosphorylation regulates the fate of viral mRNA in rotavirus-infected cells. Arch. Virol. 147:1899-1911. [DOI] [PubMed] [Google Scholar]
- 7.Dector, M. A., P. Romero, S. Lopez, and C. F. Arias. 2002. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 3:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichwald, C., F. Vascotto, E. Fabbretti, and O. R. Burrone. 2002. Rotavirus NSP5: mapping phosphorylation sites and kinase activation and viroplasm localization domains. J. Virol. 76:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 10.Estes, M. K., E. L. Pamer, and J. F. Obijeski. 1983. Rotaviruses: a review. Curr. Top. Microbiol. Immunol. 105:123-184. [DOI] [PubMed] [Google Scholar]
- 11.Fabbretti, E., I. Afrikanova, F. Vascotto, and O. R. Burrone. 1999. Two non-structural rotaviral proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333-339. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, R. A., M. A. Torres-Vega, S. Lopez, and C. F. Arias. 1998. In vivo interactions among rotavirus nonstructural proteins. Arch. Virol. 143:981-996. (Erratum, 143:2064.) [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez, S. A., and O. R. Burrone. 1991. Rotavirus NS26 is modified by addition of single O-linked residues of N-acetylglucosamine. Virology 182:8-16. [DOI] [PubMed] [Google Scholar]
- 14.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 15.Mohan, K. V., and C. D. Atreya. 2000. Comparative sequence analysis identified mutations outside the NSP4 cytotoxic domain of tissue culture-adapted ATCC-Wa strain of human rotavirus and a novel inter-species variable domain in its C-terminus. Arch. Virol. 145:1789-1799. [DOI] [PubMed] [Google Scholar]
- 16.Mohan, K. V., T. S. Dermody, and C. D. Atreya. 2000. Mutations selected in rotavirus enterotoxin NSP4 depend on the context of its expression. Virology 275:125-132. [DOI] [PubMed] [Google Scholar]
- 17.Mohan, K. V., and C. D. Atreya. 2001. Nucleotide sequence analysis of rotavirus gene 11 from two tissue culture-adapted ATCC strains, RRV and Wa. Virus Genes 23:321-329. [DOI] [PubMed] [Google Scholar]
- 18.Mohan, K. V., I. Som, and C. D. Atreya. 2002. Identification of a peroxisomal targeting signal 1 (PTS1)-containing viral protein and its targeting to peroxisomes. J. Virol. 76:2543-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrie, B. L., H. B. Greenberg, D. Y. Graham, and M. K. Estes. 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1:133-152. [DOI] [PubMed] [Google Scholar]
- 20.Poncet, D., P. Lindenbaum, R. l'Haridon, and J. Cohen. 1997. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J. Virol. 71:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres-Vega, M. A., R. A. Gonzalez, M. Duarte, D. Poncet, S. Lopez, and C. F. Arias. 2000. The C-terminal domain of rotavirus NSP5 is essential for its multimerization, hyperphosphorylation and interaction with NSP6. J. Gen. Virol. 81:821-830. [DOI] [PubMed] [Google Scholar]
- 22.Vende, P., Z. F. Taraporewala, and J. T. Patton. 2002. RNA-binding activity of the rotavirus phosphoprotein NSP5 includes affinity for double-stranded RNA. J. Virol. 76:5291-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welch, S. K., S. E. Crawford, and M. K. Estes. 1989. Rotavirus SA11 genome segment 11 protein is a nonstructural phosphoprotein. J. Virol. 63:3974-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]