Abstract
Clostridium kluyveri is unique among the clostridia; it grows anaerobically on ethanol and acetate as sole energy sources. Fermentation products are butyrate, caproate, and H2. We report here the genome sequence of C. kluyveri, which revealed new insights into the metabolic capabilities of this well studied organism. A membrane-bound energy-converting NADH:ferredoxin oxidoreductase (RnfCDGEAB) and a cytoplasmic butyryl-CoA dehydrogenase complex (Bcd/EtfAB) coupling the reduction of crotonyl-CoA to butyryl-CoA with the reduction of ferredoxin represent a new energy-conserving module in anaerobes. The genes for NAD-dependent ethanol dehydrogenase and NAD(P)-dependent acetaldehyde dehydrogenase are located next to genes for microcompartment proteins, suggesting that the two enzymes, which are isolated together in a macromolecular complex, form a carboxysome-like structure. Unique for a strict anaerobe, C. kluyveri harbors three sets of genes predicted to encode for polyketide/nonribosomal peptide synthetase hybrides and one set for a nonribosomal peptide synthetase. The latter is predicted to catalyze the synthesis of a new siderophore, which is formed under iron-deficient growth conditions.
Keywords: butyryl-CoA dehydrogenase, electron transfer flavoproteins, genome sequence, Rnf-dependent energy conservation
Clostridium kluyveri was first enriched by H. A. Barker in 1937 from the mud of a canal in Delft, The Netherlands (1). The enrichment culture fermented ethanol to acetate, butyrate, caproate, and methane. From the enrichment two organisms growing on ethanol were isolated. One organism, named C. kluyveri, ferments ethanol and acetate to butyrate, caproate, and H2 (reaction 1); toward the end of growth also butanol and hexanol are formed (2–6). The other organism, named Methanobacillus omelianskii, fermented ethanol and CO2 to acetate and methane. M. omelianskii was later found to be a coculture of a bacterium fermenting ethanol to acetate and H2 (S-organism) and an archaeon reducing CO2 with H2 to methane [Methanobacterium bryantii (7)]. All three organisms came to fame, C. kluyveri as a model organism in the early 1950s for the study of fatty acid synthesis and fatty acid oxidation (8–10), and the S-organism and M. bryantii for the development of the concept of interspecies hydrogen transfer and of syntrophy (11).
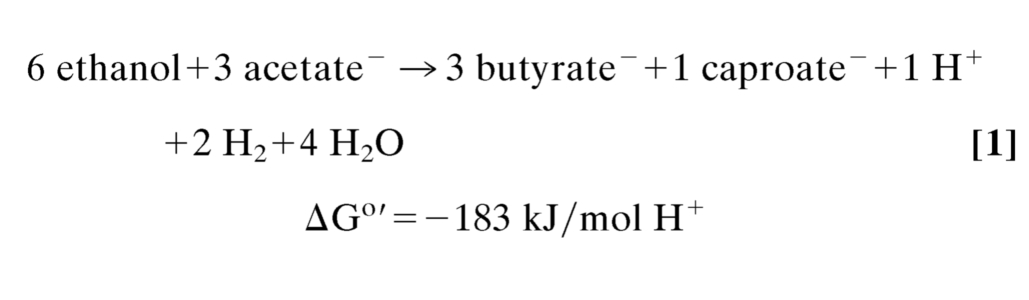 |
C. kluyveri can also ferment 2 ethanol and 2 succinate to 4 acetate, 1 butyrate, and 0.1 H2 (12) and 2 crotonate to 2 acetate, 1 butyrate, and 0.1 H2 (13, 14). The energy metabolism has been subject to many investigations (5, 6, 13), but its understanding, especially the mechanism of H2 formation via ferredoxin from NADH, has remained a challenge until today because this is thermodynamically an uphill reaction requiring energy to proceed. Likewise, carbon assimilation by this organism has been studied, and novel enzymes and pathways were discovered (15–17).
The organism has been used in the past as a source of enzymes, e.g., phosphotransacetylase for analytical purposes and enoate reductases for stereospecific hydrogenation reactions. Its ability to form caproic acid and hexanol from ethanol and butyrate has not been exploited but could be of future importance. Of further biotechnological interest are additional findings deduced from the genome sequence of C. kluyveri, such as the nonribosomal synthesis of peptide–polyketide hybrids and the presence of most of the genes required for ethanol and glycerol fermentation to 1,3-propanediol. These and certain other properties of C. kluyveri are addressed in this article on the basis of the complete genome sequence.
Results and Discussion
General Genome Features.
C. kluyveri contains one circular chromosome of 3.96 Mbp and one circular 59-kb plasmid, the latter with no obvious function (Fig. 1). The origin and terminus of replication of the chromosome were identified by GC-skew analysis and on the basis of the distinctive inflection point in the coding strand. Seventy-six percent of the 3,838 CDS are encoded on the leading strand, which is in agreement with a similarly strong coding bias observed in other clostridial [supporting information (SI) Table 1] and Bacillus genomes. The terminus of replication lies at ≈150° so that counterclockwise replication covers 210° of the chromosomal ring, which is more than in all other clostridial genomes sequenced so far. Comparative genome analysis of C. kluyveri with other clostridial genomes was carried out by using the MUMmer software package (http://mummer.sourceforge.net). Only a few regions of synteny on protein level were found, in particular with the genomes of Clostridium acetobutylicum and Clostridium tetani (SI Fig. 5). This low conservation of genome organization underlines the heterogeneity of the genus Clostridium.
Fig. 1.
Circular representation of the chromosome and plasmid of C. kluyveri. Genes encoded on the forward and reverse strands of the chromosome (outer two rings) are color-coded by red and dark blue, respectively. Predicted prophages are colored in green, and the sets of genes predicted to encode polyketide/nonribosomal peptide synthetases are indicated in yellow. The next inner ring shows the position of putative alien genes in gray. On the fourth ring the deviation from the average G+C content is plotted. Significant deviations are due to ribosomal RNA clusters (black arrows) and putative alien genes. Encoded transfer RNAs are indicated as black bars on the fifth ring. The plasmid map shows genes on the forward and reverse strands, color-coded in red and dark blue. The most inner ring shows the deviation from the average G+C content of the plasmid. Furthermore, the phage-terminase containing an intein is indicated in light blue. The intein was verified by heterologous production of the terminase in Escherichia coli (data not shown).
A search for CDS with aberrant codon usage statistics was carried out to predict possible genomic islands and “alien” genes (18), which might have been acquired horizontally from other organisms. Two hundred putative alien genes were identified in C. kluyveri (5.2% of all CDS; SI Table 1). Seventy-four of theses genes are encoded within an ≈61-kbp genomic island, which is framed by tRNA genes and which contains CDS for conserved hypothetical proteins. Thirty-four alien genes are located within a putative inserted prophage of ≈33 kbp. Compared with C. kluyveri fewer genes, an average of 42, were found in other clostridial genomes, with the exception of Clostridium difficile (249 putative alien genes; 6.6%). The latter genome has been described as highly variable (19). In addition, the genome of C. kluyveri harbors a relatively high number of possible insertion sequence elements (128) and predicted transposons or transposon fragments (56). Again, only the genome of C. difficile contains a higher number of mobile genetic elements among the sequenced clostridial genomes.
Surface-embedded and -associated as well as secreted proteins have been identified, such as 342 proteins with a predicted N-terminal signal peptide and 902 proteins possessing at least one predicted transmembrane helix, including 91 ABC transport systems, e.g., for anions, cations, and compatible solutes (SI Table 1). No CDS with the cell wall anchor LPxTG or with an N-terminal twin-arginine motif could be identified, but 17 CDS possess multiple copies of the cell wall-binding repeat 2 (PF04122), five of which contain known extracellular domains with hydrolytic activities or adhesive properties.
In the following only the CDS involved in the energy metabolism (Figs. 2 and 3 and SI Table 2) and the nonribosomal synthesis of proteins/polyketides (Fig. 4 and SI Fig. 6) in the genome of C. kluyveri will be discussed. For an overview of biosynthetic pathways starting from acetyl-CoA see SI Fig. 7. Features of endospore formation and germination are presented in SI Fig. 8.
Fig. 2.
Ethanol-acetate fermentation of C. kluyveri. For simplification purposes it was assumed that only butyrate, acetic acid, and H2 rather than caproate are formed as fermentation products. Inset shows the two membrane-associated, energy-converting enzyme complexes involved in the fermentation: ferredoxin:NAD oxidoreductase (RnfA-E) and ATP synthase (AtpA-I). For the other cytoplasmic enzymes involved see SI Table 2. The clostridial ferredoxin with two [4Fe4S] clusters accepts two electrons upon reduction (28). In the scheme it is assumed that 7 of the 10 NADH required for the reduction of 5 crotonyl-CoA to 5 butyryl-CoA (see text and reaction 5) are generated during ethanol oxidation to acetyl-CoA (gray background) and that three are generated via NAD+ reduction with Fdred2− (orange background). It has been omitted that some of the Fdred2− generated during crotonyl-CoA reduction is also used for the regeneration of NADPH, which is required for acetoacetyl-CoA reduction. The yellow area indicates the microcompartment, and the shaded area highlights the steps proposed to be involved in the catalysis of reaction 5 (see text) by the butyryl-CoA dehydrogenase complex composed of butyryl-CoA dehydrogenase (Bcd) and the electron transport flavoproteins (EtfAB). Bcd, EtfA, and EtfB each contain FAD.
Fig. 3.
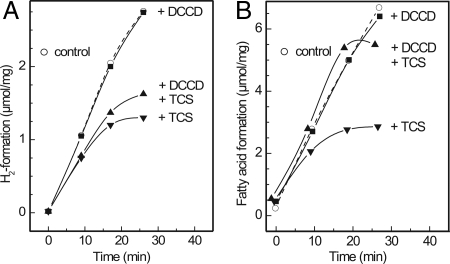
Effects of protonophore and F1Fo-ATPase inhibitor on H2 and fatty acid formation by cell suspensions of C. kluyveri. Shown is the effect of protonophore TCS and the F1Fo-ATPase inhibitor DCCD on hydrogen formation (A) and fatty acid formation (B). One mole of fatty acid corresponds to 1 mol of butyrate or 0.5 mol of caproate. For details on assay mixture composition see Materials and Methods. Open circles represent the control without TCS and DCCD (41).
Fig. 4.
CDS clusters predicted to encode nonribosomal peptide synthetase and polyketide synthase-nonribosomal peptide synthetase hybrids. A, C, Cy, E, and T are domains of nonribosomal peptide synthetases: A, adenylation domain; C, condensation domain; Cy, cyclization domain; E, epimerization domain; T, thiolation domain. ACP, AT, KR, and KS are domains of polyketide synthases: ACP, acyl carrier protein domain; AT, acyltransferase domain; KR, ketoreductase domain; KS, ketosynthase domain. M, TE, and Sfp are domains occurring in both types: M, methyltransferase domain; TE, thioesterase domain; Sfp, phosphopantetheinyl transferase domain.
C. kluyveri can fix nitrogen (20) and therefore requires molybdenum for growth. CDS for a molybdenum-dependent nitrogenase (CKL_3076-3078) as well as for a molybdate ABC transporter are present, but also for a vanadium-dependent nitrogenase (CKL_1745-1747) and an iron-only nitrogenase (CKL_0370-0372). Unexpectedly, the genome lacks CDS for both molybdopterin biosynthesis and molybdopterin-dependent enzymes.
Ethanol-Acetate Fermentation.
New insights into the exceptional fermentation strategy of C. kluyveri to convert ethanol and acetate to butyrate, caproate, and H2 (reaction 1) were gained from the genome sequence. All of the genes required for the metabolic pathway shown in Fig. 2 were found. Most genes for enzymes of the core metabolism have paralogs and are encoded in several gene clusters, some in close vicinity to energy-transferring and/or microcompartment proteins (SI Table 2).
Of the enzymes involved in the energy metabolism of C. kluyveri only two are membrane-associated proteins, i.e., the energy-converting NADH: ferredoxin oxidoreductase complex (RnfCDGEAB) and, as usual, the F1Fo-ATPase complex (AtpCDGAHFEBI). The subunits A, D, and E of the oxidoreductase and the subunits B, E, F, and I of the ATPase are integral membrane proteins with at least three transmembrane helices each.
Ethanol Dehydrogenases and Acetaldehyde Dehydrogenases in a Microcompartment.
The fermentation starts with the oxidation of ethanol via acetaldehyde to acetyl-CoA catalyzed by a NAD-dependent ethanol dehydrogenase (Adh) and an NAD(P)-dependent acetaldehyde dehydrogenase (Ald). The corresponding genes are found in a cluster (CKL_1072-1078) comprising genes for two almost identical acetaldehyde dehydrogenases, three genes for highly similar ethanol dehydrogenases, and two genes for microcompartment proteins, which are orthologs of ethanolamine using genes (eutML) of Salmonella typhimurium (SI Table 2).
From cell extracts of C. kluyveri a macromolecular complex of ethanol dehydrogenase and acetaldehyde dehydrogenase can be purified by differential manganese sulfate precipitation (21, 22), indicating that the two enzymes are located in a microcompartment similar to a carboxysome or the microcompartment harboring ethanolamine mutase plus acetaldehyde dehydrogenase in Salmonella (23). We purified the Adh/Ald complex as described above and found by mass spectroscopic analysis that all three Adh isoenzymes were present, indicating a simultaneous expression of the three adh genes in C. kluyveri (data not shown). For the ald genes this could not be determined because the two Ald proteins have identical sequences. The ethanol dehydrogenases in the microcompartment show ≈40% sequence identity to Fe-ethanol dehydrogenase from Zymomonas mobilis. The genome encodes five additional Fe-alcohol dehydrogenases, which are highly similar to each other. These could be involved in the utilization of additional substrates. For instance, C. kluyveri can grow on propanol and acetate forming propionate, butyrate, valerate, and caproate.
Enzymes Involved in Butyrate Formation.
Butyrate formation from acetyl-CoA proceeds via acetoacetyl-CoA, 3-hydroxybutyryl-CoA, crotonyl-CoA, and butyryl-CoA as intermediates. Enzymes involved are acetoacetyl-CoA thiolase (Thl), NAD- and NADP-dependent 3-hydroxybutyryl-CoA dehydrogenase (Hbd), 3-hydroxybutyryl-CoA dehydratase (Crt), and NAD-dependent butyryl-CoA dehydrogenase complex (Bcd/EtfAB). Two bcd genes can be found in the genome; one is clustered with crt, hbd, and two genes encoding electron transport proteins (etfAB). A second set of the genes crt, hbd, bcd, and etfAB is present in C. kluyveri, albeit not clustered. These CDS could have a function in caproyl-CoA formation. Butyryl- and caproyl-CoA formed by acetyl-CoA reduction react with acetate yielding fatty acids and acetyl-CoA in a reaction catalyzed by butyryl-CoA: acetate CoA transferase (Cat3), which has a broad substrate specificity (8, 10).
C. kluyveri incorporates selenium as selenomethionine (Se-Met) into its acetoacetyl-CoA thiolase (Thl) when grown in media containing normal sulfur-to-selenium ratios (24, 25). The genome sequence does not provide per se clues to understand the occurrence of Se-Met in Thl, whose gene has two highly similar paralogs encoded in direct vicinity (CKL_3696-3698). Incorporation of Se-Met is apparently not accomplished through a pathway employing a specific tRNA as in the biosynthesis of most selenocysteine-containing proteins. The sequence of the methionyl-tRNA synthetase (MetG, CKL_3766) exhibits no special features. Se-Met seems to be formed in the methionine biosynthesis pathway as a result of the interchangeability of sulfur and selenium. In this respect, an unusual feature of the methionine metabolism in C. kluyveri is the presence of three paralogous genes for S-adenosylmethionine synthetases (MetK: CKL_1633, CKL_2650, and CKL_3674). The elevated incorporation of selenium into Thl from selenite might be linked to the observation that C. kluyveri has an extremely active sulfur metabolism (24). Indeed, the clustered genes for sulfate adenylyltransferase (CKL_1795/1796), adenylylsulfate reductase (CKL_1797/1798), adenylylsulfate kinase (CKL_1799), a sulfate transport system (CKL_1800-1803), and a predicted sulfite reductase (CKL_1807) are present in the genome of C. kluyveri but absent from most other clostridial genomes.
Ferredoxin-Dependent Hydrogenase.
In the ethanol-acetate fermentation H2 is generated via a ferredoxin-dependent hydrogenase. In the genome CDS for ferredoxins and [FeFe]-hydrogenases were found. Ferredoxin (CKL_3790) from C. kluyveri is a 2× [4Fe-4S] iron sulfur protein exhibiting >60% sequence identity to the well characterized ferredoxin from Clostridium pasteurianum (26), which has a redox potential of −400 mV (27). The [FeFe]-hydrogenase from C. kluyveri (HydA) has a molecular mass of 64 kDa and shows high sequence similarity to the [FeFe]-hydrogenase 1 characterized from C. pasteurianum (28). The genome harbors additional CDS for a [FeFe]-hydrogenase (CKL_0841) and three ferredoxins (CKL_3758, CKL_0428, and CKL_3060). In addition, all three CDS for hydrogenase maturation, HydE, HydG, and HydF, were found. Interesting in this respect is the finding that C. kluyveri possesses a CDS predicted to encode a nickel CO dehydrogenase (CKL_2148). This enzyme has been demonstrated to be present in C. pasteurianum (29). Because there are four CO bound to iron in the dinuclear iron center of [FeFe]-hydrogenases, it is tempting to speculate that the nickel enzyme is involved in either the synthesis or degradation of the [FeFe]-hydrogenase.
Energetic Coupling.
A detailed analysis of the ethanol-acetate fermentation stoichiometry (reaction 1) revealed that the fermentation can be described by the coupled reactions 2–4 (5, 6).
 |
 |
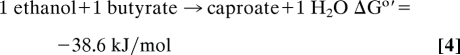 |
ATP is generated by substrate-level phosphorylation in reaction 2 (1 ATP/H+), which is highly endergonic when coupled with ADP phosphorylation (ΔG°′ = +32 kJ/mol) and therefore must be coupled to the exergonic reactions 3 and 4. The latter must also be coupled with energy conservation as deduced from the growth yields and the free energy change associated with reaction 1. How these couplings are achieved has remained a mystery, as has the question of how H2 at 105 Pa is formed from NADH regenerated during ethanol oxidation to acetyl-CoA. The steady-state intracellular NADH/NAD ratio in C. kluyveri was found at 0.3, and the redox potential E′ of the NAD/NADH couple (E°′ = −320 mV) was found to be near −300 mV and thus 120 mV more positive than E°′ = −414 mV of the H+/H2 couple (13). Molecular hydrogen formation from NADH must therefore be energy-driven, and the energy must be derived from reactions 3 and 4, most likely from the reduction of crotonyl-CoA (hexenoyl-CoA) to butyryl-CoA (caproyl-CoA) (E°′ = −10 mV) (30) with NADH (E°′ = −320 mV), which is strongly exergonic.
Thus, we propose that the reduction of ferredoxin with NADH is according to reaction 5 catalyzed by the cytoplasmic butyryl-CoA dehydrogenase complex (Bcd/EtfAB) (refs. 31 and 32; see ref. 33 for a more detailed account of this hypothesis). Reaction 5 principally resembles the coenzyme Q cycle in that twice the amount of electrons required for substrate reduction is fed into the cycle in which the flow of electrons is divided to react both with a high potential acceptor (crotonyl-CoA) and a low potential acceptor (ferredoxin). The reduced ferredoxin generated in reaction 5 is then used to regenerate NADH in reaction 6 catalyzed via a membrane-associated, energy-converting NADH: ferredoxin oxidoreductase (RnfA-G) (34, 35).
 |
It is the combination of these reactions that allows explaining H2 production by C. kluyveri and the generation of a proton motive force. The Bcd/EtfAB complex from C. kluyveri has been purified from the soluble cell fraction and was shown to catalyze reaction 5 (36). Both the butyryl-CoA dehydrogenase (Bcd) (37) and the electrontransfer proteins (EtfAB) contain FAD (38, 39). Also, the presence of the RnfA-G complex in the membrane fraction from C. kluyveri could be demonstrated. The complex was purified by us and shown to catalyze reaction 6. The mass fingerprint patterns of the subunits were found to match those predicted from the rnf CDS (data not shown). The function of RnfA-G as NADH:ferredoxin oxidoreductase was first included in a model of the energy metabolism of C. tetani (40). Purification of this membrane enzyme verified this hypothesis (E. Jayamani, C. D. Boiangiu, and W.B., unpublished observations).
The metabolic pathway shown in Fig. 2 predicts that H2 formation from ethanol depends on butyrate formation from ethanol and acetate and should be sensitive to protonophores. Indeed, it was found that in cell suspensions of C. kluyveri the formation of H2 was inhibited by low concentrations of the protonophore 3,3′,4′,5-tetrachlorosalicylanilide (TCS) (Fig. 3A). This inhibition was only slightly affected by dicyclohexylcarbodiimide (DCCD), which interferes with F1Fo-ATPase activity. Fatty acid formation was also inhibited by TCS, but this inhibition was relieved by DCCD (Fig. 3B) (41). The explanation is as follows: Addition of TCS uncouples the Rnf-catalyzed reaction (Fig. 2 Inset), resulting in increased NAD reduction. Thus, reduced ferredoxin becomes less available for H2 production. In addition, the ATP pool collapses, as does the acetyl-CoA pool via acetate kinase (Ack) and phosphotransacetylase (Pta). Consequently, buytrate and caproate syntheses are slowed down and finally stopped. The addition of DCCD prevents ATP hydrolysis and indirectly also acetyl-CoA hydrolysis; fatty acids are produced, but H2 production, because the Rnf-catalyzed reaction remains uncoupled, is inhibited.
C. kluyveri is therefore a model organism for those numerous anaerobes that oxidize ethanol and couple this oxidation with the reduction of unsaturated acids or other high potential electron acceptors. The operation of the Bcd/Etf cycle or of an analogous cycle allows the generation of reduced ferredoxin from NADH. This then may give rise to H2 production or to the generation of a proton motive force via RnfA-G or analogous complexes.
Crotonate Fermentation.
Crotonate disproportionation to acetate and butyrate involves crotonyl-CoA, 3-hydroxybutyryl-CoA, 3-ketobutyryl-CoA, acetyl-CoA, and acetyl-phosphate as intermediates in the oxidative part and crotonyl-CoA and butyryl-CoA in the reductive part. Enzymes predicted to be required are acyl-CoA: acetate CoA transferase, 3-hydroxybutyryl-CoA dehydratase, 3-hydroxybutyryl-CoA dehydrogenase, 3-ketothiolase, phosphotransacetylase, acetate kinase (oxidative part), and the butyryl-CoA dehydrogenase complex (reductive part), for all of which CDS have been found (SI Table 2).
Ethanol-Succinate Fermentation.
Ethanol-succinate fermentation to acetate and butyrate involves succinyl-CoA, succinate semialdehyde, 4-hydroxybutyrate, and 4-hydroxybutyryl-CoA as intermediates, which are not involved in ethanol-acetate or crotonate fermentations. Two specific CoA transferases (Cat1 and Cat2), NAD-dependent succinate semialdehyde dehydrogenase (SucD), NAD-dependent 4-hydroxybutyrate dehydrogenase (4Hbd), and 4-hydroxybutyryl-CoA dehydratase/isomerase (AbfD) have been characterized (42). The genes for these enzymes are clustered (SI Table 2), and their expression is induced by succinate (43, 44).
Enoate Reduction.
It was shown that cell extracts of C. kluyveri catalyze the stereospecific reduction of a wide variety of α,β-unsaturated fatty acids with H2. In the genome of C. kluyveri nine CDS for enoate reductases are found, only one of which (CKL__0743) has been characterized (45, 46). The physiological function of the enoate reductases is not known with certainty. A hypothesis is that in some ecological niches enoates such as cinnamate (47) could be present that could serve as terminal electron acceptor for the oxidation of ethanol to acetate, thus allowing the synthesis of ATP via substrate level phosphorylation.
Glycerol Utilization.
A surprising finding is the presence of a large cluster of genes predicted to be involved in glycerol dehydration to 3-hydroxypropionaldehyde including CDS for microcompartment proteins. The genome also contains two CDS for propanediol dehydrogenases (CKL_2405 and CKL_3456). The reduction of glycerol to propanediol with ethanol is strongly exergonic. It is coupled with the synthesis of 1 ATP by substrate-level phosphorylation. However, all attempts to grow C. kluyveri DSM555, the strain whose genome was sequenced, on ethanol and glycerol failed. The reason is that the gene for the subunit B of the coenzyme B12-dependent glycerol dehydratase complex is missing. In this respect it is of interest that glycerol dehydratase activity was detected in C. pasteurianum and Clostridium butyricum (48). All genes for the synthesis of coenzyme B12, the cofactor of glycerol dehydratase, can be found in the genome of C. kluyveri. This important coenzyme was first discovered in Clostridium tetanomorphum by Barker et al. (49).
Genes for Polyketide Synthases/Nonribosomal Peptide Synthetases.
Unexpectedly, because normally not present in anaerobic bacteria, the genome of C. kluyveri was found to contain four clusters of CDS predicted to code for enzymes involved in the nonribosomal synthesis of peptides and mixed polyketides/peptides (Fig. 4). Clusters 2, 3, and 4 comprising CKL_1523-1527, CKL_1730-1735, and CKL_2354-2357, respectively, are predicted to encode polyketide synthase–nonribosomal peptide synthetase multienzyme hybrid complexes, the products of which are presently unpredictable in structure and function. In contrast, cluster 1, comprising CKL_1503-1511, contains components of a nonribosomal peptide synthetase assembly line that is predicted to be involved in the synthesis of an aryl-capped siderophore containing salicylate and four thiazoli(di)ne heterocycles derived from cysteine. The stand-alone aryl acid adenylation domain (CKL_1504) is a typical feature of aryl-capped siderophore assembly lines (50). Furthermore, CKL_1505 and CKL_1511 show high sequence similarities to HMWP2 and PchF involved in the synthesis of the aryl-capped siderophores yersiniabactin and pyochelin in Yersinia and Pseudomonas, respectively (51, 52).
Yersiniabactin and Pyochelin Have a Function in Fe(III) Transport into the Cells Under Oxic Conditions.
Under anoxic conditions Fe(III) is generally reduced to Fe(II) outside the cell and then transported into the cell. It was therefore a surprise to find a gene cluster for the synthesis of a yersiniabactin/pyochelin-like siderophore in C. kluyveri, which is known to grow only under strictly anoxic conditions. A function of the putative siderophore in Fe(III) transport is supported by the finding that the genome harbors a gene cluster predicted to encode a siderophore transporter (CKL_1512-1513). Based on this information, we tested whether C. kluyveri synthesizes siderophores. Therefore, cells were grown in the presence of various iron concentrations. A siderophore activity assay demonstrated the presence of at least one efficient ferric iron chelator in the supernatant of the cell culture grown without iron supplementation (SI Fig. 6).
Materials and Methods
Sequencing Strategy.
A Sanger/pyrosequencing hybrid approach was used for whole-genome sequencing of C. kluyveri DSM555. Total genomic DNA of C. kluyveri was extracted by using Master Pure DNA Purification Kit (Epicentre Biotechnologies) and nebulized for Sanger sequencing approach. A shotgun library was constructed by using 1.5- to 4-kb size fractions. The fragments were cloned into vector pCR4-TOPO (Invitrogen). Insert ends of the recombinant plasmids were sequenced by using dye terminator chemistry with MegaBACE 1000 and 4000 (GE Healthcare) and ABI Prism 3730XL (Applied Biosystems) automated DNA sequencers. Sequences were processed with Phred and assembled into contigs by using the Phrap assembly tool (www.phrap.org). Second total genomic DNA of C. kluyveri was pyrosequenced by using a fraction of nebulized chromosomal DNA and a Roche GS20 sequencer (MWG Biotech). The 454 reads were assembled into 8,095 contigs with an average length of 433 bp and a maximum length of 7,000 bp by using the Newbler Assembler (454; Life Science). Sequence editing of shotgun sequences and 454 sequences was done by using GAP4 as part of the Staden software package (53); 6.8-fold coverage was obtained after assembly of 76,219 sequences of the 454 and Sanger sequencing approach. To solve problems with misassembled regions caused by repetitive sequences and to close remaining sequence gaps, PCR, combinatorial multiplex PCR, and primer walking with recombinant plasmids were used.
Gene Prediction and Annotation.
The initial gene prediction was accomplished by using YACOP (54). The output was verified and edited manually by using criteria such as the presence of a ribosome binding site, GC frame plot analysis, and similarity to known protein-encoding sequences (CDS). Annotation was done by using the ERGO tool from Integrated Genomics with a two-step approach. Initially, all proteins were screened against Swiss-Prot data and publicly available protein sequences from other completed clostridial genomes by using FASTA3. All predictions were verified and modified manually by comparing the protein sequences with the Pfam, GenBank, ProDom, COG, and Prosite public databases. All coding sequences were searched for similarities to protein families and domains using CD-Search (55). TMpred was used to predict transmembrane helices within the CDS (www.ch.embnet.org/software/TMPRED_form.html). The SIGI tool (56) was used for score-based identification of genomic islands, referred to as alien CDS.
Effects of Protonophore and F1Fo-ATPase Inhibitor on H2 and Fatty Acid Formation by Cell Suspensions of C. kluyveri.
Cells of C. kluyveri were harvested in the exponential growth phase under strictly anoxic conditions, washed, and resuspended in 50 mM Mops/KOH (pH 7), 5 mM MgCl2, and 0.1 mM Ti(III)citrate. The protonophore TCS and the F1Fo-ATPase inhibitor DCCD were used. The 10-ml assay mixture in 120-ml serum bottles contained 50 mM Mops/KOH (pH 7), 5 mM MgCl2, 0.1 mM Ti(III)citrate, 50 mM ethanol, and 10 mM sodium acetate. After the addition of 4.5 mg of cells (dry mass), TCS (2.2 nmol/mg of cells), and DCCD (5.6 nmol/mg of cells) the cell suspension was incubated at 0°C for 10 min; subsequently, the reaction was started by increasing the temperature to 37°C. DCCD and TCS were added in ethanolic solutions (10 mM and 2 mM, respectively). At the time indicated gas and liquid samples were taken. Hydrogen was analyzed with a Varian Aerograph Series 1400, whereas buyrate and caproate production were determined with a Carlo Erba GC 6000 (41).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Kathleen Gollnow, Jörg Kahnt, Katharina Nau, Frauke Meyer, and Stefanie Offschanka for technical assistance. We thank the Max-Planck-Gesellschaft, the Deutsche Forschungsgemeinschaft, the Niedersächsisches Ministerium für Wissenschaft und Kultur, the Fonds der Chemischen Industrie, and the Bundesministerium für Forschung und Bildung for generous support.
Footnotes
The authors declare no conflict of interest.
Data deposition: The C. kluyveri genome sequence has been deposited in the GenBank database [accession nos. CP000673 (chromosome) and CP000674 (plasmid)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0711093105/DC1.
References
- 1.Barker HA. The production of caproic and butyric acids by the methane fermentation of ethyl alcohol. Arch Mikrobiol. 1937;8:415–421. [Google Scholar]
- 2.Bornstein BT, Barker HA. The nutrition of Clostridium kluyveri. J Bacteriol. 1948;55:223–230. doi: 10.1128/jb.55.2.223-230.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker HA, Taha SM. Clostridium kluyverii, an organism concerned in the formation of caproic acid from ethyl alcohol. J Bacteriol. 1942;43:347–363. doi: 10.1128/jb.43.3.347-363.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thauer RK, Jungermann K, Wenning J, Decker K. Characterization of crotonate grown Clostridium kluyveri by its assimilatory metabolism. Arch Mikrobiol. 1968;64:125–129. doi: 10.1007/BF00406971. [DOI] [PubMed] [Google Scholar]
- 5.Thauer RK, Jungermann K, Henninger H, Wenning J, Decker K. The energy metabolism of Clostridium kluyveri. Eur J Biochem. 1968;4:173–180. doi: 10.1111/j.1432-1033.1968.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 6.Schoberth S, Gottschalk G. Considerations on the energy metabolism of Clostridium kluyveri. Arch Mikrobiol. 1969;65:318–328. doi: 10.1007/BF00412211. [DOI] [PubMed] [Google Scholar]
- 7.Bryant MP, Wolin EA, Wolin MJ, Wolfe RS. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch Mikrobiol. 1967;59:20–31. doi: 10.1007/BF00406313. [DOI] [PubMed] [Google Scholar]
- 8.Stadtman ER. The coenzyme A transphorase system in Clostridium kluyveri. J Biol Chem. 1953;203:501–512. [PubMed] [Google Scholar]
- 9.Barker HA. Bacterial Fermentations. New York: Wiley; 1956. pp. 28–56. [Google Scholar]
- 10.Stadtman ER. Functional group of coenzyme A, its metabolic relations, especially in the fatty acid cycle: Discussion. Fed Proc. 1953;12:692–693. [PubMed] [Google Scholar]
- 11.Wolin MJ, Miller TL. Interspecies hydrogen transfer: 15 years later. Am Soc Microbiol News. 1982;48:561–565. [Google Scholar]
- 12.Kenealy WR, Waselefsky DM. Studies on the substrate range of Clostridium kluyveri: The use of propanol and succinate. Arch Microbiol. 1985;141:187–194. [Google Scholar]
- 13.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bader J, Simon H. The activities of hydrogenase and enoate reductase in two Clostridium species, their interrelationship and dependence on growth conditions. Arch Microbiol. 1980;127:279–287. doi: 10.1007/BF00427205. [DOI] [PubMed] [Google Scholar]
- 15.Gottschalk G, Barker HA. Synthesis of glutamate and citrate by Clostridium kluyveri: An new type of citrate synthase. Biochemistry. 1966;5:1125–1133. doi: 10.1021/bi00868a003. [DOI] [PubMed] [Google Scholar]
- 16.Li F, Hagemeier CH, Seedorf H, Gottschalk G, Thauer RK. Re-citrate synthase from Clostridium kluyveri is phylogenetically related to homocitrate synthase and isopropylmalate synthase rather than to Si-citrate synthase. J Bacteriol. 2007;189:4299–4304. doi: 10.1128/JB.00198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thauer RK, Rupprecht E, Jungermann K. The synthesis of one-carbon units from CO(2) via a new ferredoxin dependent monocarboxylic acid cycle. FEBS Lett. 1970;8:304–307. doi: 10.1016/0014-5793(70)80293-9. [DOI] [PubMed] [Google Scholar]
- 18.Waack S, et al. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics. 2006;7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sebaihia M, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 20.Kanamori K, Weiss RL, Roberts JD. Ammonia assimilation pathways in nitrogen-fixing Clostridium kluyverii and Clostridium butyricum. J Bacteriol. 1989;171:2148–2154. doi: 10.1128/jb.171.4.2148-2154.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillmer P, Gottschalk G. Particulate nature of enzymes involved in the fermentation of ethanol and acetate by Clostridium kluyveri. FEBS Lett. 1972;21:351–354. doi: 10.1016/0014-5793(72)80200-x. [DOI] [PubMed] [Google Scholar]
- 22.Lurz R, Mayer F, Gottschalk G. Electron microscopic study on the quaternary structure of the isolated particulate alcohol-acetaldehyde dehydrogenase complex and on its identity with the polygonal bodies of Clostridium kuyveri. Arch Microbiol. 1979;120:255–262. [Google Scholar]
- 23.Penrod JT, Roth JR. Conserving a volatile metabolite: A role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sliwkowski MX, Stadtman TC. Incorporation and distribution of selenium into thiolase from Clostridium kluyveri. J Biol Chem. 1985;260:3140–3144. [PubMed] [Google Scholar]
- 25.Hartmanis MGN, Stadtman TC. Isolation of a selenium-containing thiolase from Clostridium kluyveri: Identification of the selenium moiety as selenomethionine. Proc Natl Acad Sci USA. 1982;79:4912–4916. doi: 10.1073/pnas.79.16.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertini I, Donaire A, Luchinat C, Rosato A. Paramagnetic relaxation as a tool for solution structure determination: Clostridium pasteurianum ferredoxin as an example. Proteins. 1997;29:348–358. [PubMed] [Google Scholar]
- 27.Smith ET, et al. A totally synthetic histidine-2 ferredoxin: Thermal stability and redox properties. Biochemistry. 1991;30:11669–11676. doi: 10.1021/bi00114a009. [DOI] [PubMed] [Google Scholar]
- 28.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 29.Diekert GB, Graf EG, Thauer RK. Nickel requirement for carbon-monoxide dehydrogenase formation in Clostridium pasteurianum. Arch Microbiol. 1979;122:117–120. [Google Scholar]
- 30.Crane FL, Mii S, Hauge JG, Green DE, Beinert H. On the mechanism of dehydrogenation of fatty acyl derivatives of coenzyme A. I. The general fatty acyl coenzyme A dehydrogenase. J Biol Chem. 1956;218:701–706. [PubMed] [Google Scholar]
- 31.Djordjevic S, Pace CP, Stankovich MT, Kim JJ. Three-dimensional structure of butyryl-CoA dehydrogenase from Megasphaera elsdenii. Biochemistry. 1995;34:2163–2171. doi: 10.1021/bi00007a009. [DOI] [PubMed] [Google Scholar]
- 32.Hetzel M, et al. Acryloyl-CoA reductase from Clostridium propionicum: An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur J Biochem. 2003;270:902–910. doi: 10.1046/j.1432-1033.2003.03450.x. [DOI] [PubMed] [Google Scholar]
- 33.Herrmann G, Jayamani E, Mai G, Buckel W. (November 26, 2007) Energy conservation via electron transferring flavoprotein (Etf) in anaerobic bacteria. J Bacteriol. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmehl M, et al. Identification of a new class of nitrogen-fixation genes in Rhodobacter capsulatus: A putative membrane complex involved in electron-transport to nitrogenase. Mol Gen Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 35.Boiangiu CD, et al. Sodium ion pumps and hydrogen production in glutamate fermenting anaerobic bacteria. J Mol Microbiol Biotechnol. 2005;10:105–119. doi: 10.1159/000091558. [DOI] [PubMed] [Google Scholar]
- 36.Li F, et al. (November 9, 2007) Coupled ferredoxin- and crotonyl-CoA reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JJP, Miura R. Acyl-CoA dehydrogenases and acyl-CoA oxidases: Structural basis for mechanistic similarities and differences. Eur J Biochem. 2004;271:483–493. doi: 10.1046/j.1432-1033.2003.03948.x. [DOI] [PubMed] [Google Scholar]
- 38.Sato K, Nishina Y, Shiga K. Purification of electron-transferring flavoprotein from Megasphaera elsdenii and binding of additional FAD with an unusual absorption spectrum. J Biochem (Tokyo) 2003;134:719–729. doi: 10.1093/jb/mvg199. [DOI] [PubMed] [Google Scholar]
- 39.Sato K, Nishina Y, Shiga K, Tanaka F. Hydrogen-bonding dynamics of free flavins in benzene and FAD in electron-transferring flavoprotein upon excitation. J Photochem Photobiol B. 2003;70:67–73. doi: 10.1016/s1011-1344(03)00056-3. [DOI] [PubMed] [Google Scholar]
- 40.Brüggemann H, et al. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc Natl Acad Sci USA. 2003;100:1316–1321. doi: 10.1073/pnas.0335853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeiff B. Marburg, Germany: Philipps University; 1992. MS thesis. [Google Scholar]
- 42.Scherf U, Söhling B, Gottschalk G, Linder D, Buckel W. Succinate-ethanol fermentation in Clostridium kluyveri: Purification and characterisation of 4-hydroxybutyryl-CoA dehydratase/vinylacetyl-CoA delta 3-delta 2-isomerase. Arch Microbiol. 1994;161:239–245. doi: 10.1007/BF00248699. [DOI] [PubMed] [Google Scholar]
- 43.Söhling B, Gottschalk G. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J Bacteriol. 1996;178:871–880. doi: 10.1128/jb.178.3.871-880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerhardt A, Cinkaya I, Linder D, Huisman G, Buckel W. Fermentation of 4-aminobutyrate by Clostridium aminobutyricum: Cloning of two genes involved in the formation and dehydration of 4-hydroxybutyryl-CoA. Arch Microbiol. 2000;174:189–199. doi: 10.1007/s002030000195. [DOI] [PubMed] [Google Scholar]
- 45.Tischer W, Bader J, Simon H. Purification and some properties of a hitherto-unknown enzyme reducing the carbon-carbon double bond of alpha, beta-unsaturated carboxylate anions. Eur J Biochem. 1979;97:103–112. doi: 10.1111/j.1432-1033.1979.tb13090.x. [DOI] [PubMed] [Google Scholar]
- 46.Rohdich F, Wiese A, Feicht R, Simon H, Bacher A. Enoate reductases of Clostridia: Cloning, sequencing, and expression. J Biol Chem. 2001;276:5779–5787. doi: 10.1074/jbc.M008656200. [DOI] [PubMed] [Google Scholar]
- 47.Dickert S, Pierik AJ, Linder D, Buckel W. The involvement of coenzyme A esters in the dehydration of (R)-phenyllactate to (E)-cinnamate by Clostridium sporogenes. Eur J Biochem. 2000;267:3874–3884. doi: 10.1046/j.1432-1327.2000.01427.x. [DOI] [PubMed] [Google Scholar]
- 48.Macis L, Daniel R, Gottschalk G. Properties and sequence of the coenzyme B12-dependent glycerol dehydratase of Clostridium pasteurianum. FEMS Microbiol Lett. 1998;164:21–28. doi: 10.1111/j.1574-6968.1998.tb13062.x. [DOI] [PubMed] [Google Scholar]
- 49.Barker HA, Weissbach H, Smyth RD. A coenzyme containing pseudovitamin B12. Proc Natl Acad Sci USA. 1958;44:1093–1097. doi: 10.1073/pnas.44.11.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quadri LE. Assembly of aryl-capped siderophores by modular peptide synthetases and polyketide synthases. Mol Microbiol. 2000;37:1–12. doi: 10.1046/j.1365-2958.2000.01941.x. [DOI] [PubMed] [Google Scholar]
- 51.Quadri LE, Keating TA, Patel HM, Walsh CT. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: In vitro reconstitution of aryl-4, 2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry. 1999;38:14941–14954. doi: 10.1021/bi991787c. [DOI] [PubMed] [Google Scholar]
- 52.Gehring AM, Mori I, Perry RD, Walsh CT. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry. 1998;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 53.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 54.Tech M, Merkl R. YACOP: Enhanced gene prediction obtained by a combination of existing methods. In Silico Biol. 2003;3:441–451. [PubMed] [Google Scholar]
- 55.Marchler-Bauer A, Bryant SH. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merkl R. SIGI: Score-based identification of genomic islands. BMC Bioinformatics. 2004;5:22. doi: 10.1186/1471-2105-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.