Abstract
Positive-strand RNA virus replication complexes are universally associated with intracellular membranes, although different viruses use membranes derived from diverse and sometimes multiple organelles. We investigated whether unique intracellular membranes are required for viral RNA replication complex formation and function in yeast by retargeting protein A, the Flock House virus (FHV) RNA-dependent RNA polymerase. Protein A, the only viral protein required for FHV RNA replication, targets and anchors replication complexes to outer mitochondrial membranes in part via an N-proximal sequence that contains a transmembrane domain. We replaced the FHV protein A mitochondrial outer membrane-targeting sequence with the N-terminal endoplasmic reticulum (ER)-targeting sequence from the yeast NADP cytochrome P450 oxidoreductase or inverted C-terminal ER-targeting sequences from the hepatitis C virus NS5B polymerase or the yeast t-SNARE Ufe1p. Confocal immunofluorescence microscopy confirmed that protein A chimeras retargeted to the ER. FHV subgenomic and genomic RNA accumulation in yeast expressing ER-targeted protein A increased 2- to 13-fold over that in yeast expressing wild-type protein A, despite similar protein A levels. Density gradient flotation assays demonstrated that ER-targeted protein A remained membrane associated, and in vitro RNA-dependent RNA polymerase assays demonstrated an eightfold increase in the in vitro RNA synthesis activity of the ER-targeted FHV RNA replication complexes. Electron microscopy showed a change in the intracellular membrane alterations from a clustered mitochondrial distribution with wild-type protein A to the formation of perinuclear layers with ER-targeted protein A. We conclude that specific intracellular membranes are not required for FHV RNA replication complex formation and function.
A universal feature of positive-strand RNA viruses is the involvement of host intracellular membranes in RNA replication complex formation and function. This conclusion is based primarily on four observations. First, immunofluorescence and immunoelectron microscopy have localized viral replicase proteins and nascent viral RNA synthesis to intracellular membranes (16, 18, 24, 28, 29, 33, 42, 47, 51, 53, 56). Second, in vitro viral RNA-dependent RNA polymerase (RdRp) activity cofractionates with cellular membranes (6, 8, 12, 51, 58). Third, detergents suppress, and in some instances, phospholipids enhance the in vitro activities of viral replicase proteins (2, 8, 57, 58). Fourth, lipid synthesis inhibitors (20, 30, 36) and mutations in lipid synthesis genes (26) inhibit viral RNA replication. Recent results also show that at least some positive-strand RNA viruses use membrane rearrangements to create virus-specific, membrane-bounded compartments in which RNA replication occurs (51).
Despite these observations, many fundamental questions remain about the interaction of viral replication factors with host intracellular membranes and the specific roles of membranes in viral RNA replication complex formation and function. Moreover, while most viruses assemble their replication complexes on a specific membrane or membranes, different viruses use different membranes. For various positive-strand RNA viruses, membranes derived from the endoplasmic reticulum (ER) (26, 29, 35, 42, 46-48, 51), Golgi apparatus (47), lysosomes (18, 24, 28, 47), endosomes (18, 24), vacuoles (52), mitochondria (10, 15, 32, 33), perixosomes (9, 10), and chloroplasts (14) have all been implicated in viral RNA replication complex formation and function. The significance of this diversity of intracellular membranes used by different viruses is unknown.
The wide variety of intracellular membrane compartments used by different positive-strand RNA viruses, and their specific targeting, suggests that individual viruses may have unique host factor requirements supplied by specific intracellular membranes. Alternatively, many or all host intracellular membranes could provide the functions necessary for viral RNA replication complex formation and function, and the specific intracellular localization of individual viruses may be related to other steps in the viral life cycle, such as viral protein translation or encapsidation. Results that help distinguish between these competing hypotheses potentially have significant therapeutic implications, as antiviral drugs designed to block viral RNA replication complex formation or function on a specific intracellular membrane compartment will be ineffective if alternative membranes can be used efficiently.
To study intracellular viral RNA replication complex targeting, we use Flock House virus (FHV), an alphanodavirus that has been used as a model to investigate viral capsid formation and RNA packaging (49, 50, 59), viral RNA replication and subgenomic synthesis (3, 5, 22, 27), and RNA replication complex assembly and function (32, 33, 57, 58). FHV replicates in a wide variety of cells (4), including the yeast Saccharomyces cerevisiae (37-39). FHV contains a 4.5-kb bipartite genome, in which the larger RNA segment (RNA1) encodes protein A, the FHV RdRp (4, 13). Protein A is the only viral protein needed for FHV RNA replication (3, 21, 38), and it contains the sequences necessary for the intracellular targeting of FHV RNA replication complexes to outer mitochondrial membranes (32, 33). The protein A mitochondrial outer membrane-targeting sequence is located in the N-terminal 46 amino acids and contains a 19-amino-acid transmembrane domain (TMD) (32). The presence of a single essential viral protein, a defined N-terminal targeting sequence, and the intracellular localization of viral RNA replication complexes to a single organelle all make FHV an attractive model to investigate whether viral RNA replication complexes can be retargeted to an alternative intracellular membrane.
In this report, we show that FHV RNA replication complexes can be retargeted from the mitochondria to the ER via the introduction of chimeric sequences into the protein A N-terminal region. In addition, FHV RNA replication complexes retargeted to the ER are functional in vivo and in vitro and have enhanced RNA synthesis activity compared to mitochondrially targeted replication complexes.
MATERIALS AND METHODS
Yeast strain and plasmids.
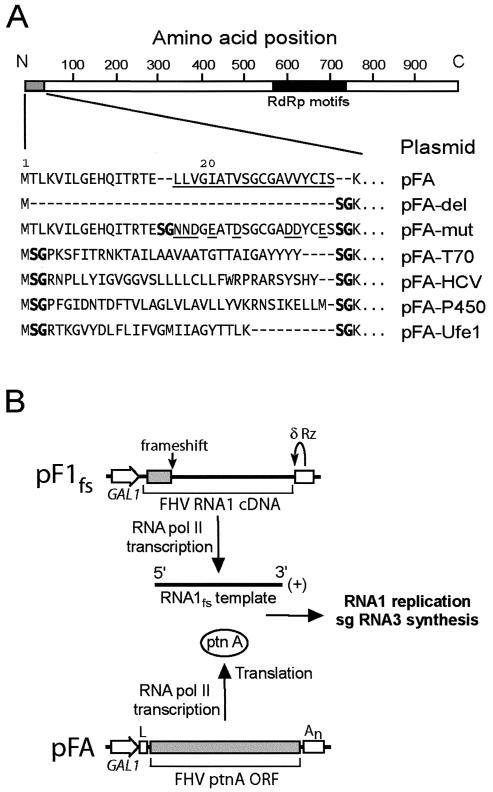
The haploid yeast strain BY4742 (MATα his3Δ1 leu2Δ0 ura3Δ0) was used for all experiments. Yeast cells were transformed, cultured, and induced with galactose as previously described (32) unless otherwise indicated. Standard molecular biology procedures were used for all cloning steps, and all products generated by PCR or with synthesized oligonucleotides were verified by automated sequencing. Plasmids pFA and pF1fs have been previously described (27, 32, 38). For the present experiments, we changed pF1fs from a HIS3-selectable yeast 2μm plasmid to a centromeric plasmid based on the pRS313 backbone (laboratory designation, pFHV1fs) to provide better control of plasmid copy number and stability. Plasmid pFA-del was generated by PCR with oligonucleotides designed to delete protein A amino acids 2 to 35 and insert a unique BspEI site. We generated chimeric protein A expression plasmids from pFA-del using mutually primed extension of overlapping nucleotides with flanking BspEI sites. Inserted sequences and plasmid designations of chimeric protein A expression plasmids are shown in Fig. 1A and described in Results below.
FIG. 1.
(A) Chimeric FHV protein A sequences and plasmid designations. A schematic of protein A with the viral RdRp motif region represented by the solid bar is shown on top for reference. The core predicted protein A TMD from Leu 17 to Ser 35 is underlined (32). The hydrophobic-to-hydrophilic amino acid mutations in pFA-mut are also underlined. The unique BspEI site used to generate the chimeric protein A constructs introduced a Ser-Gly at the insertion sequence junctions (shown in boldface). The dashes indicate no amino acids and are included for alignment purposes. (B) Schematic of plasmid-directed FHV RNA replication in yeast. RNA1 templates with authentic viral 5′ and 3′ termini are generated from pF1fs through precise placement of the GAL1 promoter start site and a hepatitis δ ribozyme (Rz), respectively, and the frameshift at the indicated location disrupts translation. The GAL1 leader (L) and CYC1 polyadenylation signal (An) flanking the protein A open reading frame (ORF) in pFA or derivatives thereof disrupt its activity as a viral RNA replication template but enhance its RNA polymerase II-directed transcription and translation. pFA-derived protein A (ptnA) subsequently directs RNA1 replication and subgenomic (sg) RNA3 synthesis from an RNA1 template transcribed from pF1fs.
Antibodies.
Mouse monoclonal antibodies (MAbs) against cytochrome oxidase subunit III (CoxIII), mitochondrial porin, dolichyl-phosphate β-d-mannosyltransferase (DPM), 3-phosphoglycerate kinase (PGK), vacuolar ATPase, and Oregon Green 488-labeled concanavalin A were from Molecular Probes (Eugene, Oreg.). All secondary antibodies for immunofluorescence and immunoblotting were from Jackson Immunoresearch (West Grove, Pa.). Rabbit polyclonal antisera against FHV protein A has been described previously (33). Mouse MAbs against FHV protein A were generated from mice immunized with a protein A-enriched insoluble pellet fraction from lysates of Escherichia coli BL21-Codon Plus (RIL) cells transformed with the expression vector pET-FHVPA (33). Hybridoma supernatants were screened for protein A reactivity by enzyme-linked immunosorbent assay using lysates from mock- and FHV-infected Drosophila S2 cells, and MAb specificity was confirmed by immunoblot and immunofluorescence analyses. The protein A epitope specificities of three hybridomas were further characterized by immunoblotting with lysates from yeast cells expressing protein A deletion mutants (32). The epitope for clones 2-2.2.2.5.3 and 3-4.1.1.2 was located between protein A amino acids 99 and 230, whereas the epitope for clone 2-1.1.2.4.8 was located between amino acids 230 and 399. Pooled ascites fluid from all three clones was used for immunoblot analyses.
RNA and protein analyses.
Total RNA isolation, Northern blot analyses and quantitation, protein isolation, and immunoblot analyses and quantitation were done as previously described (32). Cell lysates were fractionated by flotation in Nycodenz gradients as previously described (32) with several modifications. To avoid rapid FHV protein A and RNA degradation during spheroplast preparation, yeast cells were disrupted by mechanical shearing with glass beads for 1 min at 4°C in the presence of a yeast protease inhibitor cocktail (Sigma, St. Louis, Mo.) and the RNase inhibitor RNasin prior to equilibrium density gradient centrifugation. Two equal-volume fractions were taken—one from the visible membrane layer at the upper 5 to 35% Nycodenz interface and the other from the lower region where the cell lysate was loaded—and designated the low-density (LD) and high-density (HD) fractions, respectively (32). The total protein concentration of each fraction was determined by Bradford assay using bovine serum albumin as the standard, and total protein was equalized for immunoblot and in vitro RdRp analyses. In vitro RdRp assays were done as previously described (58) with several modifications. Reaction mixtures containing LD or HD fractions plus 50 mM Tris (pH 8.0); 50 mM potassium acetate; 15 mM magnesium acetate; 5 μg of actinomycin D/ml; 40 U of RNasin/μl; 1 mM (each) ATP, GTP, and CTP; 50 μM UTP; and 10 μCi of [32P]UTP in a 25-μl total volume were incubated for 3 h at 26°C, extracted once with phenol-chloroform, desalted with Sephadex G-50 columns, and analyzed in 1.2% nondenaturing agarose gels.
Immunofluorescence and EM.
Confocal immunofluorescence and transmission electron microscopy (EM) analyses were done as previously described (32, 51).
RESULTS
Retargeting sequence selection and FHV RNA replication in yeast.
We previously found that the N-terminal 46 amino acids of FHV protein A functioned as a mitochondrial outer membrane-targeting sequence (32). To investigate whether protein A and FHV RNA replication complexes could be retargeted to the ER, we constructed a series of chimeric protein A expression plasmids (Fig. 1A). Previous mapping studies showed that deletion of amino acids 9 to 45 disrupted protein A in vivo RdRp activity (32). Thus, we replaced the N-proximal region located between the initiator Met and Lys 36, the amino acid immediately downstream of the protein A TMD (32). For controls, we constructed pFA-del, in which amino acids 2 to 35 were deleted; pFA-mut, in which the protein A TMD hydrophobic amino acids Leu, Val, and Ile were mutated to the hydrophilic amino acids Asn, Asp, and Glu, respectively; and pFA-T70, in which wild-type (wt) protein A amino acids 2 to 35 were replaced with the outer mitochondrial membrane-targeting sequence from the yeast import receptor protein Tom70 (31). We initially searched for well-characterized ER-targeting sequences that contained an N-proximal TMD and were ∼30 to 40 amino acids in length, similar in size to the deleted wt protein A region in pFA-del. Few sequences fulfilled these criteria, and thus, we expanded the search to include inverted C-proximal TMDs, based on the observation that membrane targeting of some yeast proteins depends on the TMD length and composition but not the precise sequence (40). We chose three ER-targeting sequences: (i) inverted amino acids 561 to 591 from the C terminus of the hepatitis C virus (HCV) NS5B protein (48), (ii) amino acids 1 to 34 from the N terminus of yeast NADP cytochrome P450 oxidoreductase (54), and (iii) inverted amino acids 326 to 346 from the C terminus of the yeast t-SNARE Ufe1p as modified by Rayner and Pelham (40). These sequences were inserted between wt protein A amino acids 1 and 36 to generate plasmids pFA-HCV, pFA-P450, and pFA-Ufe1, respectively (Fig. 1A).
For all experiments, unless otherwise indicated, yeast cells were cotransformed with pFA or derivatives thereof to provide protein A expression and with pF1fs to provide an RNA1 template for FHV RNA replication in trans (Fig. 1B) (27, 32, 38). All plasmids expressed protein A or RNA1 from the galactose-inducible, glucose-repressible GAL1 promoter. The RNA1 transcribed from the plasmid pF1fs contains authentic FHV 5′ and 3′ termini and thus can serve as a template for viral RNA replication but cannot be translated due to a 4-nucleotide frameshifting insertion in the 5′ coding region (38). During RNA replication via a full-length genomic negative-sense (−)RNA1 intermediate, FHV produces a subgenomic, positive-sense (+)RNA3 that is colinear with the 3′ end of genomic (+)RNA1 (4). Thus, both genomic (−)RNA1 and subgenomic (+)RNA3 syntheses provide quantitative measurements of protein A-dependent FHV RNA replication.
Retargeted protein A chimeras localize to the ER.
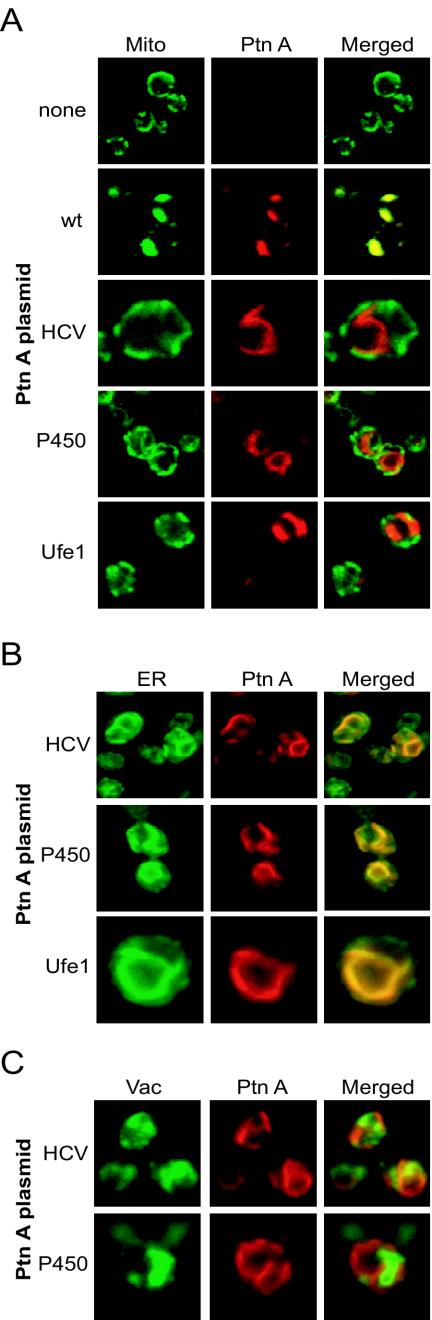
We used confocal immunofluorescence microscopy to examine the intracellular localization of protein A in yeast (Fig. 2). Consistent with previous observations (32), mitochondria from control yeast showed a peripheral distribution (Fig. 2A, top row), whereas mitochondria from yeast expressing pFA showed a clustered distribution that colocalized with protein A immunofluorescence (Fig. 2A, second row). Mitochondria from yeast expressing pFA-T70 also showed a clustered distribution that partially colocalized with protein A immunofluorescence, although the clustering was often not as prominent as that seen with wt protein A (data not shown). In contrast to pFA and pFA-T70, mitochondria from yeast expressing pFA-HCV showed a normal peripheral distribution that was distinct from the protein A immunofluorescence pattern (Fig. 2A, third row). The protein A distribution in yeast expressing pFA-HCV was predominantly centralized and perinuclear, similar to the distribution of the ER-targeted brome mosaic virus 1a and 2a replicase proteins in yeast (11, 41). A similar, largely perinuclear-type protein A distribution was seen in yeast expressing pFA-P450 (Fig. 2A, fourth row) or pFA-Ufe1 (Fig. 2A, bottom row). Immunofluorescence microscopy using a yeast ER marker confirmed that chimeric protein A localized to the ER in yeast expressing pFA-HCV (Fig. 2B, top row), pFA-P450 (Fig. 2B, second row), or pFA-Ufe1 (Fig. 2B, bottom row). As a control for other intracellular membrane compartments, immunofluorescence microscopy showed no significant localization of chimeric protein A to vacuoles in yeast expressing pFA-HCV (Fig. 2C, top row), pFA-P450 (Fig. 2C, bottom row), or pFA-Ufe-1 (data not shown). Thus, chimeric FHV protein A with specifically selected targeting sequences derived from the HCV NS5B polymerase, the yeast NADP cytochrome P450 oxidoreductase, or the yeast t-SNARE Ufe1p localized to the ER in yeast.
FIG. 2.
Chimeric protein A is retargeted to the ER. (A) Yeasts expressing pF1fs plus a control plasmid lacking the protein A open reading frame (top row), pFA (second row), pFA-HCV (third row), pFA-P450 (fourth row), or pFA-Ufe1 (bottom row) were immunostained with rabbit anti-protein A (Ptn A) and mouse anti-CoxIII for mitochondria (mito), followed by Texas red-labeled goat anti-rabbit and fluorescein isothiocyanate-labeled goat anti-mouse secondary antibodies. Representative images for CoxIII (left; green), protein A (middle; red), and merged signals (right) are shown. The merged images represent a digital superimposition of red and green signals in which areas of fluorescence colocalization are yellow. The images for pFA-HCV (third row) are shown at approximately twice the magnification of the other immunofluorescence images. (B) Yeast cells expressing pF1fs plus pFA-HCV (top row), pFA-P450 (middle row), or pFA-Ufe1 (bottom row) were immunostained with rabbit anti-protein A and Oregon Green 488-labeled concanavalin A, followed by Texas red-labeled goat anti-rabbit secondary antibodies. Representative confocal images for conconavalin A (left; green), protein A (middle; red), and merged signals (right) are shown. The images for pFA-Ufe1 (bottom row) are shown at approximately twice the magnification of the other immunofluorescence images. Concanavalin A is a lectin that selectively binds α-mannopyranosyl and α-glucopyranosyl residues. Initial experiments demonstrated that the fluorescence pattern of concanavalin A reactivity in yeast colocalized with the immunofluorescence pattern of Kar2p, a yeast ER-resident protein (data not shown). (C) Yeast cells expressing pF1fs plus pFA-HCV (top row) or pFA-P450 (bottom row) were immunostained with rabbit anti-protein A and mouse anti-vacuolar ATPase for vacuoles (Vac), followed by Texas red-labeled goat anti-rabbit and fluorescein isothiocyanate-labeled goat anti-mouse secondary antibodies. Representative confocal images for vacuolar ATPase (left; green), protein A (middle; red), and merged signals (right) are shown. The images for pFA-P450 (bottom row) are shown at approximately twice the magnification of the other immunofluorescence images.
ER-targeted protein A chimeras increase FHV RNA synthesis in vivo.
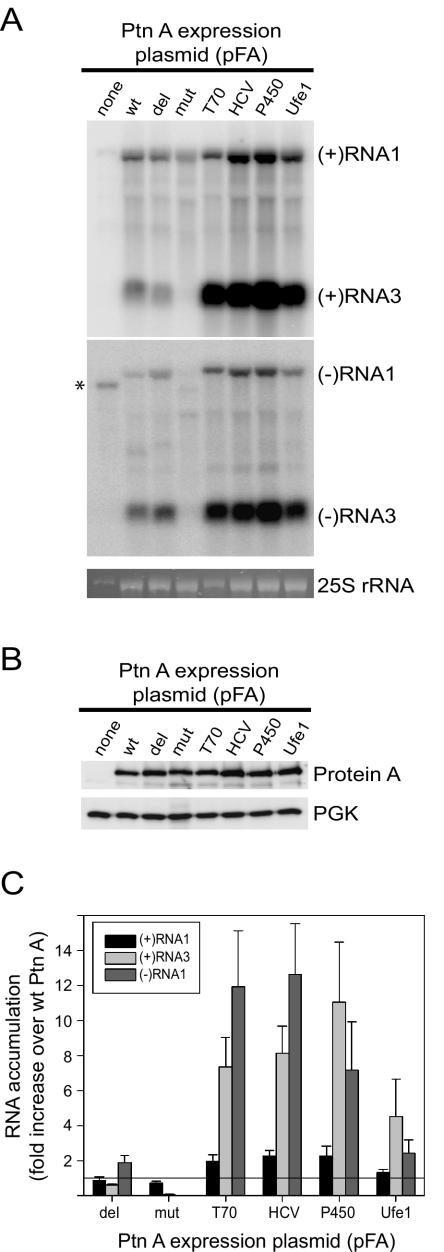
We analyzed FHV-specific RNA accumulation by Northern blot analyses to compare FHV RNA replication in yeast expressing wt protein A and in ER-targeted chimeras (Fig. 3). Both subgenomic (+)RNA3 (Fig. 3A, top blot) and (−)RNA1 template (Fig. 3A, bottom blot) accumulations were increased in yeast cells expressing pFA-T70, pFA-HCV, pFA-P450, and pFA-Ufe1. The increase in viral RNA accumulation was not due to increased FHV RNA polymerase expression, as protein A accumulations were similar in all constructs (Fig. 3B). Compared quantitatively to yeast expressing pFA, (+)RNA3 accumulation increased 5- to 11-fold and (−)RNA1 accumulation increased 2- to 12-fold in yeast cells expressing pFA-HCV, pFA-P450, and pFA-Ufe1 (Fig. 3C). The largest increases in FHV RNA accumulation were seen with pFA-T70, pFA-HCV, and pFA-P450, whereas pFA-Ufe1 showed a smaller, although still significant, increase. In contrast to subgenomic (+)RNA3 and genomic template (−)RNA1 accumulation, genomic (+)RNA1 accumulation was either unchanged or only slightly higher in yeast cells expressing pFA-T70, pFA-HCV, pFA-P450, and pFA-Ufe1 (Fig. 3C). Genomic (+)RNA1 accumulation reflects both FHV RNA replication and replication-independent yeast RNA polymerase II-directed transcription from plasmid DNA (Fig. 1B). We concluded from these data, in conjunction with the confocal-immunofluorescence results described above (Fig. 2), that FHV RNA replication complexes retargeted to the ER were active in vivo. Thus, FHV RNA replication complexes do not require a unique intracellular membrane for in vivo activity in yeast.
FIG. 3.
ER-targeted protein A chimeras increase FHV RNA accumulation in vivo. (A) Viral RNA accumulation in yeasts expressing pF1fs plus the indicated protein A (Ptn A) expression plasmids. The lane labels correspond to the plasmid designations shown in Fig. 1A. Total RNAs from equivalent numbers of cells were separated by electrophoresis and blotted with 32P-labeled complementary riboprobes that corresponded to nucleotides 2718 to 3064 from FHV RNA1 (38). Note that RNA3 is colinear with the 3′ end of RNA1 (13, 17) and that the probe sequence is present in both RNAs. Control Northern blots comparing known amounts of RNA1 and RNA3 in vitro transcripts showed that, with the probes and the transfer and hybridization conditions used, there was no significant difference between the detection efficiencies of RNA1 and RNA3. Thus, the band intensities reflect the molar ratios of RNA1 and RNA3. The riboprobes were either in the antisense or sense orientation and detected (+)RNA1 and (+)RNA3 (upper blot) or (−)RNA1 and (−)RNA3 (lower blot), respectively. The positions of RNA1 and RNA3 are shown on the right. The ethidium bromide-stained band of 25S rRNA is shown below the blots as a loading control. The asterisk indicates the position of a background band seen prominently in yeast expressing template only but also present to a lesser extent in all samples. This band was not present in total RNA preparations from FHV-infected Drosophila cells, whereas the (−)RNA1-labeled band was present (not shown). Subgenomic (−)RNA3 is produced in yeast replicating FHV RNA and in FHV-infected Drosophila cells and may function as a template for subgenomic (+)RNA3 synthesis (27, 38). (B) FHV protein A accumulation in yeast expressing pF1fs plus protein A expression plasmids. Total protein from equivalent numbers of cells was separated by electrophoresis and blotted with rabbit anti-protein A antiserum. The anti-PGK immunoblot is shown as a loading control. (C) Quantitative analysis of (+)RNA1, (+)RNA3, and (−)RNA1 accumulation. Northern blots were quantitated by phosphorimager analysis, and the results are expressed as the increase (n-fold) over yeast expressing pF1fs plus pFA. The horizontal line at 1 is placed for reference to the wt control. Averages and standard errors of the mean of at least three independent experiments are shown.
The increased RNA replication in yeast cells expressing pFA-T70, pFA-HCV, pFA-P450, and pFA-Ufe1 was not simply due to a change in the protein A TMD, as genomic (−)RNA1 and subgenomic (+)RNA3 accumulations in yeast expressing pFA-mut were negligible (Fig. 3). We changed the hydrophobic amino acids within the protein A TMD to hydrophilic amino acids in pFA-mut based on the hypothesis that a hydrophobic TMD was necessary for in vivo protein A RdRp activity. However, (+)RNA3 accumulation unexpectedly decreased only 40% in yeast expressing pFA-del compared to that in yeast expressing wt protein A, whereas (−)RNA1 accumulation increased almost twofold (Fig. 3). This suggests that the protein A N-terminal 35 amino acids are not essential for RdRp activity and that potential changes in secondary or tertiary structure may have been responsible for the absent in vivo RdRp activity with pFA-mut. Confocal immunofluorescence studies showed that protein A in yeast expressing pFA-del had a more diffuse distribution that did not colocalize with mitochondria (data not shown), similar to previously described protein A mutants with N-proximal deletions (32). Nevertheless, some protein A mutants with N-proximal deletions that include the TMD still maintain 40 to 60% membrane association (32). Thus, the retained in vivo RdRp activity of pFA-del could be due to either residual membrane association or membrane-independent FHV RNA replication complex activity. The latter hypothesis is consistent with the observation that FHV negative-strand templates can be produced in vitro in the absence of membranes or phospholipids (57, 58). The observation that pFA-del retained some in vivo RdRp activity despite the lack of clear mitochondrial localization supports the conclusion that FHV RNA replication complexes do not require a unique intracellular membrane in yeast.
Effects of yeast growth and induction kinetics on viral RNA synthesis in yeast expressing chimeric protein A.
To assess whether the increased FHV RNA synthesis in yeast expressing chimeric protein A might have been due to a slowing of yeast growth, thus allowing more time for viral RNA accumulation per cell before cell division, we measured doubling times over the entire 24-h induction period. When grown in selective medium with galactose, yeast expressing pFA plus pF1fs doubled every 9.7 ± 0.4 h (mean ± standard error of the mean), compared to 9.3 ± 1.2 h for control yeast. Doubling times were not significantly different in yeast cells expressing pFA-del (9.5 ± 0.7 h), pFA-mut (10.7 ± 0.8 h), pFA-HCV (8.5 ± 0.2 h), pFA-P450 (9.4 ± 0.2 h), or pFA-Ufe1 (9.3 ± 0.5 h) when compared to those of yeast cells expressing pFA. In contrast, the doubling time in yeast expressing pFA-T70 was increased almost twofold to 17.7 ± 1.6 h (P < 0.0004 compared to pFA), which may explain the increased (+)RNA3 and (−)RNA1 accumulation seen in these yeast cells (Fig. 3). However, differences in yeast growth kinetics did not explain the increased FHV RNA accumulation in yeast expressing ER-localized pFA-HCV, pFA-P450, or pFA-Ufe1.
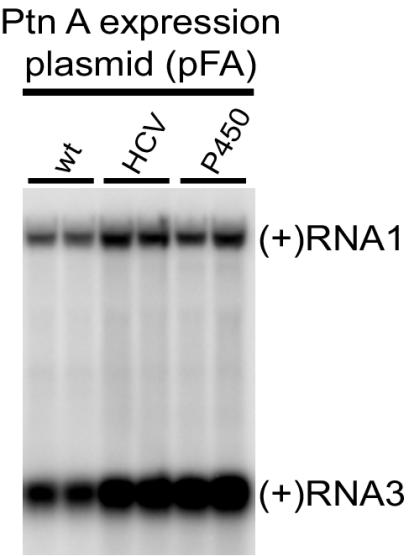
We also explored the impact of the induction period on FHV RNA accumulation in yeast cells expressing pF1fs plus pFA, pFA-HCV, or pFA-P450. Expression of FHV protein A and RNA1 replication templates was induced in selective medium with galactose, and the yeast cells were passed every 24 h into new media to maintain exponential growth and analyzed 72 h after induction (Fig. 4). FHV RNA accumulation after a 72-h induction was still increased in yeast expressing pFA-HCV or pFA-P450 compared to that in yeast expressing pFA, but the increase was less than that seen after the shorter 24-h induction period. Subgenomic (+)RNA3 accumulation in yeast expressing pFA-HCV or pFA-P450 was increased 2.5- ± 0.2- or 3.0- ± 0.2-fold, respectively, over yeast expressing pFA. Thus, enhanced FHV RNA synthesis kinetics or RNA stability may explain part of the increased FHV RNA accumulation in yeast expressing retargeted protein A.
FIG. 4.
ER-targeted protein A (Ptn A) chimeras maintain increased FHV RNA accumulation in vivo after 72 h of induction. Total RNA from equivalent numbers of cells was separated by electrophoresis and blotted with a 32P-labeled complementary riboprobe that detected (+)RNA1 and (+)RNA3. Duplicate representative samples from yeast cells expressing pF1fs plus pFA, pFA-HCV, or pFA-P450 are shown.
ER-targeted FHV protein A chimeras are membrane associated and have increased in vitro RdRp activities.
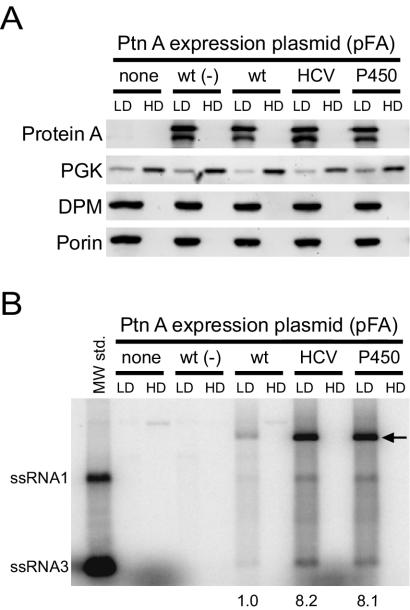
FHV protein A is membrane associated in infected Drosophila cells (33) and in yeast transformed with protein A expression vectors (32). To determine whether the ER-targeted protein A chimeras were also membrane associated, we used equilibrium density gradient centrifugation to examine the flotation behavior of protein A from yeast expressing pF1fs plus pFA, pFA-HCV, or pFA-P450 (Fig. 5A). As an additional control for subsequent in vitro RdRp assays, we also examined yeast cells expressing pFA alone, which produce protein A but do not support FHV RNA replication (27, 32, 38). Protein A from yeast cells expressing pFA, pFA-HCV, and pFA-P450 fractionated almost exclusively into the membrane-enriched LD fraction (Fig. 5A). The ER membrane protein DPM and the mitochondrial outer membrane porin protein were also present exclusively in the LD fraction, whereas the soluble cytosolic protein PGK partitioned predominantly into the membrane-depleted HD fraction. Thus, ER-targeted protein A chimeras were membrane-associated, consistent with the presence of TMDs in the sequences chosen to retarget protein A to the ER (Fig. 1A).
FIG. 5.
ER-targeted protein A (Ptn A) chimeras are membrane-associated and have enhanced in vitro FHV RdRp activities. (A) Equilibrium density gradient fractionation of lysates from yeast cells expressing pF1fs alone (none), pFA alone [wt (−)], or pF1fs plus pFA (wt), pFA-HCV, or pFA-P450. Equal amounts of total protein from both LD and HD fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with mouse MAbs to protein A, PGK, DPM, or porin. Loading equal amounts of total protein resulted in an overrepresentation of individual proteins in the LD fraction relative to the HD fraction, thus exaggerating the residual PGK signal in the LD fraction. Protein A appears as a doublet in immunoblots of equilibrium gradient fractions, where the lower band represents a C-terminal degradation product (32). (B) In vitro RdRp assay of equilibrium density gradient fractions. Equal amounts of total proteins from both LD and HD fractions were incubated with [32P]UTP and unlabeled ribonucleotides, and the reaction products were separated by nondenaturing agarose gel electrophoresis. The positions of in vitro-transcribed ssRNA1 and ssRNA3 are shown on the left. The major reaction products corresponding to ssRNA1, ssRNA3, and presumed replicative intermediate dsRNA1 (arrow) were quantitated by phosphorimager analysis in three independent experiments, and the numbers represent the increases (n-fold) in total radiolabeled products relative to the LD fraction of yeast expressing pF1fs plus pFA.
To assess the in vitro activities of ER-targeted protein A chimeras, LD and HD fractions from flotation gradients were equalized for total protein content and used for in vitro RdRp assays (Fig. 5B). No exogenous FHV RNA template was added to the RdRp reactions, and thus, any in vitro activity in the gradient fractions represented functional replication complexes that contained protein A, template FHV RNA, and any accompanying host factors. Gradient fractions from yeast expressing either pF1fs or pFA alone showed only faint high-molecular-weight reaction products, predominantly in the HD fractions. In contrast, the LD fractions from yeast expressing pF1fs plus pFA, pFA-HCV, or pFA-P450 showed distinct reaction products that comigrated with in vitro-transcribed single-stranded (ss) FHV RNA1 and RNA3. The most prominent RdRp product migrated more slowly than ssRNA1 (Fig. 5B, arrow) and likely represented double-stranded (ds) RNA1, which is a replicative intermediate and the primary reaction product previously described in in vitro FHV RdRp assays (57, 58).
The in vitro RdRp activities of LD fractions from yeast cells expressing pFA-HCV and pFA-P450 were significantly greater than that of yeast cells expressing pFA. When expressed as an increase over pFA, the production levels of ssRNA1, ssRNA3, and the presumed replicative intermediate dsRNA1 in LD fractions from yeast cells expressing pFA-HCV were increased 6.4- ± 1.4-, 9.8- ± 1.9-, and 8.5- ± 1.1-fold, respectively. For yeast expressing pFA-P450, these increases were 5.7- ± 1.4-, 8.6- ± 1.8-, and 9.3- ± 1.3-fold. The increased in vitro RdRp activity was not due to increased protein A accumulation, as LD fractions from yeast cells expressing pFA, pFA-HCV, and pFA-P450 contained similar levels of protein A (Fig. 5A, upper blot). Thus, the in vitro RdRp activities of ER-targeted protein A chimeras mirrored their increased in vivo activities (Fig. 3).
FHV replication complexes targeted to mitochondria or the ER induce distinct ultrastructural membrane alterations in yeast.
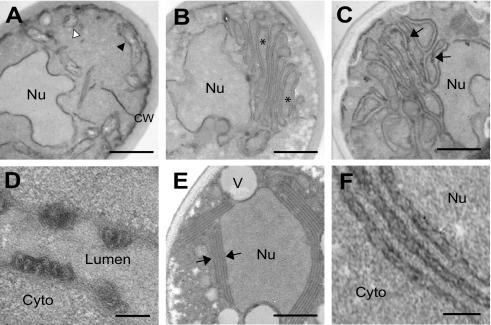
FHV infection of Drosophila cells induces distinct mitochondrial alterations, including mitochondrial clustering and the formation of 40- to 60-nm-diameter membrane-bounded spherical invaginations into the mitochondrial intermembrane space (33). Similar spherical membrane structures have been seen with a number of other positive-strand RNA viruses (7, 15, 18, 19, 24, 28, 51), and the localization of both viral replicase proteins and nascent viral RNA synthesis to these structures indicates that they are the intracellular sites of viral RNA replication complexes. To determine whether ultrastructural membrane changes similar to those in FHV-infected Drosophila cells (33) were also present in yeast cells replicating FHV RNA and to examine potential membrane changes induced by ER-targeted protein A chimeras, we used transmission EM to analyze yeast expressing pFA alone, pF1fs plus pFA, or pFA-HCV (Fig. 6). Compared to control yeast (Fig. 6A), yeast expressing pFA alone showed a proliferation of intracellular membranes and a marked clustering of membrane-bounded organelles (Fig. 6B). These structures were often located asymmetrically within cells and adjacent to nuclei, and normal-appearing mitochondria were not readily visible. A similar proliferation and ultrastructural appearance of intracellular membranes has been described in yeast expressing the mitochondrial-targeted Carnation Italian ringspot virus (CIRV) 36-kDa replicase protein (43). Yeast cells expressing pF1fs plus pFA had a similar appearance, with clustered membrane-bounded organelles (Fig. 6C). In addition, electron-dense structures were often visible along the clustered membranes (Fig. 6C), which at higher magnification appeared as tightly compressed membrane-bounded circular or ellipsoid structures (Fig. 6D). These structures protruded into the lumen of the organelle and had diameters of 30 to 50 nm, similar to the mitochondrial spherules located in the intermembrane spaces of FHV-infected Drosophila cells (33). However, the double external membrane and internal cristae that are characteristics of mitochondria were not readily apparent in the wt-protein A-induced clustered membrane structures in yeast. Nevertheless, the absence of normal mitochondria, the confocal immunofluorescence results demonstrating the localization of wt protein A to mitochondria in yeast (32) (Fig. 2A), and the similar ultrastructural appearances of FHV-infected Drosophila cells (33) and yeast expressing the CIRV 36-kDa replicase protein (43) suggest that the clustered membrane-bounded organelles in yeast expressing wt FHV protein A were mitochondria.
FIG. 6.
FHV protein A expression and RNA replication induces distinct ultrastructural membrane alterations in yeast. (A) Electron micrograph of yeast expressing pF1fs only, showing normal nucleus (Nu), cell wall (CW), and mitochondria seen in longitudinal (black arrowhead) and transverse (white arrowhead) sections. (B) Electron micrograph of yeast expressing pFA only, showing clustering of membrane-bounded organelles (asterisks). (C) Electron micrograph of yeast expressing pF1fs plus pFA, showing clustered membrane-bounded organelles plus electron-dense structures (arrows). (D) Electron micrograph (higher magnification) of membrane-bounded structures projecting into the organelle lumen of yeast expressing pF1fs plus pFA. Cyto, cytoplasm. (E) Electron micrograph of yeast expressing pF1fs plus pFA-HCV, showing perinuclear membrane layers (arrows). V, vacuole. (F) Electron micrograph (higher magnification) of perinuclear membrane layers in yeast expressing pF1fs plus pFA-HCV, showing an irregular appearance. Bars = 500 (A, B, C, and E), 100 (D), and 150 (F) nm.
The ultrastructural appearance of the membrane alterations in yeast expressing pF1fs plus pFA-HCV were significantly different than that in yeast cells expressing wt protein A. Membrane proliferation was localized primarily to the perinuclear region (Fig. 6E), consistent with the confocal-immunofluorescence results that showed localization of chimeric protein A to the perinuclear ER (Fig. 2B). The number of membrane layers adjacent to the nucleus varied from three to more than six, and they often almost completely surrounded the nucleus. Although spherule-like structures were not readily evident, high magnification often demonstrated an irregular appearance of the perinuclear membrane layers (Fig. 6F), suggesting the presence of possible underlying structures not readily visible under the fixation conditions used. Similar perinuclear layers have been observed under some conditions for yeast replicating brome mosaic virus RNA (M. D. Schwartz, J. Chen, W. M. Lee, and P. Ahlquist, unpublished data). Thus, the EM ultrastructural studies were consistent with the confocal-immunofluorescence results (Fig. 2) and suggested that structural differences between mitochondrial- and ER-targeted FHV replication complexes, or their associated membranes, may have contributed to the functional differences observed both in vivo (Fig. 3) and in vitro (Fig. 5).
DISCUSSION
In this report, we investigated the roles of alternative intracellular membranes in the formation and function of viral RNA replication complexes by retargeting FHV protein A to the ER. We drew three main conclusions. First, the introduction of engineered sequences from the HCV NS5B polymerase, the yeast NADP cytochrome P450 oxidoreductase, or the yeast t-SNARE Ufe1p into the N terminus of protein A retargeted the FHV RNA polymerase from the mitochondria to the ER. Second, ER-targeted FHV RNA replication complexes were active in vivo and in vitro. Third, ER-targeted FHV RNA replication complexes had enhanced RNA synthesis activity compared to replication complexes targeted to mitochondria. These observations indicate that unique intracellular membranes are not required for FHV RNA replication complex formation and function and suggest that any membrane-associated host functions required for FHV RNA replication complexes are provided either by a factor or factors present on multiple membranes or by distinct, membrane-specific factors with similar functions. The ability of FHV to replicate its RNA on two different intracellular-membrane compartments in yeast makes it a potentially valuable tool to investigate the contributions of host membranes and other host factors to RNA replication complex assembly and function. The natures and functions of known host factors involved in positive-strand RNA virus genome replication have recently been reviewed (1).
The demonstration that FHV RNA replication complexes can be retargeted is consistent with the results of a previous study that suggested tombusvirus RNA replication complexes could be retargeted (10). Tombusviruses are positive-strand RNA viruses of plants whose infections are associated with the formation of membranous cytoplasmic inclusions called multivesicular bodies. These structures form on mitochondria or peroxisomes, respectively, in plants infected with the tombusviruses CIRV and cymbidium ringspot virus (45). Multivesicular bodies are surrounded by multiple 80- to 150-nm-diameter vesicles that contain RNA, as detected by RNase susceptibility (15), and viral replicase proteins (9), and thus they are thought to represent the sites of tombusvirus RNA replication. Multivesicular-body localization is dependent on determinants encoded by the 5′-terminal region of the tombusvirus genome. Replacement of 600 nucleotides from the 5′ region of the CIRV genome with a similar region from the cymbidium ringspot virus genome changes the localization of multivesicular bodies from mitochondria to peroxisomes (10). FHV and CIRV show several similarities, including the normal mitochondrial localization of viral RNA replication complexes and the formation of outer mitochondrial membrane spherules (15, 33) and the presence of TMDs within the mitochondrial localization sequence of the replicase protein responsible for intracellular targeting (32, 44, 55). In addition, both FHV and CIRV can replicate viral RNA in yeast and induce the formation of similar intracellular-membrane structures (34, 37-39, 43) (Fig. 6). Further retargeting studies with FHV, CIRV, and other positive-strand RNA viruses may identify broadly applicable principles that define the interactions between viral replicase proteins and host intracellular membranes that are necessary for positive-strand RNA virus replication complex formation and function.
We retargeted FHV RNA replication complexes to the ER because that membrane compartment is used for viral RNA replication complex formation by a number of positive-strand RNA viruses (26, 29, 35, 42, 46-48, 51). The effectiveness of inverted C-terminal targeting sequences from the HCV NS5B polymerase and the yeast t-SNARE Ufe1p in retargeting FHV RNA replication complexes to the ER was consistent with the observation that the TMD amino acid composition and length, rather than the specific sequence, are important determinants for membrane localization (40). However, our results are not consistent with the results of the study by Kanaji et al. that showed a correlation between TMD hydrophobicity and intracellular-membrane localization, in which TMD hydrophobicities of <2.15 were associated with a mitochondrial localization while TMD hydrophobicities of >2.30 resulted in ER or Golgi localization (23). There was no correlation between TMD hydrophobicity and intracellular-membrane localization with the protein A chimeras used in this study. The wt protein A TMD has a hydrophobicity of 1.96, whereas the inverted HCV NS5B, P450 oxidoreductase, and inverted Ufe1p TMDs have average hydrophobicities of 2.27, 1.19, and 2.18, respectively. One potential explanation for this apparent discrepancy is that the Lys residue immediately downstream of the protein A TMD was present in all chimeric constructs (Fig. 1A), and the presence of charged residues surrounding TMDs can impact intracellular localization (23, 25). Alternatively, we cannot exclude the possibility that the ER was the default intracellular membrane target when protein A was no longer targeted specifically to the mitochondrial outer membrane, as we did not attempt to retarget FHV replication complexes to an alternative intracellular membrane other than the ER.
Previous studies that have examined FHV RNA replication in yeast (27, 38) have demonstrated significantly higher subgenomic RNA3/genomic RNA1 ratios in yeast expressing wt protein A than we found in this report (Fig. 3A and 4). Two differences in experimental design potentially contribute to this observation. We used the yeast strain BY4742, similar to the mapping study that identified the FHV protein A TMD as an important mitochondrial localization domain (32), whereas Price et al. (38) and Lindenbach et al. (27) used the yeast strain YPH500, which at 24 h after induction produces a higher subgenomic RNA3/genomic RNA1 ratio than BY4742 (P. McDowell and P. Ahlquist, unpublished data). Possibly due to the higher growth rate of BY4742, similar differences in subgenomic viral RNA accumulation between these yeast strains have been previously observed with brome mosaic virus (D. Kushner and P. Ahlquist, unpublished data). In addition, previous studies used a high-copy-number 2μm plasmid for FHV RNA1 template expression (27, 38), whereas we used a low-copy number centromeric plasmid to increase plasmid stability and reduce potential differences in DNA-dependent FHV RNA template expression levels between yeast cells expressing wt and ER-targeted protein A.
The observation that retargeting FHV protein A to ER membranes resulted in enhanced in vivo and in vitro RdRp activities compared to mitochondrial membrane-associated replication complexes was surprising, as nodavirus RNA replication is normally extremely robust. For example, in Drosophila cells infected with black beetle virus, an alphanodavirus whose RNA1 sequence shows 99% identity with FHV (4), viral RNA synthesis accounts for almost 50% of total RNA synthesis (17). Infectious-virion production in yeast spheroplasts transfected with FHV RNA is similar to that in FHV-infected Drosophila cells (39), and FHV RNA1 levels per microgram of total RNA are similar in infected Drosophila cells and yeast expressing pF1 (38). These observations suggest that FHV RNA replication efficiencies with wt protein A are similar in yeast and insect cells. The mechanisms responsible for the increased RNA accumulation observed in vivo (Fig. 3) and RNA synthesis observed in vitro (Fig. 5) with ER-targeted FHV replication complexes are unknown but could involve increased speed or efficiency of replication complex assembly, template RNA recruitment, or substrate utilization or increased stability of the template or product RNA. The reduced magnitude of the differences in (+)RNA3 accumulation between yeast cells expressing wt and ER-targeted protein A after prolonged induction (Fig. 4) implies that these differences reflect greater shifts in the early kinetics rather than in the final accumulation of RNA replication products. The observed differences in in vitro RdRp activities (Fig. 5B) and membrane ultrastructures (Fig. 6) also suggest the possibility that FHV RNA replication complexes targeted to mitochondrial or ER membranes may differ in accessibility to the cytoplasm for importing nucleotides or exporting product RNA, in host factor composition, or in other characteristics. Further experiments are being pursued to explore the underlying mechanisms responsible for the unexpected and interesting differences revealed by the retargeting of FHV RNA replication complexes.
Acknowledgments
We thank Kathleen Wessels, Pricilla McDowell, and Aaron Eichhorn for assistance. We performed the confocal immunofluorescence microscopy and transmission EM at the Keck Neural Imaging Laboratory and the Medical School Electron Microscopy Facility at the University of Wisconsin—Madison, respectively.
This work was supported by National Institutes of Health grants K08 AI01770-1 and GM35072. P.A. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola, T., A. Lampio, P. Auvinen, and L. Kaariainen. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 18:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, L. A. 1995. Requirements for the self-directed replication of Flock House virus RNA 1. J. Virol. 69:720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, L. A., and K. L. Johnson. 1998. Nodaviruses of insects, p. 225-267. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Publishing Corp., New York, N.Y.
- 5.Ball, L. A., and Y. Li. 1993. cis-acting requirements for the replication of Flock House virus RNA 2. J. Virol. 67:3544-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1991. Solubilization and immunoprecipitation of alphavirus replication complexes. J. Virol. 65:1496-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bashiruddin, J. B., and G. F. Cross. 1987. Boolarra virus: ultrastructure of intracytoplasmic virus formation in cultured Drosophila cells. J. Invertebr. Pathol. 49:303-315. [Google Scholar]
- 8.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bleve-Zacheo, T., L. Rubino, M. T. Melillo, and M. Russo. 1997. The 33K protein encoded by cymbidium ringspot tombusvirus localizes to modified peroxisomes of infected cells and of uninfected transgenic plant. J. Plant Pathol. 79:179-202. [Google Scholar]
- 10.Burgyan, J., L. Rubino, and M. Russo. 1996. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J. Gen. Virol. 77:1967-1974. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu, P. W. G., and E. G. Westaway. 1992. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch. Virol. 125:177-191. [DOI] [PubMed] [Google Scholar]
- 13.Dasmahapatra, B., R. Dasgupta, A. Ghosh, and P. Kaesberg. 1985. Structure of the black beetle virus genome and its functional implications. J. Mol. Biol. 182:183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Graaff, M., L. Coscoy, and E. M. Jaspars. 1993. Localization and biochemical characterization of alfalfa mosaic virus replication complexes. Virology 194:878-881. [DOI] [PubMed] [Google Scholar]
- 15.Di Franco, A., M. Russo, and G. P. Martelli. 1984. Ultrastructure and origin of cytoplasmic multivesicular bodies induced by carnation Italian ringspot virus. J. Gen. Virol. 65:1233-1237. [Google Scholar]
- 16.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friesen, P. D., and R. R. Rueckert. 1982. Black beetle virus: messenger for protein B is a subgenomic viral RNA. J. Virol. 42:986-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimley, P. M., I. K. Berezesky, and R. M. Friedman. 1968. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J. Virol. 2:1326-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guinea, R., and L. Carrasco. 1990. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 9:2011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, K. L., and L. A. Ball. 1997. Replication of Flock House virus RNAs from primary transcripts made in cells by RNA polymerase II. J. Virol. 71:3323-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, K. L., and L. A. Ball. 1999. Induction and maintenance of autonomous Flock House virus RNA1 replication. J. Virol. 73:7933-7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanaji, S., J. Iwahashi, Y. Kida, M. Sakaguchi, and K. Mihara. 2000. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151:277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda, R., T. Ikenoue, M. Honsho, S. Tsujimoto, J. Y. Mitoma, and A. Ito. 1998. Charged amino acids at the carboxyl-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J. Biol. Chem. 273:31097-31102. [DOI] [PubMed] [Google Scholar]
- 26.Lee, W. M., M. Ishikawa, and P. Ahlquist. 2001. Mutation of host Δ9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach, B. D., J. Y. Sgro, and P. Ahlquist. 2002. Long-distance base pairing in Flock House virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 76:3905-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magliano, D., J. A. Marshall, D. S. Bowden, N. Vardaxis, J. Meanger, and J. Y. Lee. 1998. Rubella virus replication complexes are virus-modified lysosomes. Virology 240:57-63. [DOI] [PubMed] [Google Scholar]
- 29.Mas, P., and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maynell, L. A., K. Kirkegaard, and M. K. Klymkowsky. 1992. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 66:1985-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McBride, H. M., D. G. Millar, J. M. Li, and G. C. Shore. 1992. A signal-anchor sequence selective for the mitochondrial outer membrane. J. Cell Biol. 119:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, D. J., and P. Ahlquist. 2002. Flock House virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock House virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pantaleo, V., L. Rubin, and M. Russo. 2003. Replication of carnation Italian ringspot virus defective interfering RNA in Saccharomyces cerevisiae. J. Virol. 77:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez, L., R. Guinea, and L. Carrasco. 1991. Synthesis of Semliki Forest virus RNA requires continuous lipid synthesis. Virology 183:74-82. [DOI] [PubMed] [Google Scholar]
- 37.Price, B. D., P. Ahlquist, and L. A. Ball. 2002. DNA-directed expression of an animal virus RNA for replication-dependent colony formation in Saccharomyces cerevisiae. J. Virol. 76:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional Flock House virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price, B. D., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rayner, J. C., and H. R. Pelham. 1997. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 16:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Restrepo-Hartwig, M., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubino, L., A. Di Franco, and M. Russo. 2000. Expression of a plant virus non-structural protein in Saccharomyces cerevisiae causes membrane proliferation and altered mitochondrial morphology. J. Gen. Virol. 81:279-286. [DOI] [PubMed] [Google Scholar]
- 44.Rubino, L., and M. Russo. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252:431-437. [DOI] [PubMed] [Google Scholar]
- 45.Russo, M., A. Di Franco, and G. P. Martelli. 1987. Cytopathology in the identification and classification of tombusviruses. Intervirology 28:134-143. [DOI] [PubMed] [Google Scholar]
- 46.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 49.Schneemann, A., and D. Marshall. 1998. Specific encapsulation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J. Virol. 72:8738-8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneemann, A., W. Zhong, T. M. Gallagher, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66:6728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 52.van der Heijden, M. W., J. E. Carette, P. J. Reinhoud, A. Haegi, and J. F. Bol. 2001. Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J. Virol. 75:1879-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venkateswarlu, K., D. C. Lamb, D. E. Kelly, N. J. Manning, and S. L. Kelly. 1998. The N-terminal membrane domain of yeast NADPH-cytochrome P450 (CYP) oxidoreductase is not required for catalytic activity in sterol biosynthesis or in reconstitution of CYP activity. J. Biol. Chem. 273:4492-4496. [DOI] [PubMed] [Google Scholar]
- 55.Weber-Lotfi, F., A. Dietrich, M. Russo, and L. Rubino. 2002. Mitochondrial targeting and membrane anchoring of a viral replicase in plant and yeast cells. J. Virol. 76:10485-10496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Westaway, E. G., A. A. Khromykh, and J. M. Mackenzie. 1999. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology 258:108-117. [DOI] [PubMed] [Google Scholar]
- 57.Wu, S. X., P. Ahlquist, and P. Kaesberg. 1992. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. USA 89:11136-11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, S. X., and P. Kaesberg. 1991. Synthesis of template-sense, single-strand Flockhouse virus RNA in a cell-free replication system. Virology 183:392-396. [DOI] [PubMed] [Google Scholar]
- 59.Zhong, W., R. Dasgupta, and R. R. Rueckert. 1992. Evidence that the packaging signal for nodaviral RNA2 is a bulged stem-loop. Proc. Natl. Acad. Sci. USA 89:11146-11150. [DOI] [PMC free article] [PubMed] [Google Scholar]