Abstract
Macrophagetropic R5 human immunodeficiency virus type 1 (HIV-1) isolates often evolve into dualtropic R5X4 variants during disease progression. The structural basis for CCR5 coreceptor function has been studied in a limited number of prototype strains and suggests that R5 and R5X4 Envs interact differently with CCR5. However, differences between unrelated viruses may reflect strain-specific factors and do not necessarily represent changes resulting from R5 to R5X4 evolution of a virus in vivo. Here we addressed CCR5 domains involved in fusion for a large set of closely related yet functionally distinct variants within a primary isolate swarm, employing R5 and R5X4 Envs derived from the HIV-1 89.6PI quasispecies. R5 variants of 89.6PI could fuse using either N-terminal or extracellular loop CCR5 sequences in the context of CCR5/CXCR2 chimeras, similar to the unrelated R5 strain JRFL, but R5X4 variants of 89.6PI were highly dependent on the CCR5 N terminus. Similarly, R5 89.6PI variants and isolate JRFL tolerated N-terminal CCR5 deletions, but fusion by most R5X4 variants was markedly impaired. R5 89.6PI Envs also tolerated multiple extracellular domain substitutions, while R5X4 variants did not. In contrast to CCR5 use, fusion by R5X4 variants of 89.6PI was largely independent of the CXCR4 N-terminal region. Thus, R5 and R5X4 species from a single swarm differ in how they interact with CCR5. These results suggest that R5 Envs possess a highly plastic capacity to interact with multiple CCR5 regions and support the concept that viral evolution in vivo results from the emergence of R5X4 variants with the capacity to use the CXCR4 extracellular loops but demonstrate less-flexible interactions with CCR5 that are strongly dependent on the N-terminal region.
Macrophagetropic human immunodeficiency virus type 1 (HIV-1) strains that use CCR5 for entry (R5 strains) are responsible for most person-to-person transmission events and for the early and maintenance phases of infection, while later stages of disease are characterized by the frequent emergence of variants that use CXCR4 in addition to or instead of CCR5 (dualtropic R5X4 or T-tropic X4 strains). Acquisition of the ability to utilize CXCR4 is an important event, since it is strongly associated with accelerated disease progression (21, 22) and may play a causative role (2, 6, 17). Thus, the structural basis for the R5-to-R5X4 transition in vivo has important implications for pathogenesis.
Naturally occurring HIV-1 isolates require that Env bind to CD4 prior to interacting with the coreceptor. CD4 binding leads to structural changes in gp120 that create or expose a previously obscure region, termed the bridging sheet, which is a principal site of contact with the chemokine receptor (23). In addition, the gp120 hypervariable regions largely determine which coreceptor can be utilized by each Env. Conversely, multiple regions of the chemokine receptors have been implicated in their association with gp120, and the molecular interactions underlying CCR5-gp120 interactions are complex (reviewed in reference 15). Importantly, a few mapping studies compared the R5 and R5X4 strains and suggest that these variants may utilize distinct regions of CCR5. Based on mutant and chimeric CCR5 molecules, both the N-terminal domain and the extracellular loops of CCR5 appear to function independently to mediate viral entry, while, in contrast, R5X4 strains appear to be more constrained in their ability to tolerate structural variations in CCR5 (3, 12, 28, 30). These observations have led to a hypothesis that multiple sites of interaction occur between CCR5 and R5 strains, and as a variant acquires the ability to interact with CXCR4 it relinquishes some sites of interaction with CCR5 (25).
A limitation of these coreceptor mapping studies, however, is that they have relied mainly on a small number of unrelated HIV-1 isolates. Thus, it is uncertain whether different structural determinants were identified because different viral strains were tested or whether they are actually due to the R5-to-R5X4 phenotypic changes that emerge within an infected individual. To address this question, we examined the structural determinants of CCR5 use among a set of closely related but genetically and functionally distinct env variants that we cloned from the viral quasispecies of the dualtropic primary isolate 89.6PI (31). This viral swarm, from which the prototype 89.6mc proviral molecular clone was obtained (9), contains both R5 and R5X4 env species that display a high degree of genetic relatedness (97% amino acid homology) and likely represent different stages of virus evolution in vivo.
MATERIALS AND METHODS
Molecular cloning of env gene variants.
The 89.6PI primary isolate was obtained from blood of an individual with AIDS and amplified in seronegative peripheral blood mononuclear cells for approximately 2 weeks before a genomic library was obtained. The full-length proviral molecular clone 89.6MC, widely used as a dualtropic R5X4 prototype, was generated from this library by lambda phage cloning (9). High-fidelity PCR was then used to make full-length (2.5-kb) functional env clones from the remaining 89.6PI genomic library (31). To ensure that each env gene represented a distinct proviral molecule, separate aliquots of template DNA were amplified in independent PCRs, and only one env gene from each amplification reaction was utilized. env clones were ligated into pCR-Blunt (Invitrogen, Carlsbad, Calif.) downstream of the T7 promoter. For clarity, the primary isolate swarm is referred to as 89.6PI, the prototype R5X4 env gene from the full-length infectious molecular clone is referred to as 89.6, and each independent env clone is designated by a number. The V1-C5 sequences of each env gene were determined by automated sequencing of both strands and sequence analysis carried out using MacVector software (Accelrys Inc., San Diego, Calif.).
CCR5 chimeras and mutants.
Reciprocal chimeras between CCR5 and CXCR2 were used in which the N-terminal (NT) and extracellular loop (ECL) domains were exchanged at the conserved Cys20 as described previously (12). CCR5 N-terminal deletion mutants were utilized that lack the N-terminal 4 (referred to here as −4; previously called C25-17), 8 (−8; previously called C25-18), 12 (−12; previously called C25-19) or 16 (−16; previously called C25-20) amino acids (30). A triple substitution mutant involving the first, third, and fourth extracellular domains was tested (CCR5 11/197/276) in which Ala was substituted for Asp 11, Lys 197, and Asp 276 as previously described (12). A panel of chimeras in which extracellular regions were exchanged between CXCR4 and CXCR2 has been previously described (25). Of note, in the context of previous studies evaluating prototype HIV-1 strains, these chemokine receptor constructs have been shown to be expressed in QT6 cells at levels similar to those for the wild-type chemokine receptors (12, 25, 30). We also used a panel of chimeras generated between human and rat CXCR4, which has been described as well (5).
Coreceptor fusion analysis.
Effector 293T cells were infected with T7 polymerase-expressing recombinant vaccinia virus vP11T7gene1 (1) and then transfected with plasmids encoding env genes under control of the T7 promoter. Wild-type or mutant CCR5 expression plasmids were cotransfected along with CD4 and a T7-driven luciferase reporter plasmid into target quail fibrosarcoma QT6 cells. The cells were incubated overnight in the presence of rifampin and Ara-C to inhibit vaccinia virus replication, following which effector and target cells were mixed. Six hours later the cells were lysed, and luciferase activity was measured in cell lysates as an indication of cell-cell fusion. Details of this cell-cell fusion assay have been published previously (12, 31). To ensure that coreceptors were expressed at equal levels for each Env being evaluated, target cells were transfected in bulk and then distributed among the Env-expressing cells. Negative controls in each experiment included effector cells transfected with empty vector instead of env and target cells transfected with CD4 but no chemokine receptor.
RESULTS
Role of CCR5 N-terminal and extracellular loop regions for R5 and R5X4 89.6PI variants.
Previous studies have addressed the CCR5 domains utilized for fusion by R5 and R5X4 strains (16, 28), but comparisons between a limited number of prototype isolates cannot distinguish strain-specific differences from those linked to phenotypic evolution in infected individuals. In order to compare naturally occurring, genetically related Envs with distinct phenotypes that reflect the range of coreceptor tropism associated with evolution in vivo, we analyzed a set of env genes cloned from the 89.6PI primary isolate viral swarm. This isolate was derived from an individual with AIDS (9) and contains a spectrum of variants with R5 and R5X4 phenotypes (14, 31). To assess the relative importance of the CCR5 NT and ECL regions for fusion by these Envs, we used a pair of reciprocal chimeras generated between CCR5 and CXCR2, which does not support fusion by HIV-1 Env glycoproteins. These chimeras retain the conserved Cys residue in the N terminus (residue 20) in order to maintain maximal structural integrity (12). The ability of each Env variant to fuse with the chimeric chemokine receptor/CD4 complex was assessed in a cell-cell fusion assay. Efficient fusion with a mutant coreceptor was defined as ≤50% compared with that for wild-type CCR5 for each particular Env.
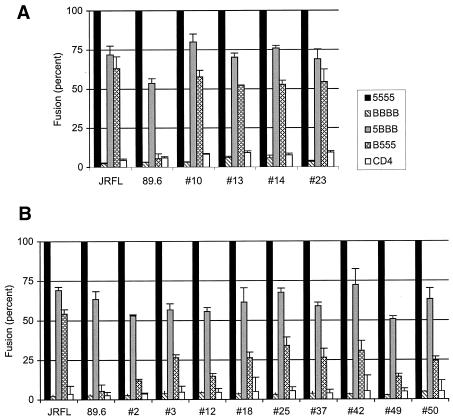
All Envs including the prototype R5 isolate JRFL, the 89.6 R5X4 prototype, and 89.6PI-derived variants fused with cells coexpressing CD4 and CCR5 but not CXCR2. As shown in Fig. 1, when the CCR5/CXCR2 recombinants were tested, JRFL fused efficiently if either the N-terminal CCR5 domain was combined with all three CXCR2 ECL in 5BBB or all three ECL of CCR5 were present with a CXCR2-derived N terminus in B555. This result suggests that JRFL can independently utilize determinants in the NT and ECL regions of CCR5 when placed in the background of CXCR2 and that neither alone is essential. In contrast, the R5X4 89.6 prototype was absolutely dependent on the N-terminal domain of CCR5, since it fused only with wild-type CCR5 and a chimera containing the CCR5 N terminus (5BBB) but not with a molecule containing intact CCR5 ECL regions but a CXCR2-derived NT (B555). This result is similar to previous reports suggesting more restricted NT-dependent CCR5 determinants for R5X4 than R5 variants (12).
FIG. 1.
Fusion of 89.6PI Env variants with CCR5/CXCR2 chimeras. Plasmids encoding R5 (A) and R5X4 (B) env genes derived from the 89.6PI quasispecies were transfected into QT6 cells and then mixed with 293T cells that were transfected with CD4 and the chimeric chemokine receptor. Fusion was determined by luciferase reporter gene expression as an indication of cytoplasmic mixing as described in Materials and Methods. The prototype R5X4 env derived from the 89.6 molecular clone was tested in parallel, along with the unrelated R5 prototype strain JRFL. The CCR5/CXCR2 recombinants were generated at the conserved Cys at residue 20, so that 5BBB contains the CCR5 N terminus on the background of CXCR2 while B555 contains the CXCR2 N terminus on the background of CCR5. Values represent luciferase reporter gene expression as a percentage of that seen with wild-type CCR5 for each env gene and represent means ± standard error of the mean for three independent experiments.
We then addressed the phenotypically divergent variants of 89.6PI. All four R5 Envs fused efficiently with both 5BBB and B555 (Fig. 1A), like JRFL but distinct from 89.6, which indicated that they also can use either NT or ECL determinants within CCR5. In contrast, the R5X4 variants analyzed resembled the prototype R5X4 89.6 Env, since the CCR5 NT domain was required for efficient fusion when placed in the context of CXCR2 (5BBB), and the CCR5 ECL regions combined with a CXCR2-derived N terminus (B555) did not support efficient fusion (Fig. 1B). Thus, despite their close genetic relatedness, R5 variants of 89.6PI more closely resemble JRFL in their redundant use of the NT or ECL region of CCR5, while R5X4 variants are highly dependent on the NT CCR5 domain.
Of note, while the R5X4 env genes were clearly distinct from the R5 variants, some heterogeneity was evident, since levels of fusion with chimera B555 ranged from approximately 10 to 30% of the level seen with wild-type CCR5. Thus, although there exists a clear distinction between the phenotypes, some R5X4 Envs exhibited slightly more flexibility than the prototype R5X4 89.6 Env. We found no correlation between the degree of B555 use and the relative efficiency of CXCR4-mediated fusion (Table 1).
TABLE 1.
CCR5 structural determinants and sequence patterns among 89.6P1 Env variantsa
| Clone | WT CCR5 | N-terminal deletion
|
Chimera
|
11/197/276 mutant | V3 charge | V1/V2 length (aa) | gp120 glycos | Relative X4 useb | ||
|---|---|---|---|---|---|---|---|---|---|---|
| −12 | −16 | 5BBB | B555 | |||||||
| R5 variants | ||||||||||
| JRFL | +++ | +++c | +++ | +++ | +++ | +++ | 4 | +2 | =d | |
| 10 | +++ | +++ | +++ | +++ | +++ | +++ | 7 | = | = | |
| 13 | +++ | +++ | +++ | +++ | +++ | +++ | 7 | = | = | |
| 14 | +++ | +++ | +++ | +++ | +++ | +++ | 7 | −3 | −1 (V1) | |
| 23 | +++ | +++ | +++ | +++ | +++ | +++ | 7 | |||
| R5X4 variants | ||||||||||
| 89.6 | +++ | − | − | +++ | − | − | 7 | = | = | 1.03 |
| 1 | +++ | + | + | − | 7 | +2 | = | 1.00 | ||
| 2 | +++ | − | − | +++ | − | − | 7 | = | −1 (V1) | 0.98 |
| 3 | +++ | +++ | +++ | +++ | + | − | 7 | = | = | 1.24 |
| 4 | +++ | + | + | + | 7 | = | = | 1.09 | ||
| 5 | +++ | + | + | + | 7 | = | = | 1.01 | ||
| 11 | +++ | + | + | − | 7 | = | = | 2.05 | ||
| 12 | +++ | + | + | +++ | − | − | 7 | = | = | 2.87 |
| 16 | +++ | + | + | + | 7 | = | = | 1.82 | ||
| 17 | +++ | + | + | + | 7 | = | −1 (V1) | 1.49 | ||
| 18 | +++ | + | + | +++ | + | + | 6 | +2 | = | 2.37 |
| 20 | +++ | + | + | − | 7 | = | −1 (V1) | 2.24 | ||
| 25 | +++ | +++ | + | +++ | + | + | 7 | = | = | 1.68 |
| 27 | +++ | + | − | − | 7 | = | = | 2.05 | ||
| 30 | +++ | + | + | − | 7 | = | = | 1.60 | ||
| 31 | +++ | + | + | − | 7 | +2 | = | 0.43 | ||
| 32 | +++ | + | + | + | 7 | +2 | = | 0.50 | ||
| 33 | +++ | +++ | + | + | 7 | = | = | 0.81 | ||
| 35 | +++ | +++ | + | + | 7 | = | = | 0.77 | ||
| 36 | +++ | + | + | − | 7 | +2 | = | 0.56 | ||
| 37 | +++ | +++ | +++ | +++ | + | +++ | 7 | +2 | = | 1.20 |
| 38 | +++ | + | + | − | 3 | = | −2 (V2 and V3) | 0.82 | ||
| 40 | +++ | +++ | + | +++ | 7 | = | = | 0.38 | ||
| 41 | +++ | + | + | − | 7 | +2 | = | 0.95 | ||
| 42 | +++ | + | + | +++ | + | + | 7 | = | = | 2.06 |
| 44 | +++ | − | − | − | 7 | = | = | 1.06 | ||
| 45 | +++ | − | − | − | 7 | = | = | 0.49 | ||
| 47 | +++ | + | − | − | 7 | = | −1 (V1) | 1.85 | ||
| 49 | +++ | − | − | +++ | − | − | 7 | = | −1 (C2) | 0.83 |
| 50 | +++ | + | + | +++ | − | + | 7 | = | −1 (V1) +1(C1) | 0.58 |
At the left is the clone designation (numbers) along with the related R5X4 89.6 prototype and unrelated R5 prototype JRFL. Fusion with wild-type CCR5 for each Env is designated as 100%. Structural determinants are shown as relative fusion with CCR5. Only the CCR5 mutants found to be discriminatory are shown, including −12 and −16 N-terminal deletions, chimeras containing either the N terminus (5BBB) or three extracellular loops (B555) of CCR5 on the background of CXCR2, or a mutant CCR5 with Ala substituted at positions 11/197/276. The predicted amino acid sequence is compared with the prototype 89.6 sequence in V3 loop charge, V1/V2 region length, and differences in predicted N-linked glycosylation (glycos) sites along with location of glycosylation site divergence. aa, amino acids.
Relative fusion using CXCR4 and CD4 compared with CCR5 and CD4.
Fusion relative to wild-type CCR5 defined as the following: +++, ≥50%; +, 25 to 50%; −, <25%.
=, no difference in V1/V2 length or V1-C5 glycosylation site pattern compared with the 89.6 prototype Env.
Utilization of CCR5 deletion mutants by 89.6PI R5 and R5X4 variants.
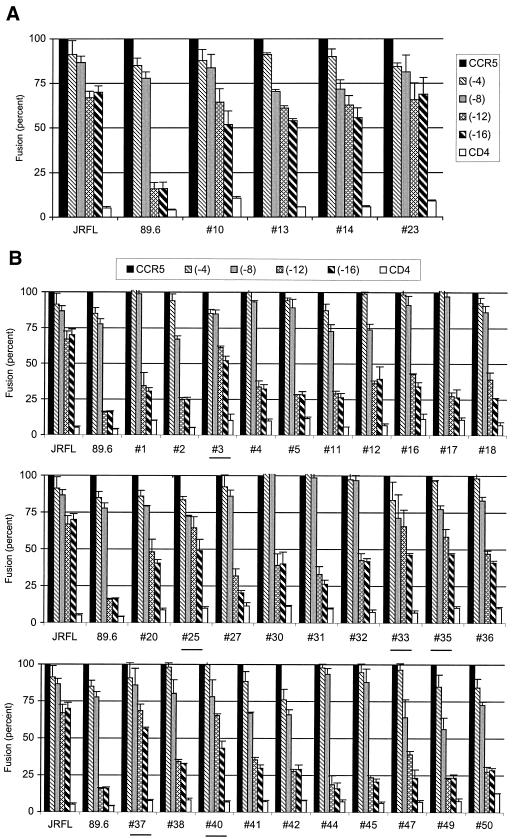
The results with CCR5/CXCR2 chimeras suggested that R5 variants of 89.6PI could fuse if either the NT or the ECL domain of CCR5 was present, while R5X4 variants absolutely required the CCR5 NT. However, chimeric coreceptors cannot distinguish between regions that are dispensable for function (e.g., the CCR5 NT for R5 variants) and those that share sufficient structural homology between parental molecules to enable function. Therefore, to more directly address the relative requirement for the CCR5 N terminus, we tested a series of truncated molecules in which 4, 8, 12, or 16 residues were deleted from the CCR5 N terminus (Fig. 2). We selected this set of mutants because in prior studies, prototype R5 Envs like JRFL, ADA, and SF162 appeared to tolerate deletion of up to 16 N-terminal amino acids for fusion, while the R5X4 89.6 prototype was unable to utilize CCR5 that lacked 12 or more amino acids (25, 30).
FIG. 2.
Fusion of 89.6PI Env variants with CCR5 N-terminal deletion mutants. The 4 R5 Env variants from the 89.6PI swarm (A) and all 30 R5X4 Env variants derived from the swarm (B) were tested in a cell-cell fusion assay with wild-type CCR5 and a panel of deletion mutants that lacked the first 4, 8, 12, or 16 N-terminal amino acids. Fusion is represented by luciferase expression as a percentage of that seen with wild-type CCR5 for each env gene and represent means ± standard error of the mean for three independent experiments. For clarity, R5X4 Envs that showed fusion ≥50% of that seen with wild-type CCR5 when tested with the deletion mutant are underlined.
Eliminating up to 16 N-terminal residues of CCR5 had little effect on fusion mediated by the prototype R5 Env JRFL (Fig. 2). In contrast, the 89.6 R5X4 prototype tolerated truncation of up to eight amino acids, but luciferase expression was reduced nearly to baseline if 12 or more residues were deleted. We then tested the naturally occurring variants from the 89.6PI swarm. For this analysis, we assessed the full panel of full-length fusion-competent R5X4 and R5 variants cloned from the 89.6PI swarm. As shown in Fig. 2A, the four R5 Envs all retained most efficient fusion even when 12 or 16 N-terminal amino acids were deleted from CCR5. This tolerance of CCR5 NT truncation resembled the unrelated JRFL R5 Env and differed from the related but functionally distinct R5X4 prototype 89.6 Env. In contrast, the majority of R5X4 variants were similar to 89.6, since fusion with CCR5 was greatly impaired by N-terminal deletions (Fig. 2B). Some Envs displayed little more than background reporter gene expression with the −12 and −16 CCR5 mutants, while others exhibited low levels of fusion (20 to 40% of control). However, a few of the 30 R5X4 variants exhibited relatively preserved fusion capacity even when most of the N terminus was eliminated (#3, #25, #33, #35, #37, and #40). Thus, the results with NT truncations are concordant with the chimeric coreceptor data in showing that the R5X4 variants were generally similar in being highly dependent on the CCR5 N terminus and distinct from the R5 variants, which were capable of utilizing CCR5 based on determinants elsewhere in the molecule.
The similarity among the related and unrelated R5 species suggests that they share a structural basis for their interaction with CCR5 and that the ability to use both NT and ECL regions of CCR5 is a characteristic of R5 variants. Similarly, the fact that most R5X4 variants require the NT domain of CCR5 indicates a common structural basis for CCR5 use that requires more-specific points of interaction with CCR5 and is linked to the dual coreceptor phenotype. At the same time, however, the observation that some R5X4 variants tolerate N-terminal deletions fairly well indicates that reliance on the CCR5 N terminus is typical but not an absolute requirement for variants that also use CXCR4.
Effect of CCR5 extracellular domain mutations on 89.6PI R5 and R5X4 variants.
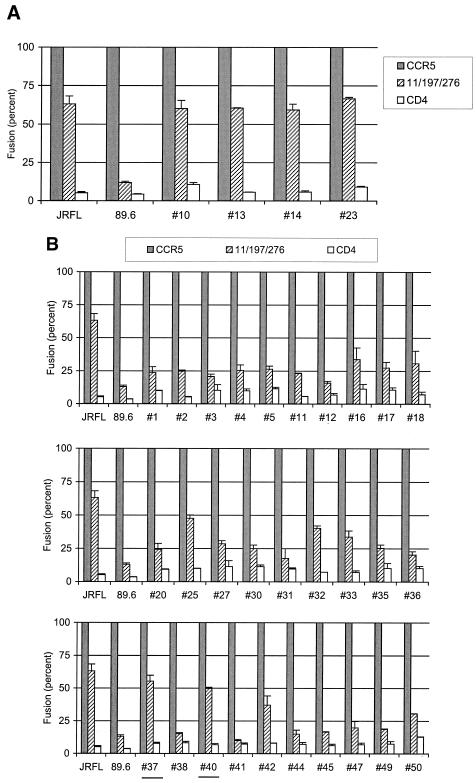
Since the above observations suggested that R5 Envs within the swarm exhibit more flexibility towards CCR5 structure than R5X4 variants, we wished to examine their tolerance of other mutations that might affect coreceptor structure. Previous studies identified several residues that are widely dispersed by primary sequence but which together appear to provide a critical determinant that distinguished between R5 and R5X4 prototypes. In particular, a D11A substitution in the N terminus combined with K197A and D276A substitutions in the second and third extracellular loops, respectively, each had little effect on fusion by the R5X4 89.6 prototype but together abrogate fusion almost completely, while fusion by several different but unrelated R5 prototypes (JRFL, BAL, and SF162) was minimally affected (12).
Consistent with this previous report, the R5 prototype JRFL was minimally affected by the CCR5 D11A/K197A/D276A mutation, while fusion by the R5X4 prototype 89.6 was greatly impaired (Fig. 3). The R5 89.6PI variants resembled JRFL and differed from 89.6 in that they retained efficient fusion when these substitutions were introduced (Fig. 3A). In contrast, nearly all of the R5X4 variants showed dramatically reduced fusion with the D11A/K197A/D276A mutant (Fig. 3B). These three residues, therefore, appear to define a structural motif critical for fusion by R5X4 variants but not for R5 variants within this swarm. Combined with results from the CCR5/CXCR2 chimeras and NT deletions, R5 members of the 89.6PI swarm consistently exhibit greater flexibility towards CCR5 structural changes than R5X4 members of the swarm.
FIG. 3.
Fusion of 89.6PI variants with a triple substitution CCR5 mutant. R5 variants (A) and R5X4 variants (B) from the 89.6PI swarm were tested for the ability to fuse with a CCR5 substitution mutant (11/197/276) containing changes in the N-terminal (D11A), second extracellular loop (K197A), and third extracellular loop (D276A) regions. Data indicate luciferase reporter gene expression as a percentage of that measured with wild-type CCR5 for each env gene and represent means ± standard error of the mean for three independent experiments. For clarity, R5X4 Envs that showed fusion ≥50% of that seen with wild-type CCR5 when tested with the substitution mutant are underlined.
Of note, 2 of 30 R5X4 variants (#37 and #40) retained ≥50% wild-type fusion with this mutant. Both of these Envs were also among the subset of R5X4 Envs that were relatively tolerant of CCR5 N-terminal truncations (Fig. 2B), suggesting that they are indeed more flexible in their interactions with CCR5. Thus, while R5X4 variants are in general quite limited in their tolerance of CCR5 structural disruption, some spectrum of tolerance does exist, indicating that rigidly constrained interaction with CCR5 is not an absolute requirement for dual CCR5 and CXCR4 utilization.
Use of CXCR4 determinants by 89.6PI variants.
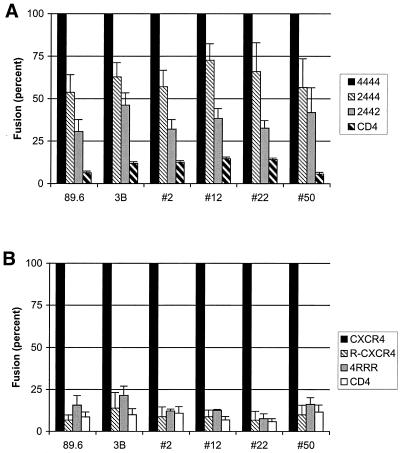
Since R5X4 variants in the swarm appeared to be particularly dependent for fusion on the N-terminal domain of CCR5, we determined which regions of CXCR4 might be important for fusion. Previous studies have suggested that the ECL domains appear to have a dominant role in the context of human-murine CXCR4 and human CXCR4/CXCR2 or CXCR4/CCR5 chimeras (4, 13, 25, 27), although both the NT and ECL regions are important in some circumstances (5, 8, 20, 29). In addition, while there are strain-dependent differences in structural determinants of CXCR4 use (29), patterns linked to the X4 versus R5X4 phenotypes have not been identified (20). We employed a series of chimeras in which equivalent regions of CXCR4 were exchanged for sequences from CXCR2, which is not used by HIV-1 for fusion (Fig. 4A). Chimera 2444 contains the N-terminal domain of CXCR2 in the background of CXCR4, while 2442 contains the first and fourth extracellular domains of CXCR2. All of these molecules were expressed at similar levels based on mean fluorescence intensity and percent positive cells by flow cytometry using the CXCR4 monoclonal antibody 12G5, which recognizes the extracellular loop regions (25). We were unable to achieve efficient expression of a molecule containing the CXCR4 N terminus introduced into CXCR2 (4222; data not shown), and so we tested another chimera that had been generated between human and rat CXCR4 (5). This molecule (4RRR) contains the N terminus of human CXCR4 on the background of the rat molecule. This chimera was also expressed at a level similar to that for wild-type CXCR4 based on fluorescence-activated cell sorter staining with 4G10, which recognizes the N-terminal domain of CXCR4 (7) (data not shown). To analyze these chimeras we selected three R5X4 variants from the 89.6PI swarm (#2, #12, and #50). We also analyzed Env #22 as the most “X4-like” of the R5X4 variants, since in initial studies with coreceptor-transfected QT6 cell line targets it appeared to be restricted to CXCR4, although subsequently it was found use CCR5 in primary cells (31, 32). In parallel we tested the X4 prototype Env 3B along with the 89.6 R5X4 prototype.
FIG. 4.
Fusion of R5X4 89.6PI variants with CXCR4 chimeras. (A) Chimeras were generated between CXCR4 and CXCR2 that introduced the N-terminal first (2444) or first and fourth (2442) extracellular domains of CXCR2 into CXCR4. (B) Chimeras were generated between human and rat CXCR4 that introduced the N-terminal first extracellular domain of human CXCR4 onto the rat molecule (4RRR). The unrelated X4 prototype strain 3B was tested in parallel. Data indicate luciferase reporter gene expression as a percentage of that measured with wild-type CXCR4 for each env gene and represent means ± standard error the mean for three independent experiments.
As shown in Fig. 4A, all of the R5X4 89.6PI Env variants retained ≥50% of wild-type fusion when the N-terminal CXCR4 domain was replaced with CXCR2 sequences (2444). Replacing both the N-terminal and fourth extracellular domains with CXCR2 sequences reduced fusion to less than 50%, but it was still detectable above background levels (2442). Similar patterns were seen for 3B and for the 89.6 prototype. This result indicates that the N terminus of CXCR4 is not essential for fusion by these isolates. We then tested the human-rat CXCR4 chimera (Fig. 4B). Introduction of the human CXCR4 N terminus onto the rat molecule (4RRR) did not confer fusion capacity for any of the Envs, indicating that it is not the principal determinant of fusion for R5X4 or X4 HIV-1 Envs and supporting the importance of the ECL domain of CXCR4 for these variants.
Genetic basis for CCR5 utilization among 89.6PI-related variants.
Multiple sequence determinants have been linked to HIV-1 tropism, the syncytium-inducing (SI)/non-syncytium-inducing (NSI) phenotype, coreceptor choice, and/or disease progression in vivo. These determinants include polymorphisms in V1/V2, charge-altering changes in V3, sequences in C4-V5, and glycosylation site alterations. In a previous analysis we found that coreceptor choice was not associated with any specific V3 sequence or charge pattern among these env genes (31). Here we determined the full V1-C5 sequences of all env clones and examined whether specific patterns could be associated with the structural requirements for CCR5 interaction.
As shown in Table 1, the 89.6 R5X4 prototype Env displays a relatively high positive V3 charge that is characteristic of CXCR4-using strains. There was relatively little variation in V3 charge among these clones, and no V3 charge pattern was linked either to relative dependence on the CCR5 N terminus, sensitivity to extracellular domain mutations, or other features (Table 1). Similarly, there were only modest differences in N-linked potential glycosylation sites, and these did not appear to be related to specific structural determinants of CCR5 use. Interestingly, eight of the clones exhibited a two-amino-acid insertion in the V2 region. Differences in V1/V2 length have been associated with disease progression and viral phenotypic evolution in vivo, although longer V2 sequences have been linked to both nonprogressor/NSI status (26) and, conversely, to the NSI-SI transition and disease progression (19). In our study, the V2 length polymorphism was seen in a similar proportion of R5 (25%) and R5X4 variants (23%), and there was no association with relative dependence on the CCR5 NT or sensitivity to structural modification of the extracellular regions. Thus, neither V3 charge pattern, gain or loss of specific N-linked glycosylation sites, nor V2 length polymorphism appeared to directly define the structural basis of CCR5 utilization among these envelopes.
DISCUSSION
The evolution of HIV-1 coreceptor tropism in vivo from NSI/R5 to SI/R5X4 is a critical event in pathogenesis, as it is closely associated with (10, 21, 22) and may be a cause of (2, 6, 17) accelerated disease progression. As a result, the molecular anatomy of coreceptor evolution has important implications for pathogenesis. In this study we took advantage of a set of closely related but functionally distinct Env variants cloned from an HIV-1 primary isolate swarm in order to examine the relationship between coreceptor phenotype and relative dependence on specific CCR5 structural elements. This viral swarm contains both R5 and R5X4 variants that display a high degree of genetic relatedness (97% amino acid homology) and likely represent different stages of virus evolution in vivo (31). Here we show that R5 variants are uniformly flexible in their use of CCR5, similar to unrelated R5 HIV-1, and can utilize elements in either the NT or ECL domains of the molecule. In contrast, the related R5X4 variants from the same swarm are largely dependent on an intact N-terminal region and are generally much more restricted in their ability to tolerate disruptions of CCR5 structure. Thus, both related and unrelated R5 variants appear to share a highly plastic ability to interact with CCR5, while the acquisition of CXCR4 use is associated with more-constrained interactions with CCR5 that are especially dependent on the NT region. By analyzing functionally distinct Envs from a single infected individual, our results suggest that these patterns reflect an evolution of Env-coreceptor interactions that occur in vivo during viral evolution from R5 to R5X4.
The CCR5 and CXCR4 domains that interact with gp120 and contribute to fusion and entry are complex, and multiple points of contact appear to be involved (reviewed in reference 15). Further complicating the issue is the fact that different types of mapping studies have been done, including studies with recombinants between human and nonhuman molecules, recombinants between human chemokine receptors that differ in their ability to function as a coreceptor, substitutions, deletions, and other mutations. Importantly, the determinants identified differ depending on the type of mutation introduced or chimeric partner, and each type of analysis has important limitations. Chimeras cannot distinguish between regions that are not involved in fusion and those that are involved but share sufficient structural homology to support fusion, while other mutations may disrupt overall structure and affect sites distant from the specific change introduced. For that reason we chose to study a large panel of Env variants and examine their overall patterns of coreceptor domain utilization and relative flexibility towards changes in CCR5 structure, employing a combination of chimeras, deletions, and substitution mutations.
Despite the complexity and limitations of chemokine receptor mapping, studies employing genetically unrelated strains have suggested that R5 and R5X4 Envs differ in their reliance on specific CCR5 regions. In particular, R5 variants seem to have redundant CCR5 determinants involving both the NT and ECL regions, while R5X4 variants are less flexible and require the CCR5 NT but can interact with the ECL domain of CXCR4 (12, 25, 30). This led to the suggestion that R5-to-R5X4 evolution results from changes in how Env interacts with CCR5, in that R5 variants interact with multiple CCR5 regions and strong interactions with the CCR5 NT are retained while interactions with the CCR5 ECL domain are replaced with the ability to utilize the ECL of CXCR4 (25). Our findings here lend support to this hypothesis, since naturally occurring R5 variants of 89.6 were similar to unrelated R5 strains in exhibiting flexible and redundant NT and ECL-dependent CCR5 interactions, while their related R5X4 species demonstrated more-restricted NT-dependent CCR5 interactions but ECL-dependent interactions with CXCR4.
We found a critical role of the CCR5 NT for R5X4 Envs, but this result does not imply that other regions of CCR5 are not involved in coreceptor function for these Envs or that the CCR5 NT is completely dispensable for R5 variants. For example, while the CCR5 NT is sufficient to confer function when placed into the background of CXCR2 in these studies, it alone is not sufficient when placed into the background of murine CCR5 (3). Similarly, even though the R5 variants tolerated deletion of up to 16 N-terminal amino acids, it has been shown that N-terminal peptides of CCR5 bind to R5 gp120/CD4 complexes and can inhibit CCR5 utilization by these variants (11). Furthermore, the D11A/K197A/D276A triple mutation disrupted fusion by R5X4 89.6PI variants, but even a D11A mutation in the CCR5 NT region had a significant effect only when combined with K197A/D276A mutations in the ECL 2 and 3 domains (12). Thus, the ECL domains clearly interact with the NT region to support coreceptor function as well. Nevertheless, our data show that these related variants differ in their relative dependence on these regions and in overall tolerance of structural disruption.
The R5X4 variants in this swarm were particularly dependent on the ECL regions of CXCR4 for function and did not require the CXCR4 NT in the context of CXCR4/CXCR2 chimeras. This was opposite from their use of CCR5 determinants, which were critically dependent on the NT in the background of the same chimeric partner. However, other studies have demonstrated that the CXCR4 NT plays a role in fusion and entry by some X4 and R5X4 Env variants (5, 20, 24, 29). Whether our finding that the CXCR4 NT plays a minor role in the context of CXCR2 chimeras reflects structural homology between the NT of these coreceptors or other factors, it remains clear that the CXCR4 ECL domains play a critical role for fusion by each of these variants. Of note, whether evolution from R5X4 to X4 is associated with systematic changes in the basis for CXCR4 utilization patterns is unknown, since strain-specific differences in CXCR4 use have not been linked to coreceptor phenotype (5, 29). The absence of pure X4 variants within our swarm preclude using this panel of Envs for such an analysis.
Although the R5X4 variants were generally much less tolerant of CCR5 structural changes, a small minority were relatively flexible in their use of CCR5. This result indicates that highly stringent CCR5 structural determinants are typical of R5X4 variants but it is not an absolute requirement for an Env to interact with both principal coreceptors. Given the high degree of diversity that develops in vivo, the fact that highly stringent CCR5 requirements are usually associated with the acquisition of CXCR4 use suggests that other factors in addition to coreceptor interactions are impacting on Env evolution. Whether these factors reflect other aspects of Env function that are not evident in in vitro analysis, or immunological pressures that might put some Env configurations at a disadvantage, remains to be determined.
A limitation of our data is that only one isolate from a single time point was available from the individual from whom 89.6PI was obtained. We hypothesize that the R5 Envs reflect variants from earlier stages of infection and the R5X4 Envs represent variants that evolve later in disease. However, the limited genetic divergence among these env genes does not enable construction of phylogenetic trees with strong evolutionary significance (31). Thus, while the association between coreceptor determinants and coreceptor phenotype is clear, a definitive link with evolution in vivo will require the use of sequential isolates from individuals followed over time.
We were unable to correlate specific Env sequences among these variants with the structural basis of CCR5 utilization. The Env determinants of coreceptor choice have been extensively studied, but there is limited information regarding the determinants that regulate which regions of the chemokine receptor are utilized. One study using a set of naturally occurring R5 variants identified a specific amino acid change at the tip of the V3 crown that determined whether an Env required the CCR5 NT or could utilize either the NT or ECL domains in the context of a chimeric chemokine receptor (18), but no similar motif was present among our variants. Using chimeric Envs and coreceptors, the V1/V2 and V3 regions of the R5X4 prototype DH12 were shown to interact with distinct regions of CXCR4 (24). We found a relatively frequent two-residue insertion in the V2 regions. Contradictory data have been reported on the relationship between V2 length polymorphisms and coreceptor use or disease status (19, 26), and we did not find an association between the V2 polymorphism and either R5 versus R5X4 phenotype or with the structural basis for CCR5 utilization. Thus, the molecular determinants within the 89.6PI Envs that regulate how CCR5 is utilized remain to be determined.
In summary, R5 and R5X4 variants from the 89.6PI primary isolate swarm differ in how they interact with CCR5. The R5 Envs possess a highly plastic capacity to interact with multiple CCR5 regions, while the R5X4 variants are less tolerant of CCR5 structural changes and more dependent on the CCR5 NT and interact with the CXCR4 ECL domains. These results lend support to the hypothesis that HIV-1 evolution in vivo results from the emergence of R5X4 variants with the capacity to use CXCR4 extracellular loops but less flexible interactions with CCR5 that are strongly dependent on the N-terminal region.
Acknowledgments
We thank A. Wade and J. Cutilli for technical assistance; S. Peiper, R. Doms, M. Alizon, and A. Brelot for mutant coreceptors; C. Broder and J. Hoxie for monoclonal antibodies; and R. Doms for critical reading of the manuscript.
This work was supported by NIH grants AI 35502, NS 27405, and MH 61139.
REFERENCES
- 1.Alexander, W. A., B. Moss, and T. R. Fuerst. 1992. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J. Virol. 66:2934-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz, R. D., S. Alexander, and J. M. McCune. 2000. Causal relationships between HIV-1 coreceptor utilization, tropism, and pathogenesis in human thymus. AIDS Res. Hum. Retrovir. 16:1039-1045. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D., R. A. Fridell, I. Aramori, S. S. G. Ferguson, M. G. Caron, and B. R. Cullen. 1997. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 16:2599-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brelot, A., N. Heveker, M. Montes, and M. Alizon. 2000. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J. Biol. Chem. 275:23736-23744. [DOI] [PubMed] [Google Scholar]
- 5.Brelot, A., N. Heveker, O. Pleskoff, N. Sol, and M. Alizon. 1997. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J. Virol. 71:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camerini, D., H. P. Su, G. Gamez-Torre, M. L. Johnson, J. A. Zack, and I. S. Chen. 2000. Human immunodeficiency virus type 1 pathogenesis in SCID-hu mice correlates with syncytium-inducing phenotype and viral replication. J. Virol. 74:3196-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chabot, D. J., and C. C. Broder. 2000. Substitutions in a homologous region of extracellular loop 2 of CXCR4 and CCR5 alter coreceptor activities for HIV-1 membrane fusion and virus entry. J. Biol. Chem. 275:23774-23782. [DOI] [PubMed] [Google Scholar]
- 8.Chabot, D. J., P. F. Zhang, G. V. Quinnan, and C. C. Broder. 1999. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J. Virol. 73:6598-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collman, R., J. W. Balliet, S. A. Gregory, H. Friedman, D. L. Kolson, N. Nathanson, and A. Srinivasan. 1992. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J. Virol. 66:7517-7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cormier, E. G., M. Persuh, D. A. D. Thompson, S. W. Lin, T. P. Sakmar, W. C. Olson, and T. Dragic. 2000. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 97:5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doranz, B. J., Z. H. Lu, J. Rucker, T. Y. Zhang, M. Sharron, Y. H. Cen, Z. X. Wang, H. H. Guo, J. G. Du, M. A. Accaviti, R. W. Doms, and S. C. Peiper. 1997. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J. Virol. 71:6305-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doranz, B. J., M. J. Orsini, J. D. Turner, T. L. Hoffman, J. F. Berson, J. A. Hoxie, S. C. Peiper, L. F. Brass, and R. W. Doms. 1999. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J. Virol. 73:2752-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 15.Dragic, T. 2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol. 82:1807-1814. [DOI] [PubMed] [Google Scholar]
- 16.Farzan, M., H. Choe, L. Vaca, K. Martin, Y. Sun, E. Desjardins, N. Ruffing, L. J. Wu, R. Wyatt, N. Gerard, C. Gerard, and J. Sodroski. 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol. 72:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glushakova, S., J. C. Grivel, W. Fitzgerald, A. Sylwester, J. Zimmerberg, and L. B. Margolis. 1998. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nat. Med. 4:346-349. [DOI] [PubMed] [Google Scholar]
- 18.Hu, Q. X., J. O. Trent, G. D. Tomaras, Z. X. Wang, J. L. Murray, S. M. Conolly, J. M. Navenot, A. P. Barry, M. L. Greenberg, and S. C. Peiper. 2000. Identification of ENV determinants in V3 that influence the molecular anatomy of CCR5 utilization. J. Mol. Biol. 302:359-375. [DOI] [PubMed] [Google Scholar]
- 19.Jansson, M., E. Backstrom, G. Scarlatti, A. Bjorndal, S. Matsuda, P. Rossi, J. Albert, and H. Wigzell. 2001. Length variation of glycoprotein 120 V2 region in relation to biological phenotypes and coreceptor usage of primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 17:1405-1414. [DOI] [PubMed] [Google Scholar]
- 20.Kajumo, F., D. A. Thompson, Y. Guo, and T. Dragic. 2000. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by negatively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology 271:240-247. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson, A., K. Parsmyr, E. Sandström, E. M. Fenyö, and J. Albert. 1994. MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J. Clin. Microbiol. 32:364-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koot, M., I. P. M. Keet, A. H. V. Ros, R. E. Y. de Goede, M. T. L. Roos, R. A. Coutinho, F. Miedema, P. T. A. Schellekens, and M. Tersmette. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681-688. [DOI] [PubMed] [Google Scholar]
- 23.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, M. K., J. Heaton, and M. W. Cho. 1999. Identification of determinants of interaction between CXCR4 and gp120 of a dual-tropic HIV-1DH12 isolate. Virology 257:290-296. [DOI] [PubMed] [Google Scholar]
- 25.Lu, Z. H., J. F. Berson, Y. H. Chen, J. D. Turner, T. Y. Zhang, M. Sharron, M. H. Jenks, Z. X. Wang, J. Kim, J. Rucker, J. A. Hoxie, S. C. Peiper, and R. W. Doms. 1997. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc. Natl. Acad. Sci. USA 94:6426-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masciotra, S., S. M. Owen, D. Rudolph, C. Yang, B. Wang, N. Saksena, T. Spira, S. Dhawan, and R. B. Lal. 2002. Temporal relationship between V1V2 variation, macrophage replication, and coreceptor adaptation during HIV-1 disease progression. AIDS 16:1887-1898. [DOI] [PubMed] [Google Scholar]
- 27.Parolin, C., A. Borsetti, H. Y. Choe, M. Farzan, P. Kolchinsky, M. Heesen, Q. Ma, C. Gerard, G. Palú, M. E. Dorf, T. Springer, and J. Sodroski. 1998. Use of murine CXCR-4 as a second receptor by some T-cell-tropic human immunodeficiency viruses. J. Virol. 72:1652-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picard, L., G. Simmons, C. A. Power, A. Meyer, R. A. Weiss, and P. R. Clapham. 1997. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J. Virol. 71:5003-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picard, L., D. A. Wilkinson, A. McKnight, P. W. Gray, J. A. Hoxie, P. R. Clapham, and R. A. Weiss. 1997. Role of the amino-terminal extracellular domain of CXCR-4 in human immunodeficiency virus type 1 entry. Virology 231:105-111. [DOI] [PubMed] [Google Scholar]
- 30.Rucker, J., M. Samson, B. J. Doranz, F. Libert, J. F. Berson, Y. Yi, R. J. Smyth, R. G. Collman, C. C. Broder, G. Vassart, R. W. Doms, and M. Parmentier. 1996. Regions in the β-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell 87:437-446. [DOI] [PubMed] [Google Scholar]
- 31.Singh, A., and R. G. Collman. 2000. Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary isolate quasispecies. J. Virol. 74:10229-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh, A., Y. Yi, S. N. Isaacs, D. L. Kolson, and R. G. Collman. 2001. Concordant utilization of macrophage entry coreceptors by related variants within an HIV type 1 primary isolate viral swarm. AIDS Res. Hum. Retrovir. 17:957-963. [DOI] [PubMed] [Google Scholar]