Abstract
Although several groups have demonstrated that RNA interference, induced by transfection of small interfering RNA (siRNA) duplexes, can protect cells against a viral challenge in culture, this protection is transient. Here, we describe lentivirus expression vectors that can stably express siRNAs at levels sufficient to block virus replication. We have used these vectors to stably express siRNAs specific for the essential human immunodeficiency virus type 1 (HIV-1) Tat transcription factor or specific for a cellular coreceptor, CCR5, that is required for infection by the majority of primary HIV-1 isolates. These lentivirus vectors are shown to protect cells, including primary macrophages, against HIV-1 infection in culture by inducing selective degradation of their target mRNA species. These data suggest that it should be possible to block the expression of specific viral or cellular genes in vivo by using viral vectors to stably express the appropriate siRNAs.
RNA interference (RNAi) was first described in nematodes as a specific mechanism of posttranscriptional gene silencing induced by introduction of long double-stranded RNA (dsRNA) molecules homologous to the target mRNA (9). During RNAi, these long dsRNA molecules are cleaved into ∼21-bp dsRNAs, termed small interfering RNA (siRNA) duplexes, by a cellular enzyme called dicer (13, 16, 20, 22). One strand of this duplex is then incorporated into the RNA induced silencing complex (RISC), where it acts as a guide RNA that specifically targets RISC proteins to homologous mRNAs (13, 28, 40). Once RISC has bound an mRNA bearing a perfectly matched sequence, the mRNA is cleaved by an unknown endonuclease component. After release of the RISC, the mRNA is degraded by cellular exonucleases, thus resulting in specific posttranscriptional silencing of the target gene (14, 17, 30).
Although RNAi was first described in Caenorhabditis elegans, it quickly became apparent that RNAi is closely related to similar phenomena previously described in plants and certain fungi and it is now clear that the ability to mount an RNAi response is conserved among all higher eukaryotes (14, 17, 30). RNAi and related phenomena have been found to play important roles in the regulation of gene expression during development and in defense of the genome against random mutagenesis induced by transposable elements (14, 17, 21, 30, 45).
An additional activity mediated by RNAi, at least in plants, is antiviral defense (7, 37). Although it remains unclear whether RNAi also has a normal role in antiviral defense in vertebrates, the induction of an RNAi response can confer specific protection against several viral pathogens in culture, including human immunodeficiency virus type 1 (HIV-1), hepatitis C virus, influenza virus, poliovirus, respiratory syncytial virus, and hepatitis B virus (2, 5, 11, 12, 15, 18, 19, 23, 33, 36, 41, 44, 50). There has therefore been considerable interest in the development of RNAi as a possible treatment for virus-induced diseases.
Although RNAi can be effectively induced in many lower eukaryotes by the simple introduction of long dsRNAs, this is not possible in vertebrate cells because of activation of the interferon response, which causes a global inhibition of mRNA translation and frequently leads to apoptosis (27). However, a gene-specific RNAi response can be induced by direct transfection of ∼21-bp siRNA duplexes or, alternatively, by introduction of plasmids that encode short RNA hairpins that can be processed by dicer to give siRNA duplexes (3, 8, 34, 43, 51). Because induction of the interferon response requires dsRNAs of >30 bp (27), this strategy allows specific gene silencing by RNAi in the absence of the global gene silencing noted upon the introduction of long dsRNAs.
Although protection against HIV-1 after transfection of cells with siRNA duplexes or siRNA expression plasmids has been reported by several groups (5, 15, 18, 23, 33, 44), this strategy has several problems, including the transient nature of the antiviral effect that is produced and the difficulty of introducing nucleic acids into primary cells. In addition, targeting of a virus that displays the sequence variability of HIV-1 by using a highly sequence-specific reagent such as an siRNA might be predicted to rapidly lead to the selection of resistant viral variants. To address these concerns, we have identified an siRNA that can effectively block the expression of the cellular protein CCR5, a key coreceptor for both initial infection and subsequent dissemination of primary HIV-1 isolates in humans (38). We have also developed a lentivirus vector that can express siRNAs, targeted either to CCR5 or the essential HIV-1 tat gene, at a level sufficient to confer a stable antiviral phenotype on both cell lines and primary macrophages. These data suggest that it may be possible to confer a stable phenotype of virus resistance on cells and tissues in vivo by targeting either cellular genes that serve as essential viral cofactors or highly conserved viral RNA sequences.
MATERIALS AND METHODS
Plasmid construction.
The mammalian expression plasmids pSuper, pHIV/Tat, pcTat, pcRev, pBC12/CMV/β-gal, and pHIT/G have been described previously (3, 5, 10, 26). The pHIV/SynTat expression plasmid is identical to pHIV/Tat, except that the wild-type tat gene has been replaced with a previously described synthetic tat gene predicted to encode the same protein product (1, 46). In addition, the HIV-1 proviral clones pNL-ADA, pNL-Luc-ADA, pNL-Luc-HXB, and pNL-Luc-1549 and plasmids expressing human CD4 (pCMV5/CD4), CXCR4 (pCMV5/CXCR4), and CCR5 (pCMV5/CCR5) have also been described previously (6, 48).
A 1,471-bp fragment of pNL-Luc-ADA encoding vpu, the first exon of tat and rev, and part of env and vpr was deleted by digestion with SalI and NheI. The sequence encoding the Vpr carboxy terminus was then restored by ligating a PCR-derived DNA fragment into the SalI and NheI sites. The PCR primers used also inserted unique SacII and XmaI sites immediately downstream of vpr. Most of the 3′ long terminal repeat (LTR) U3 region (positions −418 to −18) was then deleted by a recombinant PCR with primers that introduced unique ClaI and XbaI sites. A cytomegalovirus immediate-early promoter linked to the selectable blasticidin resistance (blr) marker was then inserted between unique BamHI and XhoI sites to generate pNL-SIN-CMV-BLR. These additional manipulations removed further segments of the viral env, rev, and nef genes, as well as the luc indicator gene present in pNL-Luc-ADA. However, the HIV-1 Rev response element was left intact (Fig. 1A).
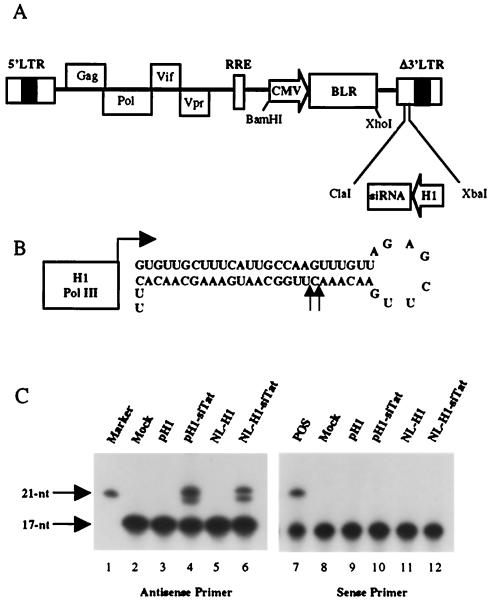
FIG. 1.
Expression of siRNAs by a lentivirus vector. (A) Schematic representation of the NL-SIN-CMV-BLR lentivirus vector. The blasticidin resistance (blr) gene was introduced as a selectable marker. Unique restriction enzyme sites are indicated. An siRNA expression cassette containing the human H1 promoter can be introduced in place of most of the 3′ LTR U3 region. (B) Predicted stem-loop structure of the Tat siRNA precursor encoded by the pNL-H1-siTat lentivirus vector. Arrows indicate the 5′ ends of the mature siRNAs encoded by this vector. (C) Primer extension analysis. pH1, cells transfected with 1 μg of pSuper; pH1-siTat, cells transfected with 1 μg of pH1-siTat; NL-H1, cells transduced with the NL-H1 lentivirus vector; NL-H1-siTat, cells transduced with NL-H1-siTat. A probe that detects the 3′ or antisense strand was used for lanes 2 to 6, and a probe specific for the 5′ or sense strand was used for lanes 7 to 12. Lane 1 contained a labeled 21-nt RNA marker, while lane 7 contained a synthetic 21-nt RNA identical to the first 21 nt of the 5′ arm of the siRNA precursor as a positive (POS) control. Mock, mock-transfected wild-type 293T cells.
Oligonucleotides encoding siRNAs directed against residues 84 to 111 in the HIV-1 tat open reading frame (ORF) (5′-AAGTGTTGCTTTCATTGCCAAGTTTGTT-3′) or residues 953 to 979 in the human ccr5 ORF (5′-AACGCTTCTGCAAATGCTGTTCTATTT-3′) were cloned into pSUPER essentially as previously described (3) to generate pH1-siTat and pH1-siCCR5, respectively. DNA fragments containing the H1 promoter (31) and sequences encoding the Tat- and CCR5-specific siRNA precursors were excised from pH1-siTat and pH1-siCCR5 by digestion with ClaI and XbaI and subcloned into the same sites in pNL-SIN-CMV-BLR to generate pNL-H1-siTat and pNL-H1-siCCR5, respectively. A similar lentivirus vector containing the H1 promoter but lacking any siRNA precursor, termed pNL-H1, was generated in parallel as a negative control.
Primary monocyte culture.
Peripheral blood mononuclear cells from healthy, HIV-1-negative donors were isolated by Ficoll-Hypaque gradient centrifugation. The cells were then resuspended in Dulbecco's modified Eagle medium and plated at 2 × 106 per well in 24-well tissue culture plates. After 3 h of culture, adherent cells were washed extensively with phosphate-buffered saline (PBS) and then cultured in Dulbecco modified Eagle medium supplemented with 10% human AB serum (Sigma) and 1,000 U of macrophage colony-stimulating factor (R&D Systems) for 1 week to allow differentiation into monocyte-derived macrophages (MDM).
Preparation of virus stocks and transduced cells.
Human 293T cells were maintained as previously described (5). To prepare HIV-1 stocks, 293T cells were transfected with 2 μg of pNL-Luc-ADA, pNL-Luc-HXB, or pNL-Luc-1549 as previously described (48). Stocks of vesicular stomatitis virus glycoprotein G (VSV-G)-pseudotyped HIV-1 virions were produced by transfection of 293T cells with 1.5 μg of pNL-ADA and 0.5 μg of pHIT/G. HIV-1-based lentivirus vectors expressing siRNAs were prepared by cotransfection of 293T cells with 1.8 μg of the lentivirus vector, 0.1 μg of pcRev, 0.1 μg of pcTat, and 50 ng of pHIT/G. The culture medium was replaced 16 h later, and supernatant media were harvested ∼40 h posttransfection and passed through a 0.45-μm-pore-size filter. Virus yields were measured by p24 Gag antigen capture enzyme-linked immunosorbent assay (NEN Life Science).
RNA and protein expression analysis.
Total RNA was prepared from 293T cells transfected with a pSUPER-derived plasmid (pH1 plasmids), or from 293T cells that had been transduced with a lentivirus vector, by using Trizol in accordance with the manufacturer's (Gibco BRL) instructions. Primer extension reactions were performed with 10 μg of total RNA with the primer extension system-avian myeloblastosis virus reverse transcriptase (Promega) in accordance with the manufacturer's instructions. Extension products were analyzed by electrophoresis through denaturing polyacrylamide gels and visualized by autoradiography.
Northern analysis was performed essentially as previously described (5). Total RNA (30 μg) was separated through a 1% agarose gel containing formaldehyde and transferred to a Hybond-N membrane (Amersham). RNA was fixed by UV cross-linking with a Stratalinker (Stratagene). HIV-1, CCR5, and CD4 mRNAs were detected with 32P-labeled random primed DNA probes.
Western analysis of protein expression in transfected 293T cells was performed as previously described (5). Briefly, cell lysates prepared from transfected 293T cells were boiled for 5 min in sodium dodecyl sulfate loading buffer containing 10% 2-mercaptoethanol. Proteins were separated by gel electrophoresis and transferred to a nitrocellulose membrane (Amersham). Western analysis was performed with a rabbit polyclonal antiserum directed against the HIV-1 Tat protein (1:1,000) or a mouse monoclonal antibody directed against β-galactosidase (β-gal; 1:2,000; Promega). The anti-Tat serum used was raised with recombinant full-length Tat and has been provided to the AIDS Research and Reference Reagent Program (catalog no. 705). Bound antibodies were detected with horseradish peroxide-conjugated anti-rabbit or anti-mouse secondary antibodies (Amersham) at a 1:3,000 dilution and visualized by chemiluminescence.
For quantitation of cell surface CCR5, transduced 293T cells were transfected with pCMV5/CCR5 and pEGFP. At 48 h after transfection, cells were stained with a monoclonal anti-CCR5 serum conjugated with Cy-Chrome (BD PharMingen) on ice for 30 min. The cells were then washed extensively with PBS, fixed with 4% formaldehyde in PBS, and then analyzed by fluorescence-activated cell sorting (FACS) on a FACScan.
Luciferase reporter virus assay.
Lentivirus vector-transduced 293T cells were transfected with 100 ng of pCMV5/CD4 and 20 ng of pCMV5/CCR5 or pCMV5/CXCR4 by using calcium phosphate. In some experiments, cells were additionally transfected with 10 ng of pHIV/Tat or pHIV/SynTat or with the parental pBC12/CMV plasmid as a negative control. At 48 h after transfection, the cells were infected with 20 ng of p24 Gag antigen of the NL-Luc-ADA virus for pCMV5/CCR5-transfected cells or the NL-Luc-HXB virus for pCMV5/CXCR4-transfected cells. After 48 h, cells were lysed in 200 μl of lysis buffer (Promega) and luciferase activities were determined with a Lumat LB9501 luminometer.
For infection of macrophages, 7-day-old cultures of MDM were infected overnight with 100 ng of p24 antigen from one of the NL-H1 lentivirus vectors. On the next day, cells were washed with PBS and placed in fresh medium. The cells were allowed to recover for 1 further day and then subjected to a second round of infection with a lentivirus vector. Two days after the second infection, the transduced MDM were infected with 20 ng of p24 antigen from the luciferase reporter virus NL-Luc-ADA or NL-Luc-1549. The cells were harvested 72 h later for luciferase assay as described above.
RESULTS
Construction of a lentivirus siRNA expression vector.
Although we and others have shown that transfection of siRNAs, or of siRNA expression plasmids, can effectively protect cells against HIV-1 (5, 15, 18, 23, 33, 44), this protection is only temporary. To address this concern, we constructed a lentivirus vector, pNL-SIN-CMV-BLR, that can be used to stably transduce cultured or primary cells with an siRNA expression cassette (Fig. 1A).
pNL-SIN-CMV-BLR was derived from pNL-Luc-ADA (48) by deletion of part or all of the HIV-1 vpu, tat, rev, env, and nef genes, as well as almost all of the U3 region in the 3′ LTR. The viral gag, pol, vif, and vpr genes, as well as the 5′ LTR and the Rev response element, were, however, left intact, as were all of the cis-acting sequences required for efficient genome packaging, reverse transcription, and integration. In place of the deleted sequences, we inserted a cytomegalovirus immediate-early promoter driving the blr selectable marker, as well as unique ClaI and XbaI sites in the 3′ LTR U3 region. These can be used to insert an siRNA expression cassette consisting of the RNA polymerase III-dependent H1 promoter (3, 31) linked to a sequence designed to form a short RNA stem-loop sequence that can be processed by endogenous dicer to yield an siRNA duplex (3, 34) (Fig. 1). This cassette replaces much of the 3′ LTR U3 region and will therefore be duplicated during reverse transcription to replace the promoter and enhancer sequences in the U3 region of the 5′ LTR, thus making this a self-inactivating double-copy lentivirus expression vector.
The initial siRNA target sequence chosen was derived from nucleotides (nt) 84 to 111 of the HIV-1 tat ORF and is shown in Fig. 1B. The sequence inserted into the pSuper siRNA expression plasmid (3) is predicted to give rise to a 26-bp dsRNA stem flanked by an 8-nt terminal loop and a 2-nt 3′ overhang derived from the transcription termination sequence for polymerase III. This expression cassette was excised from the resultant pH1-siTat plasmid by cleavage with XbaI and ClaI and then inserted into the pNL-SIN-CMV-BLR lentivirus vector to give pNL-H1-siTat (Fig. 1A). The lentivirus vector encoded by pNL-H1-siTat was packaged by cotransfection into 293T cells along with plasmids expressing HIV-1 Tat and Rev cDNAs and VSV-G. Packaged virions (50 ng of p24 Gag) were used to infect 106 293T cells, and transductants were selected with 10 μg of blasticidin per ml. Generally, >60% of the infected cells were successfully transduced with this protocol.
Primer extension was used to measure the production of siRNAs from either the initial pH1-siTat expression plasmid in transfected 293T cells or the lentivirus NL-H1-siTat vector in pooled, transduced 293T cells (Fig. 1C). The antisense or 3′ arm of the predicted stem-loop precursor (Fig. 1B) was found to give rise to two overlapping siRNAs 21 or 22 nt in length, measured from the base of the stem, and equivalent levels of each siRNA were observed in both transfected and transduced 293T cells (Fig. 1C, lanes 4 and 6). These two products presumably arise from processing by dicer at two adjacent cleavage sites in the predicted RNA hairpin (Fig. 1B). In contrast, the 5′ or sense arm did not give rise to any detectable siRNA product in vivo, although a synthetic 21-nt RNA homologous to the predicted siRNA did give rise to a readily detectable extension product (Fig. 1C, lane 7). Therefore, the RNA stem-loop structure depicted in Fig. 1B behaves like a micro-RNA stem-loop precursor in that only one arm of the precursor appears to have been stably included in the RISC (14, 17).
Stable expression of an siRNA specific for HIV-1 tat.
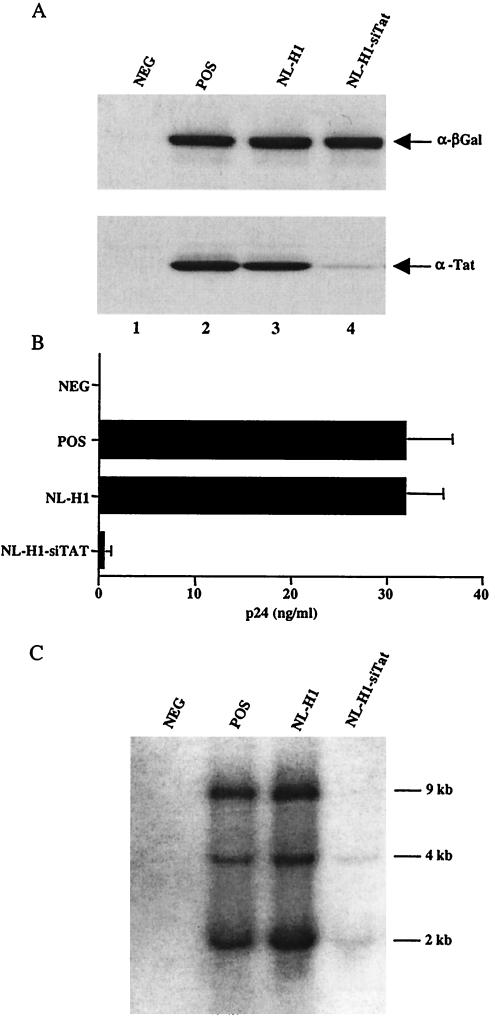
Previously, we have shown that transfection of an siRNA duplex specific for the HIV-1 tat gene can effectively block HIV-1 replication in culture (5). To test whether expression of a Tat-specific siRNA from a lentivirus vector would exert a similar phenotype, we transfected 293T cells transduced with NL-H1-siTat or with NL-H1, which is not predicted to express any siRNA, with expression vectors encoding HIV-1 Tat and β-gal. As shown in Fig. 2A, we noted a significant drop in Tat expression, compared to that of the β-gal control, in NL-H1-siTat-transduced cells. To test whether the Tat siRNA would have any effect on HIV-1 replication, we next infected control and transduced 293T cells with HIV-1 virions pseudotyped with VSV-G and then quantified virus replication by measuring the level of secreted p24 Gag protein produced by the infected cells. As shown in Fig. 2B, the NL-H1-siTat-transduced cells were largely incapable of giving rise to progeny virions while control NL-H1-transduced cells or nontransduced 293T cells gave rise to high levels of progeny virions, as measured by p24 enzyme-linked immunosorbent assay of the culture supernatants. As shown in the Northern analysis presented in Fig. 2C, the lack of progeny virion production in the NL-H1-siTat-transduced cells coincided with a large drop in the expression of all HIV-1 mRNA species after infection, as would be predicted if Tat function was largely blocked.
FIG. 2.
Inhibition of HIV-1 replication with an siRNA expressed from a lentivirus vector. (A) Normal 293T cells (lane 2) or cells transduced by lentivirus vectors (lanes 3 and 4) were cotransfected with 25 ng each of pBC12/CMV/β-gal and pHIV/Tat. At 48 h posttransfection, protein expression levels were assayed by Western analysis. Mock-transfected 293T cells served as the negative control (NEG). (B) Specific inhibition of HIV-1 replication in 293T cells transduced with the NL-H1-siTat lentivirus vector. 293T cells were infected overnight with VSV-G-pseudotyped HIV-1, and viral replication was measured by p24 Gag antigen production at 48 h after infection. 293T cells infected with wild-type, i.e., nonpseudotyped, HIV-1 served as the negative control (NEG), while transfected, nontransduced 293T cells served as the positive control (POS). The results shown are the average of three experiments with the standard deviation. (C) Total RNA was extracted from 293T cells used in panel B and subjected to Northern analysis with an HIV-1 LTR-specific probe. The approximate sizes of the three classes of HIV-1 transcript are indicated.
To demonstrate that the observed inhibition of HIV-1 replication was indeed specific, we next wished to demonstrate that mutation of the siRNA target site in the HIV-1 tat gene would rescue virus replication. Unfortunately, introduction of several point mutations into this sequence in the context of a full-length HIV-1 proviral clone resulted in an intrinsically defective virus, even though these mutations were designed not to affect the sequence of the encoded Tat protein (data not shown).
As an alternative approach to the demonstration of specificity, we therefore investigated whether HIV-1 proviral transcription could be rescued by cotransfection of an expression plasmid encoding a tat gene in which the siRNA target sequence was mutated, but not by a similar vector encoding wild-type Tat. The expression plasmid chosen, pHIV/SynTat, is identical to pHIV/Tat except that it encodes a synthetic tat gene encoding a number of nucleic acid sequence changes that do not, however, affect either the sequence or the function of the encoded Tat protein (1, 46). Importantly, the tat gene carried by pHIV/SynTat differs at three positions within the chosen siRNA target sequence (Fig. 3A) and therefore would be predicted to be largely or entirely resistant to RNAi induced by pNL-H1-siTat.
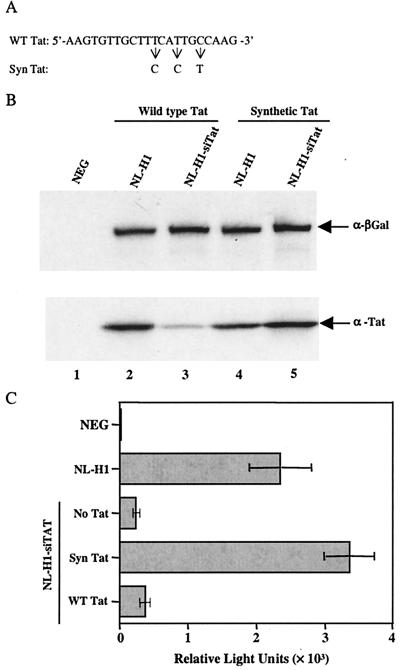
FIG. 3.
Inhibition of HIV-1 gene expression by NL-H1-siTat is specific. (A) Sequences of the wild-type (WT) and mutant synthetic (Syn) forms of the Tat siRNA target sequence. (B) 293T cells transduced with NL-H1 (lanes 2 and 4) or NL-H1-siTat (lanes 3 and 5) were transfected with 25 ng each of pBC12/CMV/β-gal and either wild-type pHIV/Tat (lanes 2 and 3) or the mutant pHIV/SynTat expression plasmid (lanes 4 and 5). Western analysis was performed as described in the legend to Fig. 2A. (C) 293T cells transduced with NL-H1 or NL-H1-siTat were transfected with expression plasmids encoding CD4 and CCR5 and, where indicated, 10 ng of either pHIV/Tat or pHIV/SynTat. At 48 h after transfection, the transduced, transfected cells were infected with the NL-Luc-ADA indicator virus and induced luciferase levels were assayed a further 48 h later. NEG, mock-transfected, NL-H1-transduced cells. The average of three experiments and the standard deviation are indicated. NEG, negative control.
To confirm this prediction, we first transfected NL-H1-siTat-transduced or control NL-H1-transduced cells with either pHIV/Tat (encoding wild-type Tat) or pHIV/SynTat (carrying the modified tat gene), as well as with pBC12/CMV/β-gal as an internal control. As can be seen in Fig. 3B, Tat expression from the pHIV/SynTat vector, unlike Tat expression from pHIV/Tat, was indeed resistant to the siRNA expressed in the NL-H1-siTat-transduced cells.
To determine whether HIV-1 proviral transcription could be selectively rescued by the mutant tat gene carried by pHIV/SynTat, we transfected NL-H1-siTat-transduced cells with expression plasmids encoding CD4 and CCR5 and 10 ng of either pHIV/Tat or pHIV/SynTat or a negative control plasmid. NL-H1-transduced cells transfected with vectors encoding CD4 and CCR5 only served as a positive control, while nontransfected NL-H1 cells served as a negative control. At 48 h later, these cultures were infected with the previously described NL-Luc-ADA indicator virus containing the luc indicator gene substituted for nef. Importantly, the luc mRNA encoded by this virus is expressed in the absence of Rev function and is not predicted to contain the tat siRNA target sequence. As shown in Fig. 3C, transfection with the pHIV/SynTat plasmid, bearing a mutated siRNA target sequence, indeed fully rescued luciferase expression by the infecting indicator virus, while the wild-type Tat expression plasmid pHIV/Tat had little or no positive effect. We therefore concluded that the observed inhibition of HIV-1 replication and gene expression was indeed due to the specific degradation of viral mRNAs bearing the tat siRNA target sequence.
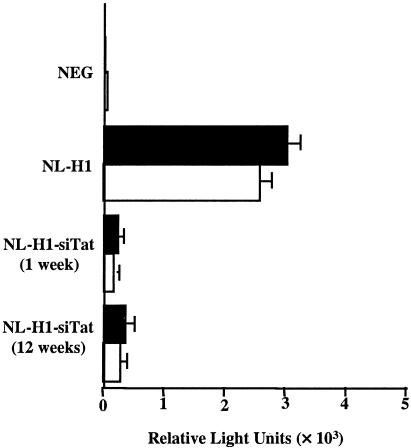
An important goal of this research was to confer a stable, virus-resistant phenotype on cells transduced with the lentivirus siRNA expression vector, rather than the transient resistance characteristic of cells transfected with siRNAs. To test whether resistance to HIV-1 replication was indeed stable, we compared the virus resistance of NL-H1-siTat-transduced cells after 1 week versus 12 weeks in culture. In this experiment, the transduced 293T cells were first transfected with expression plasmids encoding CD4 and either CCR5 or CXCR4. After 48 h, we then infected the transduced, transfected 293T cells with the previously described R5-tropic NL-Luc-ADA indicator virus or the X4-tropic NL-Luc-HXB indicator virus (6, 48). As shown in Fig. 4, 293T cells transduced with NL-H1-siTat remained nonpermissive for productive HIV-1 infection by either indicator virus after 12 weeks in culture, in contrast to the control NL-H1-transduced cells. Primer extension analyses revealed that the level of expression of the siRNA encoded by the NL-H1-siTat provirus was essentially the same after 1 week or 12 weeks in culture (data not shown). We therefore concluded that the antiviral state induced by the lentivirus NL-H1-siTat siRNA expression vector was stable over the period tested.
FIG. 4.
Stable inhibition of HIV-1 gene expression in NL-H1-siTat-transduced cells. 293T cells transduced with NL-H1-siTat were cultured under selection for either 1 week or 12 weeks. Transduced cells were then transfected with expression plasmids encoding CD4 and CCR5 or CD4 and CXCR4 and infected with the R5-tropic NL-Luc-ADA or X4-tropic NL-Luc-HXB indicator virus, and induced luciferase activities were determined 2 days later. The average of three experiments and the standard deviation are indicated. NEG, negative control.
Inhibition of CCR5 expression with a lentivirus siRNA expression vector.
Although the NL-H1-siTat lentivirus vector can potently inhibit HIV-1 replication (Fig. 2 to 4), this represents an idealized system in that the siRNA is designed to target the sequence of a cloned HIV-1 provirus. However, siRNA function can be severely compromised by even a small number of mismatches with the mRNA target sequence (Fig. 3). Given both the known variability of primary HIV-1 gene sequences and the mutability of these sequences in the face of selective pressure, it appears unlikely that siRNAs targeted against HIV-1 would be able to suppress virus replication over the long term in vivo.
To address this issue, we designed siRNAs targeted to the CCR5 coreceptor. CCR5 is critical for HIV-1 transmission between humans and is necessary for infection by the majority of primary HIV-1 isolates (38). Although important for HIV-1 infection, CCR5 is dispensable for human health and well-being, as shown by the existence of individuals who are homozygous for a point mutation that totally blocks CCR5 expression (25).
We identified an siRNA, directed against residues 953 to 979 of the human ccr5 ORF, that was highly effective at inhibiting CCR5 expression by cotransfection (data not shown). This sequence was incorporated into the stem of a predicted RNA stem-loop structure, expressed under the control of the H1 promoter and then introduced into the pNL-SIN-CMV-BLR lentivirus vector to give pNL-H1-CCR5, as shown in Fig. 1. This lentivirus siRNA expression vector was then packaged in 293T cells and used to infect other 293T cells, and transductants were selected by culture in blasticidin.
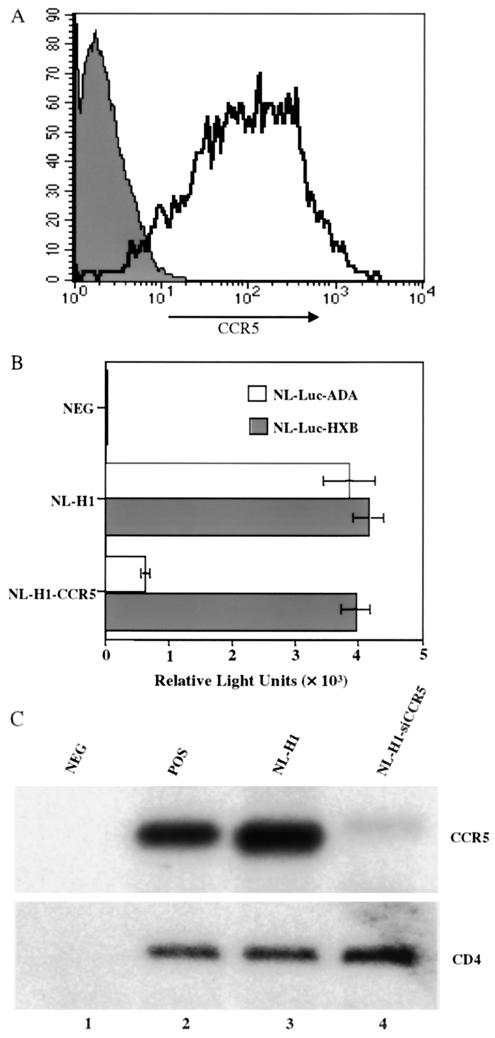
To test whether the siRNA expressed from the lentivirus vector would indeed exert an inhibitory effect on CCR5 expression, we transfected pooled 293T cells transduced with NL-H1-siCCR5, with NL-H1 as a control, with plasmids expressing green fluorescent protein (GFP), and with CCR5. At 48 h after transfection, cells were stained with a monoclonal antibody specific for CCR5 and subjected to FACS to quantitate the level of cell surface CCR5 expression on GFP-expressing cells. Although the levels of GFP expression in the different transduced cells were similar (data not shown), the level of CCR5 expression observed was dramatically lower in the cells transduced with NL-H1-siCCR5 than in the control NL-H1-transduced cells (Fig. 5A).
FIG. 5.
Inhibition of CCR5 expression by a lentivirus-expressed siRNA. (A) Transduced 293T cells were transfected with plasmids expressing CCR5 and GFP. At 48 h after transfection, the level of cell surface CCR5 expression on GFP-positive cells was analyzed by FACS with an anti-CCR5 monoclonal antibody. CCR5 expression on NL-H1-siCCR5-transduced cells is represented by shading, and that on NL-H1-transduced cells is represented by the open area. (B) Transduced 293T cells were transfected with expression plasmids encoding CD4 and CCR5 or CD4 and CXCR4. At 48 h after transfection, CD4+ CCR5+ cells were infected with NL-Luc-ADA while CD4+ CXCR4+ cells were infected with NL-Luc-HXB. Induced luciferase levels were determined 48 h after infection and are indicated. NEG, nontransfected 293T cells. The average of three experiments and the standard deviation are indicated. (C) Transduced or control 293T cells were transfected with CCR5 and CD4 expression plasmids. At 48 h after transfection, total RNA was extracted and subjected to Northern analysis to measure CCR5 mRNA (top) and CD4 mRNA (bottom) expression. Mock-transfected 293T cells served as a negative control (NEG), while wild-type 293T cells transfected with CCR5 and CD4 served as a positive control (POS).
To test whether the siRNA expressed from the NL-H1-siCCR5 lentivirus vector would selectively inhibit infection by R5-tropic HIV-1, we next transfected transduced 293T cells with plasmids expressing CD4 and CCR5 or CD4 and CXCR4. Mock-transfected 293T cells served as a negative control. Two days later, we infected the transfected 293T cells with the R5-tropic indicator virus NL-Luc-ADA or the X4-tropic indicator virus NL-Luc-HXB (48). At 48 h later, the cells were lysed and induced luciferase expression levels were quantitated. As shown in Fig. 3B, NL-Luc-ADA and NL-Luc-HXB were able to infect the control NL-H1-transduced cells with comparable efficiencies, as shown by the similar levels of expression of the virus-encoded luciferase. In contrast, while the NL-H1-siCCR5-transduced cells remained fully permissive for infection by the X4-tropic NL-Luc-HXB indicator virus, infection by the R5-tropic NL-Luc-ADA indicator virus was strongly reduced (Fig. 5B).
If the siRNA expressed in the NL-H1-siCCR5-transduced cells is indeed functional, then it should induce the selective degradation of CCR5 mRNA (14, 17, 30). To test this hypothesis, we prepared total RNA from cultures transfected as described above and performed Northern analysis for either the CCR5 or the CD4 mRNA expressed from the cotransfected plasmids. As shown in Fig. 5C, we observed a marked and specific inhibition of the level of CCR5 mRNA expression in the NL-H1-siCCR5-transduced cells compared to that in control NL-H1-transduced cells, while the level of CD4 mRNA expression remained constant. Together, these data indicate that the CCR5-specific siRNA produced by the NL-H1-siCCR5 lentivirus vector is able to specifically reduce not only CCR5 mRNA and protein expression but also infection by R5-tropic HIV-1.
Inhibition of HIV-1 replication in primary macrophages by siRNAs expressed from lentivirus vectors.
The data presented thus far were all obtained with the human cell line 293T, and we were therefore anxious to confirm that the lentivirus siRNA expression vectors would also function in primary cells. We therefore used the lentivirus vectors to transduce primary MDM. It has previously been shown by several groups that these nondividing cells can be effectively infected by HIV-1 or by HIV-1-derived lentivirus vectors (4, 24, 32, 49).
To obtain good levels of infection without selection and without having to concentrate the lentivirus vector preparations and to see if we could observe functional synergy between two distinct lentivirus siRNA expression vectors, we transduced each MDM culture twice over a 2-day interval (Fig. 6). Two days after the second transduction, the MDM were infected with aliquots of the R5-tropic NL-Luc-ADA or the X4-tropic NL-Luc-1549 indicator virus. The env gene used in NL-Luc-1549 is derived from a primary X4-tropic HIV-1 strain that, unlike laboratory X4-tropic HIV-1 isolates, is able to infect primary macrophages with exclusively endogenous CXCR4 as a coreceptor (48).
FIG. 6.
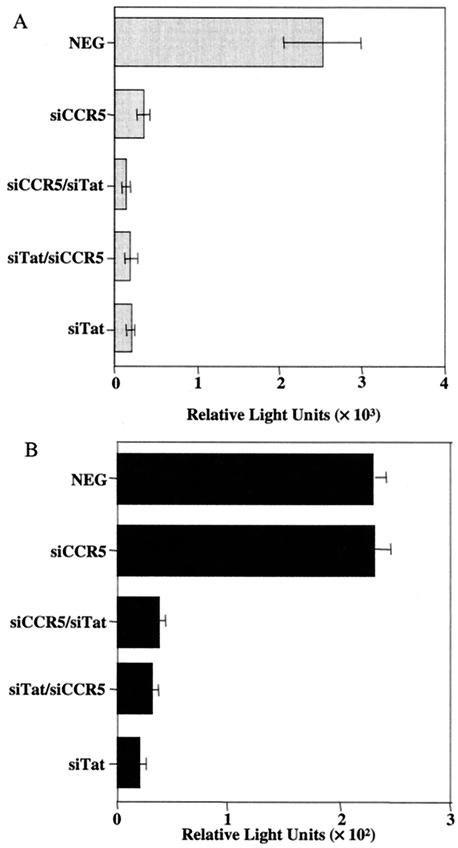
Single-cycle assay of HIV-1 replication in primary macrophages. MDM were cultured for 7 days in the presence of macrophage colony-stimulating factor and then transduced twice with a lentivirus vector stock over a 2-day period. The order of vector infection is shown; e.g., siCCR5 denotes MDM transduced twice with NL-H1-siCCR5, while siCCR5/siTat denotes MDM transduced first with NL-H1-siCCR5 and then with NL-H1-siTat. Two days later, the transduced MDM were infected with the R5-tropic reporter virus NL-Luc-ADA (A) or X4-tropic reporter virus NL-Luc-1549 (B) and induced luciferase levels were measured a further 48 h later. Doubly NL-H1-transduced MDM served as the negative control (NEG). The average of three experiments and the standard deviation are indicated.
As shown in Fig. 6A, we observed efficient inhibition of MDM infection by the R5-tropic NL-Luc-ADA indicator virus after transduction with NL-H1-siTat, NL-H1-siCCR5, or both lentivirus vectors. The efficiency of inhibition by NL-H1-siTat was such that it was not possible to see any synergy with NL-H1-siCCR5 under these assay conditions. In contrast to the R5-tropic NL-Luc-ADA indicator virus, infection of primary macrophages by the X4-tropic NL-Luc-1549 virus was entirely unaffected by transduction of the cells with NL-H1-siCCR5, as predicted. However, this virus remained fully susceptible to inhibition by NL-H1-siTat (Fig. 6B). We therefore concluded that these lentivirus siRNA expression vectors are capable of expressing siRNAs in primary macrophages at levels sufficient to effectively block functional expression of the cellular ccr5 or viral tat gene product.
DISCUSSION
While there has been considerable excitement over the potential of RNAi as a treatment for virus-induced diseases, this potential will only be achievable if the relevant nucleic acids can be effectively delivered to the appropriate cells in vivo, if an effective level of expression of the siRNAs can be maintained over a long period, and if the target virus is not able to rapidly undergo selection for variants that are resistant to the siRNAs used.
While we and others have previously reported excellent protection against HIV-1 and other viruses in cell lines and primary cells in culture (2, 5, 11, 12, 15, 18, 19, 23, 33, 36, 41, 44, 50), the approach used, i.e., primarily transfection of siRNA duplexes, gives rise to only transient inhibition. To address this concern, we have developed a simple lentivirus vector that can effectively and stably express siRNAs at a level sufficient to block HIV-1 replication in culture (Fig. 1). Although the pNL-SIN-CMV-BLR vector may not be sufficiently defective to be suitable for use in humans, these data nevertheless provide proof of principle for the concept that stable expression of siRNAs, and hence a stable antiviral phenotype, can be achieved with lentivirus vectors or other viral vectors.
A second concern noted above is the issue of viral variability and the resultant ability of viruses to escape inhibition by siRNAs targeted to even highly conserved regions in the viral genome. While it might, in principle, be possible to design siRNAs targeted to multiple critical viral sequences that, in combination, would provide long-term protection, a possible alternative strategy is to target a cellular gene product that is essential for virus replication but dispensable to the host. The human ccr5 gene provides an ideal example of such a target, as it is critical for infection by most primary HIV-1 isolates yet apparently of little or no importance to the human host (25, 38).
With the lentivirus vector siRNA expression strategy, we were indeed able to effectively and specifically reduce CCR5 protein and mRNA expression in transduced cells (Fig. 5). More importantly, this approach allowed us to selectively inhibit infection by R5-tropic, but not X4-tropic, HIV-1 isolates of not only cell lines but also primary macrophages (Fig. 5B and 6). These data therefore confirm and extend the recent report by Qin et al. (35) showing that a lentivirus-expressed siRNA targeted to CCR5 can inhibit infection of primary T cells by R5-tropic HIV-1. Together, these reports suggest that this strategy has the potential to block HIV-1 infection in vivo in a manner that cannot be overcome simply by a 1- or 2-nt change in the viral siRNA target sequence (12). Of course, HIV-1 might still be able to escape this inhibitory effect by switching to another coreceptor, such as CXCR4 (25, 38). While this certainly represents a serious concern, we note that it may also be possible to simultaneously knock down CXCR4 expression in adult patients with RNAi without serious side effects for the host and to thereby preclude this avenue of viral escape (29).
Although the focus of this report has been the potential of lentivirus siRNA expression vectors for rendering cells nonpermissive for HIV-1 replication, these vectors also have the potential to induce the stable knockdown of important cellular genes unrelated to the HIV-1 life cycle in either cultured cells or experimental animals (39, 42, 47). We therefore intend to make the pNL-SIN-CMV-BLR lentivirus vector, as well as a variant expressing gfp in place of blr, available to academic researchers on request.
Acknowledgments
M.-T.M.L. and G.A.C. contributed equally to this report.
We thank Timothy M. Clay and Michael A. Morse for assistance in the isolation of primary human macrophages.
This work was supported in part by grant AI42538 from the National Institute of Allergy and Infectious Diseases and by a travel grant from the Burroughs Wellcome Fund, both to B.R.C.
REFERENCES
- 1.Adams, S. E., I. D. Johnson, M. Braddock, A. J. Kingsman, S. M. Kingsman, and M. Edwards. 1988. Synthesis of a gene for the HIV transactivator protein TAT by a novel single stranded approach involving in vivo gap repair. Nucleic Acids Res. 16:4287-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitko, V., and S. Barik. 2001. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubei, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. VPR is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 7.Covey, S. N., N. S. Al-Kaff, A. Langara, and D. S. Turner. 1997. Plants combat infection by gene silencing. Nature 385:781-782. [Google Scholar]
- 8.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 9.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 10.Fouchier, R. A. M., B. E. Meyer, J. H. M. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge, Q., M. T. McManus, T. Nguyen, C.-H. Shen, P. A. Sharp, H. N. Eisen, and J. Chen. 2003. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. USA 100:2718-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 13.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-295. [DOI] [PubMed] [Google Scholar]
- 14.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 15.Hu, W.-Y., C. P. Myers, J. M. Kilzer, S. L. Pfaff, and F. D. Bushman. 2002. Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol. 12:1301-1311. [DOI] [PubMed] [Google Scholar]
- 16.Hutvágner, G., J. McLachlan, A. E. Pasquinelli, É. Bálint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 17.Hutvágner, G., and P. D. Zamore. 2002. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 12:225-232. [DOI] [PubMed] [Google Scholar]
- 18.Jacque, J.-M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapadia, S. B., A. Brideau-Andersen, and F. V. Chisari. 2003. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. USA 100:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ketting, R. F., T. H. Haverkamp, H. G. van Luenen, and R. H. Plasterk. 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNase D. Cell 99:133-141. [DOI] [PubMed] [Google Scholar]
- 22.Knight, S. W., and B. L. Bass. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in C. elegans. Science 293:2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M.-J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 19:500-504. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 8:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 26.Malim, M. H., J. Hauber, R. Fenrick, and B. R. Cullen. 1988. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335:181-183. [DOI] [PubMed] [Google Scholar]
- 27.Manche, L., S. R. Green, C. Schmedt, and M. B. Mathews. 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12:5238-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez, J., A. Patkaniowska, H. Urlaub, R. Lührmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563-574. [DOI] [PubMed] [Google Scholar]
- 29.Martinez, M. A., A. Gutierrez, M. Armand-Ugon, J. Blanco, M. Parera, J. Gomez, B. Clotet, and J. A. Este. 2002. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS 16:2385-2390. [DOI] [PubMed] [Google Scholar]
- 30.McManus, M. T., and P. A. Sharp. 2002. Gene silencing in mammals by short interfering RNAs. Nat. Rev. 3:737-747. [DOI] [PubMed] [Google Scholar]
- 31.Myslinski, E., J.-C. Amé, A. Krol, and P. Carbon. 2001. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 29:2502-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naldini, L., and I. M. Verma. 2000. Lentiviral vectors. Adv. Virus Res. 55:599-609. [DOI] [PubMed] [Google Scholar]
- 33.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S.-K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 34.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin, X.-F., D. S. An, I. S. Y. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Randall, G., A. Grakoui, and C. M. Rice. 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. USA 100:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ratcliff, F., B. D. Harrison, and D. C. Baulcombe. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558-1560. [DOI] [PubMed] [Google Scholar]
- 38.Ross, T. M., P. D. Bieniasz, and B. R. Cullen. 1999. Role of chemokine receptors in HIV-1 infection and pathogenesis. Adv. Virus Res. 52:233-267. [DOI] [PubMed] [Google Scholar]
- 39.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, M. Zhang, M. T. McManus, F. B. Gertler, M. L. Scott, and L. Van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz, D. S., G. Hutvágner, B. Haley, and P. D. Zamore. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10:537-548. [DOI] [PubMed] [Google Scholar]
- 41.Shlomai, A., and Y. Shaul. 2003. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology 37:764-770. [DOI] [PubMed] [Google Scholar]
- 42.Stewart, S. A., D. M. Dykxhoorn, D. Palliser, H. Mizuno, E. Y. Yu, D. S. An, D. M. Sabatini, I. S. Y. Chen, W. C. Hahn, P. A. Sharp, R. A. Weinberg, and C. D. Novina. 2003. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9:493-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sui, G., C. Soohoo, E. B. Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Surabhi, R. M., and R. B. Gaynor. 2002. RNA interference directed against viral and cellular targets inhibits human immunodeficiency virus type 1 replication. J. Virol. 76:12963-12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 46.Tiley, L. S., P. H. Brown, and B. R. Cullen. 1990. Does the human immunodeficiency virus Tat trans-activator contain a discrete activation domain? Virology 178:560-567. [DOI] [PubMed] [Google Scholar]
- 47.Tiscornia, G., O. Singer, M. Ikawa, and I. M. Verma. 2003. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. USA 100:1844-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tokunaga, K., M. I. Greenberg, M. A. Morse, R. I. Cumming, H. K. Lyerly, and B. R. Cullen. 2001. Molecular basis for cell tropism of CXCR4-dependent human immunodeficiency virus type 1 isolates. J. Virol. 75:6776-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinberg, J. B., T. J. Matthews, B. R. Cullen, and M. H. Malim. 1991. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 174:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson, J. A., S. Jayasena, A. Khvorova, S. Sabatinos, I. G., Rodrigue-Gervais, S. Arya, F. Sarangi, M. Harris-Brandts, S. Beaulieu, and C. D. Richardson. 2003. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. USA 100:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng, Y., E. J. Wagner, and B. R. Cullen. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9:1327-1333. [DOI] [PubMed] [Google Scholar]