Abstract
A physics-based method, aimed at determining protein structures by using NOE-derived distance constraints together with observed and computed 13Cα chemical shifts, is applied to determine the structure of a 20-residue all-β peptide (BS2). The approach makes use of 13Cα chemical shifts, computed at the density functional level of theory, to derive backbone and side-chain torsional constraints for all of the amino acid residues, without making use of information about residue occupancy in any region of the Ramachandran map. In addition, the torsional constraints are derived dynamically—i.e., they are redefined at each step of the algorithm. It is shown that, starting from randomly generated conformations, the final protein models are more accurate than existing NMR-derived models of the peptide, in terms of the agreement between predicted and observed 13Cβ chemical shifts, and some stereochemical quality indicators. The accumulated evidence indicates that, for a highly flexible BS2 peptide in solution, it may not be possible to determine a single structure (or a small set of structures) that would satisfy all of the constraints exactly and simultaneously because the observed NOEs and 13Cα chemical shifts correspond to a dynamic ensemble of conformations. Analysis of the structural flexibility, carried out by molecular dynamics simulations in explicit water, revealed that the whole peptide can be characterized as having liquid-like behavior, according to the Lindemann criterion. In summary, a β-sheet structure of a highly flexible peptide in solution can be determined by a quantum-chemical-based procedure.
Keywords: protein structure determination, validation, refinement, protein flexibility, molecular dynamics
We recently introduced a new physics-based method that exploits distance constraints derived from nuclear Overhauser effects (NOEs) and 13Cα chemical shifts to determine the structure of a 76-residue all-α-helical protein (the Bacillus subtilis acyl carrier protein) at a high level of accuracy (1) without resorting to other experimental data (such as vicinal coupling constants, backbone residual dipolar couplings, etc.) or knowledge-based information (for example, from automated chemical-shift predictors, side-chain rotamer libraries, etc.). This methodology (2), validated on 139 conformations of the human protein ubiquitin, enabled us to offer a new criterion for an accurate assessment of the quality of NMR-derived protein conformations and to examine whether x-ray or NMR-solved structures are better representations of the observed 13Cα chemical shifts in solution. A detailed analysis (2) of the disagreement between observed and density functional theory (DFT)-computed 13Cα chemical shifts in these ubiquitin conformations illustrated the accuracy of the calculations and, more importantly, demonstrated that these disagreements reflect the dynamic nature of the protein rather than inaccuracies of the method. Our methodology has also been used (3) to show that neutral, rather than charged, basic and acidic groups are a better approximation of the observed 13Cα chemical shifts of a protein in solution. Furthermore, the results obtained (3) indicated that side-chain flexibility influences the computed 13Cα chemical shifts in ubiquitin and, hence, revealed the importance of a proper consideration of side-chain conformations for an accurate refinement of protein structures. Because automated servers are widely used for prediction of backbone torsional angles using observed chemical shifts for a given protein sequence, we evaluated the performance of our method compared with that of automated servers (2). In particular, we considered a problem inverse to structure prediction—i.e., we tested the sensitivity of these methods to significant differences in protein conformation (in terms of DFT-computed and observed chemical shifts). As a result, the servers appeared to be much less accurate than our methodology, which indicates that results obtained by using automated servers may not be able to provide enough guidance in selecting the most accurate conformations during protein-structure determination.
Evidence obtained from the probability-based secondary structure identification method of Wang and Jardetzky (4) suggests that the reliability to distinguish an α-helix from a statistical coil based on chemical-shift information follows the ranking 13Cα > 13C′ > 1Hα > 13Cβ > 15N > 1HN, whereas a different trend (1Hα > 13Cβ > 1HN ∼ 13Cα ∼ 13C′ ∼ 15N) was found for the corresponding reliability to distinguish a β-strand conformation from a statistical coil. This trend raises the question as to whether a mainly 13Cα-driven methodology can be used to predict high-quality all-β-sheet structures and, if so, how well the corresponding 13Cβ chemical-shift predictions would be. Our physics-based method (1, 3) relies on the hypothesis that an accurate protein structure prediction can be carried out by simply identifying a set of conformations that simultaneously satisfies two sets of constraints: (i) computed torsional constraints for all amino acid residues in the sequence (obtained from a comparison of computed 13Cα chemical shifts with the experimental ones), and (ii) a fixed set of experimental NOE-derived distance constraints. This approach makes use of 13Cα chemical shifts, computed at the density functional level of theory, to obtain torsional constraints for all backbone and side-chain torsional angles for each residue without assuming the occupancy of any region of the Ramachandran map (1). The method used in this work makes use of 100% of the observed 13Cα chemical shifts to derive torsional constraints for all of the residues in a protein, in contrast to the traditional methods that use the 13Cα chemical shifts to identify only those portions of the backbone of the molecule that correspond to well defined secondary structure, thereby making use of only up to ≈40% of the residues in proteins (5).
A 20-residue peptide capable of forming a three-stranded antiparallel β-sheet in aqueous solution—i.e., the BS2 peptide with the sequence TWIQNDPGTKWYQNDPGTKIYT (Fig. 1), for which both a complete set of 13Cα chemical shifts and a reduced number of NOEs were reported (6)—was chosen to determine whether our method, previously shown to be able to compute α-helical structures (1), could also succeed in computing a β-sheet structure (and at the same time predict observed 13Cβ chemical shifts). The BS2 peptide is one of three designed 20-residue peptides—namely, BS1, BS2, and BS3—discussed by Santiveri et al. (6). The three peptides share the same sequence except for the two turn-regions (residues 6, 7 and 14, 15 in Fig. 1). The BS2 peptide represents an improved β-sheet model (compared with the BS1 peptide) with the highest population of residues in the three-stranded antiparallel β-sheet conformation in aqueous solution (6). The stabilization of the β-sheet structure was achieved by substitution of the Gly and Ser turn residues of BS1 by d-proline and Gly in BS2. In contrast, replacement of the d-proline residues of BS2 by l-proline (in the BS3 peptide) destabilizes the BS3 structure to a statistical coil (6).
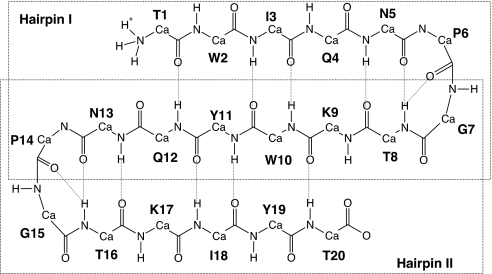
Fig. 1.
Schematic representation of the BS2 peptide (6), with ionized N- and C-terminal groups, shown in an ideal three-stranded antiparallel β-sheet motif. All residues are named by using the single-letter code and a number designating the position of each residue in the sequence. Dashed red lines indicate the hydrogen bonds observed in MD simulations (see MD Simulations). The two β-hairpins, I and II, forming the observed conformation of the BS2 peptide are indicated by two large dashed boxes.
Experimental structure determination of small peptides—e.g., those containing <25 residues, which are able to fold as monomers and do not contain disulfide bonds—is very valuable because such determinations can provide important information for force-field development (7) and evaluation (8). Moreover, small proteins can also be useful for the design and improvement of search algorithms aimed at an efficient exploration of the conformational space (9, 10). It should be noted that both of these applications require knowledge of high-quality conformations representing the native state. Until now, this approach has been confined mainly to x-ray-derived structures rather than to NMR-derived ones because it is often assumed that most of the NMR structures do not achieve the accuracy of high-quality x-ray structures (11, 12), although NMR-derived structures for ubiquitin are better representations of the 13Cα chemical shifts in solution than the x-ray structure (2).
The goal of this work was twofold. First, to determine whether it is possible to obtain an accurate set of conformations that simultaneously satisfies the NOE-derived distance constraints and the 13Cα-derived torsional constraints for the BS2 peptide in solution (6). To carry out such an analysis, two sets of conformations were generated with our physics-based method—namely, by using the observed 13Cα chemical shifts together with either a full set or a subset of NOE-derived distance constraints. Our second goal was to obtain atomic-level information about the structure and flexibility of the BS2 peptide in solution and, hence, to provide cross-validation of the results obtained from the 13Cα-derived analysis and NOE-derived distance violations. For this second goal, we carried out 20-ns MD simulations with explicit water starting from four structures selected (arbitrarily) from the final ensemble of conformations derived by using our new methodology (see Materials and Methods). Characterization of the structural flexibility of molecules in solution is of fundamental importance for the study of biological function, stability, and folding, and is a field of active experimental (13, 14) and theoretical research (15, 16).
Results and Discussion
Assessment of the Structural Quality of the BS2 Peptide.
In the following subsections, we present the results of the analysis of different ensembles of conformations of the BS2 peptide—namely, the Santiveri set (6), the Run_1 set, and the Run_2 set, with 20, 10, and 20 conformations, respectively. The Run_1 and Run_2 sets were both determined by using the 13Cα-derived torsional constraints but with a different number of NOE-derived constraints (see Materials and Methods).
Analysis in terms of the computed 13Cα and 13Cβ chemical shifts.
Fig. 2 shows a bar diagram of the root-mean-square deviations (rmsds) between the computed and observed 13Cα chemical shifts for each of the conformations from the following sets: Santiveri (as red bars), Run_1 (black bars), and Run_2 (green bars). Analysis based on the individual rmsds (indicated by bars in Fig. 2) or on the conformationally averaged rmsd (ca-rmsd) (indicated by dashed horizontal lines in Fig. 2) shows the importance of considering torsional-angle constraints derived from the computed 13Cα chemical shifts for the purpose of structure determination. Thus, although traditional methods and our method make use of NOE-derived distance constraints, the use of computed torsional-angle constraints for all residues in the sequence, not only those in secondary structure elements, led to lower ca-rmsds for the ensembles obtained with both the full set of NOE constraints and its subset, as shown in Table 1. The correlation coefficients (17), R, for the 13Cβ chemical shifts, shown in Table 1, are also consistent with this conclusion.
Fig. 2.
Bars indicate the rmsd between computed and observed 13Cα chemical shifts for each conformation from the following sets: Santiveri (red bars), Run_1 (black bars), and Run_2 (green bars). Dashed horizontal lines (see values in Table 1) designate the ca-rmsd values computed for each of these three sets as described in Materials and Methods; the color used for each horizontal line matches the set from which it was derived.
Table 1.
Results for the BS2 peptide
| Conformation set* | 13Cβ correlation coefficient,†R | ca-rmsd,‡ ppm | Maximum distance violation,§ Å | Number of abnormally short interatomic distances¶ |
|---|---|---|---|---|
| Santiveri (20) | 0.97 | 4.6 | 2.36 | 7.79 ± 1.99 (∼0.39) |
| Run_1 (10) | 0.98 | 3.5 | 0.88 | 10.10 ± 2.47 (∼0.50) |
| Run_2 (20) | 0.99 | 2.2 | 2.62 | 0.16 ± 0.37 (∼0.01) |
Computed for each set of conformations listed in column 1; the number of conformations in each set is indicated, in column 1, in parentheses.
*Santiveri denotes the original set of 20 conformations obtained by Santiveri et al. (6). The Run_1 and Run_2 sets were generated as explained in Materials and Methods.
†The correlation coefficient (17), R, (or Pearson coefficient) between computed and observed 13Cβ chemical shifts for each of the sets in column 1.
‡Values computed as explained in Materials and Methods.
§From the NOE-classified intensities (6).
¶Computed by using WHAT IF (19) as an average for the conformations of Santiveri, Run_1, and Run_2 sets. An abnormally short interatomic distance is defined (19) as the distance between two atoms that is shorter than the sum of their van der Waals radii minus 0.4 Å. In parentheses is shown the per-residue number of abnormal short interatomic distances.
Analysis in terms of NOE-derived distances and torsional angles.
Analysis of the distance violations indicated that the Santiveri set shows similar distributions of NOE violations for the 20 conformations as that of Run_2, although both sets show significantly higher maximum violations than that of Run_1 [see supporting information (SI) Fig. 4]. Thus, as shown in Table 1, the maximum distance violations are comparable for the Santiveri set (∼2.4) and Run_2 set (∼2.6), whereas they are significantly lower (∼0.9 Å) for the Run_1 set.
Some large (>2 Å) NOE-distance violations exist for the Santiveri set (Table 1). This analysis was carried out by using the regularized geometry of the conformations from the Santiveri set—i.e., all residues of the 20 conformations were replaced with the standard ECEPP/3 geometry (18). The conformations resulting from this regularization procedure are quite close, but not identical, to the original ones, with all-heavy-atom rmsd values ranging up to ∼0.2 Å. SI Figs. 5 and 6 and the related discussion (see SI Text) demonstrate that the maximum violations shown in Table 1 for the Santiveri set result from deficient orientations of the side chains in model 15 [out of 20 models reported by Santiveri et al. (6)] rather than the regularization. It should be mentioned that large (>2 Å) NOE-distance violations were also obtained for the structures from Run_2 (Table 1) because only a subset of NOE-derived distance constraints was used to derive these conformations.
An analysis in terms of violations of the torsional-angle constraints used during the last step of the structure-determination procedure was carried out for the φ, ψ, and χ torsional angles (86 angles) of all of the residues of the Santiveri, Run_1, and Run_2 sets. The selected set of φ, ψ, and χ torsional angles belongs to the minimal-rmsd model (2) in which the 13Cα chemical shift of each residue individually best matched the experimental one. This analysis does not consider the ω torsional angles because the departure of the peptide unit from the planar trans conformation, except for proline, is <10° (19). The percentage of agreement (within a 30° tolerance range) obtained for Run_1 (56%), Run_2 (50%), and the Santiveri (42%) sets indicates that the latter ensemble of conformations possesses a higher dispersion of the backbone and side-chain torsional angles than that of Run_1 or Run_2 set (see Fig. 3). This property is important because it has been recognized for a long time (20) that a high-quality structure determination should show a small rmsd among all conformers.
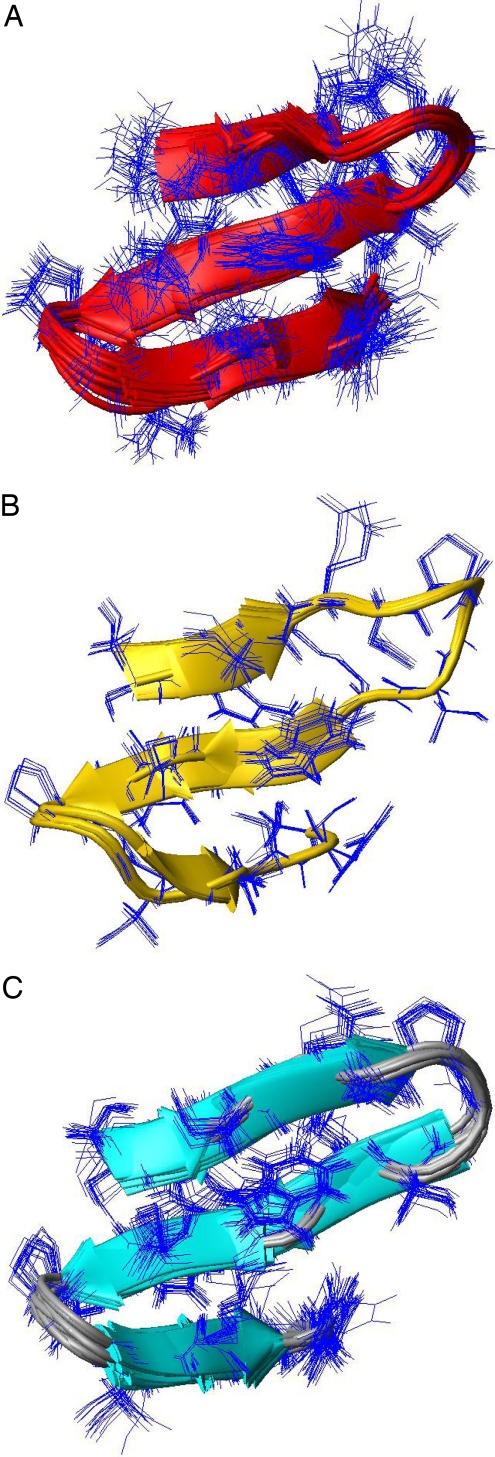
Fig. 3.
Sets of conformations of the BS2 peptide. (a) Superposition of 20 NMR-derived conformations (represented by ribbon diagrams) of the BS2 peptide obtained by Santiveri et al. (6). Side chains are represented by thin blue lines. (b) Same as in a for the 10 NMR-derived conformations after Run_1. (c) Same as in a for the 20 NMR-derived conformations after Run_2.
A comparison of some stereochemical quality indicators.
The conformations from the Santiveri, Run_1, and Run_2 sets were analyzed by using the PROCHECK server (11). The results reported in SI Table 2 reveal very similar distributions—i.e., within the standard deviation—of the residues in the most favored and additional allowed regions of the Ramachandran map. All of these ensembles of conformations contain no residues in the generously allowed and disallowed regions of the Ramachandran map.
Regarding the standard deviation of the ω values, which obey a Gaussian distribution with an average of ∼178° and a standard deviation of ∼5.5° (19), only the Run_2 set (5.90°) is neither as tightly constrained as the Santiveri set (0.02°) nor as underconstrained as the Run_1 set (7.16°).
There is a significant difference between the Santiveri, Run_1, and Run_2 sets in terms of the per-residue average number of abnormally short interatomic distances (19)—namely, ∼0.39, ∼0.50, and ∼0.01, respectively (as shown in Table 1). A similar analysis carried out for seven small NMR-derived proteins (2) also revealed a very large number (∼0.8 per residue) of abnormally short interatomic distances. However, the result obtained for the Run_2 ensemble (0.01 per residue) is close to the ideal value of 0.0 that characterizes x-ray-derived structures. Consistent with these analyses, the computed nonbonded ECEPP/3 energy for all conformations in Run_2 is significantly lower (by at least two orders of magnitude) than those computed from conformations of Run_1 or Santiveri set, respectively. This indicates that, for a highly flexible peptide in solution such as BS2, it might not be feasible to find a small set of conformations showing distance violations of <0.9 Å (as with the Run_1 set of conformations) and simultaneously good agreement between observed and predicted 13Cα chemical shifts and without steric clashes (as with the Run_2 set of conformations). Existence of atomic clashes, as in the Santiveri and Run_1 sets, prevents the use of these conformations for unconstrained MD analysis and, hence, a set (arbitrarily selected) of conformations from the Run_2 was chosen for this purpose in the next section.
Analysis of the Flexibility of the BS2 Peptide.
13Cα conformational shifts analysis.
Protein flexibility can be estimated (15) based on analysis of so-called conformational shifts, defined as the deviations of the observed 13Cα and 13Cβ chemical shifts from the statistical-coil values (21, 22). The conformational shifts were shown to reflect a wide range of conformational changes (23). Thus, the upper limit of the time scale of the motions affecting chemical shifts varies from microseconds to milliseconds for 13C and 15N nuclei and from hundreds of nanoseconds to hundreds of microseconds for protons. In contrast, the lower time limits of conformational changes affecting 1H, 13C, and 15N chemical shifts were shown to be on the picosecond time scale. The more flexible regions of a protein are expected to possess smaller average conformational shifts than the less flexible ones (15). This leads to the idea that changes in conformational shifts (CS) would be inversely proportional to the amplitude of backbone motions (15).
The CS for the BS2 peptide were computed as the deviation (24) of the conformationally averaged 13Cα chemical shifts from the corresponding statistical-coil values, for each amino acid residue μ from the 10 and 20 conformations of the Run_1 and Run_2 sets, respectively. They were used to calculate the average of the absolute value of the CS per strand (〈CS〉) for the N-terminal (residues 2–5), central (residues 8–13), and C-terminal (residues 16–19) strands. The results from the Run_1 set indicate that the C-terminal strand (〈CS〉 = 2.0 ppm) is more flexible than either the N-terminal strand (〈CS〉 = 2.9 ppm) or the central strand (〈CS〉 = 3.1 ppm). Further analysis of the 〈CS〉 from the Run_2 set gives qualitatively similar results (1.8, 2.5, and 1.0 ppm for the N-terminal, central, and C-terminal strands, respectively), hence indicating that the C-terminal strand is the most flexible one. However, the most flexible part of the BS2 peptide appears to be the turns—namely, (DPro-6, Gly-7) and (DPro-14, Gly-15) showing an average, over both turns, of 〈CS〉 ∼ 0.6 and 1.5 ppm for the Run_1 and Run_2 sets, respectively. The conformational shift analysis does not suffer from limitations of 15N relaxation measurements, such as peak overlap or broadening, poor signal intensity, insensitivity to internal fluctuations (that are slower than overall tumbling), etc. (16). However, values computed by using the conformational shift analysis rely heavily on the set of statistical-coil chemical shifts used, which are usually determined from oligopeptides in solution and, in some cases such as alanine, are a topic of debate. These oligopeptides display a predominantly flexible backbone, although the side chains frequently adopt a nonstatistical-coil arrangement (25, 26), which might significantly influence the 13Cα chemical shifts (3, 27). Accurate determination of molecular flexibility based only on 13Cα conformational shifts may be biased by the preferential side-chain orientation observed in oligopeptides.
MD analysis.
To study flexibility of the BS2 peptide in solution, we also carried out ∼20-ns MD runs starting from each of the four arbitrarily selected conformations (models 7, 9, 10, and 11 shown in SI Fig. 7) chosen from the 20 structures generated in Run_2. Because we are interested in studying the near-equilibrium dynamics, only the snapshots from the first 7 ns of each MD run (SI Fig. 8) were selected for further analysis. Details of the simulations are given in Materials and Methods.
SI Table 3 contains the atomic fluctuations of the backbone atoms computed per residue for the four BS2 models. If we consider the average fluctuations for each of the three β-strands of the peptide, the largest average fluctuation (0.79 Å) over the four MD runs takes place for residues 16–19 pertaining to the C-terminal strand, which indicates a larger relative flexibility of this part of the molecule. The average atomic fluctuations per strand are similar for the N-terminal and the central strands (0.69 and 0.63, respectively). This result is in qualitative agreement with the conclusion drawn from the analysis of the 13Cα conformational shifts.
We carried out an analysis of the values of the generalized Lindemann parameter (ΔL), which provides information about solid vs. liquid-like behavior of the system (28). The characteristic ΔL ≈ 0.15 corresponds to the transition between solid-like (ΔL < 0.15) and liquid-like (ΔL > 0.15) behavior. The BS2 peptide in solution can be considered as a predominantly liquid-like system because the ΔL value computed for all heavy atoms and for the backbone heavy atoms are similar and ∼0.24 [average over the four models used for the MD runs; see SI Text (Details of the Methods)]. It is interesting to compare this result with the Lindemann ΔL values obtained for ubiquitin (29), which are 0.14 and 0.29 for the heavy atoms of the backbone and those of the side chains, respectively. Furthermore, there is a wide dispersion of the ΔL values among the residues with the lowest values of ∼0.13–0.15 depending on the model.
Lifetimes of the backbone hydrogen bonds, which can be considered as an additional indicator of conformational changes, were computed for each of the four models (see SI Table 4). All of the backbone hydrogen bonds observed in the MD simulations of the BS2 peptide are shown in Fig. 1. In general, the lifetimes of the hydrogen bonds between strands 1 and 2 within hairpin I are similar to those of strands 2 and 3 within hairpin II. The only significant difference appears for the much shorter (13–88%) lifetime of the hydrogen bond between residues Thr-20 and Lys-9 compared with the lifetime of the bond between residues Gln-12 and Thr-1 (99%). This observation leads to the conclusion that the C terminus of the BS2 peptide appears to be more flexible than the N terminus.
The results of the hydrogen-bond analysis also indicate that the amide hydrogens of residue Thr-8 can form two backbone hydrogen bonds with the carbonyl oxygen of Asn-5 and DPro-6, and likewise Thr-16 with the carbonyl oxygen of Asn-13 and DPro-14. The interstrand hydrogen bonds (Thr-8 … Asn-5 and Thr-16 … Asn-13) exist for almost the entire length of the simulation (70–90%), whereas the lifetimes of the hydrogen bonds between threonines (8 and 16) and d-prolines (6 and 14) are ≈20% (see SI Table 4). As seen from the MD runs, each turn as a whole fluctuates with respect to the average structure (see SI Fig. 9), leading to significantly larger atomic fluctuations for these parts of the peptide compared with the β-strands (as shown in SI Fig. 9). The average atomic fluctuations for the two turns and three strands are 1.01 and 0.64 Å, respectively, in agreement with conclusions derived from the 〈CS〉 analysis of Run_1 and Run_2 sets.
Conclusion
In this work, we demonstrated that an accurate all-β-sheet structure can be determined by simply identifying a set of conformations that simultaneously satisfy a set of constraints—namely, 13Cα-dynamically derived torsional-angle constraints for all amino acid residues in the sequence—and a fixed set of NOE-derived distance constraints. In particular, two sets of conformations for the BS2 peptide were determined here by using different numbers of NOE-derived distance constraints. As expected, use of the 13Cα-derived torsional constraints led to noticeably lower ca-rmsds for both sets compared with the Santiveri models. Analysis of the accuracy of these sets, as a measure of the closeness with which the calculations reproduce the structure in solution, in terms of the NOE-derived distance violations, the 13Cβ chemical shifts, and some stereochemical quality factors, indicates that our self-consistent physics-based method is able to produce a more accurate set of conformations than that obtained with the traditional methods. Further, the results suggests that, for a flexible molecule in solution, it may not be possible to determine a single structure (or a small set of structures) that would satisfy all of the constraints exactly and simultaneously. This is a consequence of the well known fact (30) that NMR parameters, such as the observed NOE-derived distances and the 13Cα chemical shifts, correspond to a dynamic ensemble of conformations and, therefore, may not be reproduced exactly by a limited set of static structures (2, 31).
Further analysis of the per-residue average 13Cα conformational shifts from the Run_1 and Run_2 sets enabled us to conclude that the third, C-terminal strand in the β-sheet of the BS2 peptide is the most flexible strand although less flexible than the turns. In line with these results, the MD simulations carried out for the BS2 peptide yielded a plausible atomic description of the motion of this peptide in solution, as revealed by both the pattern of hydrogen bonds and the generalized Lindemann parameter, and also provided additional evidence for greater flexibility of the C-terminal strand. The fact that the observed 13Cα chemical shifts, supplemented only by NOE-derived distance constraints, provide accurate information for validation and refinement of protein structures, and site-specific information about the flexibility of the molecule in solution, may be very useful for NMR spectroscopists and theoreticians interested in analyses of the stability and protein-folding mechanism.
Although the present method is more CPU-time-demanding than traditional methods to solve protein structure, such as the one used by Santiveri et al. (6), the higher computational cost does not constitute a real problem because of the increasing availability of computer clusters with large numbers of faster processors. Conceivably, advances in computational capabilities will enable us to estimate, at the quantum-chemical level, the weight factor of each conformation of the ensemble, and hence, convert the ca-rmsd into a Boltzmann-average rmsd. This development will constitute a significant advance in the interpretation of the ensemble-averaged experimental quantities.
Materials and Methods
Sequence of the BS2 Peptide.
The BS2 peptide studied here has the following sequence: TWIQNDPGTKWYQNDPGTKIYT (Fig. 1), where DP denotes d-proline. Replacement of the d- by l-proline (the BS3 peptide) leads to a complete destabilization of the β-sheet motif (6).
NMR Data for the BS2 Peptide.
A total of 130 NOE-derived distances for which NOE classified intensities are provided [from table ST4 of Santiveri et al. (6)], and 20 13Cα and 18 13Cβ chemical shifts [referenced to 3-(trimethylsilyl) propionate sodium salt (TSP)] [from table ST1 of Santiveri et al. (6)], were used in this work. Only the experimental data for aqueous solution (at pH 3.5 and t = 10°C) reported by Santiveri et al. (6) were used here. We assumed that there was no assignment error in any of the constraints, and that 100% of the NOE-derived distances had zero distance-constraint errors. The latter assumptions were adopted because the accuracy of the structure determination is very sensitive to NOE-derived distance errors (31).
Existing Set of Conformations for the BS2 Peptide.
A set of 20 conformations of the BS2 peptide was originally derived by Santiveri et al. (6), using traditional NMR methods. This ensemble of conformations was used here only for comparison purposes and is referred to as the Santiveri set. Whether the use of traditional methods, combined with chemical-shift-based torsional-angle constraints derived from automated server predictors, could lead to better results than those obtained by Santiveri et al. goes beyond the current analysis.
Run_1 and Run_2 Set of Conformations.
A full set of 130 NOE-derived distances (6) was used to determine 10 conformations of Run_1. A subset of 118 NOE-derived distances—i.e., after removing the last 12 from the full list of 130—was used to determine 20 conformations of Run_2. This subset of 12 NOEs was chosen because it does not significantly affect the β-sheet twist (6). In both runs, the 20 13Cα chemical shifts were used, as explained below under Protein Structure Determination. We did not study the influence of the selection of the subsets of NOE-derived distances on the results obtained for the Run_2 set because such analysis goes beyond our current computational capacity.
Conformational Shifts.
The 13Cα conformational shifts for each amino acid in the sequence was computed as the difference between the observed (or the computed conformationally averaged) 13Cα chemical shifts and their corresponding statistical-coil value, as reported by Wishart et al. (24). The reported statistical-coil value (24) (63.3 ppm for Pro) was adopted for d-Pro.
Protein Structure Determination.
A recently introduced physics-based method (1), aimed at determining protein structures in solution, is used here to obtain the most probable set of conformations of the 20-residue BS2 peptide that satisfies both the observed 13Cα chemical shifts and a set of NOE-derived distance constraints. The procedure used to determine Run_1 and Run_2 sets of conformations consists of the following steps.
The variable-target-function (VTF) approach with a simplified soft-sphere potential function (32) was used to generate an ensemble of conformations at random that simultaneously satisfy a set of distance constraints derived from the experimental NOEs and the torsional constraints derived from the 13Cα conformational shifts. A clustering procedure was carried out to select a small subset of the total number of the VTF-derived set of conformations—namely, those possessing a maximum NOE-derived distance violation lower than 1 Å—by using the minimal spanning tree (MST) method (33).
The 13Cα and 13Cβ chemical shifts were computed at the DFT level for each conformation of the set obtained in step i. The DFT procedure was applied to each amino acid X in the sequence by treating X as a terminally blocked tripeptide with the sequence Ac-GXG-NMe in the conformation of each generated peptide structure. The 13Cα and 13Cβ chemical shifts for each amino acid residue X were computed (26) at the B3LYP/6–311+G(2d,p) level of theory, whereas the remaining residues in the tripeptide were treated at the B3LYP/3–21G level of theory—i.e., by using the locally dense approach (34). All ionizable residues were considered neutral during the quantum-chemical calculations (3). The isotropic shielding values, calculated with the Gaussian 98 package (35), were referenced with respect to a tetramethylsilane (TMS) 13Cα chemical-shift scale, as described in ref. 26. Conversion of the computed TMS-referenced values for the 13Cα chemical shifts to a TSP reference was carried out by adding 1.25 ppm in place of 1.82 ppm (36), as discussed by Vila et al. (2). Examination of the chemical shifts of each residue of all of the clustered conformations considered here enabled us to identify a new minimal-rmsd model (2), in which the 13Cα chemical shift of each residue individually best matched the experimental one, thereby providing a new set of φ, ψ, and χ torsional-angle constraints.
Only one conformation among all of the selected conformations described in step i was selected. This conformation possessed the lowest rmsd between the computed and observed 13Cα chemical shifts. The selected conformation was used as a starting one in a conformational search with Monte Carlo with minimization (MCM) (37) carried out with two types of constraints: the original fixed set of NOEs and the new set of φ, ψ, and χ torsional angles derived in step ii. This time, instead of using a simplified soft-sphere potential function, we used a complete force-field containing the following terms: (a) the internal potential energy, as described by the ECEPP/3 force field (18); (b) the solvent free energy calculated by using a solvent-accessible surface area model (38); and (c) additional energy terms aimed at penalizing violations of the distance and torsional-angle constraints (39). Finally, a clustering procedure was carried out to select a small subset of the total number of the MCM-derived set of conformations by using the MST method (33) and assuming a specific rmsd cutoff for all heavy atoms.
Steps ii and iii were repeated iteratively by using the set of conformations obtained in step iii and, hence, allowing us to obtain an updated set of φ, ψ, and χ torsional-angle constraints. At any stage of the procedure, a tolerance range Λ, with 20° ≤ Λ ≤ 35°, for the torsional constraints was adopted. Variation of the torsional angles within a tolerance range Λ is considered acceptable and hence is not subject to energetic penalties. Among all of the conformations generated in the final use of step iii, only one conformation is selected, because it is characterized by the lowest rmsd between the computed 13Cα chemical shifts and the observed ones. Thus, the procedure of step iii, applied to such a conformation, led to a new set of structures. The final number of conformations in this set is determined by the cutoff rmsd value adopted for the clusterization procedure in step iii, that is 0.3 and 0.4 Å for Run_1 and Run_2, respectively. As a consequence, the Run_1 set (10 conformations) is tighter than that of Run_2 (20 conformations), as seen in Fig. 3 b and c.
NOE Analysis.
The evaluation of the total number of violations and maximum violations (shown in SI Fig. 4) for the Santiveri, Run_1, and Run_2 sets, respectively, was carried out only with the full set of 130 NOEs.
CPU Time for the Quantum-Chemical Calculations.
See SI Text (Details of the Methods).
13Cα Chemical Shifts in the Presence of Conformational Averaging.
A new scoring function (ca-rmsdα) called the conformationally averaged rmsd was proposed recently (2) as a criterion to assess the quality of protein models. For details, see SI Text (Details of the Methods).
MD Simulations.
All MD simulations were carried out by using the AMBER 8.0 package (40) and the AMBER parm99 force field. For details of the MD simulations and trajectory analysis, see SI Text (Details of the Methods).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. M. A. Jiménez for providing the coordinates and the NMR experimental data for the BS2 peptide, and Professors D. A. Case and G. T. Montelione for helpful comments on this article. This work was supported by National Institutes of Health Grants GM-14312 and GM-24893, and National Science Foundation Grant MCB05–41633. Support was also received from the Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina (FONCyT-ANPCyT Grant PAV 22642/22672) and the Universidad Nacional de San Luis, Argentina (Grant P-328501). This work was conducted by using the resources of a Beowulf-type cluster located at the Baker Laboratory of Chemistry and Chemical Biology, Cornell University, and the National Science Foundation Terascale Computing System at the Pittsburgh Supercomputer Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711022105/DC1.
References
- 1.Vila JA, Ripoll DR, Scheraga HA. Use of 13Cα chemical shifts in protein structure determination. J Phys Chem B. 2007;111:6577–6585. doi: 10.1021/jp0683871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vila JA, Villegas ME, Baldoni HA, Scheraga HA. Predicting 13Cα chemical shifts for validation of protein structures. J Biomol NMR. 2007;38:221–235. doi: 10.1007/s10858-007-9162-x. [DOI] [PubMed] [Google Scholar]
- 3.Vila JA, Scheraga HA. Factors affecting the use of 13Cα chemical shifts to determine, refine, and validate protein structures. Proteins. 2007 doi: 10.1002/prot.21726. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Jardetzky O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 2002;11:852–861. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X-P, Case DA. Automated prediction of 15N, 13Cα, 13Cβ, and 13C′ chemical shifts in proteins using a density functional database. J Biomol NMR. 2001;21:321–333. doi: 10.1023/a:1013324104681. [DOI] [PubMed] [Google Scholar]
- 6.Santiveri CM, Santoro J, Rico M, Jiménez MA. Factors involved in the stability of isolated β-sheets: Turn sequence, β-sheet twisting, and hydrophobic surface burial. Protein Sci. 2004;13:1134–1147. doi: 10.1110/ps.03520704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jang S, Kim E, Pak Y. Free energy surfaces of miniproteins with a ββα motif: Replica exchange molecular dynamics simulation with an implicit solvation model. Proteins. 2006;62:663–671. doi: 10.1002/prot.20771. [DOI] [PubMed] [Google Scholar]
- 8.Zhou R. Free energy landscape of protein folding in water: Explicit vs. implicit solvent. Proteins. 2003;53:148–161. doi: 10.1002/prot.10483. [DOI] [PubMed] [Google Scholar]
- 9.Mohanty S, Hansmann UHE. Folding of proteins with diverse folds. Biophys J. 2006;91:3573–3578. doi: 10.1529/biophysj.106.087668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Höfinger S, Almeida B, Hansmann UHE. Parallel tempering molecular dynamics folding simulation of a signal peptide in explicit water. Proteins. 2007;68:662–669. doi: 10.1002/prot.21268. [DOI] [PubMed] [Google Scholar]
- 11.Laskowski RA, MacArthur MW, Moss DS, Thornton J. PROCHECK—A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 12.Williamson MP, Kikuchi J, Asajura T. Application of 1H-NMR chemical-shifts to measure the quality of protein structures. J Mol Biol. 1995;247:541–546. doi: 10.1006/jmbi.1995.0160. [DOI] [PubMed] [Google Scholar]
- 13.Korzhnev DM, Orekhov VY, Arseniev AS. Model-free approach beyond the borders of its applicability. J Magn Reson. 1997;127:184–191. doi: 10.1006/jmre.1997.1190. [DOI] [PubMed] [Google Scholar]
- 14.Palmer AG., III NMR characterization of the dynamics of biomacromolecules. Chem Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 15.Berjanskii M, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 16.Berjanskii M, Wishart DS. The RCI server: Rapid and accurate calculation of protein flexibility using chemical shifts. Nucleic Acids Res. 2007;35:W531–W537. doi: 10.1093/nar/gkm328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Press HW, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in FORTRAN 77: The Art of Scientific Computing. Cambridge, UK: Cambridge Univ Press; 1992. pp. 630–633. [Google Scholar]
- 18.Némethy G, et al. Energy parameters in polypeptides. 10. Improved geometrical parameters and nonbonded interactions for use in the ECEPP/3 algorithm, with application to proline-containing peptides. J Phys Chem. 1992;96:6472–6484. [Google Scholar]
- 19.Vriend GJ. WHAT IF—A molecular modeling and drug design program. J Mol Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 20.Güntert P, Wüthrich K. Improved efficiency of protein structure calculations from NMR data using the program DIANA with redundant dihedral angle constraints. J Biomol NMR. 1991;1:447–456. doi: 10.1007/BF02192866. [DOI] [PubMed] [Google Scholar]
- 21.Grathwohl C, Wüthrich K. 13C NMR of protected tetrapeptides TFA-Gly-Gly-L-X-L-Ala-OCH3, where X stands for 20 common amino-acids. J Magn Reson. 1974;13:217–225. [Google Scholar]
- 22.Spera S, Bax A. Empirical correlation between protein backbone conformation and Cα and Cβ 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- 23.Berjanskii M, Wishart DS. Application of the random coil index to studying protein flexibility. J Biomol NMR. 2007;40:31–48. doi: 10.1007/s10858-007-9208-0. [DOI] [PubMed] [Google Scholar]
- 24.Wishart DS, et al. 1H, 13C and 15N random coil NMR chemical-shifts of the common amino-acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 25.Bundi A, Wüthrich K. 1H-NMR parameters of the common amino-acid residues measured in aqueous-solutions of the linear tetrapeptides H-Gly-Gly-X-L-Ala-OH. Biopolymers. 1979;18:285–297. [Google Scholar]
- 26.Vila JA, Ripoll DR, Baldoni HA, Scheraga HA. Unblocked statistical-coil tetrapeptides and pentapeptides in aqueous solution: A theoretical study. J Biomol NMR. 2002;24:245–262. doi: 10.1023/a:1021633403715. [DOI] [PubMed] [Google Scholar]
- 27.Villegas ME, Vila JA, Scheraga HA. Effects of side-chain orientation on the 13C chemical shifts of antiparallel β-sheet model peptides. J Biomol NMR. 2007;37:137–146. doi: 10.1007/s10858-006-9118-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Vitkup D, Karplus M. Native proteins are surface-molten solids: Application of the Lindemann criterion for the solid versus liquid state. J Mol Biol. 1999;285:1371–1375. doi: 10.1006/jmbi.1998.2374. [DOI] [PubMed] [Google Scholar]
- 29.Lindorff-Larsen K, et al. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 30.Constantine KL, et al. Structural and dynamic properties of a β-hairpin-forming linear peptide. 1. Modeling using ensemble-averaged constraints. J Am Chem Soc. 1995;117:10841–10854. [Google Scholar]
- 31.Zhao D, Jardetzky O. An assessment of the precision and accuracy of protein structures determined by NMR—Dependence on distance errors. J Mol Biol. 1994;239:601–607. doi: 10.1006/jmbi.1994.1402. [DOI] [PubMed] [Google Scholar]
- 32.Vásquez M, Scheraga HA. Variable-target-function and buildup procedures for the calculation of protein conformation—Application to bovine pancreatic trypsin-inhibitor using limited simulated nuclear magnetic resonance data. J Biomol Struct Dyn. 1988;5:757–784. doi: 10.1080/07391102.1988.10506426. [DOI] [PubMed] [Google Scholar]
- 33.Kruskal JB., Jr On the shortest spanning subtree of a graph and the traveling salesman problem. Proc Am Math Soc. 1956;7:48–50. [Google Scholar]
- 34.Chesnut DB, Moore KD. Locally dense basis-sets for chemical-shift calculations. J Comput Chem. 1989;10:648–659. [Google Scholar]
- 35.Frisch MJ, et al. Vol. 98. Pittsburgh: Gaussian; 1998. Gaussian. Revision A.7. [Google Scholar]
- 36.Wishart DS, et al. 1H, 13C and 15N chemical-shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Scheraga HA. Monte-Carlo-minimization approach to the multiple-minima problem in protein folding. Proc Natl Acad Sci USA. 1987;84:6611–6615. doi: 10.1073/pnas.84.19.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ooi T, Oobatake M, Nemethy G, Scheraga HA. Accessible surface-areas as a measure of the thermodynamic parameters of hydration of peptides. Proc Natl Acad Sci USA. 1987;84:3086–3090. doi: 10.1073/pnas.84.10.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ripoll DR, Ni F. Refinement of the thrombin-bound structure of a hirudin peptide by a restrained electrostatically driven Monte Carlo method. Biopolymers. 1992;32:359–365. doi: 10.1002/bip.360320411. [DOI] [PubMed] [Google Scholar]
- 40.Case DA, et al. Vol. 8. San Francisco: Univ of California; 2004. AMBER. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.