Abstract
The 5′ long terminal repeat (LTR) of spleen necrosis virus (SNV) contains a unique posttranscriptional control element that facilitates Rev/Rev-responsive element-independent expression of unspliced human immunodeficiency virus type 1 (HIV-1) gag reporter RNA. HIV-1 Gag expression is eliminated when SNV LTR is repositioned to the 3′ untranslated region or when the RU5 region is positioned in the antisense orientation. RU5 corresponds to the 5′ RNA terminus, and results presented here indicate that Gag production is sustained upon introduction of transcribed spacers that reposition SNV RU5 35 to 200 nucleotides downstream. Concordant results of deletion and point mutagenesis identified two functionally redundant and synergistic motifs (designated A and C) that are necessary and sufficient for SNV RU5 activity. Enzymatic analysis of SNV RU5 RNA structure determined that A and C correspond to stem-loop structures. Quantitative RNA and protein analysis of A and C mutants revealed that the structural integrity of A and C is necessary for protein production, and loss of function correlates with little change in steady-state level, splicing efficiency, or cytoplasmic accumulation of HIV-1 gag reporter RNA. Instead, the structural mutations eliminate cytoplasmic utilization as an mRNA template for Gag protein production. Point mutations of unpaired loop-and-bulge nucleotides that maintain the structure of A eliminate activity. The results show that the unpaired UUGU loop and U-rich bulges function together and are candidate SNV RU5 binding sites for the host cell protein(s) that directs cytoplasmic utilization of unspliced HIV-1 reporter RNA.
Structured RNA elements in divergent retroviruses facilitate various posttranscriptional steps in expression of unspliced viral RNA. The 3′ untranslated regions (UTRs) of Mason-Pfizer monkey virus (MPMV) and the related simian retrovirus type 1 contain a constitutive transport element (CTE) that directs nuclear export of unspliced RNA in conjunction with host proteins Tap and NXT1/p15 (10, 14, 26, 27). The 3′ UTRs of avian leukosis virus and the related Rous sarcoma virus contain one and two copies, respectively, of a direct repeat (DR) element that is necessary for cytoplasmic accumulation and stability of unspliced viral RNA and for assembly of progeny virions (1, 22-24, 29, 30, 33). The 5′ long terminal repeats (LTRs) of spleen necrosis virus (SNV) and MPMV contain a unique posttranscriptional control element that facilitates reporter gene expression from unspliced human immunodeficiency virus type 1 (HIV-1) gag reporter RNA independently of Rev/Rev-responsive element (RRE), and also nonviral luc RNA (6, 15, 28).
Extensive mutagenesis studies of CTE and DR have mapped necessary and redundant structural motifs that present unpaired nucleotides for interaction with cellular posttranscriptional modulators. The MPMV and SRV-1 CTEs share 92% sequence homology and are 154-nucleotide (nt) orientation- and position-dependent RNA elements (27, 32). The RNA structure of the CTE that is predicted by M-Fold software (20, 35) is a stable stem-loop structure (ΔG = −39.4 kcal/mol) that consists of a degenerate repeat of ∼70 nt, a 9-nt terminal loop, and two 16-nt internal loops that are rotated 180° relative to each other (11, 32). The M-Fold prediction was validated by RNA structure analysis and extensive mutagenesis and functional analysis by a Gag reporter gene assay (11, 32). Point mutations designed to disrupt the double-stranded stem regions adjacent to the loops eliminated HIV-1 gag reporter gene activity (11). Compensatory mutations to restore base pairing of the stem rescued Gag production, confirming that the double-stranded stem structure is essential for activity. The stem has been postulated to orient the single-stranded loop nucleotides into the functionally correct position. Mutagenesis experiments by Tabernero et al. (32) indicated that the sequence of the internal loops, an AAGA bulge, and the stem structure between the loops are essential for CTE activity. Collectively, the mutagenesis data indicate that the secondary structure of CTE RNA and primary sequence of the unpaired nucleotides in the internal loops are necessary for CTE activity.
The DR is a 135-nt redundant stem-loop structure that also functions in an orientation-dependent manner (33). Each DR consists of two subelements referred to as DR1 and DR2 (29). Introduction of substitution mutations into predicted internal and terminal loop sequences in DR1 of the replication-competent Prague Rous sarcoma virus C reduces replication by a factor of 10 to 20 and reduces genomic RNA packaging by a factor of 10 (1). Two separate point mutations at the predicted stem-loop junction of one of the internal loops of the DR reduced expression of unspliced cat transcripts to between 10 and 40% of wild-type expression (24). Similar to the mutational analyses of CTE, these mutagenesis results demonstrated the functional importance of predicted unpaired nucleotides in the internal loops of DR. The role of the predicted double-stranded stem in DR remains to be verified, but it is projected to correctly position the unpaired nucleotides for interaction with functionally relevant cellular proteins.
The posttranscriptional control element in the 5′ SNV LTR was initially identified by its ability to facilitate Rex/Rex-responsive element-independent expression of bovine leukemia virus RNA (2). Subsequently, the SNV LTR was determined to facilitate Rev/RRE-independent expression of unspliced HIV-1 gag reporter RNA (6). Rev/RRE-independent Gag expression is eliminated when the RU5 region of the LTR is positioned in the antisense orientation or the LTR is repositioned to the 3′ UTR (6, 8). SNV RU5 does not function by derepressing inhibitory sequences located in the HIV-1 gag gene (28). Ribosomal sedimentation and ribosome profile analysis established that SNV RU5 augments polysome loading onto HIV-1 gag reporter RNA and also nonviral luciferase (luc) reporter RNA (6, 28). Experiments with bicistronic luc reporter plasmids determined that SNV RU5 does not function as an internal ribosome entry sequence (28). Together these results indicate that the RU5 region of the SNV LTR functions as a unique, orientation-dependent RNA element that enhances translation. Sequence or structural motifs necessary for activity remain to be defined.
Here, results of RNA and protein assays on SNV RU5 deletion and substitution mutants and enzymatic mapping of SNV RU5 RNA identified sequence and structural motifs necessary for activity. The experiments determined that SNV RU5 contains two functionally redundant stem-loops that are necessary for Rev/RRE-independent Gag production. Our genetic and biochemical results indicate that SNV RU5 stem-loop structures present unpaired nucleotides for functional interaction with a cellular posttranscriptional modulator(s). Quantitative RNA analysis by RNase protection assay (RPA) indicates that the mutations do not diminish the steady-state level or cytoplasmic accumulation of gag RNA. Instead, the mutations preclude efficient translational utilization of the cytoplasmic transcripts.
MATERIALS AND METHODS
Plasmid construction.
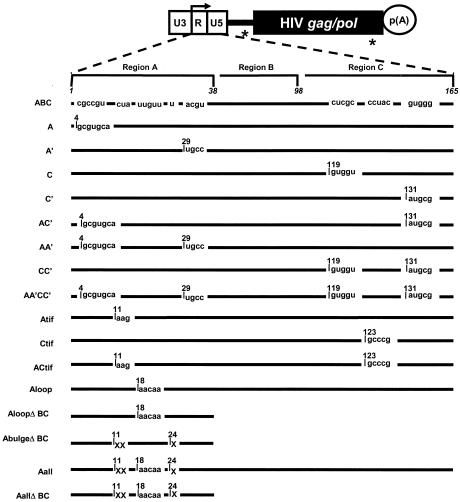
SNV RU5 deletion mutants were created by PCR with primer pairs that flank the region to be deleted and with the ExSite PCR-based site-directed mutagenesis kit and manufacturer protocol (Stratagene). SNV RU5 point mutants were generated by PCR-based site-directed mutagenesis using the QuikChange site-directed mutagenesis kit and manufacturer protocol (Stratagene). Primer sequences are available upon request, and the substitution mutations are summarized in the text. The deletion coordinates within the RU5 sequence are base pairs 4 to 42 (ΔA), 46 to 115 (ΔB), and 115 to 165 (ΔC) or combinations of these (ΔAB, ΔBC, and ΔAC). The PCR template for mutagenesis was pYW100 (6) with the following exceptions. ΔAB was generated from PCR template ΔB. ΔAC was generated from PCR template ΔA. AA′ was created from PCR template A. CC′ was created from PCR template C. AC′ was created from PCR template A. AA′CC′ was created from PCR template AA′C′. ACtif was created from PCR template Atif. AloopΔBC was created from PCR template ΔBC. AallΔBC was created from PCR template ΔBC. Antisense CBA was created by digesting pYW207 (6) with BamHI and ApaI to excise the antisense CBA region, which was inserted into pYW205 (6) that had been digested with BamHI and ApaI. The intermediate plasmid pTR170 was generated by site-directed mutagenesis of pYW100 to create a ClaI site between regions B and C of RU5. Subsequently, mutant ABA was created by digestion of pTR170 with ClaI and BamHI to excise the C region and ligation with complementary linkers that contain the A region.
SNV RU5 spacer mutants pTR162, pTR163, pTR143, and pTR200 contain the indicated inserts that were ligated at a unique AvaI site at position 2. pTR162 was created by introduction of a 35-bp synthetic linker with AvaI termini into the AvaI site in pYW100. pTR163 was created by insertion of a 35-bp synthetic linker with XhoI and SalI termini into an XhoI site within the linker in pTR162. pTR143 was created by introduction of 100 bp of pUC19 (coordinates 896 to 1,000 [New England Biolabs]) with AvaI termini into the AvaI site of pYW100. pTR200 was created by introduction of 194 bp of pGFP-N1 (coordinates 944 to 1139 [Clontech]) with AvaI termini into the AvaI site of pYW100. All mutant plasmids were verified by extensive restriction digestion and sequence analysis and lack an AUG translation initiation codon.
DNA transfection and reporter protein analysis.
Reporter gene assays were performed on 293 cells transfected by a CaCl2 protocol (6) in three replicate 100- or 33-mm-diameter plates. The cells were harvested 48 h posttransfection in phosphate-buffered saline, centrifuged at 2,000 × g for 3 min, and resuspended in ice-cold lysis buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% NP-40). Gag levels were quantified by a Gag enzyme-linked immunosorbent assay (ELISA) (Coulter Corp.) and normalized to Luc activity (relative light units) measured with a Lumicount luminometer (Packard); Luc was expressed from cotransfected pGL3 (Promega).
RNA preparation and analysis.
For preparation of nuclear and cytoplasmic RNAs, cells were harvested 48 h posttransfection and resuspended in 400 μl of hypotonic buffer (10 mM HEPES-KOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol). The cell suspensions were chilled on ice for 10 min and vortexed for 10 s, and nuclei were sedimented by centrifugation at 350 × g for 1 min at 4°C. Isolation of nuclear and cytoplasmic RNA from the pellet and supernatant, respectively, was performed with TriReagent by using the manufacturer's protocol (Molecular Research Center, Cincinnati, Ohio). The RNA was treated twice with DNase for 30 min at 37°C, phenol extracted, and ethanol precipitated. For RPAs, 25 μg of total RNA, 15 μg of nuclear RNA, or 25 μg of cytoplasmic RNA was analyzed as previously described (6). Antisense runoff α-32P-labeled RNA transcripts were synthesized with MAXIscript T7 polymerase (Ambion) according to the manufacturer's instructions. HIV-1 template pGEM (140 to 440) was digested with NotI, and pGAPDH was digested with NcoI. The HIV probe is 344 nt in length and complementary to the HIV 5′ UTR and the 5′ 106 nt of the gag open reading frame (6). Expected sizes of protected fragments are 303 nt for the unspliced product and 146 nt for spliced gag RNA. Spacer mutant T7 templates were generated as PCR products by using primers that amplify a region including 66 nt of the U3 promoter through the first 144 nt of the HIV 5′ UTR. The in vitro-transcribed RNAs were isolated by gel elution, and the RPAs were performed with RPAIII (Ambion) according to the manufacturer's protocol. RNA was ethanol precipitated with 3 × 105 cpm of HIV-1 gag probe and 1 × 104 cpm of a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene probe. RPA products were separated on acrylamide gels and were visualized by PhosphorImager (Molecular Dynamics) analysis, and RNA signals were quantified by ImageQuant version 4.2 software (Molecular Dynamics).
RNA secondary structure mapping.
MAXIscript T7 polymerase (Ambion) was used to transcribe the SNV RNAs from PCR products that contain the T7 promoter. The RNAs were dephosphorylated and 5′ end labeled with [γ-32P]ATP, and the full-length transcript was purified from a denaturing polyacrylamide gel. Target RNA was used at 250 ng per mapping reaction. For alkaline hydrolysis reactions, the target RNA was incubated in alkaline hydrolysis buffer (50 mM sodium carbonate [pH 9.2], 1 mM EDTA) with 3 μg of tRNA at 95°C for 5 min. For RNase T1 sequencing, target RNA was incubated in RNA sequencing buffer (20 mM sodium citrate [pH 5], 1 mM EDTA, 7 M urea) with 1 μg of tRNA at 50°C for 15 min, cooled to room temperature, and incubated with RNase T1 (Ambion) for 15 min at room temperature. For RNase T1 structure mapping, target RNA was incubated in RNA structure buffer (100 mM Tris [pH 7], 1 M KCl, 100 mM MgCl2) with 1 μg of tRNA and RNase T1 for 15 min at room temperature. For RNase A structure mapping, target RNA was incubated in RNA structure buffer with 1 μg of tRNA and RNase A (Ambion) for 15 min at room temperature. For RNase V1 structure mapping, target RNA was incubated in structure buffer with 1 μg of RNA and RNase V1 (Ambion) for 15 min at room temperature. All reactions were terminated by ethanol precipitation. Digested products were separated on 10 or 20% denaturing acrylamide gels and visualized with a PhosphorImager (Molecular Dynamics).
RESULTS
Two functionally redundant, synergistic regions are necessary for SNV RU5 activity.
SNV RU5 mutations were assayed for Rev/RRE-independent HIV-1 Gag production in the context of reporter plasmid pYW100, which was used in a previous characterization of SNV RU5 activity (Fig. 1A) (6). Deletion mutagenesis was used to define sequence boundaries necessary for SNV RU5 activity. SNV RU5 was arbitrarily divided into three regions, designated A (coordinates 4 to 42), B (46 to 115), and C (115 to 165), which were deleted individually and in combination. The mutants were transiently transfected into human 293 cells, and HIV-1 Gag production was measured by ELISA. As reported previously, SNV RU5 positioned in the sense orientation (ABC), but not the antisense orientation (antisense CBA), facilitated Rev/RRE-independent Gag production (Table 1) (8). Individual deletion of A, B, or C (ΔA, ΔB, or ΔC) and combined deletion of A and B (ΔAB) or B and C (ΔBC) reduced but did not eliminate Gag production. By contrast, combined deletion of A and C (ΔAC) eliminated Gag production. Because the presence of either A or C is sufficient for partial activity, we sought to determine whether two A regions act in a functionally redundant manner to restore Gag production to near-wild-type levels. To test this, we created a mutant that contains two copies of A (ABA). We postulated that introduction of two copies of A would either restore activity or produce altered base pairing that sterically hinders efficient translation. The results were consistent with the second possibility and showed that two copies of A do not act in a redundant manner to restore SNV RU5 activity.
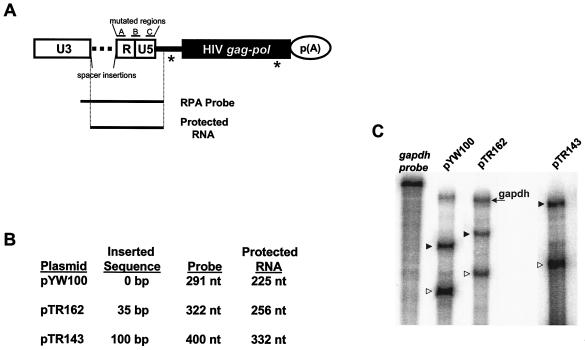
FIG. 1.
Reporter plasmid and RPA analysis. (A) Representation of parental SNV/HIV-1 reporter plasmid pYW100 showing the relative positions of mutated A, B, and C regions and the location of spacer insertions, the RPA probe, and protected RNA species. White rectangles, SNV LTR; horizontal dotted line, position of inserted spacer sequences; black line, HIV-1 5′ UTR; black rectangle, HIV-1 gag-pol genes; circle, polyadenylation signal. Splice sites are indicated by an asterisk. (B) List of plasmids that lack or contain spacer insertions, with the length of the inserted sequence, the length of the RPA probe, and the predicted size of protected RNA band after RNase digestion. (C) Representative RPA of spacer mutant RNA. HIV-1 Gag reporter plasmids were transfected into 293 cells. At 48 h posttransfection, total RNA was isolated and treated with DNase, and 20 μg of RNA was subjected to RPA with the indicated probe and GAPDH RNA probe. Transcripts were subjected to polyacrylamide gel electrophoresis and RNA signals were quantified by phosphorimager analysis. Undigested GAPDH RNA probe is labeled in italics, lanes are labeled with the designated reporter RNA. An RNA sample from pTR143 was electrophoresed separately to facilitate better resolution and the GAPDH RNA probe was omitted due to overlapping sizes of the GAPDH RNA protected band and the pTR143 probe. The position of protected GAPDH RNA is indicated on the right. ▸, position of spacer probes; ▹, position of protected RNA.
TABLE 1.
Effect of SNV RU5 deletion mutations on Gag production
| RU5 region | Relative Gag levela |
|---|---|
| ABC | 1.0 |
| Antisense CBA | <MD |
| ΔA | 0.3 ± 0.14 |
| ΔB | 0.4 ± 0.14 |
| ΔC | 0.5 ± 0.13 |
| ΔAB | 0.3 ± 0.14 |
| ΔBC | 0.4 ± 0.08 |
| ΔAC | <MD |
| ABA | <MD |
HIV-1 Gag reporter protein levels in representative triplicate transfections of 293 cells were standardized to Luc activity from cotransfected pGL3. A value of 1.0 is equivalent to 40 ng of Gag protein per ml. Values are means ± standard deviations. <MD, less than the minimum detectable level, 0.01 ng/ml.
In summary, results of deletion mutagenesis indicate that A or C is sufficient for Rev/RRE-independent Gag production and that A and C act in a functionally redundant manner. Together, A and C act synergistically to support wild-type levels of Gag production.
Location at the 5′ terminus is not essential for SNV RU5 activity.
Retroviral LTRs are composed of three regions: U3, R, and U5. U3 contains the viral promoter, and the first base pair of R corresponds to the RNA start site. Previous results determined that repositioning SNV RU5 distal to the RNA start site eliminates Rev/RRE-independent Gag production and stimulation of luciferase production (6, 28). Here we sought to assess the requirement of the 5′-proximal position of SNV RU5 for Rev/RRE-independent HIV-1 Gag production. We introduced spacer sequences of various lengths (35, 70, 100, and 200 bp) that lack ATGs between SNV U3 and SNV RU5 (Fig. 1A). RPAs on total cellular RNA were performed with homologous uniformly labeled 5′ RNA probes (Fig. 1A and B). The results verified that the spacer sequences are transcribed (Fig. 1C). Results of multiple independent transfection assays performed in triplicate indicate that the spacer sequences did not significantly reduce Gag production (Table 2). These results indicate that SNV RU5 activity is sustained when RU5 is repositioned 35 to 200 nt downstream of U3 and imply that precise proximity to the RNA start site is not necessary for activity.
TABLE 2.
Effect of spacer insertions between SNV U3 and RU5 on Gag production
| Plasmid | Spacer length (bp)a | Relative Gag levelb |
|---|---|---|
| pYW100 | 0 | 1.0 |
| pTR162 | 35 | 1.19 ± 0.19 |
| pTR163 | 70 | 1.21 ± 0.09 |
| pTR143 | 100 | 0.77 ± 0.03 |
| pTR200 | 200 | 0.94 ± 0.06 |
Number of base pairs inserted between U3 and RU5. All inserts do not contain an ATG.
HIV-1 Gag reporter protein levels in triplicate transfections of 293 cells were standardized to Luc activity from cotransfected pGL3. A value of 1.0 is equivalent to 40 ng of Gag protein per ml. Values are mean ± standard deviations.
A and C functional motifs are stem-loop structures.
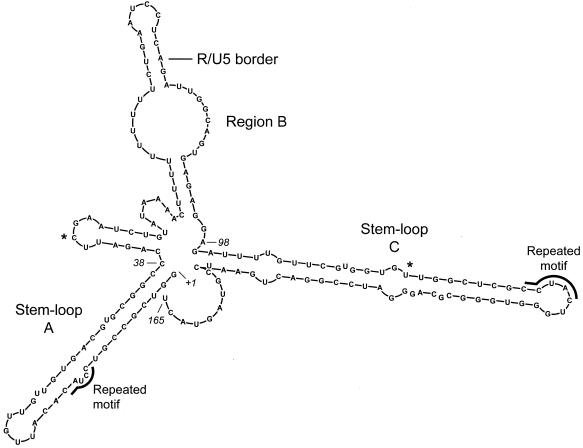
The Zuker M-Fold program (20, 35), which accurately predicted the secondary structure of the CTE (11, 32) and DR (33), was used to predict the secondary structure of SNV RU5. The most likely structure contains two stem-loop structures that correspond roughly to the A and C regions and a central spacer region that corresponds to B (Fig. 2). M-Fold computations, which are executed at fixed conditions of 37°C with 1 M NaCl and no divalent cations, predicted seven potential structures. The structures contain similar stems and terminal loop regions in A and C. The variation common to all predictions is the structural character of the B region. When folded independently, the functionally necessary A and C regions are predicted to maintain the stable stem-loop structures that they form in full-length SNV RU5. The predicted free energy of the most stable predicted SNV RU5 structure is −54 kcal/mol, which is comparable to those of the DR (−47 kcal/mol) and CTE (−39 kcal/mol) (11, 33).
FIG. 2.
Secondary structure of 165 nucleotide SNV RU5 predicted by the M-Fold software (20, 35). This structure depicts the most stable of seven predicted structures. The position of the R/U5 border is shown. Asterisks indicate positions of original deletions between A and B and between B and C. The predicted free energy (ΔG) is −54 kcal/mol.
Enzymatic mapping of SNV RU5 RNA was performed to analyze the secondary structure, and the data were evaluated against the structural predictions produced by M-Fold. In denaturing buffer, RNase T1 is expected to cut at every G residue. In structure buffer, RNase T1 is expected to cut at unpaired G residues, RNase A is expected to cut at unpaired C and U residues, and RNase V1 is expected to cut at double-stranded nucleotides (4, 11, 16, 33). Nucleotides predicted to be particularly prone to cleavage by RNases A and T1 are those located in the terminal loop regions. The 38-base A-region RNA and 58-base C-region RNA were transcribed in vitro, labeled at the 5′ end with [γ-32P]ATP, and subjected to digestion with each RNase. The RNase concentrations were titrated, and the profiles of digestion products were separated on denaturing sequencing gels and visualized by phosphorimager analysis. Alkaline hydrolysis of A-region and C-region RNAs was used to generate a ladder representing each nucleotide of the RNA. The assays were repeated multiple times with independently transcribed RNAs, and digestion patterns did not change.
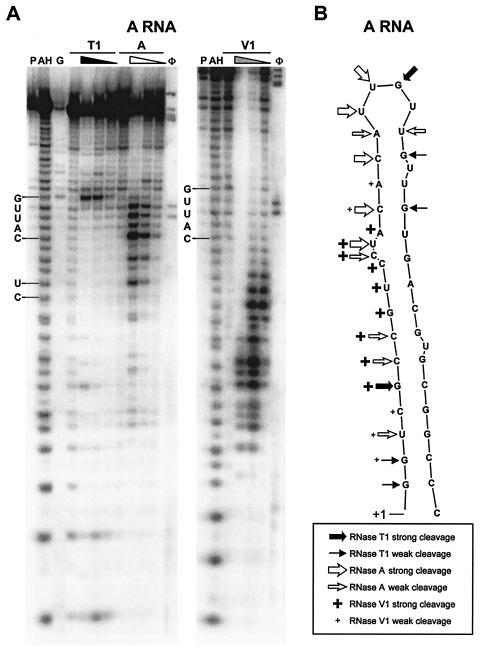
The RNase titrations clearly show sites that are readily accessible to cleavage (unpaired loop nucleotides) as opposed to less accessible sites (stem-and-bulge nucleotides). A region RNA showed strong RNase A and T1 cleavage products in the predicted terminal loop region and RNase A cleavage of the CU bulge (Fig. 3). The observation that strong cleavage begins 2 nt upstream of the terminal loop predicted by M-Fold suggests fluidity between a terminal loop of UUGU and a second predicted structure that contains a terminal loop of CAUUG. The RNase V1 digestions verify the predicted double-stranded stem.
FIG. 3.
Enzymatic digestion of A region RNA. (A) After transcription in vitro, the A region RNA was labeled at the 5′ end with [γ-32P]ATP and digested with various concentrations of RNase T1, A, and V1. A representative of four gels is shown. Lanes: P, probe; AH, alkaline hydrolysis ladder; G, RNase T1 sequencing ladder; φ, φX174 ladder. Four lanes are included for each RNase digestion. The first lane contains only buffer. For RNase T1, samples were incubated with 0, 1, 0.1, and 0.01 U of RNase. For RNase A, samples were incubated with 0, 10−3, 10−4, and 10−5 U of RNase. For RNase V1, samples were incubated with 0, 0.1, 0.01, and 0.001 U of RNase. The position and sequence of the terminal loop CAUUG and the CU bulge are indicated. (B) Diagram of M-Fold-predicted A structure, with areas cleaved by the RNases indicated.
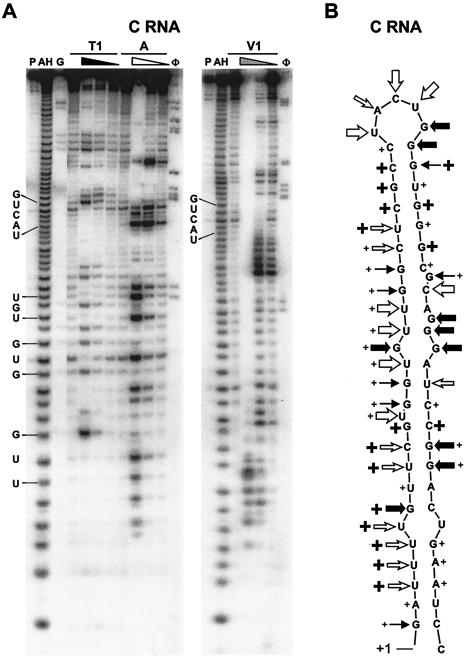
C-region RNA showed expected products of strong cleavage by the single-strand-specific RNases A and T1 that are in perfect concordance with the predicted terminal loop UACUG (Fig. 4). As expected, areas in the stem predicted to contain bulges were also cleaved by the single-strand-specific RNases A and T1. RNase V1 digestion verified the double-stranded stem structure of C RNA. Overlapping digests of stem-and-bulge regions by single-strand-specific RNases A and T1 and double-strand-specific RNase V1 suggest that these regions form stem structures but that there is a region of fluidity centered around the UGU and GGA bulges.
FIG. 4.
Enzymatic digestion of C-region RNA. (A) After transcription in vitro, the C-region RNA was labeled at the 5′ end with [γ-32P]ATP and digested with various concentrations of RNase T1, A, and V1. A representative of four gels is shown. Lanes: P, probe; AH, alkaline hydrolysis ladder; G, RNase T1 sequencing ladder; φ, φX174 ladder. Four lanes are included for each RNase digestion at the concentrations given in the legend to Fig. 3. The positions of the terminal loop UACUG and the UGU, GUG, and UUG bulges are indicated. (B) Diagram of M-Fold-predicted C structure and areas cleaved by the RNases (indicated as in Fig. 3B).
Of particular interest is the terminal loop of the C stem, which is flanked by a strong 7-nt stem that lacks bulged nucleotides. While the terminal loop is cleaved extensively by the single-strand-specific RNases, the flanking stem is cleaved solely by the double-strand-specific RNase V1. Enzymatic digestion of the contiguous SNV RU5 RNA is expected to show similar folding patterns, but mapping a 165-base substrate RNA is expected to produce a profile with lower resolution. When the full-length SNV RU5 RNA was subjected to the RNase digestion protocol, the A region contained the same digestion pattern. As expected, the C region was not resolved sufficiently under these conditions to determine the pattern of nucleotide cleavage (data not shown). In summary, the results from enzymatic mapping using three RNases with different specificities support the M-Fold-predicted structure of the A- and C-region RNAs.
A and C stem-loop structures are necessary for Gag production.
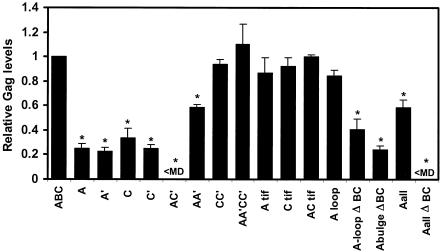
Substitution mutations were introduced into pYW100 to dissect the contribution of the secondary structure motifs and to analyze specific sequence motifs (Fig. 5). The first group of substitution mutations were designed to disrupt base pairing of either the A or C stem (A, A′, C, and C′). Each of these substitution mutations reduced but did not eliminate Gag production (Fig. 6). The levels are similar in magnitude to the levels exhibited by the ΔA or ΔC deletion mutants (Table 1). When the A and C′ substitution mutations were combined to simultaneously disrupt the predicted A and C stems (AC′), Gag production was eliminated. This loss of function is analogous to that with the ΔAC deletion mutant (Table 1) and posits the model that the A and C stem structures are necessary for SNV RU5 activity.
FIG. 5.
Summary of the SNV RU5 point mutations. Depicted at the top is the parental SNV/HIV-1 reporter plasmid. White rectangle, SNV LTR; black line, HIV-1 5′ UTR; black rectangle, HIV-1 gag/pol genes; circle, polyadenylation signal. Splice sites are indicated by an asterisk. The bent arrow indicates the RNA start site, which defines the U3/R border. Nucleotide numbers delineating regions A, B, and C are shown in italics above wild-type A, B, and C regions (not drawn to scale). Plasmid names are on the left. Nucleotide numbers correspond to the first mutated nucleotide at each mutation site and the altered sequence. Lines indicate unchanged sequence. “X” indicates deleted nucleotides.
FIG. 6.
Relative Gag levels produced in 293 cells transfected with the indicated HIV-1 Gag reporter plasmids. 293 cells were cotransfected with wild-type SNV RU5 (ABC) or mutant Gag reporter plasmids and pGL3 luciferase expression plasmid. Cell lysates were analyzed for Gag production by ELISA and for Luc activity by a chemiluminescent Luc assay. Values are from a representative triplicate experiment among more than 10 independent experiments. Gag levels are shown relative to SNV RU5 (ABC), and a value of 1.0 is equivalent to 40 ng of Gag per ml. Gag levels are standardized to cotransfected Luc. Standard deviations are shown. <MD, less than minimum detectable Gag level. *, significantly different from the value for SNV RU5 as determined by the Student t test (P < 0.005).
To test the model that these stem structures are necessary for activity, compensatory mutations were introduced singly and in combination into A and C (Fig. 5). Compensatory mutations in either the A stem (AA′) or the C stem (CC′) restored Gag production to 60 and 100%, respectively (Fig. 6). Introduction of both compensatory mutations into the nonfunctional AC′ mutant (AA′CC′) rescued Gag production from undetectable to 100%. The results of the deletion and substitution mutagenesis exhibit complete confirmation that A and C act in a functionally redundant manner to facilitate Rev/RRE-independent expression of HIV gag RNA. The rescue of Gag production by the compensatory mutations indicates that secondary structures that correspond to the A and C stem-loops are necessary for Gag production.
Nucleotides distinct from a conserved functional CTE motif are necessary for SNV RU5 activity.
Interestingly, the sequence motif CCUAC, which is repeated in the A region and the C region (Fig. 2), resembles the CCUAG motif, which is repeated in the functionally necessary unpaired loop regions of the CTE (32). Base substitutions were introduced individually and in combination to change CCUAC to CAAGC or GCCCG (Atif, Ctif, and ACtif) (Fig. 5). In all three cases, Gag production remained between 85 and 100% of wild-type levels, indicating that maintenance of the CCUAC motifs is not necessary for activity of SNV RU5 (Fig. 6).
Unpaired nucleotides in A are necessary for SNV RU5 activity.
To investigate nucleotides within A that are necessary for activity, base substitutions were introduced into the A terminal loop sequences that change UUGUU to AACAA (Aloop and AloopΔBC) (Fig. 5). In addition, unpaired nucleotides in the stem (CU and U bulges) were deleted (AbulgeΔBC). These mutations were predicted by M-Fold to maintain the stem structure while mutating the unpaired regions. When the A terminal loop sequence was mutated in wild-type SNV RU5 (Aloop), Gag production was unchanged (compare ABC to Aloop in Fig. 6). Compared to the parental deletion mutant ΔBC (Table 1), the AloopΔBC mutant exhibited partial activity of 40% (Fig. 6). Deletion of the single nucleotide bulges independently of the loop mutations (AbulgeΔBC) produced a minor but statistically significant reduction, from 40 to 25% (P = 0.04). The A loop-and-bulge mutations were combined to examine the possibility that these unpaired nucleotides act together to convey function (Aall and AallΔBC) (Fig. 5). Compared to ABC, the Aall mutant exhibited a reduction in Gag production to 60% (Fig. 6), a trend that is similar to that seen with the ΔA deletion mutant (Table 1) and verified the earlier conclusion that C is sufficient for partial activity. The AallΔBC mutant exhibited complete elimination of Gag production. These data indicate that individual mutation of unpaired loop-and-bulge regions in A do not abolish SNV RU5 function, while combined mutations of these regions result in loss of activity.
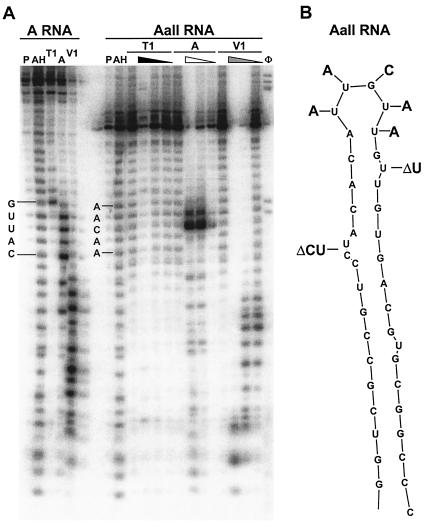
To verify that the stem-loop structure was maintained after introduction of these mutations, the RNase A, T1, and V1 digestion patterns were compared between A and AallΔBC (Fig. 7). As expected, the mutation of the terminal loop region (from UUGUU to AACAA) changed the sensitivity to RNase A. The AACAA motif was solely sensitive to RNase A cleavage at the C residue. RNase V1 digested the stem region similarly for both RNAs. The results demonstrate that, as predicted, the stem structure is maintained and that the loss of function is attributable to mutation of unpaired nucleotides.
FIG. 7.
Enzymatic digestion and position of mutations of AallΔBC. (A) After transcription in vitro, AallΔBC RNA was end labeled with [γ-32P]ATP and digested with various concentrations of RNase T1, A, and V1. Digested samples from A RNA were run in parallel for direct comparison. Lanes: P, probe; AH, alkaline hydrolysis ladder; G, RNase T1 sequencing ladder; φ, φX174 ladder. Four lanes are included for each RNase digestion and were used at the same concentrations as in Fig. 3B. Positions of the A terminal loop CAUUG and the Aall ΔBC terminal loop AACAA are shown. (B) Structure of wild-type A region, with deleted and mutated nucleotides indicated.
In summary, combined mutation of the A loop and bulge (AallΔBC) eliminated Gag production, while individual mutation reduced but did not eliminate Gag production. Collectively, these results indicate that the UUGUU sequence and the U and CU sequences function together to support optimal SNV RU5 activity. Our enzymatic digestion data on AallΔBC RNA verified that these mutations are in unpaired regions of the A stem-loop.
Loss of Gag production is not attributable to reduction in cytoplasmic gag RNA.
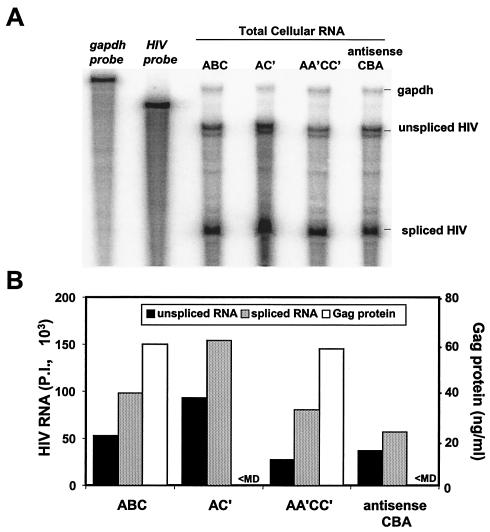
RPAs were used to assess the possibilities that the elimination of Gag protein production in response to SNV RU5 mutation is attributable to a defect in maintenance of steady-state RNA level, to a change in splicing efficiency, or to cytoplasmic RNA accumulation. The RPA probe used in the experiments spans the 3′ 194 nt of the HIV-1 UTR and the 5′ 106 nt of the gag coding region and overlaps the 5′ splice donor site (6). Total cellular RNA was evaluated from the wild-type SNV RU5 reporter (ABC), the nonfunctional AC′ and antisense SNV RU5 mutants, and the functional AA′CC′ compensatory mutant. The RPAs revealed that similar steady-state levels of unspliced HIV-1 gag RNA are sustained despite loss of Gag production in response to the AC′ and antisense mutations (Fig. 8). Similar ratios of spliced to unspliced reporter transcript are observed. Furthermore, the rescue of Gag production by the AA′CC′ compensatory mutation is not attributable to an increased steady-state gag RNA level. The data indicate that elimination of Gag production by SNV RU5 mutation is not attributable to a reduced steady-state RNA level or a deleterious effect on RNA splicing efficiency.
FIG. 8.
Quantification of HIV-1 reporter transcripts in total cellular RNA by RPA and Gag protein by ELISA. (A) Wild-type SNV RU5 (ABC) and selected mutant plasmids were transfected into 293 cells. At 48 h posttransfection, total cellular RNA was isolated and treated with DNase, and 25 μg was subjected to RPA with an HIV-specific probe complementary to the HIV-1 5′ UTR and a GAPDH RNA probe to control for RNA loading. Transcripts were subjected to polyacrylamide gel electrophoresis, and RNA signals were quantified by phosphorimager analysis. Undigested probes are labeled in italics, lanes are labeled with the reporter RNA, and RNase protection products are indicated on the right. RNA signals were quantified by ImageQuant version 4.2 software (Molecular Dynamics). (B) Unspliced and spliced RNA phosphorimager units (P.I.) were normalized to GAPDH RNA levels, and Gag levels were measured by ELISA.
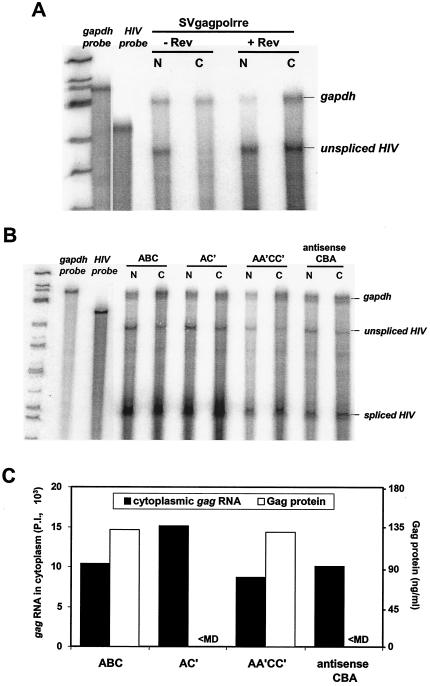
To evaluate the possibility that elimination of Gag production is attributable to elimination of cytoplasmic accumulation of gag RNA, nuclear and cytoplasmic RNA was prepared and subjected to RPA analysis. As a control to establish lack of nuclear contamination of cytoplasmic fractions, gag RNA was analyzed from 293 cells transfected with the Rev-dependent control plasmid pSVgagpolrre in the absence and presence of cotransfected Rev (3). Consistent with appropriate fractionation, unspliced gag RNA was observed in the cytoplasm solely in the presence of Rev (Fig. 9A). The protocol was used to evaluate wild-type SNV RU5 (ABC), nonfunctional AC′ and antisense CBA, and functional compensatory AA′CC′ mutant RNA. Results of five independent experiments consistently demonstrated similar levels of unspliced gag RNA in the nucleus and cytoplasm (Fig. 9B and C). The observation in this particular experiment that ABC and AC′ exhibit higher levels of spliced RNA than AA′CC′ and antisense CBA was not consistently observed. The data are in agreement with the RPA results on total cellular RNA and indicate that the nonfunctional mutants exhibit little change in steady-state RNA level or cytoplasmic accumulation of gag RNA. While Gag protein is produced from wild-type SNV RU5 and the AA′CC′ compensatory mutant, Gag protein production from the AC′ mutant or antisense CBA is less than the minimum detectable (Fig. 9C). The data indicate that despite similar levels of unspliced gag RNA in the cytoplasm, the translational efficiency of ABC and AA′CC′ is significantly greater than that of either AC′ or antisense CBA. Our results indicate that structural features in A and C are necessary for Gag protein synthesis.
FIG. 9.
Quantification of nuclear and cytoplasmic levels of HIV-1 reporter RNA by RPA. HIV-1 Gag reporter plasmids were transfected into 293 cells. At 48 h posttransfection, nuclear and cytoplasmic RNAs were isolated and treated with DNase, and 15 μg of nuclear or 25 μg of cytoplasmic RNA was subjected to RPA with the HIV-1 5′ UTR and a GAPDH RNA probe. Transcripts were subjected to polyacrylamide gel electrophoresis, and RNA signals were quantified by phosphorimager analysis. Undigested probes are labeled in italics, lanes are labeled with the reporter RNA, and RNase protection products are indicated on the right. (A) RPA of nuclear (N) and cytoplasmic (C) RNA from 293 cells transfected with pSVgagpolrre in the absence (−) or presence (+) of Rev. (B) RPA of nuclear (N) and cytoplasmic (C) RNA from 293 cells transfected with wild-type SNV RU5 (ABC) and selected mutants (6). (C) Summary of cytoplasmic gag RNA and Gag protein expressed from SNV RU5 and selected mutants. The RNA signals were quantified from the RPA by ImageQuant version 4.2 software (Molecular Dynamics) and normalized to GAPDH RNA levels. P.I., phosphorimager units. Gag levels were measured by ELISA. <MD, less than minimum detectable levels.
DISCUSSION
The concordant results of our deletion and substitution mutagenesis experiments indicate that SNV RU5 A and C regions act in a functionally redundant and synergistic manner to facilitate Rev/RRE-independent expression of unspliced HIV-1 gag reporter RNA. Structure mapping experiments by enzymatic RNA digestion demonstrate that A and C exist as stable stem-loop structures. The rescue of Gag production by compensatory mutations in A and C demonstrates that the structural character of the A and C regions is necessary for SNV RU5 activity. Point mutation of unpaired sequences in the A region elucidated their necessary role in SNV RU5 activity. The data indicate that the stem structure correctly positions unpaired nucleotides for interaction with a cellular effector protein(s). The RPA results indicate that the loss of Gag protein production is attributable not to loss of available gag RNA in the cytoplasm but to loss of gag RNA that is a productive mRNA template for protein synthesis.
Despite elimination of protein production, the AC′ and antisense CBA reporter plasmids exhibit similar levels of unspliced gag RNA in the cytoplasm. Previous data from our lab showed that HIV-1 gag reporter plasmids that contain a deletion of SNV RU5, but maintain the U3 promoter region, also exhibit similar levels of unspliced gag RNA in the cytoplasm despite reduction of HIV-1 Gag protein production (6). Control experiments with the Rev-dependent HIV-1 gag reporter pSVgagpolrre eliminated the possibility that the signal is attributable to nuclear contamination. A speculative explanation is that the SNV U3 promoter region, which is common to all the mutants, directs recruitment of ribonucleoprotein (RNP) components that modulate nuclear export. Recent mechanistic evaluation of pre-mRNA processing in Saccharomyces cerevisiae has shown that RNP components, which are recruited cotranscriptionally in conjunction with RNA polymerase II transcription machinery, effect pre-mRNA processing and export events (9, 17-19). The necessity of de novo transcription for activity of SNV RU5 has already been demonstrated (8) and supports the possibility that SNV U3 recruits a nuclear protein(s) that affects nuclear export.
The finding that SNV RU5 structural motifs modulate translational utilization of gag RNA indicates that a candidate SNV RU5-interactive cellular protein is the nuclear protein Sam68, because Sam68 has recently been shown to enhance cytoplasmic utilization of unspliced CTE-containing HIV-1 gag RNA (7). Our preliminary data demonstrate that while Sam68 enhances expression from SNV RU5-containing pYW100 18-fold in 293 cells, expression of the nonfunctional SNV RU5 antisense mutant is also facilitated (T. M. Roberts and K. Boris-Lawrie, unpublished results) (31). These results indicate that SNV RU5 is not necessary for the positive effect of Sam68 on this reporter RNA and do not support the notion that Sam68 mediates SNV RU5 activity.
Another possible SNV RU5-interactive cellular protein is an RNA helicase that facilitates hnRNP decondensation, similar to the role postulated for Dbp5 during mRNA export (12, 34). Once the hypothetical helicase is in the cytoplasm, its activity facilitates translational efficiency by neutralization of structural barriers to ribosome scanning. In retroviral RNA, a collection of structural barriers is imposed by the packaging signal and other necessary replication motifs throughout the extensive 5′ UTR (13, 21, 25). An intriguing possibility is that the SNV RU5-interactive protein neutralizes these structures and facilitates translation at the expense of viral RNA packaging. The protein would therefore act as a molecular switch to define the cytoplasmic fate of virion RNA (5). In this model, SNV RU5 would facilitate cytoplasmic utilization of unspliced viral RNA as mRNA template for protein synthesis at the expense of available genomic RNA for packaging into progeny virions. Experiments that characterize the cellular SNV RU5-interactive proteins will test these possibilities and are also expected to illuminate molecular mechanisms of eukaryotic posttranscriptional control.
Acknowledgments
We thank Alan Cochrane for the gift of SAM68 and SAM68ΔC reporter plasmids. We thank Stacey Hull and members of the Center for Retrovirus Research for comments on the manuscript and stimulating discussions.
This work was supported by grants from the American Cancer Society, Ohio Division, the National Institute of Allergy and Infectious Diseases (R29A140851), and the National Cancer Institute (P30CA16058).
REFERENCES
- 1.Aschoff, J. M., D. Foster, and J. M. Coffin. 1999. Point mutations in the avian sarcoma/leukosis virus 3′ untranslated region result in a packaging defect. J. Virol. 73:7421-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boris-Lawrie, K., and H. M. Temin. 1995. Genetically simpler bovine leukemia virus derivatives can replicate independently of Tax and Rex. J. Virol. 69:1920-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray, M., S. Prasad, J. W. Dubay, E. Hunter, K. T. Jeang, D. Rekosh, and M. L. Hammarskjold. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. USA 91:1256-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunel, C., and P. Romby. 2000. Probing RNA structure and RNA-ligand complexes with chemical probes. Methods Enzymol. 318:3-21. [DOI] [PubMed] [Google Scholar]
- 5.Butsch, M., and K. Boris-Lawrie. 2002. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 76:3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butsch, M., S. Hull, Y. Wang, T. M. Roberts, and K. Boris-Lawrie. 1999. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J. Virol. 73:4847-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyle, J. H., B. W. Guzik, Y. C. Bor, L. Jin, L. Eisner-Smerage, S. J. Taylor, D. Rekosh, and M. L. Hammarskjold. 2003. Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/BRK. Mol. Cell. Biol. 23:92-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dangel, A. W., S. Hull, T. M. Roberts, and K. Boris-Lawrie. 2002. Nuclear interactions are necessary for translational enhancement by spleen necrosis virus RU5. J. Virol. 76:3292-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dantonel, J. C., K. G. Murthy, J. L. Manley, and L. Tora. 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389:399-402. [DOI] [PubMed] [Google Scholar]
- 10.Ernst, R. K., M. Bray, D. Rekosh, and M. L. Hammarskjold. 1997. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol. Cell. Biol. 17:135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst, R. K., M. Bray, D. Rekosh, and M. L. Hammarskjold. 1997. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA 3:210-222. [PMC free article] [PubMed] [Google Scholar]
- 12.Gatfield, D., H. Le Hir, C. Schmitt, I. C. Braun, T. Kocher, M. Wilm, and E. Izaurralde. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11:1716-1721. [DOI] [PubMed] [Google Scholar]
- 13.Geballe, A. P., and M. K. Gray. 1992. Variable inhibition of cell-free translation by HIV-1 transcript leader sequences. Nucleic Acids Res. 20:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzik, B. W., L. Levesque, S. Prasad, Y. C. Bor, B. E. Black, B. M. Paschal, D. Rekosh, and M. L. Hammarskjold. 2001. NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol. Cell. Biol. 21:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hull, S., and K. Boris-Lawrie. 2002. RU5 of Mason-Pfizer monkey virus 5′ long terminal repeat enhances cytoplasmic expression of human immunodeficiency virus type 1 gag-pol and nonviral reporter RNA. J. Virol. 76:10211-10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knapp, G. 1989. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 180:192-212. [DOI] [PubMed] [Google Scholar]
- 17.Lei, E. P., H. Krebber, and P. A. Silver. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15:1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei, E. P., and P. A. Silver. 2002. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev. 16:2761-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Licatalosi, D. D., G. Geiger, M. Minet, S. Schroeder, K. Cilli, J. B. McNeil, and D. L. Bentley. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101-1111. [DOI] [PubMed] [Google Scholar]
- 20.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 21.Miele, G., A. Mouland, G. P. Harrison, E. Cohen, and A. M. Lever. 1996. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 70:944-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogert, R. A., and K. L. Beemon. 1998. Mutational analysis of the Rous sarcoma virus DR posttranscriptional control element. J. Virol. 72:3407-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogert, R. A., L. H. Lee, and K. L. Beemon. 1996. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J. Virol. 70:3834-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paca, R. E., R. A. Ogert, C. S. Hibbert, E. Izaurralde, and K. L. Beemon. 2000. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J. Virol. 74:9507-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkin, N. T., E. A. Cohen, A. Darveau, C. Rosen, W. Haseltine, and N. Sonenberg. 1988. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 7:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paschal, B. M., and L. Gerace. 1995. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J. Cell Biol. 129:925-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasquinelli, A. E., R. K. Ernst, E. Lund, C. Grimm, M. L. Zapp, D. Rekosh, M. L. Hammarskjold, and J. E. Dahlberg. 1997. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 16:7500-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts, T. M., and K. Boris-Lawrie. 2000. The 5′ RNA terminus of spleen necrosis virus stimulates translation of nonviral mRNA. J. Virol. 74:8111-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson, S. B., W. Guo, S. C. Winistorfer, R. C. Craven, and C. M. Stoltzfus. 1998. The upstream, direct repeat sequence of Prague A Rous sarcoma virus is deficient in mediating efficient Gag assembly and particle release. Virology 247:86-96. [DOI] [PubMed] [Google Scholar]
- 30.Simpson, S. B., L. Zhang, R. C. Craven, and C. M. Stoltzfus. 1997. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J. Virol. 71:9150-9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soros, V. B., H. V. Carvajal, S. Richard, and A. W. Cochrane. 2001. Inhibition of human immunodeficiency virus type 1 Rev function by a dominant-negative mutant of Sam68 through sequestration of unspliced RNA at perinuclear bundles. J. Virol. 75:8203-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabernero, C., A. S. Zolotukhin, A. Valentin, G. N. Pavlakis, and B. K. Felber. 1996. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J. Virol. 70:5998-6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, J., and B. R. Cullen. 1999. Structural and functional analysis of the avian leukemia virus constitutive transport element. RNA 5:1645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao, J., S. B. Jin, B. Bjorkroth, L. Wieslander, and B. Daneholt. 2002. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 21:1177-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.