Abstract
Vaccinia virus is the prototypic orthopoxvirus and was the vaccine used to eradicate smallpox, yet the expression profiles of many of its genes remain unknown. Using a genome tiling array approach, we simultaneously measured the expression levels of all 223 annotated vaccinia virus genes during infection and determined their kinetics. For 95% of these genes, significant transcript levels were detected. Most remarkably, classification of the genes by their expression profiles revealed 35 genes exhibiting immediate-early expression. Although a similar kinetic class has been described for other virus families, to our knowledge, this is the first demonstration of its existence in orthopoxviruses. Despite expression levels higher than for genes in the other three kinetic classes, the functions of more than half of these remain unknown. Additionally, genes within each kinetic class were spatially grouped together in the genome. This genome-wide picture of transcription alters our understanding of how orthopoxviruses regulate gene expression.
Keywords: gene transcription, genome tiling array, microarray, vaccinia virus
The orthopoxviruses are a closely related group that share high sequence similarity and many aspects of basic biology (1). This group includes two significant human pathogens, namely, variola virus, the causative agent of smallpox, and monkeypox virus, which is the current cause of emerging disease in central and West Africa. Vaccinia virus (VACV) is the prototypic orthopoxvirus and was the live vaccine used to eradicate smallpox. VACV is now in use as a vector for recombinant vaccines, some of which are in clinical trials.
Like other orthopoxviruses, VACV has a linear, double-stranded DNA genome that is nearly 200 kb long. The most common laboratory strain of VACV, Western Reserve (VACWR), is predicted to encode 223 ORFs. This number includes 12 ORFs (VACWR-001 through VACWR-012 and VACWR-218 through VACWR-207) that are repeated at each end of the genome, leaving a total of 211 unique ORFs.
VACV has a broad cellular tropism in vitro and potential host range in vivo, but there is no clearly identified animal reservoir for the virus in nature. As a result of an exclusively cytoplasmic life cycle, VACV encode its own enzymes and proteins required for gene transcription, genome replication, virion production, and morphogenesis and, for the most part, does not depend on host cell proteins for these processes. In addition, VACV infection induces a rapid and massive shutdown of host gene expression that acts at several levels (1–4).
Expression kinetics have been described for a variety of VACV genes. These studies, and work done to define promoters and transcription complexes, have led to the definition of four temporal gene classes and three distinct promoter types. The promoters have been named early, intermediate, and late, with each promoter associated with one gene class (5). In addition, some genes have elements of early and late promoters in their upstream region, giving rise to a fourth class referred to as early/late.
The virion contains early transcription complexes bound to DNA in position to promote transcription of early genes upon entry into host cells (6, 7). Despite analogy with other large, double-stranded DNA viruses, such as herpes viruses, and some use of the term in the literature (8), an immediate-early class of genes has not come to be included in the established model of poxvirus gene expression (5, 9, 10).
In terms of broad functions, besides proteins required for expression of the subsequent gene classes, early genes tend to encode virulence factors for modulation of host responses (11–14). Proteins encoded by late genes are generally required for virion morphogenesis and structure or early gene transcription factors packaged in newly made viral cores (15–17).
Although there is a wealth of information in the literature regarding VACV gene expression, this knowledge is highly fragmented with respect to experimental settings and methodologies used, and substantial gaps exist. Experimental data showing expression kinetics are only available for approximately two-thirds of the annotated ORFs, leaving predictions based on canonical promoter motifs as the only guide for 46 ORFs (www.poxvirus.org). For 23 ORFs lacking prototypic promoter sequences, even predictions are not possible.
Although microarrays have been used to show viral gene expression (18, 19), those arrays did not allow resolution between the ORFs and the untranslated regions. Tiling arrays, however, provide a comprehensive and unbiased sampling of transcriptional activity by using overlapping probes covering the entire region of interest (20). In this study, we simultaneously measured transcription of the entire VACV genome during infection and derived a complete picture of gene expression. The close relationship between the orthopoxviruses makes it reasonable to assume that the findings apply across the group (10).
Results and Discussion
The Genome Tiling Array Allows for Simultaneous Measurements of VACV Gene Transcription Throughout Infection.
We aimed to determine the temporal transcription profile of all 223 annotated VACV ORFs during infection. HeLa cells were chosen as hosts because they support productive infection and are commonly used for growing VACV stocks (21). Cells were infected at high density and with a high multiplicity (MOI = 10) of VACWR to give synchronous infection. A tiling array containing 25-mer DNA probes interrogated both strands of the VACV genome at a 4-nt resolution. To facilitate the comparison of probe signal intensity among the different arrays, background subtraction and normalization steps were required (see Materials and Methods).
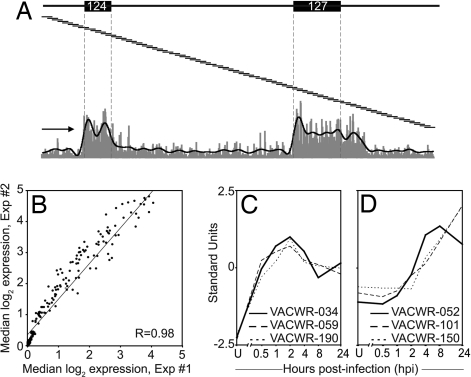
An overview of the tiling array technique is presented in Fig. 1A. It shows probe signal intensity from a sample taken 2 h postinfection (hpi) covering ≈7 kb of the forward strand including VACWR-124 and -127. The signal intensities of probes lying completely within an ORF are pooled to calculate the median signal intensity and used for significance testing. Visual examination of the data provided confidence in the power of the method to resolve adjacent ORFs and also revealed unexpected complexities in VACV transcription. For example, a stretch of high signal continues well past the end of VACWR-027, indicating that this early transcript has an uncharacteristically long 3′ untranslated region (Fig. 1A).
Fig. 1.
Validation of the genome tiling array technology. (A) Shown is an overview of the genome tiling array technique including probe signal intensity of a sample from 2 hpi, covering ≈7 kb of the forward strand. ORFs VACWR-124 and VACWR-127 are indicated. The 25-mer probes (not to scale) spanning this region are represented as overlapping lines. A fluorescence signal is obtained from each individual probe (vertical gray bars at the bottom) and all probes lying completely within an ORF (bounded by vertical dashed lines) are pooled. The black line is a result of smoothing the signal over 25 probes. The arrow indicates transcriptional orientation. (B) RNA was prepared in two different experiments at 2 hpi, and median probe signal intensities for each gene were compared. (C) Shown are relative expression levels (standard units) for a set of known early or (D) late genes over time course.
Resolving transcription of adjacent ORFs is likely to be most difficult at late times after infection because the inefficient termination of late genes may result in “run-on” transcription of downstream genes (22). Although our analysis alone does not discriminate between run-on and “true” transcription, the tiling array allows for comparisons of transcription associated with ORFs to that of intergenic regions. When comparing the signal intensities of adjacent ORFs and the intergenic regions at 8 hpi, we found that although some run-on transcription is apparent, the median signal intensity for the downstream ORF is generally higher than that of the upstream intergenic region, indicative of true transcription [supporting information (SI) Fig. 6A].
The reproducibility of the technique was assessed by using biological replicate RNA samples from 1, 2, 4, and 8 hpi. Pearson correlation coefficients (R) for median ORF expression levels were between 0.93 and 0.99. A representative scatter plot from two independent 2-hpi RNA samples is shown in Fig. 1B. In addition, analyzing RNA extracted from uninfected HeLa cells detected no VACV gene expression (SI Table 1), demonstrating that there was no significant cross-hybridization between the VACV probes and human RNA. Finally, we benchmarked our results against data from the literature for a set of genes with well established kinetics (three early and three late). To compare expression profiles between individual ORFs, the expression values were standardized. In concordance with their known expression profile, the expression of the three early genes (VACWR-034, -059, and -190) (23–25) was initiated at 0.5 hpi and peaked at 2 hpi (Fig. 1C). In contrast, expression of the three late genes (VACWR-052, -101, and -150) (26–28) was initiated at ≈2–4 hpi and peaked or was still rising at 8 or 24 hpi (Fig. 1D). Taken together, these results support our experimental approach and illustrate its high throughput, reproducibility, and general agreement with established methods.
The Vast Majority of Annotated VACV ORFs Are Expressed During the Viral Cycle.
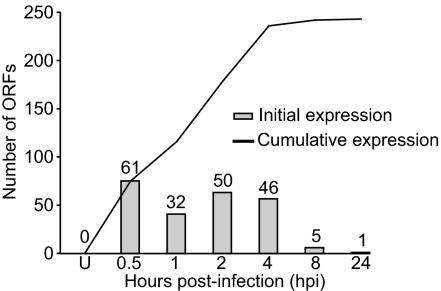
Median expression values and associated P values were determined for all annotated ORFs (SI Table 1). In terms of overall VACV transcription, within 30 min after infection 61 genes were transcribed, and by 1 hpi another 32 genes were transcribed (Fig. 2). By 24 hpi, 197 genes (93%) had been transcribed at some point during infection. Transcripts from 14 ORFs were never detected in this analysis. Seven of these were reported to be expressed, but 4 (VACWR-092, -097, -134, and -162) were detected by using other cell lines (29–31). The study demonstrating expression of VACWR-064 used a higher viral dose (MOI = 50) than what was used in our analysis. In fact, at 4 hpi there was border-line expression of VACWR-064 (P = 2.5 × 10−5) in our analysis, suggesting that the lack of expression might be due to a lower sensitivity in our assay. For VACWR-074 and VACWR-100 we have no explanation for the inconsistencies observed.
Fig. 2.
The vast majority of annotated ORFs for VACV are expressed during infection. Shown are the number of ORFs whose transcription is initiated (bars) in uninfected cells (U) and at 0.5–24 hpi and the cumulative number of genes whose expression was detected by each time point (line).
Cluster Analysis of Expression Profiles Revealed an Immediate-Early Gene Class.
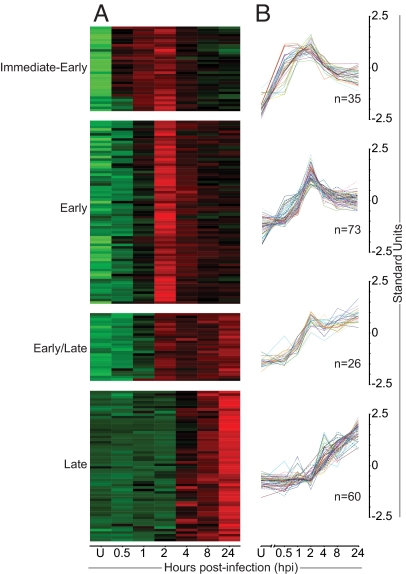
Having genome-wide information about VACV transcription, we wanted to assess whether these data would fit the existing model with early, intermediate, and late gene classes, with a fourth class resulting from combined early/late expression. To group the expression profiles in an unbiased manner, a hierarchical clustering analysis was performed based on relative expression levels. It was found that four clusters provided the most reproducible grouping of genes (data not shown). The resulting gene clusters are shown as heatmaps, going from green to red with increasing expression levels (Fig. 3A) and corresponding line plots (Fig. 3B).
Fig. 3.
Cluster analysis of expression profiles revealed an immediate-early class of genes. Genes were divided into four groups based on hierarchical clustering of kinetic expression data. (A) A heat-map representation of ORF expression is shown. Colors indicate the probe intensity level for each gene in uninfected cells (U) and at 0.5–24 hpi; green and red represent lower and higher expression, respectively. (B) Relative expression levels are shown throughout the same time course as in A.
The most striking finding of this analysis was the clear division of early gene expression into two classes. In keeping with nomenclature established for other virus families (32), we refer to these as immediate-early and early. The immediate-early class comprised 35 genes and was characterized by expression initiated by 0.5–1 hpi, peaking at 2 hpi before declining (Fig. 3B). The early class was the largest by number, containing 73 genes with the onset of transcription at 1–2 hpi and maximum level of transcription at 2 hpi. A late class of 60 genes was also identified generally expressed from 4 hpi, with levels increasing over time. Finally, an early/late class was found with 26 genes exhibiting onset of transcription typical of early genes but with sustained expression more similar to late genes. Three ORFs (VACWR-033, -063, and -086) did not fit well into any of the classes and were excluded from this part of the analysis.
No “immediate-early/late” gene class was identified, suggesting that immediate-early genes are distinct from the early genes. When early promoter sequences (either verified or putative) for the two classes were compared, only minor differences were seen in the consensus promoter sequences, primarily just upstream of the transcription start site. Thus, although only one type of early promoter has been identified, it is possible that minor changes in promoter sequences or other as-yet-unrecognized sequences allow transcription complexes to preferentially assemble on the promoters of immediate-early genes in virions. Another possibility is that hitherto-unrecognized enhancer elements exist for the immediate-early genes. This is not unreasonable because the alternative would be that the 134 genes with early promoter elements will be transcriptionally active upon initiation of infection, giving the virus relatively poor control over gene expression in the early stages of infection.
We would also like to point out that there is a difference between defining “functional” versus “kinetic” classes of viral genes. Early poxvirus genes are traditionally defined functionally because they are expressed before viral DNA replication. The term immediate-early used here is a kinetic description of preferential expression before the early genes, and no functional distinction has been made between these classes. Either way, our identification of an immediate-early class of genes forces a revision of the current paradigm of poxvirus gene expression.
A second surprise of the cluster analysis was the lack of an intermediate gene class. A few genes in the late class were expressed at maximum levels at 4 hpi, which may suggest that they are intermediate genes. However, even when the analysis was biased in favor of detecting an intermediate class, it was never identified with statistical significance. Genes reported to belong to an intermediate class or having intermediate promoters were either distributed among the immediate-early (VACWR-072), early (VACWR-077), and late (VACWR-119 and VACWR-120) classes, or did not fit well into any class (VACWR-086) (SI Table 2). Some genes with intermediate promoters also have additional promoter elements in their upstream regions (29, 33). The complexity introduced by promoter combinations might explain why they fail to form a distinct cluster. It is also possible that shorter time intervals would be required for distinction between intermediate and late genes (34).
As a test of the biological relevance of the cluster analysis, we examined whether the expression of genes in the late class depended on viral DNA replication. The ORF expression levels were determined at 8 hpi in the presence and absence of Cytosine beta-arabinofuranoside (AraC), a nucleoside analog commonly used to block DNA replication. Of the 60 late genes, the transcription of 52 were either completely (n = 33) or partially (reduced by ≥30%, n = 19) blocked upon AraC treatment (SI Table 2). In contrast, transcription was not inhibited by AraC for any of the 134 genes from the other classes (SI Fig. 7). These results confirm that late gene transcription by VACV depends, for the most part, on DNA replication and strengthen the validity of the clustering analysis.
Tiling Array Analysis Is Largely Unaffected by Run-On Transcription.
The current data processing does not distinguish between run-on and true transcription. Having assigned genes into kinetic classes, the impact of run-on transcription from upstream genes was assessed at 8 hpi. ORFs were divided into four groups based on the expression kinetics of their upstream neighboring ORF: Early ORFs located after early ORFs, early after late, late after early, and late after late. Median transcript levels for each ORF were compared with those of the 100-nt untranslated regions on either end (SI Fig. 6B). ORFs lacking flanking untranslated regions were not included. This showed that irrespective of the kinetic class of the upstream neighbor, ORF transcripts were, on average, much more abundant than transcripts from the flanking regions. Nevertheless, these analyses will be further refined to perform de novo transcript mapping of the virus. That will help to discover novel transcripts and more precisely determine the boundaries of transcription initiation and termination.
Functions of Genes in the Different Kinetic Classes.
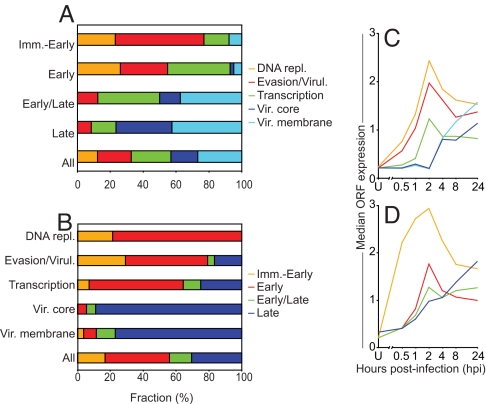
After the clustering of the genes by their expression profiles, a detailed analysis of gene functions within each kinetic class was performed. Each gene was assigned to one of five functional categories based on literature annotations and predicted functions (SI Table 3): DNA replication, immune evasion/virulence, transcription, virion core proteins, and virion membrane proteins (SI Table 2). Genes of unknown function and genes with functions outside of these categories were excluded. In addition, for ORFs that are fragments of a larger ancestor gene, only the ORF proximal to the promoter was placed into the corresponding category, and the downstream ORF(s) were denoted as “pseudo” and excluded. The analysis showed marked differences between genes in the two earliest classes, which were mainly involved in DNA replication, evasion/virulence, and transcription, and the early/late and late genes, which had a larger proportion of genes encoding virion core and membrane proteins (Fig. 4A). Strikingly, more than half of the immediate-early genes were of unknown function (SI Table 2), suggesting that the genes expressed first during infection are not well studied.
Fig. 4.
Function and expression levels of genes in the different classes. (A) Shown are the four gene classes with the fraction (%) of each class having a certain function indicated. (B) Shown are groups of genes associated with a specific function, with the fraction (%) of each group belonging to each kinetic class indicated. (C and D) Shown is median ORF expression at each time point for genes within each functional category (C) and kinetic class (D).
Among the earliest expressed were genes involved in DNA replication, including genes encoding a ssDNA-binding phosphoprotein and a thymidylate kinase (SI Table 2, Immediate-early genes). Genes encoding enzymes involved in replication, including the viral DNA polymerase itself, were in the early class. None of the genes involved in DNA replication were found in the early/late or late classes (Fig. 4B). The peak of expression of the early classes of genes at ≈2 hpi and the lack of genes required for replication in the late class are consistent with current knowledge of the orthopoxvirus replicative cycle.
VACV encodes a broad range of proteins that contribute to virulence in vivo, and some have established roles in the modulation of host defense (35–37). We found that at least 27 of these were expressed (SI Table 2). These genes were most frequently found in the two earliest classes (Fig. 4B) but were also present in the later classes. This implies that VACV continues to express immunomodulatory proteins throughout infection to provide a more favorable environment for viral growth in the face of the host's immune response.
Gene products important for RNA transcription were also represented in all temporal classes but were most abundant in the early class. In agreement with the literature, we found that the RNA polymerase subunits were expressed by genes of the early classes. To reconcile this observation with the presence of these proteins in virions, we looked at their expression levels later in infection and noted that although they peak at 2 hpi, levels remained relatively high at 8 hpi.
In general, genes coding for structural components of the virion core or membrane were largely expressed late, but there were exceptions. VACWR-156 (membrane phosphoglycoprotein) was immediate-early; VACWR-051 (involved in plaque and EEV formation) and VACWR-159 (transmembrane phosphoprotein) were early; and VACWR-181 (membrane glycoprotein) and VACWR-187 (EEV membrane glycoprotein) were early/late. Viral membrane proteins are often also expressed on the surface of infected cells (38). The broad expression time and cellular localization suggests that these proteins might play more than one role during infection. In fact, VACV proteins with dual functions are described in refs. 39 and 40.
Immediate-Early Genes Are More Highly Expressed than Genes of Other Classes.
To this point, our analysis has been based on relative expression levels. Next, we compared absolute median expression levels amongst the four temporal classes identified. The most striking result was that RNA levels for the immediate-early genes were significantly higher (P = 0.003–0.023) than for genes of the other three temporal classes throughout infection (Fig. 4D). Indeed, despite a dramatic decline in levels after the peak at 2 hpi, immediate-early transcript levels still exceeded those of late genes up to 24 hpi. These data further support the division of immediate-early and early genes. Looking at individual genes, there were several exhibiting expression levels clearly higher than the average in their class. Some examples are: immediate-early, VACWR-059 (double-stranded RNA-binding protein) and VACWR-184 (unknown); early, VACWR-018 (unknown); early/late, VACWR-131 (core protein); and late, VACWR-169 (unknown). Because of their exceptionally high expression levels, these genes might be of special interest for future investigations.
The signal intensities were also compared for genes possessing different functions. This showed that RNA levels were highest for genes involved in DNA replication, followed by those associated with evasion/virulence and transcription through 4 hpi (Fig. 4C). As expected, given their predominance in the late gene class, levels for virion core and membrane ORFs did not dominate until 24 hpi.
VACV Transcriptome: Filling Experimental Knowledge Gaps Regarding VACV Gene Transcription.
A comparison was made between the tiling array data and published data from the poxvirus database (SI Table 2). Of the 197 genes transcribed in HeLa cells, 20 were not previously shown to be expressed and lacked association with a typical early or late promoter. Their transcription was studied in more detail. We assume that an ORF is expressed from its promoter (and not merely due to run-on transcription) if it fulfills at least one of the following criteria: (i) ORF expression level is higher than that of the upstream region, (ii) ORF encodes known epitopes (therefore demonstrating translation), or (iii) ORF exhibits different expression kinetics than the upstream ORF. From this, it was concluded that at least 8 of the 20 ORFs exhibit true transcription.
Quantitative reverse-transcriptase PCR (qRT-PCR) was used to validate the transcription of these 20 genes and another 13 genes sampled from the four classes. This showed that all 33 genes generated detectable qRT-PCR products (SI Table 2). There was a significant correlation (P ≤ 10−7) of the fold-change in transcript levels between 2 and 8 hpi obtained with the tiling array analysis and qRT-PCR (SI Fig. 8). This suggests that tiling arrays can be used as a semiquantitative measurement of gene expression and that the signal strengths derived for individual ORFs here are a good representation of relative gene expression levels.
Of the 125 genes with kinetic expression previously described, 117 (94%) were found to be concordant with our data (SI Table 2), whereas 8 genes (VACWR-042, -071, -099, -111, -115, -142, -181, and -191) showed conflicting results. These discrepancies might be explained by methodological differences, such as the use of a different viral dose or cell type, which may alter the expression profile. Furthermore, we provide empirical evidence of the kinetic expression for 62 of the remaining ORFs, for which data were lacking. This underscores the advantages of and necessity for the simultaneous study of expression of all genes using a systematic approach.
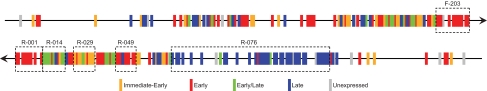
To get an overview of transcription throughout the genome, a map of the transcriptome was constructed (Fig. 5). Visual inspection supported by statistical analysis (see Materials and Methods) revealed a strong preference for colocalization of genes within the same kinetic class. For all four classes, the occurrence of a neighboring gene within the same kinetic class was significantly higher than expected (SI Table 4). Six regions were identified in which genes of a certain class were significantly over-represented with respect to their overall genomic distribution (Fig. 5).
Fig. 5.
Genes of different kinetic classes are clustered in the genome. Temporal expression is shown for each gene along the two strands of the VACV genome. The genes are positioned in their proper order, but sizes and spacing have been drawn identical for all ORFs. Boxes denote regions with an overrepresentation of genes from a certain kinetic class. F/R indicates the strand and numbers represent the VACWR name of the first ORF in each region.
A further extension of this broad gene expression approach would be to analyze the expression at the protein level and combine these data with the rapidly increasing amount of information about immunogenicity of VACV proteins [www.immuneepitope.org (41)]. This may allow us to unravel the rules that underpin immunogenicity and immunodominance in the host response to large pathogens. Last, a comprehensive study encompassing viral and host genes, especially those of immunological relevance, will help reveal the interactions between these two players in pathogenesis.
We have rendered the first complete picture of an orthopoxvirus transcriptome and demonstrated that the vast majority of annotated VACV genes are expressed. This also led to the surprising discovery of an immediate-early class of genes, more than half of which have unknown function despite being expressed at very high levels. Our study demonstrates the power of a genome-wide approach, compels a revision of the current understanding of orthopoxvirus gene regulation, and suggests many lines of investigation in orthopox virology and pathogenesis.
Materials and Methods
Viral Stocks.
The WR strain of vaccinia virus was obtained from B. Moss (National Institutes of Allergy and Infectious Diseases, Bethesda, MD).
Infection and Flow Cytometry.
HeLa cells were incubated at 107 cells per 100 μl complete media with VACWR at an MOI of 10:1. After 60 min, 30 × 106 cells were distributed to 225-cm2 flasks with 30 ml of complete media and cultured at 37°C. The method of infection was tested by using a GFP-encoding VACV42) and/or staining with an anti-VACV serum (ViroStat) and analyzed by flow cytometry. AraC was used at 40 μg/ml during infection in some samples to block viral DNA replication. Expression was considered significantly decreased when median signal intensity was ≥30% reduced. Productive infection or replication block were tested by enumerating the plaques formed in the AraC-treated samples at 2 and 24 hpi.
RNA Preparation, Labeling, and Hybridization.
Cells were harvested and resuspended in TRIzol and purified according to manufacturer's protocol. RNA clean-up was performed by using RNeasy columns. Ribosomal RNA was depleted by using the RiboMinus transcriptome isolation kit. NA samples were chemically labeled with biotin, using the ULS aRNA labeling kit, and hybridized to the arrays. Arrays were scanned by using the Affymetrix GeneChip Scanner 3000 7G and standard Affymetrix protocol as described in ref. 43.
Q-PCR.
Q-PCR was performed as described in ref. 44. For each primer set, expression levels were quantified relative to that of human 18S rRNA. Confirmation of single amplicons and lack of primer-dimers were performed by melting-curve analysis and gel electrophoresis. Samples prepared in the absence of reverse transcription were run for each primer pair to confirm specificity and to exclude that the signal came from contaminating genomic DNA.
Affymetrix GeneChip NimbleExpress Tiling Array Design.
The genome tiling array was built with NimbleGen probe synthesis technology. Packaging of the array was developed in collaboration with Affymetrix, using 25-mer probes covering both strands of the VACV genome (NCBI: AY243312.1) with a 4-nt spacing (97,334 probes). The array includes 15,308 negative control probes specific for Arabidopsis thaliana and 14,399 synthetic “antigenomic” probes with varying GC content.
Background Subtraction and Data Normalization.
Probe level data were log2-transformed to stabilize their variance. The fluorescence signal resulting from nonspecific hybridization was subtracted by using the synthetic and A. thaliana probes as empirical estimators. As a strong dependence of nonspecific signal on GC content was observed (SI Fig. 9), the median background probe signal at the corresponding GC content was subtracted from the signal of each VACV probe. Probes with GC content <3 nt were not well represented and were grouped. After removal of nonspecific background, a dependence of the specific signal on GC content was still apparent. This was minimized by mapping all probe signals to their corresponding values in the empirical distribution of probes with a GC content equal to 8. To enable the direct comparison of signal intensities across different arrays, probe signals were quantile-normalized (45). For replicate samples, quantile-normalized signals for each probe were averaged.
Data Summarization and Significance Testing.
Median quantile-normalized probe intensities for probes lying completely within each ORF were used as representative ORF signal intensity. Probes for identical ORFs (VACWR-001 through VACWR-012 and VACWR-218 through VACWR-207) were combined. The significance of signal intensity in each ORF was calculated by using the binomial distribution (20). P ≤ 10−5 was used as a threshold, because this approximately corresponds to a value of 10−3 after adjusting for the number of tests performed (n = 223).
Conversion to Standard Units.
To observe the correlations between temporal expression patterns, the data were converted to standard units: Standard Units = (S − μ)/σ. S = probe signal, μ = mean probe signal over the time course, and σ = standard deviation.
Gene Clustering and Bootstrapping Analysis.
Standardizedexpression profiles for each ORF were clustered by using the “cutHclust” function of the ClassDiscovery library (46) in the R statistical programming language (47). Data from 1–24 hpi were used. Data from uninfected cells and 0.5 hpi were excluded as their signal intensities were considerably lower and introduced excessive noise. All parameters were left at default values except for k, which was varied from 2 to 7. The “BootstrapClusterTest” function was used with values of k in the same range to determine the number of biologically relevant clusters. Other parameters included: cutHclust, metric = Pearson, and nTimes = 100. Data from 10 iterations were pooled. The robustness of clusters was measured averaging the number of times that two patterns ended up in the same cluster through 1,000 iterations on a subset of the data. A Pearson correlation test was run comparing expression values for each gene with the average for each cluster. R > 0.7 was considered as significant.
Gene Colocalization Statistics.
The cluster of each gene and the kinetics of its neighbors was noted in genomic space. The position of each gene was shuffled randomly 10,000 times while keeping track of the neighboring genes' kinetics during each iteration. Each pairwise distribution of neighboring genes was calculated in this manner. The observed frequency of colocalization was converted into a Z score (and P value), using standard deviations obtained through the randomization process.
Supplementary Material
ACKNOWLEDGMENTS.
We thank L. Giancola and M. Maybeno for VACV growth and purifications. This work was supported by National Institutes of Health Grants ROI-AI-56268 and HHSN266200400124C (to A.S.) and HHSN266200400036C (to E.J.L.), the Wenner-Gren Foundation (E.A.); the Sweden-America Foundation (M.S.), and an Australian National Health and Medical Research Council R. D. Wright Career Development Award (to D.C.T.). This is Kirin publication number 963.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711573105/DC1.
References
- 1.Becker Y, Joklik WK. Messenger RNA in cells infected with vaccinia virus. Proc Natl Acad Sci USA. 1964;51:577–585. doi: 10.1073/pnas.51.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedle S, Cooper RJ. The inhibition of HELA cell RNA synthesis following infection with vaccinia virus. J Gen Virol. 1984;65:1687–1697. doi: 10.1099/0022-1317-65-10-1687. [DOI] [PubMed] [Google Scholar]
- 3.Bablanian R, Coppola G, Scribani S, Esteban M. Inhibition of protein synthesis by vaccinia virus. IV. The role of low-molecular-weight viral RNA in the inhibition of protein synthesis. Virology. 1981;112:13–24. doi: 10.1016/0042-6822(81)90607-3. [DOI] [PubMed] [Google Scholar]
- 4.Bablanian R, Baxt B, Sonnabend JA, Esteban M. Studies on the mechanisms of vaccinia virus cytopathic effects. II. Early cell rounding is associated with virus polypeptide synthesis. J Gen Virol. 1978;39:403–413. doi: 10.1099/0022-1317-39-3-403. [DOI] [PubMed] [Google Scholar]
- 5.Broyles SS. Vaccinia virus transcription. J Gen Virol. 2003;84:2293–2303. doi: 10.1099/vir.0.18942-0. [DOI] [PubMed] [Google Scholar]
- 6.Munyon WH, Kit S. Induction of cytoplasmic ribonucleic acid (RNA) synthesis in vaccinia-infected LM cells during inhibition of protein synthesis. Virology. 1966;29:303–309. doi: 10.1016/0042-6822(66)90037-7. [DOI] [PubMed] [Google Scholar]
- 7.Kates JR, McAuslan BR. Poxvirus DNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1967;58:134–141. doi: 10.1073/pnas.58.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golini F, Kates JR. Transcriptional and translational analysis of a strongly expressed early region of the vaccinia virus genome. J Virol. 1984;49:459–470. doi: 10.1128/jvi.49.2.459-470.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moss B. Poxviridae and their replication. In: Fields BN, Knipe DN, editors. Virology. 2nd Ed. New York: Raven; 1990. pp. 2079–2111. [Google Scholar]
- 10.Condit RC, Niles EG. Regulation of viral transcription elongation and termination during vaccinia virus infection. Biochim Biophys Acta. 2002;1577:325–336. doi: 10.1016/s0167-4781(02)00461-x. [DOI] [PubMed] [Google Scholar]
- 11.Rosales R, Harris N, Ahn BY, Moss B. Purification and identification of a vaccinia virus-encoded intermediate stage promoter-specific transcription factor that has homology to eukaryotic transcription factor SII (TFIIS) and an additional role as a viral RNA polymerase subunit. J Biol Chem. 1994;269:14260–14267. [PubMed] [Google Scholar]
- 12.Rosales R, Sutter G, Moss B. A cellular factor is required for transcription of vaccinia viral intermediate-stage genes. Proc Natl Acad Sci USA. 1994;91:3794–3798. doi: 10.1073/pnas.91.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vos JC, Sasker M, Stunnenberg HG. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 1991;10:2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vos JC, Sasker M, Stunnenberg HG. Promoter melting by a stage-specific vaccinia virus transcription factor is independent of the presence of RNA polymerase. Cell. 1991;65:105–113. doi: 10.1016/0092-8674(91)90412-r. [DOI] [PubMed] [Google Scholar]
- 15.Gershon PD, Moss B. Early transcription factor subunits are encoded by vaccinia virus late genes. Proc Natl Acad Sci USA. 1990;87:4401–4405. doi: 10.1073/pnas.87.11.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broyles SS, Fesler BS. Vaccinia virus gene encoding a component of the viral early transcription factor. J Virol. 1990;64:1523–1529. doi: 10.1128/jvi.64.4.1523-1529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broyles SS, Moss B. DNA-dependent ATPase activity associated with vaccinia virus early transcription factor. J Biol Chem. 1988;263:10761–10765. [PubMed] [Google Scholar]
- 18.Kennedy PG, et al. Transcriptomal analysis of varicella-zoster virus infection using long oligonucleotide-based microarrays. J Gen Virol. 2005;86:2673–2684. doi: 10.1099/vir.0.80946-0. [DOI] [PubMed] [Google Scholar]
- 19.Stingley SW, et al. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J Virol. 2000;74:9916–9927. doi: 10.1128/jvi.74.21.9916-9927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 21.Pasmanik-Chor M, et al. Expression of mutated glucocerebrosidase alleles in human cells. Hum Mol Genet. 1997;6:887–895. doi: 10.1093/hmg/6.6.887. [DOI] [PubMed] [Google Scholar]
- 22.Mahr A, Roberts BE. Arrangement of late RNAs transcribed from a 7.1-kilobase EcoRI vaccinia virus DNA fragment. J Virol. 1984;49:510–520. doi: 10.1128/jvi.49.2.510-520.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan JR, Roberts BE. Organization of RNA transcripts from a vaccinia virus early gene cluster. J Virol. 1984;51:283–297. doi: 10.1128/jvi.51.2.283-297.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murcia-Nicolas A, Bolbach G, Blais JC, Beaud G. Identification by mass spectroscopy of three major early proteins associated with virosomes in vaccinia virus-infected cells. Virus Res. 1999;59:1–12. doi: 10.1016/s0168-1702(98)00114-2. [DOI] [PubMed] [Google Scholar]
- 25.Symons JA, Tscharke DC, Price N, Smith GL. A study of the vaccinia virus interferon-gamma receptor and its contribution to virus virulence. J Gen Virol. 2002;83:1953–1964. doi: 10.1099/0022-1317-83-8-1953. [DOI] [PubMed] [Google Scholar]
- 26.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosel JL, Earl PL, Weir JP, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol. 1986;60:436–449. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi T, Oie M, Ichihashi Y. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology. 1994;202:844–852. doi: 10.1006/viro.1994.1406. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt JF, Stunnenberg HG. Sequence and transcriptional analysis of the vaccinia virus HindIII I fragment. J Virol. 1988;62:1889–1897. doi: 10.1128/jvi.62.6.1889-1897.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doglio L, De Marco A, Schleich S, Roos N, Krijnse Locker J. The Vaccinia virus E8R gene product: a viral membrane protein that is made early in infection and packaged into the virions' core. J Virol. 2002;76:9773–9786. doi: 10.1128/JVI.76.19.9773-9786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Townsley AC, Senkevich TG, Moss B. The product of the vaccinia virus L5R gene is a fourth membrane protein encoded by all poxviruses that is required for cell entry and cell–cell fusion. J Virol. 2005;79:10988–10998. doi: 10.1128/JVI.79.17.10988-10998.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajcani J, Andrea V, Ingeborg R. Peculiarities of herpes simplex virus (HSV) transcription: An overview. Virus Genes. 2004;28:293–310. doi: 10.1023/b:viru.0000025777.62826.92. [DOI] [PubMed] [Google Scholar]
- 33.Pacha RF, Meis RJ, Condit RC. Structure and expression of the vaccinia virus gene which prevents virus-induced breakdown of RNA. J Virol. 1990;64:3853–3863. doi: 10.1128/jvi.64.8.3853-3863.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldick CJ, Jr, Moss B. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate-stage genes. J Virol. 1993;67:3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reading PC, Moore JB, Smith GL. Steroid hormone synthesis by vaccinia virus suppresses the inflammatory response to infection. J Exp Med. 2003;197:1269–1278. doi: 10.1084/jem.20022201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng A, Tscharke DC, Reading PC, Smith GL. The vaccinia virus A41L protein is a soluble 30 kDa glycoprotein that affects virus virulence. J Gen Virol. 2001;82:2095–2105. doi: 10.1099/0022-1317-82-9-2095. [DOI] [PubMed] [Google Scholar]
- 37.Kotwal GJ, Hugin AW, Moss B. Mapping and insertional mutagenesis of a vaccinia virus gene encoding a 13,800-Da secreted protein. Virology. 1989;171:579–587. doi: 10.1016/0042-6822(89)90627-2. [DOI] [PubMed] [Google Scholar]
- 38.Colamonici OR, Domanski P, Sweitzer SM, Larner A, Buller RM. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J Biol Chem. 1995;270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- 39.Webb TJ, et al. Inhibition of CD1d1-mediated antigen presentation by the vaccinia virus B1R and H5R molecules. Euro J Immunol. 2006;36:2595–2600. doi: 10.1002/eji.200636024. [DOI] [PubMed] [Google Scholar]
- 40.Herrera E, Lorenzo MM, Blasco R, Isaacs SN. Functional analysis of vaccinia virus B5R protein: essential role in virus envelopment is independent of a large portion of the extracellular domain. J Virol. 1998;72:294–302. doi: 10.1128/jvi.72.1.294-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters B, et al. A The immune epitope database and analysis resource: From vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norbury CC, et al. CD8+ T cell cross-priming via transfer of proteasome substrates. Science. 2004;304:1318–1321. doi: 10.1126/science.1096378. [DOI] [PubMed] [Google Scholar]
- 43.Lockhart DJ, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotech. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 44.Sundstrom M, Chatterji U, Schaffer L, de Rozieres S, Elder JH. Feline immunodeficiency virus OrfA alters gene expression of splicing factors and proteasome-ubiquitination proteins. Virology. 2007 doi: 10.1016/j.virol.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 46.Coombes KR. ClassDiscovery: Classes and methods for “class discovery” with microarrays or proteomics. R package, Version 1.3. 2007. Available at http://bioinformatics.mdanderson.org/software/OOMPA/classdiscoverythtml/00index.html.
- 47.Team R. R: A Language and Environment for Statistical Computing. 2006. Available at www.R-project.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.