Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) replicates in the cytoplasm of infected cells, but its nucleocapsid (N) protein localizes specifically to the nucleus and nucleolus. The mechanism of nuclear translocation and whether N associates with particular nucleolar components are unknown. In the present study, we show by confocal microscopy that the PRRSV N protein colocalizes with the small nucleolar RNA (snoRNA)-associated protein fibrillarin. Direct and specific interaction of N with fibrillarin was demonstrated in vivo by the mammalian two-hybrid assay in cells cotransfected with the N and fibrillarin genes and in vitro by the glutathione S-transferase pull-down assay using the expressed fibrillarin protein. Using a series of deletion mutants, the interactive domain of N with fibrillarin was mapped to a region of amino acids 30 to 37. For fibrillarin, the first 80 amino acids, which contain the glycine-arginine-rich region (the GAR domain), was determined to be the domain interactive with N. The N protein was able to bind to the full-length genomic RNA of PRRSV, and the RNA binding domain was identified as the region overlapping with the nuclear localization signal situated at positions 41 to 47. These results suggest that the N protein nuclear transport may be controlled by the binding of RNA to N. The PRRSV N protein was also able to bind to both 28S and 18S ribosomal RNAs. The protein-protein interaction between N and fibrillarin was RNA dependent but independent of N protein phosphorylation. Taken together, our studies demonstrate a specific interaction of the PRRSV nucleocapsid protein with the host cell protein fibrillarin in the nucleolus, and they imply a potential linkage of viral strategies for the modulation of host cell functions, possibly through rRNA precursor processing and ribosome biogenesis.
The disease associated with porcine reproductive and respiratory syndrome virus (PRRSV) impacts swine production worldwide (for a review, see reference 1). PRRSV emerged almost simultaneously in the late 1980s and early 1990s in North America and Europe, respectively, and is widespread worldwide. Although clinical signs are similar on both continents, North American and European types of PRRSV are genetically distinct, with a sequence identity of only 63% (2, 20, 38). PRRSV is a single-stranded positive-sense RNA virus belonging to the family Arteriviridae, which forms the order Nidovirales along with another family, Coronaviridae (8, 19). The PRRSV genome is structurally similar to that of coronaviruses but is much smaller: the full-length genome is ∼15,000 nucleotides (21, 38). The virion consists of five membrane-associated proteins (GP2a, GP2b, GP4, GP5, and M) and a nucleocapsid (N) protein (27). GP3 is considered a structural component in European isolates but is not found in the virions of North American strains (12, 17, 32).
The N protein of PRRSV is comprised of 123 and 128 amino acids for North American and European genotypes, respectively. It is highly immunogenic (18) and is a basic protein with an isoelectric point of 10.4 that has been reported to be a serine phosphoprotein (36). Upon translation, the N protein interacts with itself by noncovalent interactions. As the N protein migrates to the Golgi complex, which is thought to be the maturation site for nidoviruses, it becomes disulfide linked via a cysteine residue at position 23, and the disulfide-linked homo-dimers are assembled into nucleocapsids (37).
The PRRSV N protein is concentrated mainly in the perinuclear region of virus-infected cells. However, Rowland and coworkers (24) reported the accumulation of N in the nuclei and nucleoli of virus-infected cells, as well as in cells transfected with constructs that expressed the N protein tagged with the green fluorescent protein. Recently, others have reported the nucleolar localization of the N proteins of equine arteritis virus, which is another arterivirus, and coronaviruses (15, 29, 39). The presence of N in the nucleolus is a unique observation, since the entire life cycle of PRRSV and other arteriviruses is considered to be restricted to the cytoplasm. Two conserved stretches of basic amino acids have been identified in the N protein at positions 10 to 13 and 41 to 47, and these sequences resemble two classical types of nuclear localization signal (NLS), ′pat4′ and ′pat7′, respectively (24). Subsequently, the ′pat7′ NLS was shown to be functional and sufficient for translocation of N to the nucleolus. Since the Golgi apparatus is thought to be the maturation site of arteriviruses, it has been postulated that the PRRSV N protein plays dual roles during virus infection: a structural role in the cytoplasm and a nonstructural role in the nucleus and/or nucleolus.
The nucleolus is a highly structured and dynamic nuclear organelle that is involved in the transcription of rRNA and ribosome biogenesis (23, 31). Ribosomal and other nucleolar proteins are first synthesized in the cytoplasm and then transported to the nucleus through the nuclear pore complex. Since the nucleolus is not a membrane-bound organelle, nucleolar proteins diffuse through the nucleoplasm and accumulate in the nucleolus. In addition to ribosome biogenesis, the nucleolus is involved in the regulation of apoptosis and control of the cell cycle (7, 26, 33). More than 270 proteins and over 150 different species of small nucleolar RNAs (snoRNAs) have been identified in the nucleolus so far (3, 31). Fibrillarin is one of the major proteins in the nucleolus associated with snoRNAs and forms small nucleolar ribonucleoprotein particles (snoRNPs) (13). We examined whether the PRRSV N protein in the nucleolus colocalized with nucleolar proteins, and in this communication, we report that the N protein colocalizes with fibrillarin and specifically interacts with it. We also show here that the PRRSV N protein is an RNA binding protein and suggest a possible role of N as a potential competitor for fibrillarin.
MATERIALS AND METHODS
Cells and viruses.
HeLa, BSC, TK-143, and Marc-145 (a subclone of MA104 cells [16]) cells were grown as monolayers in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (CanSera). The cells were maintained at 37°C with 5% CO2. Stocks of PRRSV (North American genotype strain PA8 [38]), wild-type vaccinia virus, and recombinant vaccinia virus expressing T7 RNA polymerase (vTF7-3) (10) were prepared in Marc-145 or BSC cells.
Antibodies.
Monoclonal antibodies (MAbs) specific for N were described previously (21). The monospecific polyclonal rabbit antiserum raised to the recombinant N protein expressed in Escherichia coli was obtained from Serge Dea (Institute Armand-Frappier, Laval, Quebec, Canada). The fibrillarin-specific antiserum, originally obtained from an autoimmune patient with scleroderma (22), was kindly provided by Marvin Fritzler (University of Calgary, Calgary, Alberta, Canada).
Plasmid constructions and mutagenesis.
cDNA cloning of the PRRSV N gene to produce pGEM-N, pCITE-N, pCITE-[GST-N], the carboxy-terminal deletion mutants (pGEX-C11, pGEX-C50, pGEX-C66, pGEX-C86, and pGEX-C98), and the amino-terminal deletion mutants (pGEX-N18, pGEX-N30, pGEX-N52, and pGEX-N69) is described elsewhere (37). The human fibrillarin gene was kindly provided by John Aris (University of Florida, Gainesville, Fla.). The fibrillarin coding sequence was amplified from the parental cDNA clone (4) using a pair of primers (forward, 5′-TAAggatccCCATGAAGCCAGGATTCAGTCCC-3′, and reverse, 5′-CTCgatatcTCAGTTCTTCACCTTGGGGGG-3′, where lowercase letters indicate BamHI and EcoRV recognition sequences, respectively) and cloned into the BamHI and SmaI sites of pGEX-3X (Amersham Pharmacia). DNA manipulations, cloning, and PCR were performed according to standard procedures (25). The E. coli strains XL-1 Blue (Stratagene) and DH5α were used as hosts for generating mutant genes and for general-purpose cloning, respectively.
For construction of fibrillarin mutants, inverse PCR was conducted using the following primer sets: for Δα, 5′-TAACTCGAGTGAGATGGGAATTCATCGTG-3′ (DFαF) and 5′-CTCCTCGAGAGTTGGCCTTAATGGAAATC-3′ (DFαR); for ΔR/α, 5′-TAACTCGAGTGAGATGGGAATTCATCGTG-3′ (DFR/αF) and 5′-CTCCTCGAGCGAAATCGAGACTCTCTTCT-3′ (DFR/αR); for ΔR+α, 5′-TAACTCGAGTGAGATGGGAATTCATCGTG-3 (DFR+/αF) and 5′-CTCCTCGAGACTGGTTTCCTCTTTTTCCT-3′ (DFR+/αR); for ΔG, 5′-TAACTCGAGAACCAGTCGGGGAAGAATGT-3′ (DFGF) and 5′-CTCCTCGAGACTGAATCCTGGCTTCATGG-3′ (DFGR). PCR mutagenesis and screening of mutants was performed as described previously (35).
Immunofluorescence and image analysis.
For immunofluorescence analysis, Marc-145 cells were used for virus infection and HeLa cells were used for N gene transfection. Cells were seeded on microscope slide coverslips, which were set in 35-mm-diameter dishes, and grew to a confluence of ∼50%. The cells were either infected with PRRSV at a multiplicity of infection of 1 to 5 or transfected with 1.5 μg of pXJ40-N using Lipofectamine (Invitrogen) according to the manufacturer's instructions. At 48 h posttransfection, the cell monolayers were washed once with phosphate-buffered saline (PBS) and fixed with 3.7% formaldehyde for 10 min at room temperature. The cells were then blocked with 1% bovine serum albumin in PBS for 30 min at room temperature and permeabilized with 0.1% NP-40 for 10 min. After being washed with PBS, the cells were incubated with the N-specific MAb SDOW17 or human anti-fibrillarin antiserum for 1 h at room temperature, followed by incubation with goat anti-mouse immunoglobulin G conjugated with fluorescein isothiocyanate (FITC) or with goat anti-rabbit immunoglobulin G conjugated with Texas red dye (Molecular Probes, Eugene, Oreg.). The coverslips were washed with PBS and mounted on microscope slides in the presence of 60% glycerol in PBS with 0.1% sodium azide. Fluorescence was visualized using a laser confocal scanning microscope (model TCS SP2; Leica Microsystems GmbH, Heidelberg, Germany).
DNA transfection, metabolic labeling, and immunoprecipitation.
Cells were grown to 90% confluence and infected for 1 h at 37°C with vTF7-3 (10) at a multiplicity of infection of 10. Following infection, fresh medium containing 10% fetal bovine serum was added, and incubation continued for an additional 1 h. The cells were washed twice in OPTI-MEM (Invitrogen) and then transfected for 12 h using Lipofectamine according to the manufacturer's instructions. The transfection solution was removed, and the cells were starved for 1 h in methionine-deficient medium. The cells were labeled for 5 h in the presence of 50 μCi of EasyTag EXPRESS protein-labeling mix ([35S]methionine and [35S]cysteine; specific activity, 407 MBq/ml; Perkin-Elmer)/ml. At the end of the labeling period, the cells were harvested, washed twice with cold PBS, and lysed with RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 150 mM NaCl, 50 mM Tris-HCl [pH 7.4], 10 mM EDTA, 0.1% sodium dodecyl sulfate [SDS]) containing 1 mM phenylmethylsulfonyl fluoride (PMSF). After incubation on ice for 20 min, the cell lysates were centrifuged at 14,000 rpm for 30 min in a microcentrifuge (model 5415; Eppendorf), and the supernatants were recovered.
For immunoprecipitation, cell lysates equivalent to 1:15 volume of a 100-mm-diameter dish were adjusted with RIPA buffer to a final volume of 100 μl and incubated for 2 h at room temperature with 1 μl of a mixture of N-specific MAbs. The immune complexes were adsorbed to 7 mg of protein A-Sepharose CL-4B beads (Amersham Biosciences) for 16 h at 4°C. The beads were collected by centrifugation at 6,000 rpm in a microcentrifuge (model 5415; Eppendorf) for 2 min and washed twice with RIPA buffer and once with washing buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl). The beads were resuspended in 20 μl of SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (10 mM Tris-HCl [pH 6.8], 25% glycerol, 10% SDS, 0.12% [wt/vol] bromophenol blue) containing 10% β-mercaptoethanol, boiled for 5 min, and analyzed by SDS-12% PAGE. The gels were dried on filter paper and autoradiographed using a PhosphorImager (model SI; Molecular Dynamics).
Protein expression in E. coli and GST pull-down assay.
The proteins were expressed in E. coli as a glutathione S-transfererase (GST) fusion protein. Briefly, 100 ml of Luria-Bertani medium containing 100 μg of ampicillin/ml was inoculated with 1/100 of an overnight culture and grown to an optical density of 0.6 at 600 nm. Protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. The bacteria were collected at 6,000 × g for 10 min, and the pellet was resuspended in 5 ml of PBS and sonicated on ice three times for 30 s each time with 2-s intervals (model W-385; Ultrasonics Inc.). Triton X-100 was added to a final concentration of 1%, and the proteins were solubilized for 30 min at 4°C with constant agitation. The insoluble fraction and cell debris were removed by centrifugation at 10,000 × g for 10 min at 4°C. The supernatants were incubated with 100 μl of a 50% slurry of glutathione S-Sepharose 4B beads (Amersham Pharmacia) for 30 min at 4°C with constant agitation. The beads bound to the GST fusion proteins were collected at 1,000 × g, washed five times in PBS containing 1% Triton X-100, and resuspended in a final volume of 250 μl of binding buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 5 mM dithiothreitol, 0.5% NP-40, 1 mM PMSF, 5% glycerol), resulting in a 20% slurry for use in GST pull-down assays.
For GST pull-down assays, radiolabeled viral N protein was produced either in PRRSV-infected cells or by in vitro transcription-translation. For the authentic viral N protein, PRRSV-infected Marc-145 cells were radiolabeled 36 h postinfection (p.i.) for 8 h with 50 μCi of [35S]methionine (EasyTag EXPRESS protein-labeling mix of [35S]methionine and [35S]cysteine; specific activity, 407 MBq/ml; Perkin-Elmer)/ml. For production of radiolabeled recombinant N protein, plasmid pCITE-N was transcribed in vitro by T7 RNA polymerase using T7 mMESSAGE mMACHINE (Ambion). The in vitro transcript was translated in rabbit reticulocyte lysate (Promega) in the presence of 50 μCi of [35S]methionine (EasyTag EXPRESS)/ml according to the manufacturer's instructions.
Approximately equal amounts, as judged by Coomassie blue staining, of various GST fusion proteins complexed to glutathione-Sepharose beads in a 20% slurry were incubated with the radiolabeled N proteins in binding buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 5 mM dithiothreitol, 0.5% NP-40, 1 mM PMSF, 5% glycerol) in a final volume of 400 μl overnight at 4°C with constant agitation. The beads were rinsed five times in binding buffer and boiled for 5 min in SDS-PAGE sample buffer containing 10% β-mercaptoethanol, and the disassociated proteins were analyzed by electrophoresis in 12% polyacrylamide gels. The gels were dried and exposed to a PhosphorImager to obtain radiographic images.
RNase A treatment.
GST fusion proteins were bound to glutathione-Sepharose beads as described above and washed once with binding buffer containing 5 mM EDTA. The beads were resuspended in the same buffer containing 100 μg of RNase A/ml and incubated for 1 h at 37°C. Simultaneously, cell lysate containing radiolabeled N protein was diluted at a ratio of 1:100 in binding buffer supplemented with 5 mM EDTA containing 100 μg of RNase A/ml, followed by incubation for 1 h at 37°C. The glutathione-Sepharose beads coupled to the GST fusion protein were pelleted and resuspended in the RNase-treated cell lysate and incubated overnight at 4°C for the standard GST pull-down assay.
Western blot analysis.
Cell lysates were separated on an SDS- 12% polyacrylamide gel under reducing conditions, followed by transfer to a nitrocellulose membrane in 10 mM 3-[cyclohexamino]-1-propanesulfonic acid buffer (pH 10.4; Sigma) prepared in 20% methanol. The membrane was blocked with 5% (wt/vol) skim milk in Tris-buffered saline (TBS; 10 mM Tris HCl [pH 8], 150 mM NaCl) overnight and washed three times for 5 min each time in TBS containing 0.05% Tween 20. The membrane was then incubated with N-specific rabbit antiserum at a dilution of 1:5,000 in 1% bovine serum albumin- TBS for 2 h. The membrane was washed again and incubated with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (H+L) (Bio-Rad) at a dilution of 1:5,000 in 1% bovine serum albumin- TBS for 1 h. Color was developed with BCIP (5-bromo-6-chloro-3-indolyl phosphate; p-toluidine salt) and nitro blue tetrazolium chloride (Bio-Rad) in 100 mM Tris-HCl- 100 mM NaCl- 5 mM MgCl2 (pH 9.5) according to the manufacturer's instructions.
Mammalian two-hybrid assay.
The N gene of PRRSV was subcloned downstream of the yeast GAL4 DNA binding domain in the pM expression vector (Clontech), and the human fibrillarin gene was cloned into the pVP16 expression vector (Clontech) downstream of the activation domain of the herpes simplex virus VP16 transactivator. The resulting plasmid constructs, pM-N and pVP-fibrillarin, were cotransfected along with the luciferase reporter plasmid, p5xGAL4SV40-luc, into HeLa cells. The reporter construct p5xGal4SV40-luc, which contains the luciferase reporter gene and five copies of the GAL4 DNA binding sequence upstream of the simian virus 40 promoter, was generously provided by Milo Vassallo and Naoko Tanese (New York University School of Medicine, New York, N.Y.). The pM and pVP16 N gene constructs, along with p5xGal4SV40-luc, were cotransfected into HeLa cells at 50% confluence in a ratio of 5:5:1 and assayed for reporter gene activity 48 h posttransfection. Luciferase actvities were determined using the Dual-Luciferase Reporter Assay kit (Promega) according to the instructions of the manufacturer. In this system, Renilla luciferase was included in all transfections, and the Renilla activities were used to normalize the transfection efficiencies.
In vitro transcription and translation. To generate an RNA probe for Northwestern blot assay, the full-length cDNA clone of PA8 virus genomic RNA, including a polyadenylation tail of 72 A’s at the 3′ terminus followed by a unique NotI site, was assembled and placed under the T7 promoter. The entire cassette was cloned in pOK12, named pOK12-PA8T7 (unpublished data), which was then used as a template for synthesis of the RNA probe. The full-length cDNA clone was linearized with NotI and used as a template to generate noncapped RNA using the MEGAscript kit (Ambion, Inc.) in accordance with the manufacturer's instructions. A transcription reaction mixture (20 μl) contained 10 μCi of [α-32P]UTP (3,000 Ci/mmol; 370 MBq/ml) (Perkin-Elmer). For fibrillarin, pCIneo-fibrillarin was transcribed and translated using the rabbit reticulocyte lysate transcription-translation system (TNT; Promega) in the presence of 50 μCi of [35S]methionine (EasyTag EXPRESS)/ml according to the manufacturer's instructions.
Northwestern in vitro RNA binding assay.
The nucleocapsid proteins were isolated from PRRS virions, N-gene-transfected HeLa cells, or PRRSV-infected cells by immunoprecipitation as described above. Virions were isolated from the precleared culture medium of virus-infected Marc-145 cells by centrifugation at 100,000 × g for 60 min at 4°C. Samples were then separated by SDS-PAGE and transferred to 0.45-μm-pore-size nitrocellulose membranes by using a wet-transfer apparatus (Bio-Rad Laboratories) for 1 h at 280 mA. The membranes were washed in probe buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA, 1× Denhardt's solution) for 10 min at room temperature, followed by blocking in the same buffer containing 250 μg of of baker's yeast tRNA (Roche Molecular Biochemicals)/ml for 1 h. The RNA probe used for the Northwestern RNA binding assay corresponded to the full-length genomic sequence of PRRSV strain PA8. The RNA probe was generated by in vitro transcription of the full-length genomic cDNA clone in the presence of [α-32P]UTP (3,000 Ci/mmol; 370 MBq/ml) (Perkin-Elmer). The membranes were incubated with 32P-labeled PRRSV RNA for 1 h at room temperature in probe buffer and then washed in the same buffer three times at room temperature before exposure to a PhosphorImager.
RESULTS
Colocalization of PRRSV N and fibrillarin in the nucleolus.
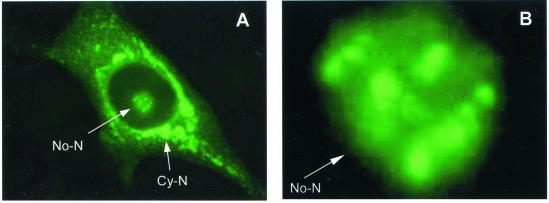
The nucleolar localization of the N protein was first examined in Marc-145 cells infected with the PRRSV strain PA8, which is a typical North American isolate. Virus-infected cells were stained with the N-specific MAb SDOW17 labeled with FITC and viewed under a fluorescence microscope. In addition to the typical cytoplasmic distribution of the N protein, the accumulation of N in the nucleolus was evident 16 h p.i. A representative photograph showing the N protein in the nucleolus of an infected cell is presented in Fig. 1A. Further magnification of the same cell showed that the N protein staining was not uniformly distributed throughout the nucleolus but rather in clusters, which were evident in regions with intense FITC staining (Fig. 1B).
FIG. 1.
Cellular distribution of the PRRSV N protein. Marc-145 cells prepared on microscope coverslips were infected with PRRSV (North American PA8 strain) and incubated for 16 h. The cells were fixed with 4% formaldehyde and permeabilized with 0.1% NP-40. The cells were then stained with the N-specific MAb SDOW17, followed by staining with goat anti-mouse antibody conjugated with FITC. (A) The cells were examined by fluorescence microscopy (magnification, ×40). (B) Nucleolar region under higher magnification (×100), showing the accumulation of FITC-MAb within regions of the nucleolus. No-N, nucleolar N; Cy-N, cytoplasmic N.
The nucleolus is structurally divided into three major subcompartments: the fibrillar center, a dense fibrillar component, and a granular component (14). Investigation of the possible association of N with specific nucleolar components began with experiments to determine if the N protein colocalized with fibrillarin, a protein whose distribution is almost exclusively restricted to the dense fibrillar component that surrounds the fibrillar centers in the mammalian nucleolus (5). Dual-label immunofluorescence staining was conducted using FITC-labeled fibrillarin-specific antiserum and Texas red-labeled anti-N-specific rabbit serum raised against the N protein. The subnucleolar distribution of FITC-labeled antibody was consistent with the localization of fibrillarin within the dense fibrillar component region of the nucleolus (Fig. 2B and E). Figure 2B and E also shows the localization of fibrillarin to a single Cajal (coiled) body. In contrast, the PRRSV N protein was widely distributed throughout the cell, including the cytoplasm and the nucleoplasm (Fig. 2A and D). In addition, the N protein was seen to accumulate in what appeared to be the dense fibrillar component and the surrounding region (Fig. 2A). The merging of the two images showed a yellow region of colocalization (Fig. 2C), which corresponded to the dense fibrillar component and the fibrillar centers within the nucleolus. Fibrillarin and the N protein did not colocalize in the nucleoplasmic region surrounding the nucleolus (Fig. 2C) or in the cytoplasm, suggesting that colocalization was restricted exclusively to the nucleolus. Interestingly, N staining was not observed in the Cajal body (see Discussion).
FIG. 2.
Colocalization of the N protein with fibrillarin in the nucleolus. Marc-145 cells prepared on microscope coverslips were infected with PRRSV and incubated for 16 h (A through C). Alternatively, HeLa cells were transfected with the N gene for 24 h (D though F). The cells were fixed with 4% formaldehyde and permeabilized with 0.1% NP-40. The cells were then costained with rabbit serum specific for N and human autoimmune serum specific for fibrillarin, followed by staining with goat anti-rabbit antibody conjugated with Texas red and goat anti-human antibody conjugated with FITC. The cells were examined by laser scanning confocal microscopy. (A, B, and C) PRRSV-infected cell costained with N-specific rabbit antiserum (red) and fibrillarin-specific human autoimmune serum (green). The intensely stained green spots correspond to the dense fibrillar component, which surrounds the fibrillar center. (D, E, and F) N gene-transfected and costained cells. The yellow regions in panels C and F are the areas where FITC and Texas red colocalized. No-N, nucleolar N; Cy-N, cytoplasmic N; Fibn, fibrillarin; Cajal, coiled body. Magnification, ×100.
To determine if the N-fibrillarin colocalization required the participation of other viral proteins, the N gene was cloned into the eukaryotic expression vector pCi-Neo and transfected into Marc-145 cells, followed by dual immunofluorescent staining (Fig. 2D and E). Similar to the results obtained for virus-infected cells, N and fibrillarin colocalized to the fibrillar region within the nucleolus (Fig. 2F), but not to the nucleoplasm or the cytoplasm. Diffused staining of N in the nucleoplasm was weaker in N gene-transfected cells (Fig. 2D) than in virus-infected cells (Fig. 2A). Previously, it was reported that the PRRSV N protein subcellular distribution was mainly nuclear and nucleolar during the early stage of infection (6 to 20 h p.i.), and by 30 to 48 h p.i., it became exclusively cytoplasmic (36). The intensity of the nucleoplasmic staining was also reflected by the accumulated N protein and the kinetics of the protein shuttling. The weaker signal observed in Fig. 2C was probably due to these factors. Nonetheless, the N protein localization in the nucleoplasm was obvious even in transfected cells. These data show that the N protein alone is able to colocalize with fibrillarin in the region of the nucleolus that corresponds to the dense fibrillar component and fibrillar center regions of the nucleolus.
The N protein interacts with fibrillarin in vivo and in vitro.
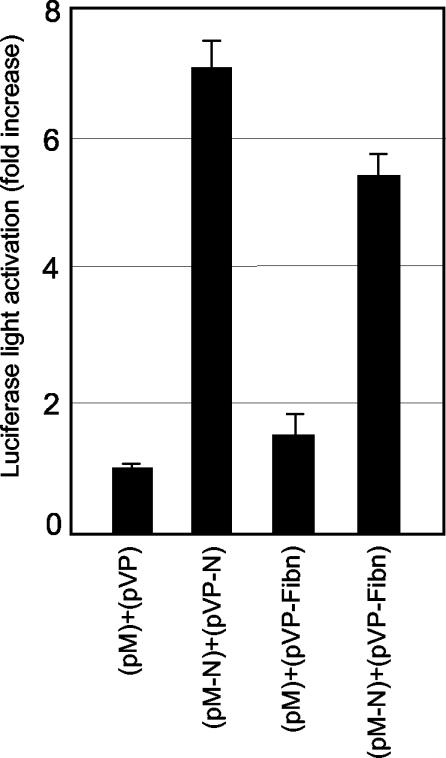
The mammalian two-hybrid assay was used to determine if N and fibrillarin formed a physical association in vivo. The N gene of PRRSV was subcloned downstream of the yeast GAL4 DNA binding domain in the pM expression vector, and the fibrillarin gene was cloned into pVP16 downstream of the activation domain of the herpes simplex virus VP16 transactivator. The resulting plasmids, pM-N and pVP-fibrillarin, were cotransfected along with a luciferase reporter plasmid, p5xGal4SV40-luc, into HeLa cells. At 48 h posttransfection, cell lysates were prepared and luciferase activity was measured. An ∼5-fold increase in luciferase activity was observed in cells cotransfected with the N and fibrillarin genes relative to lysates prepared from cells transfected with the control plasmids, pM and pVP16, or pM and pVP-fibrillarin (Fig. 3). Since the N protein itself is known to form homodimers in vivo (37), cells transfected with pM-N and pVP-N (the N gene cloned into pVP16) were included as a positive control. The level of luciferase activity was similar to that of the cells transfected with pM-N and pVP-fibrillarin. These results indicate that N and fibrillarin have the capacity to interact in vivo.
FIG. 3.
Interaction of the N protein with fibrillarin in vivo determined by the dual-luciferase reporter assay in the mammalian two-hybrid system. HeLa cells were cotransfected with pM-N and pVP-fibrillarin, along with the reporter plasmid p5xGAL4SV40-luc, in a ratio of 5:5:1. The cells were harvested 48 h posttransfection, and luciferase activities were measured. The data represent the means of three independent experiments plus 1 standard deviation after normalization using Renilla luciferase expression as an internal control.
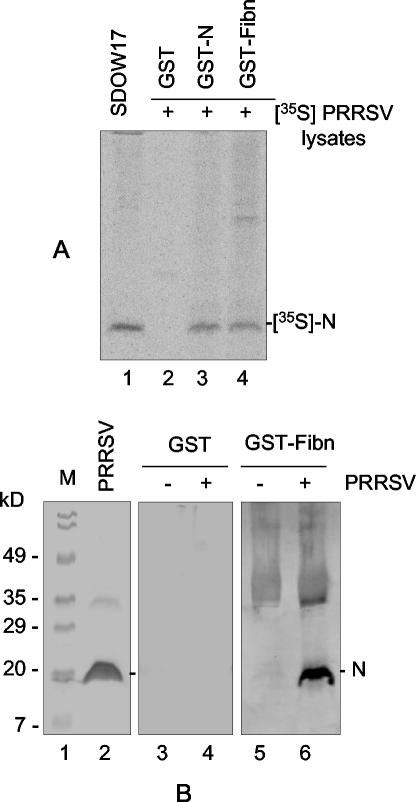
The capacity for interaction between N and fibrillarin was further determined in vitro by the GST pull-down assay. Fibrillarin was expressed as the GST-fibrillarin fusion protein in E. coli and coupled to glutathione-Sepharose beads. Cells were infected with PRRSV and radiolabeled with [35S]methionine. Labeled-cell lysates were then incubated with glutathione-Sepharose beads bound to either GST or the GST-fibrillarin fusion protein. After extensive washing, any labeled proteins bound to the GST-fibrillarin coupled beads were dissociated in the presence of β-mercaptoethanol and resolved by SDS-PAGE, followed by autoradiography (Fig. 4A). The N protein specifically immunoprecipitated from purified virions by MAb SDOW17 served as a positive control and molecular size marker (lane 1). GST alone did not precipitate any detectable amounts of radiolabeled protein from virus-infected cells (lane 2). Incubation of the GST-fibrillarin fusion protein with PRRSV-infected cell lysates resulted in the precipitation of a protein that migrated identically to the one immunoprecipitated by SDOW17 (compare lanes 1 and 4). Since N forms stable homodimers via covalent and noncovalent interactions (37), GST-N was included to precipitate radiolabeled N from virus-infected cells and, in lane 3, showed the recovery of a labeled protein identical in size to the proteins specifically precipitated with MAb SDOW17 and GST-fibrillarin.
FIG. 4.
Interaction of the N protein with fibrillarin in vitro measured by the GST pull-down assay. Human fibrillarin was expressed in E. coli as a GST-fibrillarin fusion protein. The GST-fibrillarin fusion protein was coupled to glutathione-Sepharose beads in a 20% slurry and incubated with N protein prepared as lysates from cells infected with virus in the presence of [35S]methionine. The beads were washed five times in binding buffer and dissociated by boiling them in sample buffer followed by SDS- 12% PAGE. The gel was dried and either exposed to a PhosphorImager to obtain radiographic images (A) or subjected to Western blotting using N-specific rabbit antiserum (B). (A) Bead binding assay. Lane 1, immunoprecipitation of purified virions using MAb SDOW17; lanes 2 through 4, precipitation of N by GST alone as a negative control, GST-N, or GST-fibrillarin (GST-Fibn), respectively. +, present. (B) Immunoblot assay. Lane 1, molecular mass markers; lane 2, purified virions probed with N antibody; lanes 4 and 6, PRRSV-infected cell lysates precipitated with GST or GST-fibrillarin. Lanes 3 and 5 represent lysates from uninfected cells precipitated with GST or GST-fibrillarin.
The identification of the protein precipitated in the GST pull-down assay as the N protein was further confirmed by immunoblot analysis. Following electrophoresis, the proteins on the gel were transferred to a nitrocellulose membrane and probed with N-specific rabbit antiserum (Fig. 4B). The N protein was detected in the PRRSV-infected cell lysate (lane 2) and in lane 6, where the cell lysate reacted with GST-fibrillarin. These data demonstrate that the protein specifically precipitated by GST-fibrillarin and therefore detected in the GST pull-down assay was indeed the PRRSV N protein.
Identification of interactive domains on PRRSV N and fibrillarin.
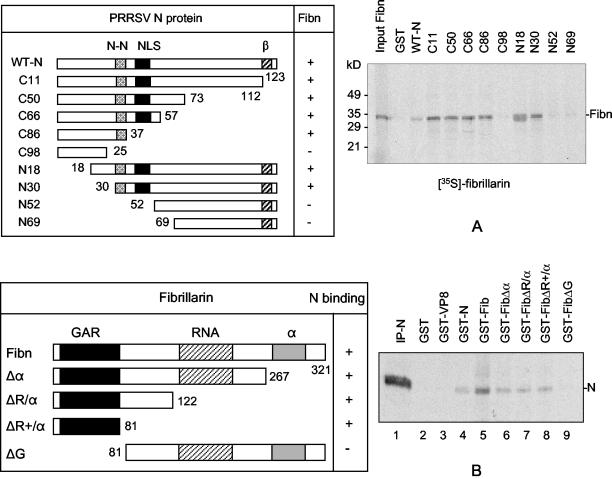
Since the N protein was found to interact with fibrillarin, a mapping study was conducted to identify the fibrillarin-interactive domain using a series of deletion mutants. The N gene was progressively deleted from either the 5′ terminus or the 3′ terminus (Fig. 5A), and each deletion construct was expressed in E. coli as a GST fusion protein. Approximately equal amounts of mutant proteins, determined by Coomassie blue staining, were individually coupled to glutathione-Sepharose beads. Fibrillarin was then synthesized by in vitro transcription translation in the presence of [35S]methionine and added as a probe to the Sepharose beads to determine the N-fibrillarin interactions by the GST pull-down assay (Fig. 5A). The C terminus N protein mutants, C11, C50, C66, and C86, and the N terminus mutants, N18 and N30, bound to fibrillarin. Correspondingly, C98, N52, and N69, which lacked the 30-to-37 amino acid region, did not interact with fibrillarin. From these results, a conclusion was made that the region between positions 30 and 37 was responsible for the interaction of N with fibrillarin.
FIG. 5.
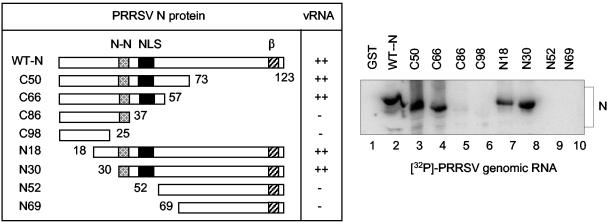
Identification of the fibrillarin (Fibn) binding domain of N (A) and the N protein-interactive domain of fibrillarin (B). The N gene was progressively deleted from either the 5′ or the 3′ terminus, and the deletion mutants were individually expressed in E. coli as GST fusion proteins. For fibrillarin, deletion constructs were generated by inverse PCR as described in Materials and Methods, and the individual mutants were expressed as GST fusion proteins in E. coli. The expressed N or fibrillarin mutant proteins were coupled to glutathione-Sepharose beads, and the Sepharose beads were complexed with radiolabeled fibrillarin synthesized in vitro by transcription translation (A) or with cell lysates prepared from PRRSV-infected, [35S]methionine-labeled Marc-145 cells (B). The beads were washed five times, and the bound protein was analyzed by SDS-PAGE and autoradiography. WT, wild-type; N-N, N protein-N protein interactive domain (37); NLS, nuclear localization signal (24); β, β-strand conserved among arteriviruses (35); GAR, glycine-arginine rich domain (4); RNA, RNA binding domain; α, α-helix domain; IP-N, immunoprecipitation of N. + and − indicate binding affinity between N and fibrillarin; the numbers indicate amino acid positions.
We also investigated the N protein-interactive domain within fibrillarin. Human fibrillarin is a 321-amino-acid protein that contains three distinct domains (4). Fibrillarin begins with a glycine- and arginine-rich (GAR) domain, followed by a stretch of amino acids resembling a domain of putative RNA binding, and the C terminus, which contains the highest potential for α-helix conformation (4). To map the N-interactive domain of fibrillarin, four constructs were made such that each of these three domains was included or excluded (Fig. 5B). The fibrillarin mutants were expressed as GST fusion proteins in E. coli, which were then each bound to glutathione-Sepharose beads. The fibrillarin-bound beads were incubated with the radiolabeled cell lysate prepared from PRRSV-infected cells and subjected to the GST pull-down assay. Of the four mutants, only mutants containing the GAR domain were able to precipitate the N protein (Fig. 5B).
RNA binding of PRRSV N.
For PRRSV, the genomic RNA and the polymerized N protein are thought to be the sole components of the viral nucleocapsid, suggesting that N has a direct interaction with viral genomic RNA. The interaction between N and genomic RNA was examined by Northwestern RNA blot analysis, and the results are shown in Fig. 6. The N protein from either virus-infected cells (Fig. 6A, lane 2) or purified virions (Fig. 6A, lane 3) was resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Since it is unknown if a particular region of the viral genomic RNA is required for N protein binding, the full-length genomic RNA was used as a probe. The radiolabeled RNA probe was synthesized in vitro by transcription of the full-length viral genomic cDNA clone placed under the T7 promoter (unpublished data). The N protein-bound membrane was then probed with the transcribed viral RNA. A single protein was hybridized to the viral RNA for both PRRSV-infected cells (Fig. 6A, lane 2) and PRRSV virions (lane 3). No signal was detected from uninfected cells (lane 1). These results demonstrate that the PRRSV N protein has the capacity to bind viral genomic RNA. RNA binding was also observed using recombinant N protein expressed in the T7-based vaccinia virus system (Fig. 6B, lane 5) or the GST-N fusion protein expressed in E. coli (Fig. 6C, lane 8). Since the E. coli-expressed GST-N protein was unphosphorylated (unpublished data), lane 8 suggests that RNA binding is independent of N protein phosphorylation.
FIG. 6.
Binding of the N protein to the viral genomic RNA. PRRS virions or PRRSV-infected cells were immunoprecipitated using MAb SDOW17 and resolved by SDS-PAGE, followed by transblotting to a nitrocellulose membrane. PRRSV full-length genomic RNA was synthesized in vitro from the full-length cDNA clone of PRRSV using T7 RNA polymerase in the presence of [α-32P]UTP. The protein-bound membrane was probed with the radiolabeled RNA transcript for 1 h at 37°C, followed by exposure to a PhosphorImager. (A) Authentic viral proteins. Marc, Marc-145 cells. (B) Recombinant N protein synthesized by T7-based vaccinia virus expression. Lane 4, T7 RNA polymerase expressing recombinant vaccinia virus (vTF7)-infected cells; lane 5, vTF7-infected and pCITE-N gene-transfected cells; lane 6, PRRS virions immunoprecipitated using MAb SDOW17. (C) E. coli-expressed GST fusion proteins. Lane 7, GST alone; lane 8, GST-N fusion protein; lane 9, GST-rotavirus VP8 fusion protein as a negative control.
To identify the RNA binding domain of N, deletion mutants of N were used for RNA blot analysis (Fig. 7). Individual N mutants were expressed in E. coli as a GST fusion protein, and approximately equal amounts of mutant proteins were loaded on a gel, followed by transfer onto a nitrocellulose membrane. The membrane was then probed with in vitro-transcribed 32P-labeled full-length genomic RNA. The genomic RNA bound efficiently to the constructs containing a region of amino acids 37 to 57 (Fig. 7, WT-N, C50, C66, N18, and N30), but the constructs lacking this region (C89, C98, N52, and N69) did not bind to the RNA. From this study, we concluded that the RNA binding domain of N lies between amino acids 37 and 57, which overlaps the ′pat7′ NLS sequence located at positions 41 to 47.
FIG. 7.
Identification of the N protein RNA binding domain. The N gene deletion mutants were individually expressed in E. coli as GST fusion proteins. The individual proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes in equal amounts. The membranes were probed with the [32P]UTP-labeled full-length PRRSV genomic RNA transcribed in vitro by T7 RNA polymerase. After being washed, the membrane was exposed to a PhosphorImager for image analysis. WT, wild-type; N-N, N protein-N protein interactive domain (37); NLS, nuclear localization signal (24); β, β-strand conserved among arteriviruses (35). The numbers indicate amino acid positions; + and − indicate binding affinity of N to the viral genomic RNA (vRNA).
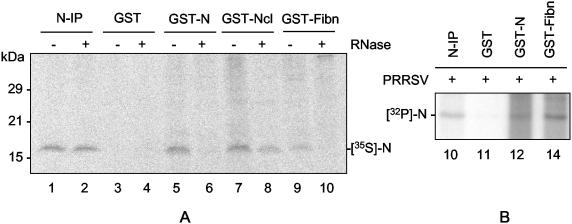
Since the PRRSV N protein is found both in the cytoplasm, where the viral RNA coexists with N, and in the nucleolus, where the viral RNA is absent but rRNA is abundant, it was of interest to examine if the PRRSV N protein was able to bind rRNA. The viral N or GST-N fusion protein was resolved by SDS-PAGE and blotted onto a nitrocellulose membrane. The membrane was then hybridized, in the presence of yeast tRNA, with radiolabeled total cellular RNA prepared from uninfected Marc-145 cells (Fig. 8A). Both the GST-N fusion protein expressed in E. coli (Fig. 8A, lane 3) and the authentic viral N protein immunoprecipitated from virions (Fig. 8A, lane 4) bound the cellular RNA. As a control, the individual viral proteins from virion particles were resolved, immobilized on nitrocellulose, and then probed with the RNA (Fig. 8A, lane 5). Only the N protein bound cellular RNA.
FIG. 8.
Binding of the N protein to rRNA. Either the GST-N fusion protein expressed in E. coli or the purified PRRS virions (A, lanes 4 and 5; B, lanes 3 and 6) was resolved by SDS-PAGE and transferred to membranes. The membranes were hybridized with the total cellular RNA (A) or with either 28S or 18S rRNA (B) extracted from uninfected Marc-145 cells. The RNAs were radiolabeled in vitro using [γ-32P]ATP and polynucleotide kinase. N-IP, N immunoprecipitated using MAb SDOW17; GST-VP8, GST fused with rotavirus VP8 protein.
N protein binding to cellular RNA was further elucidated using a probe for each subunit of rRNA (Fig. 8B). Total cellular RNA was prepared from Marc-145 cells and resolved by gel electrophoresis. Both 28S and 18S rRNAs were located by staining and purified from the gel (data not shown). The purified rRNA subunits were radiolabeled in vitro and hybridized to the membrane to which N was bound. Both the GST-N and virion N proteins were hybridized by each of the 28S (Fig. 8B, lane 2) and 18S (Fig. 8B, lane 5) rRNAs, demonstrating that PRRSV N binds to both subunits of rRNA. Minor bands were also observed for the E. coli-expressed GST-N protein (Fig. 8B, lanes 2 and 5), and the same bands were detected by the total-RNA probe (Fig. 8A, lane 3). The specificities of these bands were not further ascertained.
Involvement of RNA in the interaction of N with fibrillarin.
Fibrillarin is an RNA binding protein in the nucleolus. Fibrillarin binds to pre-rRNA and participates in the precise processing of the pre-rRNA via ribose methylation at specific sites. Since the N protein possessed the capacity to bind both viral genomic RNA and rRNA, we wished to examine whether RNA was involved in the interaction between fibrillarin and N. For this experiment, the GST-fibrillarin fusion protein was immobilized on glutathione-Sepharose beads and incubated with the radiolabeled N protein prepared from virus-infected cells in the presence of RNase (Fig. 9A). Since the N-N interaction is RNA dependent (37), the capacity of immunobilized N to pull down N was included as a control (lanes 5 and 6). In addition, we found that another nucleolar protein, nucleolin (C23), was able to bind to the PRRSV N protein (unpublished data), and therefore nucleolin as a GST fusion protein was included as another control (lanes 7 and 8). N immunoprecipitated with MAb SDOW17 was used as a marker (lanes 1 and 2). In the absence of RNase, the N protein bound GST-N (lane 5), GST-nucleolin (lane 7), and GST-fibrillarin (lane 9). After the addition of RNase A, at a concentration of 100 μg/ml, the N-[GST-N] and N[GST-fibrillarin] interactions were inhibited (lanes 6 and 10). This concentration of RNase did not affect the ability of N to bind to GST-nucleolin, demonstrating that the conditions used for RNase treatment did not affect the stability of the labeled N protein (lane 8). These data indicate that the presence of RNA is required for the N-fibrillarin interaction.
FIG. 9.
Involvement of RNA in the interaction of fibrillarin and N. (A) The GST-fibrillarin (Fibn) fusion protein was expressed in E. coli and subjected to a GST pull-down assay in the presence of 100 μg of RNase A/ml. The GST-fibrillarin-bound Sepharose beads were incubated with the [35S]methionine-labeled N protein prepared from virus-infected cells. The beads were washed, and the bound proteins were resolved by SDS-PAGE followed by autoradiography. Nucleolin as a GST fusion protein (GST-Ncl) was included as a control. N-IP, the immunoprecipitated N protein as a molecular mass marker. (B) GST pull-down assay using the phosphorylated N protein radiolabeled in vivo with inorganic [32P]orthophosphate upon virus infection. GST fusion proteins were subjected to a GST pull-down assay using the 32P-labeled N protein as the probe prepared from cells infected by PRRSV.
The binding of fibrillarin to the phosphorylated form of N was studied using the inorganic 32P-labeled N protein prepared from PRRSV-infected cells (Fig. 9B). The results showed that GST-fibrillarin bound the 32P-N protein. Together with the results shown in Fig. 6, these data show that both the phosphorylated and unphosphorylated forms of N are capable of binding fibrillarin and RNA.
DISCUSSION
We have shown that the PRRSV N protein colocalizes with fibrillarin in the nucleoli of virus-infected cells and in N-gene-transfected cells. Fibrillarin and the N protein colocalized in the fibrillar region of the nucleolus, most notably in the regions associated with the dense fibrillar component, but not in the granular component surrounding the fibrillar regions, in Cajal (coiled) bodies, or in the nucleoplasm. The specific interaction between N and fibrillarin was biochemically confirmed in vivo by the mammalian two-hybrid assay and in vitro using the GST-fibrillarin pull-down assay. In both assays, the degree of interaction of N with fibrillarin was comparable to N interacting with itself, indicating that N and fibrillarin form a relatively stable heterocomplex. The interactive domains of both N and fibrillarin were mapped using deletion constructs. For N, eight amino acids (IAQQNQSR) at positions 30 to 37 were identified as the binding domain with fibrillarin. Interestingly, this string of amino acids was previously mapped as the region which participated in the formation of the N-N interaction (37). This region corresponds to a relatively hydrophilic region of N and forms part of the conformational epitope recognized by MAb SDOW17 (35). Therefore, this domain is likely to be exposed to the aqueous environment rather than sequestered internally. Due to the high concentration of amide-containing asparagine (N) and glutamine (Q) residues, it is possible that this region mediates relatively strong hydrogen bonding for the formation of protein-protein interactions (40), such as N-N interactions and N-fibrillarin interactions. For fibrillarin, the N protein binding domain was mapped to the 80 amino acids at the N terminus. This region contains as many as 46 glycines (G) and 13 arginines (A) in a stretch of 70 residues and is referred to as the GAR domain. The biological function of the GAR domain is unclear.
The localization of viral nucleolar proteins to the nucleolus generally occurs through the interactions of basic regions on the viral protein with stretches of acidic residues on nucleolar proteins, such as B23 and nucleolin (C23) (11, 28). Upon binding viral protein in the cytoplasm or in the nucleus, B23 and nucleolin can function as shuttle proteins, directing the transport of viral proteins across the nuclear pore complex into the nucleoplasm and then to the nucleolus. The PRRSV N protein contains two domains enriched with basic amino acid residues, which may function as a potential recognition sequence for B23 and nucleolin. Unlike nucleolin and B23, however, fibrillarin lacks corresponding stretches of positively charged residues. Furthermore, it has been shown that the N protein has the ability to bind to importin-α and importin-β (unpublished data); both play essential roles in the nuclear transport of a protein through the nuclear pore complex. Therefore, it is assumed that N protein transport to the nucleolus is perhaps mediated via the importin-α/β-based pathway and/or the B23/nucleolin-based pathway rather than via the involvement of fibrillarin (Fig. 9, lanes 7 and 8 [N interacts with nucleolin]). The role of N during its interaction with fibrillarin may represent a unique function of N in the nucleolus. Fibrillarin-N protein complex formation appears to be restricted to the dense fibrillar component and nearby regions of the nucleolus and to be associated with box C/D snoRNAs to form snoRNP complexes. These snoRNAs contain an antisense element complementary to rRNAs, and the antisense snoRNAs function as the guide for snoRNPs in the dense fibrillar component. The snoRNPs then participate in the site-specific 2′-O-ribose methylation of the pre-rRNA, the precise pre-rRNA cleavage processing, and assembly of ribosomal subunits (30, 31). In fact, the crystal structure of fibrillarin revealed a methyltransferase-like domain in the C-terminal region (34). Since fibrillarin and N function as RNA binding proteins (Fig. 8) (4, 34), colocalization to the same site may reflect a common RNA binding function. We have shown that the N-fibrillarin interaction requires RNA. Since replication of the PRRSV genomic RNA is restricted to the cytoplasm and since fibrillarin exists mainly in the nucleus and nucleolus, it is unlikely that the viral genomic RNA participates in the N and fibrillarin interactions. Rather, rRNA—presumably precursor rRNA, which is abundantly present in the dense fibrillar component where the N-fibrillarin colocalization is detected—may be the RNA species participating in this interaction in the nucleolus. Indeed, we have shown that the N protein was able to bind to rRNA (Fig. 8), and both 28S and 18S rRNAs were capable of binding to N. A possibility is that N and fibrillarin may compete for the same rRNA substrate in the dense fibrillar component of the nucleolus, which may result in the modification of fibrillarin function in virus-infected cells. Alternatively, the binding of nucleolar RNAs in the fibrillar region may direct a conformational change in N sufficient to facilitate a stable interaction with other nucleolar proteins, such as fibrillarin, and subsequently to allow the N protein to counterregulate fibrillarin function, such as site-specific methylation, leading to uncontrolled cleavages of pre-rRNA. This hypothesis is further supported by the observation that the colocalization of N and fibrillarin did not occur in the Cajal body (Fig. 2B and E), where unlike in the nucleolus, rRNA is absent (6). Studies are in progress to examine a modulatory function of N for fibrillarin, such as RNA methylation.
Previously, it was shown that the phosphorylated form of N was found in both the cytoplasm and the nucleoplasm, suggesting that phosphorylation was not a determinant for the nuclear localization of N (36). In the present study, we show that the N protein binds to the viral genomic RNA via a region containing amino acids 37 to 57 (Fig. 7). This region includes the putative NLS situated at positions 41 to 47. It is postulated, therefore, that RNA binding may be a determinant for N protein nuclear localization. When the viral genomic RNA accumulates and binds to the N protein in the cytoplasm, presumably at a later stage of infection, the NLS of N is masked by the bound RNA. In this scenario, the N-RNA complex becomes a viral capsid and is utilized to constitute virions during virus assembly. When the N protein is naked, however (for example, at an early stage of infection, when the N protein concentration surpasses the genomic RNA in the molar ratio), the NLS of N is exposed and recognized by importin-α and importin-β, leading to the transport of the cargo protein through the nuclear pore complex into the nucleus. In the nucleus, importin-α and -β dissociate, and the N protein is further transported to the nucleolus, perhaps by nucleolin or B23, and more specifically to the dense fibrillar component, where N binds to fibrillarin with the help of rRNA. Through this series of interactions, N may play an important role during the early stage of virus infection to modify the function of fibrillarin toward an active production of the virus.
The nuclear and nucleolar distribution of the viral capsid protein has been observed in other members of the order Nidovirales and the families Arteriviridae (equine arteritis virus [29]) and Coronaviridae (15, 38). The nucleocapsid proteins of coronaviruses, such as infectious bronchitis virus, transmissible gastroenteritis virus, and mouse hepatitis virus, have been reported to localize to the nucleoli of infected cells (39). Among these viruses, it is of particular interest that the nucleocapsid proteins of avian infectious bronchitis virus and mouse hepatitis virus colocalize with two nucleolar antigens, fibrillarin and nucleolin (9). Thus, colocalization and specific interaction of the nucleocapsid protein with fibrillarin may be a common feature of nidoviruses. Elucidation of the role that the nidovirus N protein plays in the nucleolus may provide insight into the mechanisms of viral strategies that alter host cell functions, possibly by modulating host protein synthesis and the cell cycle during virus replication.
Acknowledgments
We are deeply indebted to John Aris for providing the human fibrillarin cDNA clone and Marvin Fritzler for providing anti-fibrillarin human antiserum for this study.
This work was supported by Ontario Pork and the Ontario Ministry of Agriculture and Food and by PHS NIH grant R15GM61319-01 and USDA NRI grant 97-35204-5071 awarded to R.R.R.
REFERENCES
- 1.Albina, E. 1997. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet. Microbiol. 55:309-316. [DOI] [PubMed] [Google Scholar]
- 2.Allende, R., T. L. Lewis, Z. Lu, D. L. Rock, G. F. Kutish, A. Ali, A. R. Doster, and F. A. Osorio. 1999. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J. Gen. Virol. 80:307-315. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. S., C. E. Lyon, A. H. Fox, A. K. L. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. [DOI] [PubMed] [Google Scholar]
- 4.Aris, J. P., and G. Blobel. 1991. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl. Acad. Sci. USA 88:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biggiogera, M., M. Malatesta, S. Abolhassani-Dadras, F. Amalric, L. I. Rothblum, and S. Fakan. 2001. Revealing the unseen: the organizer region of the nucleolus. J. Cell Sci. 114:3199-3205. [DOI] [PubMed] [Google Scholar]
- 6.Borhmann, K., J. Ferreira, N. Santama, K. Weis, and A. I. Lamond. 1995. Molecular analysis of the coiled body. J. Cell Sci. Suppl. 19:107-113. [DOI] [PubMed] [Google Scholar]
- 7.Carmo-Fonseca, M., L. Mendes-Soare, and I. Campos. 2000. To be or not to be in the nucleolus. Nat. Cell Biol. 2:E107-E112. [DOI] [PubMed] [Google Scholar]
- 8.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 9.Chen, H., T. Wurm, P. Britton, G. Brooks, and J. A. Hiscox. 2002. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 76:5233-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginisty, H., H. Sicard, B. Benoit, and P. Bouvet. 1999. Structure and functions of nucleolin. J. Cell Sci. 112:761-772. [DOI] [PubMed] [Google Scholar]
- 12.Gonin, P., H. Mardassi, C. A. Gagnon, B. Massie, and S. Dea. 1998. Nonstructural and antigenic glycoprotein is encoded by ORF3 of the IAF-Klop strain of porcine reproductive and respiratory syndrome virus. Arch. Virol. 143:1927-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jansen, R. P., E. C. Hurt, H. Kern, H. Lehtonen, M. Carmo-Fonseca, B. Lapeyre, and D. Tollervey. 1991. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol. 113:715-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordan, E. G. 1984. Nucleolar nomenclature. J. Cell Sci. 67:217-220. [DOI] [PubMed] [Google Scholar]
- 15.Hiscox, J. A., T. Wurm, L. Wilson, P. Britton, D. Cavanagh, and G. Brooks. 2001. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 75:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 17.Mardassi, H., P. Gonin, C. A. Gagnon, B. Massie, and S. Dea. 1998. A subset of porcine reproductive and respiratory syndrome virus GP3 glycoprotein is released into the culture medium of cells as a non-virion-associated and membrane-free (soluble) form. J. Virol. 72:6298-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meulenberg, J. J. M., A. P. van Nieuwstadt, A. van Essen-Zandbergen, J. N. A. Bos-de-Ruijter, J. P. M. Langeveld, and R. H. Meloen. 1998. Localization and fine mapping of antigenic sites on the nucleocapsid protein of porcine reproductive and respiratory syndrome virus with monoclonal antibodies. Virology 252:106-114. [DOI] [PubMed] [Google Scholar]
- 19.Meulenberg, J. J., M. M. Hulst, E. J. de Meijer, P. L. Moonen, A. den Besten, E. P. de Kluyver, G. Wensvoort, and R. J. Moormann. 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 192:62-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelsen, C. J., M. P. Murtaugh, and K. S. Faaberg. 1999. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 73:270-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson, E. A., J. Christopher-Hennings, T. Drew, G. Wensvoort, J. E. Collins, and D. A. Benfield. 1993. Differentiation of U. S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 31:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochs, R. L., M. A. Lischwe, W. H. Spohn, and H. Busch. 1985. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol. Cell. 54:123-133. [DOI] [PubMed] [Google Scholar]
- 23.Olson, M. O., M. Dundr, and A. Szebeni. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10:189-196. [DOI] [PubMed] [Google Scholar]
- 24.Rowland, R. R., R. Kervin, C. Kuckcleburg, A. Sperlich, and D. A. Benfield. 1999. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 64:1-12. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Scheer, U., and R. Hock. 1999. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11:385-390. [DOI] [PubMed] [Google Scholar]
- 27.Snijder, E. J., and J. M. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava, M., and H. B. Pollard. 1999. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 13:1911-1922. [PubMed] [Google Scholar]
- 29.Tijms, M. A., Y. van der Meer, and E. J. Snijder. 2002. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 83:795-800. [DOI] [PubMed] [Google Scholar]
- 30.Tollervey, D., H. Lehtonen, R. Jansen, H. Kern, and E. C. Hurt. 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-ribosomal RNA processing, pre-ribosomal RNA methylation, and ribosomal assembly. Cell 72:443-457. [DOI] [PubMed] [Google Scholar]
- 31.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9:337-342. [DOI] [PubMed] [Google Scholar]
- 32.van Nieuwstadt, A. P., J. M. Meulenberg, A. van Essen-Zanbergen, A. Petersen-den Besten, R. J. Bende, R. J. Moormann, and G. Wensvoort. 1996. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J. Virol. 70:4767-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visintin, R., and A. Amon. 2000. The nucleolus: the magician's hat trick for cell cycle tricks. Curr. Opin. Cell Biol. 12:372-377. [DOI] [PubMed] [Google Scholar]
- 34.Wang, H., D. Boisvert, K. K. Kim, R. Kim, and S. H. Kim. 2000. Crystal structure of a fibrillarin homologue from Methanococcusjannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J. 19:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wootton, S. K., G. Koljesar, L. Yang, K. J. Yoon, and D. Yoo. 2001. Antigenic importance of the carboxy-terminal beta-strand of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. Clin. Diagn. Lab. Immunol. 8:598-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wootton, S. K., R. R. Rowland, and D. Yoo. 2002. Phosphorylation of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. J. Virol. 76:10569-10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wootton, S. K., and D. Yoo. 2003. Homo-oligomerization of the porcine reproductive and respiratory syndrome virus nucleocapsid protein and the role of disulfide linkages. J. Virol. 77:4546-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wootton, S. K., D. Yoo, and D. Rogan. 2000. Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch. Virol. 145:2297-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wurm, T., H. Chen, T. Hodgson, P. Britton, G. Brooks, and J. A. Hiscox. 2002. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 75:9345-9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, D., C. J. Tsai, and R. Nussinov. 1997. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Eng. 10:999-1012. [DOI] [PubMed] [Google Scholar]