Abstract
The threat of avian influenza A (H5N1) infection in humans remains a global health concern. Current influenza vaccines stimulate antibody responses against the surface glycoproteins but are ineffective against strains that have undergone significant antigenic variation. An alternative approach is to stimulate pre-existing memory T cells established by seasonal human influenza A infection that could cross-react with H5N1 by targeting highly conserved internal proteins. To determine how common cross-reactive T cells are, we performed a comprehensive ex vivo analysis of cross-reactive CD4+ and CD8+ memory T cell responses to overlapping peptides spanning the full proteome of influenza A/Viet Nam/CL26/2005 (H5N1) and influenza A/New York/232/2004 (H3N2) in healthy individuals from the United Kingdom and Viet Nam. Memory CD4+ and CD8+ T cells isolated from the majority of participants exhibited human influenza–specific responses and showed cross-recognition of at least one H5N1 internal protein. Participant CD4+ and CD8+ T cells recognized multiple synthesized influenza peptides, including peptides from the H5N1 strain. Matrix protein 1 (M1) and nucleoprotein (NP) were the immunodominant targets of cross-recognition. In addition, cross-reactive CD4+ and CD8+ T cells recognized target cells infected with recombinant vaccinia viruses expressing either H5N1 M1 or NP. Thus, vaccine formulas inducing heterosubtypic T cell–mediated immunity may confer broad protection against avian and human influenza A viruses.
Introduction
As transmission of highly virulent avian influenza A (H5N1) viruses to humans continues to occur in large parts of the world (385 reported cases including 243 deaths as of June 2008; ref. 1), there remains real concern that such a virus might mutate or reassort with a human influenza virus and hence acquire efficient human to human transmission and cause an influenza pandemic (2).
Currently available influenza vaccines induce antibodies against the viral surface glycoproteins HA and neuraminidase (NA), with the HA being by far the more important. While neutralizing antibodies can provide sterilizing immunity by blocking infection, this protection is only specific for the immunizing and closely related strains and is ineffective against heterologous strains with serologically distinct HAs. H5N1 influenza viruses continue to evolve and change antigenically (3, 4), and the WHO has reported the emergence of H5 HAs that are antigenically distinct from the H5 HAs used for vaccine production (5). H5N1 vaccine candidates developed so far are based on the clade 1 viruses (6–9). Induction of cross-reactive neutralizing antibodies against several clade 2 H5N1 strains by an adjuvant clade 1 vaccine has been documented recently (10). However, drastic antigenic changes are inevitable if and when the H5N1 virus adapts to transmit efficiently between humans. A pandemic strain may even be of another subtype. Thus, antibody-based vaccines developed against current H5N1 strains alone are unlikely to confer adequate protection against a pandemic strain, and there is an urgent need for another vaccine strategy.
An alternative or additional approach would be to stimulate T cell–mediated immunity, particularly virus-specific CD8+ CTLs that target the highly conserved internal proteins, as well as CD4+ T cells that provide help for the generation and maintenance of CD8 memory T cells (reviewed in refs. 11, 12). While CTLs do not prevent the establishment of infection, there is good evidence in mice and humans that T cells provide partial protection against influenza by promoting viral clearance and reducing the severity of symptoms (13–15). Influenza-specific CTLs mostly target internal proteins (16–21) and can provide partial protection across heterologous strains by targeting such conserved regions (22).
In view of the annual exposure to seasonal human influenza viruses (predominantly H3N2 and H1N1), most healthy adults may possess immunological memory against influenza virus. Indeed, influenza A virus–specific and mostly cross-reactive CD4+ (23, 24) and CD8+ (24–27) memory T cell responses have been demonstrated in humans. Recognition of target cells infected with swine or avian strains by T cell cultures derived from healthy individuals has also been reported (28, 29).
Despite the evidence supporting potential heterosubtypic protection that influenza A–specific T cells may confer, our understanding of T cell–mediated immunity to influenza infection remains incomplete, particularly in the context of human infection with avian influenza A (H5N1) viruses (30). Such limitation stems largely from the technology available for earlier studies. As the frequency of circulating influenza-specific T cells decline after recovery, in vitro stimulation of T cell cultures was necessary in most studies to expand the antigen-specific populations to a detectable level. A shortcoming of this method is that T cells expanded by culture in the presence of antigen may consist of preferentially expanded populations that do not truly represent the overall memory T cell repertoire in vivo. This limitation can now be overcome by using IFN-γ enzyme-linked immunosorbent spot (ELISpot) assays, in which fresh PBMCs are stimulated with overlapping peptides representing the whole virus proteome set out in a 3-dimensional matrix system. This can provide a more comprehensive picture of the virus-specific memory T cell populations circulating in peripheral blood.
In the present study, the overall influenza A virus–specific memory T cell response and the extent of cross-reactivity to H5N1 in healthy individuals from the United Kingdom (UK) and Viet Nam (n = 48 and 42, respectively) were evaluated ex vivo using the overlapping peptides spanning the full proteome of the influenza A/New York 232/2004 (H3N2) and A/Viet Nam/CL26/2004 (H5N1) strains. The H5N1 cross-reactive T cell responses were further examined against target cells infected with the recombinant vaccinia viruses (rVACVs) expressing the matrix protein 1 (M1) or nucleoprotein (NP) of the H5N1 strain. We report here that the majority of healthy individuals possessed influenza A–specific CD4+ and CD8+ memory T cell populations that were broadly cross-reactive to H5N1 internal proteins and discuss the relevance of these observations to a pre-pandemic vaccine strategy.
Results
Ex vivo assessment of CD4+ and CD8+ T cell responses against the full proteome of the H5N1 and H3N2 influenza A viruses in healthy individuals.
In this study we examined the virus-specific CD4+ and CD8+ memory T cell responses to the human H3N2 influenza A virus proteome and determined the extent of cross-reactivity to the H5N1 virus in healthy individuals with low risk of avian influenza exposure in 2 populations, using 48 volunteers from the UK and 42 from Viet Nam (a country in which H5N1 outbreaks in poultry and avian to human transmission continue to be reported). Because participants in this study reported no previous H5N1 virus exposure (Vietnamese participants were confirmed H5 seronegative by hemagglutination inhibition assay), we reasoned that any recognition of H5N1 antigens would involve cross-reactive memory T cell populations established from previous exposure to human influenza strains.
Freshly isolated PBMCs were tested for responses to a panel of 457 overlapping peptides spanning all internal proteins of clade 1 H5N1 influenza virus (referred to as peptide matrix system 1; Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI32460DS1) using ex vivo IFN-γ ELISpot in conjunction with an additional panel of 259 peptides of the H3N2 internal proteins that differ in sequence from the equivalent regions of the H5N1 strain (peptide matrix system 2; Supplemental Table 1). Together, the 2 panels contained 716 overlapping peptides representing the complete internal protein regions of the H5N1 and H3N2 influenza A strains (83%–98% amino acid sequence identity). The extent of T cell cross-recognition of the surface glycoproteins HA and NA of the H5N1 and H3N2 strains (peptide matrix systems 1 and 3; Supplemental Table 1) was evaluated in 20 Vietnamese healthy donors (34%–39% amino acid sequence identity). All overlapping peptides were placed into one 3-dimensional and two 2-dimensional peptide matrix systems as described in Supplemental Table 1.
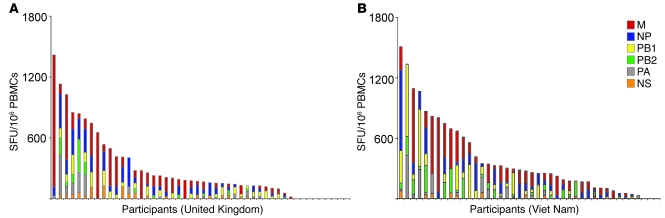
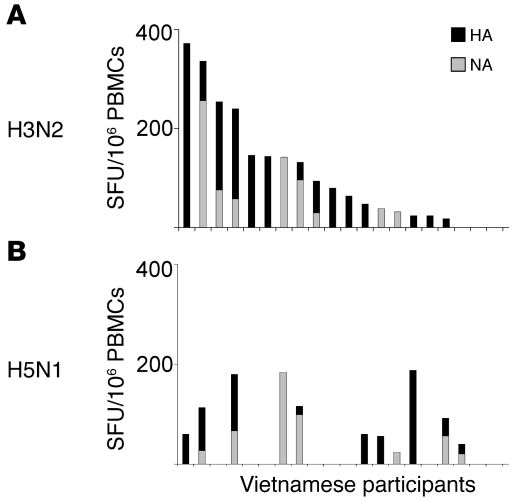
The majority of the participants (91% in the UK and 98% in Viet Nam) exhibited influenza A virus–specific memory T cell responses. Furthermore, 81% (UK) and 93% (Viet Nam) of the participants displayed broadly cross-reactive memory T cell responses against the H5N1 internal proteins (Figure 1). The distribution and magnitude of the IFN-γ responses against all H5N1 internal proteins are summarized in Figure 1. The frequencies of such cross-reactive memory T cell populations were generally comparable with those found in other acute respiratory virus infections (e.g., respiratory syncytial virus, ref. 31; adenovirus, ref. 32) and those specific for other repeatedly encountered foreign antigens (e.g., tetanus toxoid; ref. 33) but were significantly lower than those found in chronic viral infections (e.g., HIV, CMV; ref. 34). Cross-recognition of the H5N1 HA and/or NA was also detected in 11 of the 20 healthy Vietnamese individuals tested, but with considerably lower frequencies compared with the responses against the H3N2 HA and NA (Figure 2).
Figure 1. Cross-reactive memory T cell responses targeted to the internal proteins of avian influenza A (H5N1) virus in healthy individuals.
All participants (including responders and non-responders) from the UK (A; n = 48) and Viet Nam (B; n = 42) are represented on the x axis. The total magnitudes of ex vivo ELISpot IFN-γ responses to the overlapping peptide pools covering all H5N1 internal proteins are represented on the y axis. Each colored segment represents the source protein corresponding to peptide pools eliciting H5N1 cross-reactive T cell responses. M, matrix protein; PA, polymerase acidic protein.
Figure 2. Ex vivo recognition of the HA and NA of the H3N2 and H5N1 influenza A strains by healthy Vietnamese individuals.
Shown are the magnitudes of ex vivo ELISpot IFN-γ responses to the overlapping peptide pools representing HA (black) and NA (gray) of the H3N2 (A) or H5N1 (B) strains from H5-seronegative healthy Vietnamese participants (n = 20). These participants are shown on the x axis in the same order for A and B.
The breadth and magnitude of the T cell responses varied considerably between individuals (Figures 1 and 2). In a study that assessed influenza-specific T cell responses using in vitro bulk cultures derived from 3 volunteers, the memory T cell responses were detected against epitopes distributed on a wide variety of virus proteins (24). A similar broad repertoire of influenza-specific T cell responses was observed in a number of participants in this study. However, among the 90 participants, there was a wide range in the number of target regions recognized. Many donors of diverse HLA backgrounds displayed IFN-γ T cell responses distributed widely across the influenza proteome, as demonstrated in a study involving subjects with A2 and A11 haplotypes (27). Other individuals exhibited responses more specifically targeted to one or more internal proteins (examples are provided in Supplemental Figure 1). The number of viral proteins targeted by the H5N1 cross-reactive memory T cell populations ranged from 0 to 10, with the majority of responders recognizing 3 or 4 different proteins.
M1 and NP are the dominant targets of H5N1 cross-recognition.
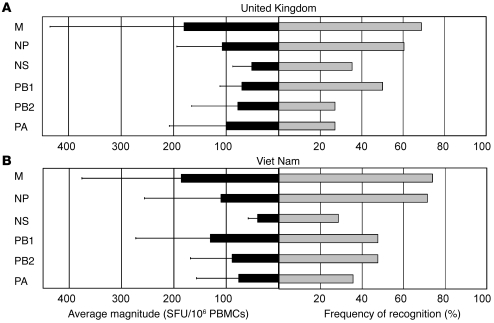
M1 and NP appeared to be the most immunogenic protein targets of H5N1 cross-reactive memory T cell responses (Figure 3). The relative dominance of these 2 proteins was similarly observed during the assessment of the overall memory T cell responses to the H3N2 strain (data not shown). The majority of the healthy participants (77%, UK; 86%, Viet Nam) displayed cross-recognition of the H5N1 M1 and/or NP. M1 elicited the highest magnitude of H5N1 cross-reactive IFN-γ responses ex vivo, followed by NP (UK) and polymerase basic protein 1 (PB1; Viet Nam).
Figure 3. Relative dominance of cross-reactive memory T cell responses to the internal proteins of avian influenza A (H5N1) virus in healthy individuals.
Gray bars represent the percentage of participants from the UK (A) and Viet Nam (B) who exhibited a detectable IFN-γ T cell response to at least one peptide pool corresponding to the specified source protein on ex vivo IFN-γ ELISpot assay. The average magnitude of IFN-γ T cell responses directed to each protein is represented by black bars. Data are mean ± SD. UK, n = 48; Viet Nam, n = 42.
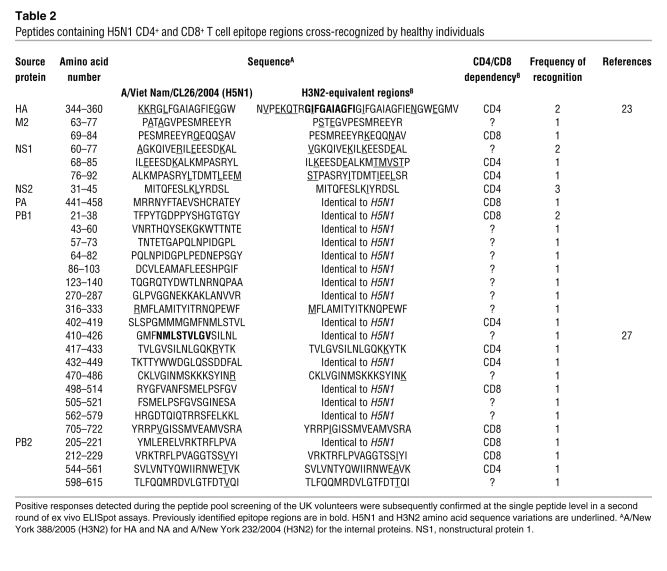
Confirmation of the H5N1 CD4+ and CD8+ T cell epitope regions.
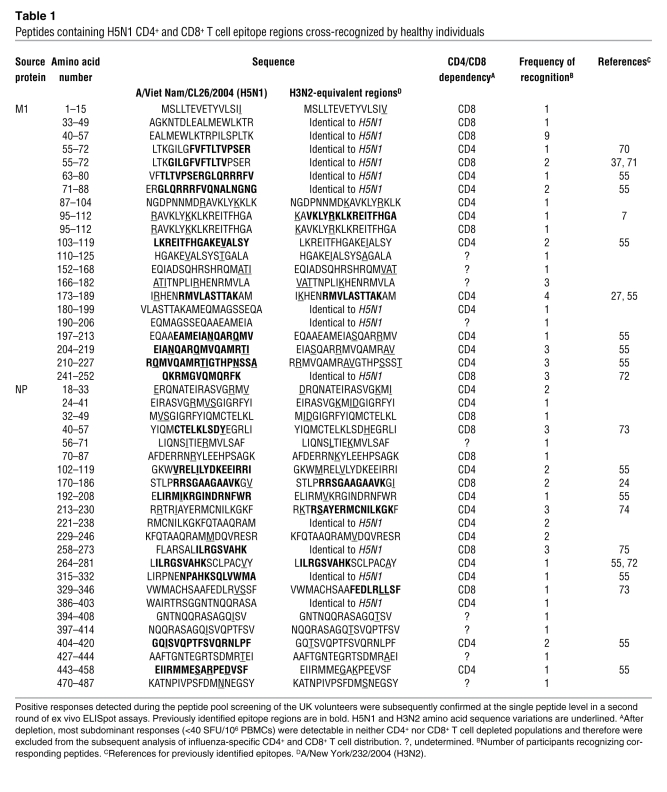
The positive responses detected during the peptide pool screening of the UK volunteers were confirmed subsequently at the single peptide level in a second round of ex vivo ELISpot assays. With increased sensitivity of ELISpot assay, it was possible to detect not only immunodominant responses but also subdominant responses involving memory T cell frequencies as low as 20 spot-forming units/106 (SFU/106) PBMCs. Direct ex vivo detection of such low-frequency T cell populations had not been possible in many earlier studies because of the limited sensitivity of assays such as Cr release assay. These assays often required in vitro proliferation of low-frequency populations in the presence of antigens, and those with low in vitro proliferation capacities often went undetected. We confirmed 73 peptides containing H5N1 T cell epitope regions recognized by healthy individuals, and of these, 28 were identical in sequence to the equivalent H3N2 regions (Tables 1 and 2). In addition, there were 62 H3N2 epitope regions whose sequences differed from their H5N1 equivalents (see Supplemental Table 2 for all confirmed epitope regions). Notably, at least 1 immunodominant or subdominant epitope region was present in each protein, but M1 and NP contained the highest number of H5N1 cross-reactive T cell epitope regions (21 and 23, respectively) as well as the majority of the most frequently recognized peptides (Table 1).
Table 1 .
Peptides containing H5N1 CD4+ and CD8+ T cell epitope regions cross-recognized by healthy individuals
Table 2 .
Peptides containing H5N1 CD4+ and CD8+ T cell epitope regions cross-recognized by healthy individuals
Distribution of influenza A–specific CD4+ and CD8+ memory T cell responses across the virus proteome.
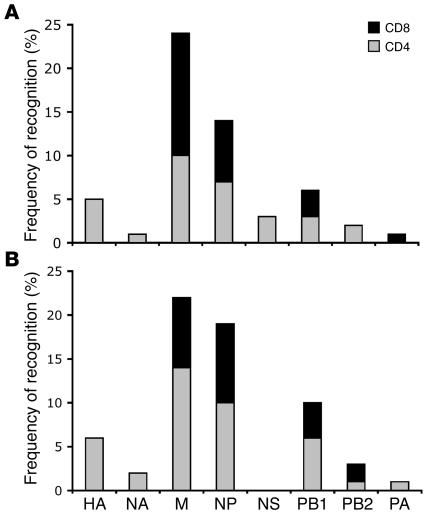
The responses detected against positive peptide pools and confirmed T cell epitope regions were characterized further for CD4+ or CD8+ T cell dependency by depletion experiments using anti-CD4 or CD8 antibodies conjugated to magnetic beads. The distribution of ex vivo CD4+ and CD8+ memory T cell responses across the whole influenza proteome was assessed: CD4+ T cell responses were more abundant (59%) than CD8+ responses (41%) and were distributed widely across all influenza proteins, whereas CD8+ T cell responses were targeted mostly against internal proteins (Figure 4). Detected responses to the HA and NA were entirely CD4+ T cell dependent.
Figure 4. Distribution of influenza A–specific CD4+ and CD8+ memory T cell responses across the virus proteome.
The total frequency of CD4+ (gray bars) and CD8+ (black bars) T cell recognition of positive peptide pools of each viral protein is shown for participants from the UK (A; n = 34) and Viet Nam (B; n = 27). The analysis involved ex vivo IFN-γ responses against the peptide pools of the H3N2 HA and NA and all H5N1 internal proteins. Responders who were unavailable to supply fresh blood samples for the depletion study were excluded from this analysis.
Recognition of target cells infected with rVACVs expressing H5N1 M1 or NP.
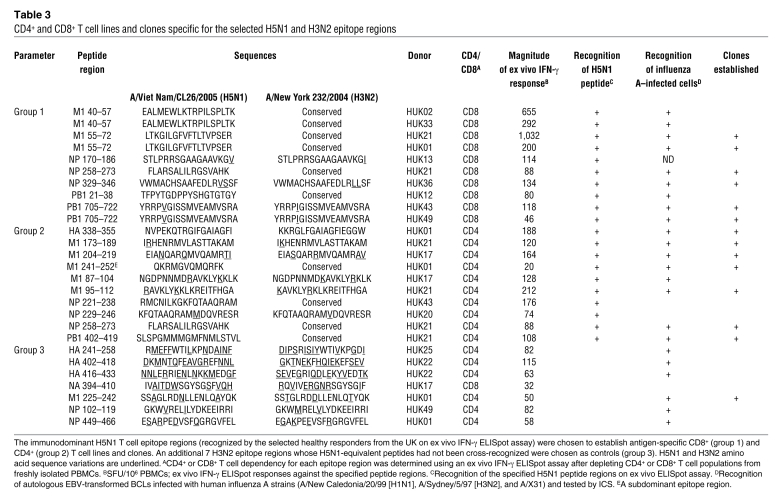
To test further the effector functions of the above T cells, 10 CD4+ and 10 CD8+ antigen-specific T cell lines were established from healthy responders in the UK using the immunodominant H5N1 T cell epitope regions cross-recognized on ex vivo ELISpot assay (Table 3). An additional 7 H3N2 epitope regions whose H5N1 equivalent peptides had not been cross-recognized were chosen as controls. The antigen specificity and CD4/CD8 dependency of each T cell line was confirmed by IFN-γ ELISpot and intracellular cytokine staining (ICS) (data not shown). In addition, T cell lines were prescreened by IFN-γ ICS for their ability to recognize autologous target cells infected with human influenza A viruses (summary provided in Table 3). Peptide-specific CD4+ and CD8+ T cell clones were generated from the majority of the established T cell lines by limiting dilution.
Table 3 .
CD4+ and CD8+ T cell lines and clones specific for the selected H5N1 and H3N2 epitope regions
In view of the relatively high peptide concentration required for ex vivo IFN-γ ELISpot assay, artificial T cell cross-recognition of a number of peptides seemed a possibility. To investigate whether T cell responses to H5N1 peptides were functional against naturally processed H5N1 virus proteins, rVACVs expressing the M1 or NP of H5N1 were constructed (H5N1 M1-VACV, NP-VACV). Autologous EBV-transformed B cell lines (BCLs) were infected by these viruses and used as targets. rVACVs expressing the M1 or NP of the human H1N1 strains (M1-VACV and NP-VACV) and H3N2 strain (NP-VACV) (19, 21, 35) were also used in parallel. The extent of cross-recognition of target cells infected with rVACVs by CD4+ and CD8+ T cell clones derived from healthy individuals were evaluated by Cr release assay, CD107a degranulation assay, and ICS (IFN-γ, TNF-α, IL-2, and IL–4).
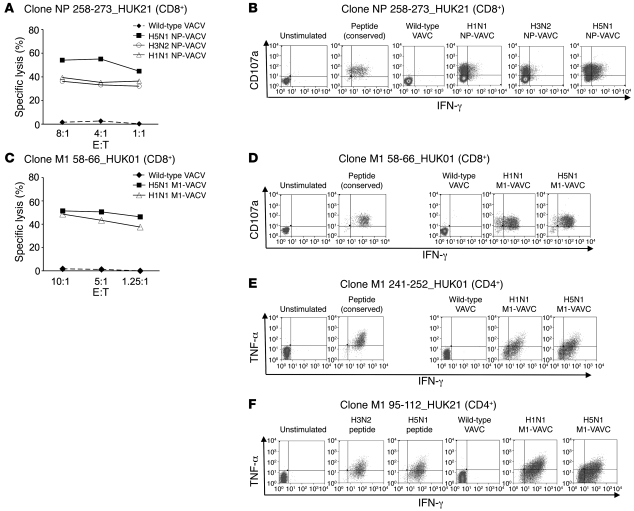
High levels of cross-recognition of target cells infected with H5N1 M1-VACV or NP-VACV were exhibited by a number of CD4+ and CD8+ T cell clones. Cells infected with wild-type VACVs were not recognized. Good cytolytic capacity was displayed by the cross-reactive CD8+ T cells even at low effector to target ratios (representative examples provided in Figure 5, A and C). These clones also appeared to be highly functional, simultaneously secreting cytokines such as IFN-γ and TNF-α coupled with upregulation of CD107a in recognition of the target cells expressing the M1 or NP of the H5N1 strain as well as those of the human strains (representative examples provided in Figure 5, B and D, and Supplemental Figure 2). Highly cross-reactive CD4+ T cell clones, such as clones M1 95–112_ HUK21 (clone specific for M1 95–112 and derived from donor HUK21) and M1 241–252_HUK01 also displayed polyfunctional capacities, producing high levels of cytokines such as IFN-γ, TNF-α, and IL-2 upon cross-recognition (Figure 5, E and F, and Supplemental Figure 2). No production of IL-4 was detected.
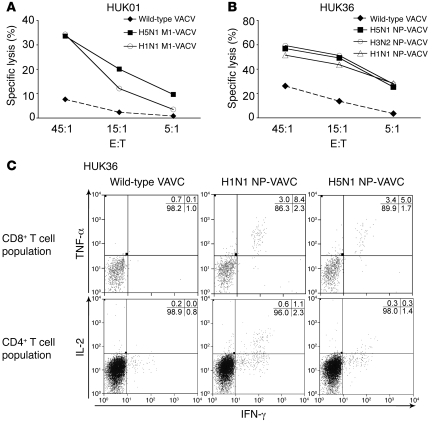
Figure 5. Effector functions displayed by CD4+ and CD8+ T cell clones upon recognition of target cells infected with the rVACVs expressing H5N1 M1 or NP.
Representative examples. (A and C) 51Cr release assay. Cross-reactive cytolytic activities against target cells infected with H5N1 NP-VACV (A) or H5N1 M1-VACV (C) displayed by CD8+ T cell clones specific for NP 258–273 (donor HUK21) (A) or for M1 58–66 (donor HUK01) (B). E:T, effector to target ratio. (B and D) ICS for effector cytokine secretion or upregulation of degranulation marker by CD8+ T cell clones specific for NP 258–273 (donor HUK21) (B) or for M1 58–66 (donor HUK01) (D) in recognition of peptide-pulsed or VACV-infected target cells. (E and F) ICS for effector cytokine secretion by CD4+ T cell clones specific for M1 241–252 (donor HUK01) (E) or for M1 95–112 (donor HUK21) (F).
Cross-reactive CTL activity against the H5N1 M1 and NP was also displayed by 7-day polyclonal virus-specific T cell cultures derived from healthy individuals in the UK (representative examples shown in Figure 6, A and B). Despite relatively low frequencies, it was possible to separately detect and analyze CD4+ and CD8+ H5N1 cross-reactive subpopulations that secreted detectable levels of cytokine(s) upon recognition of target cells expressing the H5N1 M1 or NP (representative example displayed in Figure 6C).
Figure 6. Cross-recognition of target cells infected with rVACVs expressing H5N1 M1 or NP by 7-day polyclonal T cell cultures established with human influenza A viruses.
Representative examples. (A and B) 51Cr release assay. Cross-reactive cytolytic activities displayed against target cells infected with H5N1 M1-VACV (donor HUK01) (A) or H5N1 NP-VACV (donor HUK36) (B) by the T cell cultures established with A/New Caledonia/20/99 (H1N1). (C) ICS for effector cytokine secretion (IFN-γ, TNF-α, and IL-2) by separately gated CD8+ or CD4+ population of the T cell culture established with A/New Caledonia/20/99 (H1N1) (donor HUK36). Numbers represent percentages of cells.
Discussion
Heterosubtypic T cell–mediated immunity to influenza A viruses in humans has been widely reported (24–26, 36). While cross-reactive T cells cannot prevent establishment of infection, they can enhance recovery by promoting virus clearance and reduce the severity of illness (15). It has also been demonstrated that in vitro T cell cultures derived from healthy individuals can lyse target cells infected with swine or avian influenza viruses (28, 29). Together with a large number of studies in mice that demonstrated cross-protection conferred by influenza A–specific T cells during subsequent challenges (reviewed in ref. 11), these studies suggest that vaccines stimulating cross-reactive T cell populations, potentially used in conjunction with the current antibody-based strategy, may confer broader and more effective protection against a potential pandemic strain. However, most studies using T cell cultures expanded in vitro were unable to assess accurately the overall influenza-specific memory T cell response and the extent of cross-reactivity to heterologous strains, particularly regarding the breadth, magnitude, and immunodominance hierarchy of the responses, at the population level. If a novel vaccine formula designed to stimulate cross-protective T cells is to be developed, it is crucial to address these questions first.
In the present study, we provide for what we believe to be the first time, a comprehensive ex vivo assessment of virus-specific CD4+ and CD8+ memory T cell responses against the full proteome of the human influenza A (H3N2) virus together with measurement of the extent of cross-reactivity to avian influenza A (H5N1) using freshly isolated PBMCs from 48 healthy individual from the UK and 42 healthy individuals from Viet Nam. Of 90 healthy donors with no prior exposure to H5N1, 81% and 93% of healthy donors living in the UK and Viet Nam, respectively, displayed detectable cross-reactive CD4+ and/or CD8+ memory T cell responses to the H5N1 internal proteins. In contrast, cross-recognition of the H5N1 HA and NA was exhibited by only 9 and 7, respectively, of 20 Vietnamese participants tested, and the T cell pools were of considerably lower frequencies compared with those specific for the H3N2 HA and NA. It is worth noting that HA and NA, predominantly targeted by CD4+ T cell populations, appear less immunogenic for T cell recognition than most internal proteins, regardless of the subtypes tested. Despite relatively high amino acid sequence identity (approximately 80%) between the NA of the H5N1 and human H1N1 strains, ex vivo cross-reactive T cell recognition of the NA was unremarkable. Thus, it would seem reasonable to suggest that internal protein regions, highly conserved and more frequently recognized by both CD4+ and CD8+ cross-reactive memory T cells, may serve as effective targets for vaccines designed to boost potentially cross-protective memory T cell populations.
Previous reports have identified M1 and NP as major targets of influenza-specific CTL recognition in humans (20, 37). Our ex vivo study verified M1 and NP as the most immunogenic protein targets of H5N1 cross-reactive CD4+ and CD8+ T cell responses. Moreover, CD4+ and CD8+ T cell clones specific for M1 or NP epitopes displayed cross-reactive effector functions against the target cells expressing the H5N1 M1 or NP, respectively. Transfer of NP-specific CTL clone protects mice from otherwise lethal challenges with homologous and heterologous influenza A viruses (22). Although no such direct evidence exists in humans, a good correlation has been documented between pre-existing influenza A virus–specific CTL activities and enhanced viral clearance upon nasal challenge (15). Thus, it is likely that our observations on cross-reactive influenza-specific memory T cells in healthy individuals may correlate with enhanced antiviral protection upon exposure to H5N1 influenza A viruses. Interestingly, the dominance of M1- or NP-specific responses observed at the population level during peptide pool screening was evident at the individual level when positive pool responses were confirmed at the single peptide level: M1- or NP-specific epitope regions were consistently immunodominant in a number of participants of diverse HLA backgrounds (data not shown). This held true for both CD4+ and CD8+ H5N1 cross-reactive and H3N2 strain–specific T cell epitope regions. It appears that M1 and/or NP may serve as potential universal influenza vaccine targets to provide broad protection to populations of diverse genetic backgrounds. Notably, vaccination of mice with DNA encoding NP elicited NP-specific CD4+ and CD8+ effector T cells and provided measurable protection against otherwise lethal heterosubtypic challenges (38).
Influenza A–specific T cells detectable by ex vivo IFN-γ ELISpot have been previously characterized as a subset of circulating memory T cell populations capable of immediate effector function upon antigen encounter without a need to differentiate or proliferate (39). IFN-γ secretion, an important component of antiviral protection in influenza infection (40), was detectable within 6–12 hours of stimulation with peptides. In view of the diverse tissue tropism associated with highly pathogenic H5N1 viruses (41), it is possible that circulating H5N1 cross-reactive memory T cell populations capable of immediate effector function could confer partial protection against human infection with H5N1 viruses.
Highly cross-reactive CD4+ and CD8+ effector functions against target cells infected with rVACVs expressing the H5N1 M1 or NP were detected both at the single-cell and polyclonal levels. In addition to exhibiting good cytolytic activities (CD8+ clones), cross-reactive T cell clones also displayed a high degree of polyfunctionality, simultaneously secreting IFN-γ, TNF-α, and IL-2 (CD4+ clones), together with upregulation of CD107a in recognition of H5N1 antigens. The induction of such multifunctional T cell populations in mouse and man has been associated with superior antiviral protection and vaccine efficacy (42–44). Further characterization of such broadly cross-reactive and highly functional CD4+ and CD8+ T cell responses is in progress. This includes epitope optimization and determination of HLA restrictions, not included in the comprehensive analysis reported here.
Population differences in H5N1 cross-reactive T cell responses were observed between the cohorts from the UK and Viet Nam. A higher percentage of Vietnamese participants displayed detectable H5N1 cross-reactive memory T cell responses ex vivo. In addition, although the difference did not reach statistical significance, we detected ex vivo IFN-γ T cell responses of higher magnitude to the H5N1 internal proteins from Vietnamese participants (mean overall magnitudes: 390 (Viet Nam) and 287 (UK) SFU/106; P = 0.058). Potential explanations for these differences include host genetic differences, exposure to region-specific, unrelated pathogens (an example is discussed in ref. 45), and variations in the circulating influenza virus strains in the 2 regions. Moreover, there is a clearly defined influenza season in the UK, whereas in southern Viet Nam, acute influenza cases are reported throughout the year.
Considerable variation in virus-specific T cell recognition patterns was observed between different individuals in aspects such as the magnitude and breadth of the responses, CD4 and CD8 dependence, cross-reactivity, and dominant protein targets. McMichael et al. demonstrated a correlation between pre-existing influenza A–specific CTL activity and enhanced virus clearance upon subsequent intranasal challenge with live virus even in the absence of cross-protective antibodies (15). This suggests that pre-existing influenza-specific memory T cells played a clear role in protection against disease in subsequent infection with heterologous strains. Thus, it is likely that individual variations in circulating memory T cell pools, particularly in the memory pools that are cross-reactive between subtypes, would at least partially contribute to the determination of clinical outcome of subsequent influenza infections together with other variables such as HLA haplotypes (46), age (47), antigen doses (48), and exposure history to influenza viruses and other pathogens (45, 49). It may even be possible that such variations have contributed to differential outcomes in human infection cases with highly pathogenic H5N1. We are currently investigating this possibility in Viet Nam.
In a recent retrospective epidemiological analysis, Epstein provided a good example of heterosubtypic protection against influenza A infection conferred by immunological memory established from previous natural infection: adults who had experienced natural infection with the H1N1 strain in 1957 were significantly better protected from the H2N2 pandemic strain later that year (50). In view of the absence of evidence suggesting the existence of neutralizing antibodies that are cross-reactive between the H1N1 and H2N2 strains, this observation further suggests that pre-existing cross-reactive memory T cells may be beneficial in subsequent heterosubtypic infection. However, it is worth noting that T cell memory to influenza in humans, particularly CD8+ CTL activities, is short-lived, with a half-life of 2–3 years (51), and needs regular boosting by natural infection or appropriately designed vaccines.
There is growing interest in targeting conserved regions of internal proteins, such as M1 and NP, in combination with the existing vaccine formula in preparation for a potential pandemic. In view of the 80%–90% amino acid sequence identity of these proteins between the avian and human type A strains, our data pertaining to the immunodominant protein regions recognized by healthy individuals of diverse HLA backgrounds could serve as a potent catalyst for such endeavors. For instance, the epitope regions we identified that frequently elicited highly cross-reactive CD4+ and CD8+ T cell responses merit special attention for the potential development of a universal vaccine that would generate or boost T cell responses against seasonal human influenza as well as avian influenza viruses.
Despite extensive HLA polymorphism, the majority of HLA-A and -B allelic variants can be classified into 9 supertypes based on their shared peptide-binding specificity (52, 53). Epitope-based vaccines containing conserved peptides recognized by various MHC molecules may therefore confer broad and potent protection against influenza. However, developing such vaccines that can provide broad coverage across the diverse populations remains a challenge, as different HLA types are expressed at varying frequencies in different ethnic groups. Thus, the analysis of cross-reactivity of and population coverage by (54) epitopes of interest should parallel comprehensive epitope mapping efforts (55, 56). Such endeavors may be facilitated by a centralized database, such as the Immune Epitope Database and Analysis Resource (http://www.immuneepitope.org), which provides comprehensive T and B cell epitope listing of various organisms as well as relevant analysis tools and resources (30, 57).
Currently available inactivated subunit vaccines are inefficient at boosting influenza-specific CTLs, as antigens cannot reach the cytosol and thus cannot be processed and presented as MHC-peptide complexes for T cell recognition. Stimulation of cross-reactive memory T cells could be potentially achieved by cold adapted live attenuated influenza vaccines (LAIVs) that closely mimic natural infection. Several trials have reported that LAIVs can boost virus-specific CTLs as well as mucosal and serum antibodies and provide broad cross-protection against heterologous human influenza A viruses (58, 59). While the safety and efficacy of the reassortant viruses bearing modified H5 HAs (60) in humans remains to be determined, it would be worth evaluating the extent of cross-protection against H5N1 potentially conferred by currently available seasonal human LAIVs. Memory T cell populations boosted by these vaccines may in theory cross-react and provide partial protection against H5N1 by targeting highly conserved internal virus proteins.
The aim of such T cell–based approaches would be to provide broader partial protection against overwhelming infection and help lower morbidity and mortality rather than to provide complete protection against establishment of infection. This would be a highly relevant and perhaps more realistic public health goal in a pandemic situation.
In summary, the data presented here form an important basis from which to evaluate the role of virus-specific cross-reactive T cells in broad partial protection from human infection with avian (H5N1) and human influenza A viruses.
Methods
Subjects.
Influenza-specific T cell memory responses were studied in 48 healthy volunteers in Oxford, UK (age 20–63 yr), and 42 healthy volunteers in Ho Chi Minh City, Viet Nam (age 23–51 yr), all of whom were deemed to be at low risk of H5N1 exposure. Hemagglutination inhibition assay (using horse red blood cells) confirmed the absence of H5 antibody in Vietnamese participants. Samples were collected between December 2005 and November 2006. All participants were devoid of any symptoms indicative of acute influenza infection at the time of sample collection. Informed consent was obtained from all participating individuals prior to the study.
Ethical approval was obtained from the Scientific and Ethical Committee of the Hospital for Tropical Diseases, Ho Chi Minh City, Viet Nam, and the Oxford Tropical Research Ethics Committee (OXTREC), Oxford, UK.
Synthetic peptides.
A total of 1,028 15- to 18-mer peptides overlapping by 10 amino acid residues and spanning the full proteome of the H5N1 and H3N2 influenza A viruses (151 HA, 125 NA, 58 M1, 26 M2, 121 NP, 57 nonstructural protein 1 [NS1], 31 NS2, 155 acid polymerase [PA], 135 basic polymerase [PB1], and 169 PB2) were designed using the Los Alamos National Library web-based software PeptGen (http://www.hiv.lanl.gov/content/sequence/PEPTGEN/peptgen.html) and synthesized (purity >95%; PEPscreen; Sigma-Aldrich) using the sequences of the following strains: A/Viet Nam/CL26/2004 (H5N1), A/New York 388/2005 (H3N2) (HA and NA), and A/New York 232/2004 (H3N2) (internal proteins).
Designing and deconvoluting 2- and 3-dimensional peptide matrix systems.
The overlapping peptides spanning the H5N1 internal proteins and the H3N2 HA and NA were assigned into a 3-dimensional matrix system in which each peptide was represented in 3 different peptide pools. The first dimension of the peptide matrix system was designed so that peptides from different source proteins were separated into different pools (595 peptides, peptide matrix system 1; Supplemental Table 1). (Peptide pools 15 and 16 of the peptide matrix system 1 and peptide pool 2 of the peptide matrix system 2 contained a mixture of overlapping peptides of the M1 and M2. However, the responses detected against these pools were determined to be targeted predominantly to the M1 when the positive pool responses were confirmed at the single peptide level.) The optimal peptide arrangement for the second and third dimensions of the 3-dimensional peptide matrix system was determined by a series of software simulations (using Deconvolute This! software; ref. 61), with the goal of minimizing the number of subsequent assays required to determine individual peptides containing T cell epitopes (61). The overlapping peptides spanning the H3N2 internal proteins with amino acid sequences that were different from their H5N1 equivalents were arranged into a smaller 2-dimensional peptide matrix system in which each peptide was represented in 2 different pools (295 peptides, peptide matrix system 2; Supplemental Table 1). An additional 2-dimensional matrix system for the overlapping peptides spanning the H5N1 surface glycoproteins (77 HA and 61 NA) was designed (138 peptides, peptide matrix system 3; Supplemental Table 1). The responses from positive peptide pools of the peptide matrix system 1 were deconvoluted using Deconvolute This! software (61).
Lymphocyte purification and ELISpot assays.
Six- to 12-hour IFN-γ ELISpot assays were performed using either freshly isolated or cryopreserved PBMCs as described previously (39). Influenza-specific T cells detected ex vivo using this method in humans have been previously characterized to be of memory phenotype (CD45RO+) (39). In view of the fact that little or no mRNA for IFN-γ is expressed in naïve CD4+ and CD8+ T cells (62), and that frequencies of antigen-specific naive T cell populations are very low (below the limit of direct detection by methods such as IFN-γ ELISpot assay) (63, 64), it was extremely unlikely that the ex vivo IFN-γ responses we detected were generated by naive T cell populations. All peptide pool screening assays were carried out with freshly separated PBMCs, whereas some individual peptide testing was done with cryopreserved PBMCs. No significant difference was observed between responses generated by fresh or cryopreserved PBMCs. PBMCs (250,000) were used in each ELISpot well in the final volume of 100 μl. The end concentration of peptide in each well was 2–3.5 μM when testing peptide pools and 2 μM when testing individual peptides. For negative and positive controls, PBMCs were incubated with media alone and with phytohemagglutinin (Wellcome), respectively. SFUs were counted using an automated ELISpot reader (AID). Responses were considered positive when the number of SFUs per well was greater than 3 times the average negative control value for the given assay and 4 times the mean negative control value (1.6 SFU/well) obtained from the initial peptide pool screening of all participants (median, 1 SFU/well; range, 0–7 SFU/well). Adjusted SFUs after subtracting average negative values were expressed as SFU per 106 PBMCs.
CD4+ and CD8+ T cell depletion.
Confirmed epitope-containing peptides generating more than 40 SFU/106 PBMCs (participants in the UK) and positive peptide pools generating more than 40 SFU/106 PBMCs (participants in Viet Nam) were characterized further by CD4+ and CD8+ T cell depletion of fresh PBMCs using anti-CD4 or anti-CD8 antibody–coated magnetic beads (Dynabeads; Invitrogen) according to the manufacturer’s protocol. CD4+ T cell–depleted and CD8+ T cell–depleted PBMC populations were tested in parallel to aid interpretation. After depletion, most subdominant responses (of less than 40 SFU/106 PBMCs) were detectable in neither CD4+ nor CD8+ T cell–depleted populations and therefore were excluded from the subsequent analysis of influenza-specific CD4+ and CD8+ T cell distribution.
Antigen-specific CD4+ and CD8+ T cell lines and clones.
Establishment of peptide-specific T cell lines has been previously described (65). Briefly, 3 × 106 to 5 × 106 PBMCs were pulsed as a pellet for 1 h at 37°C with 100 μM of peptides containing T cell epitope regions and cultured in R10 at 2 × 106 cells per well in a 24-well Costar plate. IL-2 was added to a final concentration of 100 U/ml on day 3. The cultures were restimulated after 14 d using autologous BCLs pulsed with the appropriate peptides. Antigen-specific CD4+ and CD8+ T cell clones were established by limiting dilution of the T cell lines as described previously (66).
Induction of influenza-specific polyclonal T cell culture.
Induction of influenza-specific short-term culture has been previously described (20, 21, 25). Briefly, freshly isolated PBMCs at 1.5 × 106 cells/ml were incubated with 4–5 HA units of influenza A/New Caledonia/20/99 (H1N1) or A/Sydney/5/97 (H3N2) in serum-free RPMI for 1 h at 37°C. FCS was then added to 10%, and the cells were incubated for 6–14 days.
Influenza A viruses.
The following virus strains were used: A/New Caledonia/20/99 (H1N1), A/Sydney/5/97 (H3N2), A/X31 (H3N2), and B/Hong Kong/330/2001 (a generous gift from Alan J. Hay, World Influenza Centre, National Institute for Medical Research, UK). Viruses were stored within allantoic fluid at –80°C.
Construction of rVACVs.
The M1 open reading frame (nucleotides 1–759) of the cDNA copy of RNA segment 7 from influenza A/VN/1194/2006 (H5N1) (kindly provided by Alan J. Hay) was amplified by PCR using the following primers designed to introduce a Kozak sequence around the initiating methionine codon (shown in bold) and unique NotI and SmaI restriction sites (underlined) for subcloning: 5′-AGAATGATATCGCGGCCGCCACCATGAGTCTTCTAACCGAGGTCGAAACGTA-3′ (5′ primer); 5′-GTGCAGATGCAGCGATTCAAGTGACCCGGGAAGGCCTAG-3′ (3′ primer).
The PCR product was purified from an agarose gel after electrophoresis, digested with EcoRV and SmaI, and cloned into a SmaI site of the VACV expression vector pSC11 (67) downstream of the 7.5K promoter, and the resultant plasmid was termed pSC11-H5N1 M1. The DNA sequence was confirmed by sequencing. A thymidine kinase–negative rVACV expressing the M1 was constructed as described (67–69) and termed H5N1 M1-VACV.
Similarly, a full-length cDNA copy of RNA segment 5 (encoding NP) from influenza A/VN/1194/2006 (H5N1) was amplified by PCR using the following primers containing NotI and SmaI restriction sites (underlined): 5′-ATAAGAATGCGGCCGCCACCATGGCGTCTCAAGGCACCAAACGA-3′ (5′ primer); 5′-AGACAATGCAGAGGAGTATGACAATTAACCCGGGAAGGCCTAG-3′ (3′ primer).
The M1 gene was removed from pSC11-H5N1 M1 by NotI and SmaI digestion, and the NP PCR product was digested with the same enzymes and cloned into the plasmid. The resultant plasmid was termed pSC11-H5N1 NP, and the fidelity of the DNA sequence was confirmed by sequencing. There was a single nucleotide difference from the published sequence (AY651498) at position 59 (A to G) that led to an amino acid change (Q20R). This plasmid was used to construct a rVACV expressing NP as for M1, and the virus was termed H5N1 NP-VACV. rVACVs expressing the M1 (21) or NP (19, 35) of the H1N1 and H3N2 human influenza A strains have been reported.
Target cell preparation.
EBV-transformed BCLs were washed 3 times and suspended in 100 μl serum-free RPMI. rVACV or influenza A virus was added at 3–10 pfu/cell. Wild-type VACV was used as a negative control. After incubating for 1.5 h at 37°C, cells were washed and resuspended in R10 medium and incubated for 4 h (influenza or VACV) or overnight (VACV). Infected cells were washed 3 times with R10 medium before being used for ICS or before being labeled with 200 μCi Na51Cr prior to cytotoxicity assays (20, 21). BCLs pulsed with appropriate peptides at 100 μM for 1 h at 37°C were also tested.
Cytotoxicity assay.
A standard 51Cr release assay was used as described previously (20, 21).
ICS and flow cytometry.
The following directly conjugated antibodies were obtained from BD Biosciences: IFN-γ (PE), TNF-α (APC), CD107a/b (FITC), IL-2 (APC), IL-4 (FITC), CD4 (perCP), and CD8 (perCP). Antigen-specific CD4+ and CD8+ T cell clones or virus-specific polyclonal short-term cultures were stimulated with virus-infected or peptide-pulsed target cells in the presence of anti-CD107a/b for 1 h and incubated with monensin (BD Biosciences) for a further 5 h at 37°C. Negative controls included unstimulated cells and target cells infected with influenza B/Hong Kong/330/2001 or wild-type VACV. CD4+ or CD8+ T cell populations producing cytokines were detected by flow cytometry (44).
Statistics.
The SPSS statistics package was used for all statistical analysis. The log-transformed mean magnitudes of total influenza-specific IFN-γ responses in the UK and Viet Nam groups were compared using the independent sample 2-tailed t test. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank all participants for providing blood samples used in this study and the medical staff of the Hospital for Tropical Medicine, Ho Chi Minh City, for their kind help with donor recruitment and sample collection. We thank Alan J. Hay for the generous gift of influenza viruses and a cDNA copy of influenza A/VN/1194/2006 (H5N1). We also thank Lisa Schimanski for kind help with molecular cloning techniques, Kati di Gleria for peptide synthesis, Ann Atzberger and Craig Waugh for cell sorting, Yash Bhasin for technical assistance with rVACV construction, Aleksandra Leligdowicz for sharing her experience of peptide matrix systems, Tim Rostron for HLA typing, and Mario Roederer for Deconvulute This! software. The study was funded by the UK Medical Research Council, the Wellcome Trust, and the Rhodes Trust (Rhodes Scholarship to L.Y.-H. Lee). G.L. Smith is a Wellcome Trust Principal Research Fellow.
Footnotes
Nonstandard abbreviations used: BCL, B cell line; ELISpot, enzyme-linked immunosorbent spot; ICS, intracellular cytokine staining; M1, matrix protein 1; NA, neuraminidase; NP, nucleoprotein; PB1, polymerase basic protein 1; rVACV, recombinant vaccinia virus; SFU, spot-forming unit.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:3478–3490 (2008). doi:10.1172/JCI32460
See the related Commentary beginning on page 3273.
References
- 1. WHO. 2008. Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. http://www.who.int/csr/disease/avian_influenza/country/cases_table_2008_06_19/en/index.html .
- 2.Abdel-Ghafar A.N., et al. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 3.Horimoto T., et al. Antigenic differences between H5N1 human influenza viruses isolated in 1997 and 2003. J. Vet. Med. Sci. 2004;66:303–305. doi: 10.1292/jvms.66.303. [DOI] [PubMed] [Google Scholar]
- 4.Li K.S., et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 5.WHO. 2007. Antigenic and genetic characteristics of H5N1 viruses and candidate H5N1 vaccine viruses developed for potential use as pre-pandemic vaccines. http://www.who.int/csr/disease/avian_influenza/guidelines/recommendationvaccine.pdf. [PubMed] [Google Scholar]
- 6.Bresson J.L., et al. Safety and immunogenicity of an inactivated split-virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: phase I randomised trial. Lancet. 2006;367:1657–1664. doi: 10.1016/S0140-6736(06)68656-X. [DOI] [PubMed] [Google Scholar]
- 7.Lin J., et al. Safety and immunogenicity of an inactivated adjuvanted whole-virion influenza A (H5N1) vaccine: a phase I randomised controlled trial. Lancet. 2006;368:991–997. doi: 10.1016/S0140-6736(06)69294-5. [DOI] [PubMed] [Google Scholar]
- 8.Treanor J.J., Campbell J.D., Zangwill K.M., Rowe T., Wolff M. Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N. Engl. J. Med. 2006;354:1343–1351. doi: 10.1056/NEJMoa055778. [DOI] [PubMed] [Google Scholar]
- 9.Leroux-Roels I., et al. Antigen sparing and cross-reactive immunity with an adjuvanted rH5N1 prototype pandemic influenza vaccine: a randomised controlled trial. Lancet. 2007;370:580–589. doi: 10.1016/S0140-6736(07)61297-5. [DOI] [PubMed] [Google Scholar]
- 10.Leroux-Roels I., et al. Broad Clade 2 cross-reactive immunity induced by an adjuvanted clade 1 rH5N1 pandemic influenza vaccine. PLoS ONE. 2008;3:e1665. doi: 10.1371/journal.pone.0001665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas P.G., Keating R., Hulse-Post D.J., Doherty P.C. Cell-mediated protection in influenza infection. Emerg. Infect. Dis. 2006;12:48–54. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rimmelzwaan G.F., Fouchier R.A.M., Osterhaus A.D.M.E. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr. Opin. Biotechnol. 2007;18:529–536. doi: 10.1016/j.copbio.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Yap K.L., Ada G.L., McKenzie I.F. Transfer of specific cytotoxic T lymphocytes protects mice inoculated with influenza virus. Nature. 1978;273:238–239. doi: 10.1038/273238a0. [DOI] [PubMed] [Google Scholar]
- 14.Lin Y.L., Askonas B.A. Biological properties of an influenza A virus-specific killer T cell clone. Inhibition of virus replication in vivo and induction of delayed-type hypersensitivity reactions. J. Exp. Med. 1981;154:225–234. doi: 10.1084/jem.154.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMichael A.J., Gotch F.M., Noble G.R., Beare P.A. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 1983;309:13–17. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- 16.Bennink J.R., Yewdell J.W., Gerhard W. A viral polymerase involved in recognition of influenza virus-infected cells by a cytotoxic T-cell clone. Nature. 1982;296:75–76. doi: 10.1038/296075a0. [DOI] [PubMed] [Google Scholar]
- 17.Townsend A.R., Skehel J.J. Influenza A specific cytotoxic T-cell clones that do not recognize viral glycoproteins. Nature. 1982;300:655–657. doi: 10.1038/300655a0. [DOI] [PubMed] [Google Scholar]
- 18.Townsend A.R., Skehel J.J. The influenza A virus nucleoprotein gene controls the induction of both subtype specific and cross-reactive cytotoxic T cells. J. Exp. Med. 1984;160:552–563. doi: 10.1084/jem.160.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yewdell J.W., Bennink J.R., Smith G.L., Moss B. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 1985;82:1785–1789. doi: 10.1073/pnas.82.6.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMichael A.J., Michie C.A., Gotch F.M., Smith G.L., Moss B. Recognition of influenza A virus nucleoprotein by human cytotoxic T lymphocytes. J. Gen. Virol. 1986;67:719–726. doi: 10.1099/0022-1317-67-4-719. [DOI] [PubMed] [Google Scholar]
- 21.Gotch F., McMichael A., Smith G., Moss B. Identification of viral molecules recognized by influenza-specific human cytotoxic T lymphocytes. J. Exp. Med. 1987;165:408–416. doi: 10.1084/jem.165.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor P.M., Askonas B.A. Influenza nucleoprotein-specific cytotoxic T-cell clones are protective in vivo. Immunology. 1986;58:417–420. [PMC free article] [PubMed] [Google Scholar]
- 23.Gelder C.M., Welsh K.I., Faith A., Lamb J.R., Askonas B.A. Human CD4+ T-cell repertoire of responses to influenza A virus hemagglutinin after recent natural infection. . J. Virol. 1995;69:7497–7506. doi: 10.1128/jvi.69.12.7497-7506.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jameson J., Cruz J., Ennis F.A. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 1998;72:8682–8689. doi: 10.1128/jvi.72.11.8682-8689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMichael A.J., Askonas B.A. Influenza virus-specific cytotoxic T cells in man; induction and properties of the cytotoxic cell. Eur. J. Immunol. 1978;8:705–711. doi: 10.1002/eji.1830081007. [DOI] [PubMed] [Google Scholar]
- 26.Biddison W.E., Shaw S., Nelson D.L. Virus specificity of human influenza virus-immune cytotoxic T cells. J. Immunol. 1979;122:660–664. [PubMed] [Google Scholar]
- 27.Gianfrani C., Oseroff C., Sidney J., Chesnut R.W., Sette A. Human memory CTL response specific for influenza A virus is broad and multispecific. Hum. Immunol. 2000;61:438–452. doi: 10.1016/S0198-8859(00)00105-1. [DOI] [PubMed] [Google Scholar]
- 28.Jameson J., Cruz J., Terajima M., Ennis F.A. Human CD8+ and CD4+ T lymphocyte memory to influenza A viruses of swine and avian species. . J. Immunol. 1999;162:7578–7583. [PubMed] [Google Scholar]
- 29.Kreijtz J., et al. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T lymphocyte populations directed to human influenza A virus. J. Virol. 2008;82:5161–5166. doi: 10.1128/JVI.02694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bui H.H., Peters B., Assarsson E., Mbawuike I., Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc. Natl. Acad. Sci. U. S. A. 2007;104:246–251. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Bree G.J., et al. Characterization of CD4+ memory T Cell responses directed against common respiratory pathogens in peripheral blood and lung. . J. Infect. Dis. 2007;195:1718–1725. doi: 10.1086/517612. [DOI] [PubMed] [Google Scholar]
- 32.Tang J., et al. Adenovirus hexon T-cell epitope is recognized by most adults and is restricted by HLA DP4, the most common class II allele. Gene Ther. 2004;11:1408–1415. doi: 10.1038/sj.gt.3302316. [DOI] [PubMed] [Google Scholar]
- 33.Mayer S., Laumer M., Mackensen A., Andreesen R., Krause S.W. Analysis of the immune response against tetanus toxoid: enumeration of specific T helper cells by the Elispot assay. Immunobiology. 2002;205:282–289. doi: 10.1078/0171-2985-00131. [DOI] [PubMed] [Google Scholar]
- 34.Papagno L., et al. Comparison between HIV- and CMV-specific T cell responses in long-term HIV infected donors. Clin. Exp. Immunol. 2002;130:509–517. doi: 10.1046/j.1365-2249.2002.02005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend A., et al. Defective presentation to class I-restricted cytotoxic T lymphocytes in vaccinia-infected cells is overcome by enhanced degradation of antigen. J. Exp. Med. 1988;168:1211–1224. doi: 10.1084/jem.168.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boon A.C., et al. Recognition of homo- and heterosubtypic variants of influenza A viruses by human CD8+ T lymphocytes. . J. Immunol. 2004;172:2453–2460. doi: 10.4049/jimmunol.172.4.2453. [DOI] [PubMed] [Google Scholar]
- 37.Gotch F., Rothbard J., Howland K., Townsend A., McMichael A. Cytotoxic T lymphocytes recognize a fragment of influenza virus matrix protein in association with HLA-A2. Nature. 1987;326:881–882. doi: 10.1038/326881a0. [DOI] [PubMed] [Google Scholar]
- 38.Ulmer J.B., et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 39.Lalvani A., et al. Rapid effector function in CD8+ memory T cells. . J. Exp. Med. 1997;186:859–865. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris A.G., Lin Y.L., Askonas B.A. Immune interferon release when a cloned cytotoxic T-cell line meets its correct influenza-infected target cell. Nature. 1982;295:150–152. doi: 10.1038/295150a0. [DOI] [PubMed] [Google Scholar]
- 41.de Jong M.D., et al. Fatal avian influenza A (H5N1) in a child presenting with diarrhea followed by coma. N. Engl. J. Med. 2005;352:686–691. doi: 10.1056/NEJMoa044307. [DOI] [PubMed] [Google Scholar]
- 42.Betts M.R., et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. . Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darrah P.A., et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 44.Duvall M.G., et al. Polyfunctional T cell responses are a hallmark of HIV-2 infection. Eur. J. Immunol. 2008;38:350–363. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goulding J., et al. Respiratory infections: do we ever recover? Proc. Am. Thorac. Soc. 2007;4:618–625. doi: 10.1513/pats.200706-066TH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boon A.C., et al. Preferential HLA usage in the influenza virus-specific CTL response. J. Immunol. 2004;172:4435–4443. doi: 10.4049/jimmunol.172.7.4435. [DOI] [PubMed] [Google Scholar]
- 47.Mbawuike I.N., Lange A.R., Couch R.B. Diminished influenza A virus-specific MHC class I-restricted cytotoxic T lymphocyte activity among elderly persons. Viral Immunol. 1993;6:55–64. doi: 10.1089/vim.1993.6.55. [DOI] [PubMed] [Google Scholar]
- 48.La Gruta N.L., et al. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. . Proc. Natl. Acad. Sci. U. S. A. 2006;103:994–999. doi: 10.1073/pnas.0510429103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selin L.K., et al. Attrition of T cell memory: selective loss of LCMV epitope-specific memory CD8 T cells following infections with heterologous viruses. Immunity. 1999;11:733–742. doi: 10.1016/S1074-7613(00)80147-8. [DOI] [PubMed] [Google Scholar]
- 50.Epstein S. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J. Infect. Dis. 2006;193:49–53. doi: 10.1086/498980. [DOI] [PubMed] [Google Scholar]
- 51.McMichael A.J., Gotch F.M., Dongworth D.W., Clark A., Potter C.W. Declining T-cell immunity to influenza, 1977–82. Lancet. 1983;2:762–764. doi: 10.1016/s0140-6736(83)92297-3. [DOI] [PubMed] [Google Scholar]
- 52.Sidney J., Grey H.M., Kubo R.T., Sette A. Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs. Immunol. Today. 1996;17:261–266. doi: 10.1016/0167-5699(96)80542-1. [DOI] [PubMed] [Google Scholar]
- 53.Sidney J., Peters B., Frahm N., Brander C., Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bui H.H., et al. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roti M., et al. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. . J. Immunol. 2008;180:1758–1768. doi: 10.4049/jimmunol.180.3.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang M., et al. CTL epitopes for influenza A including the H5N1 bird flu; genome-, pathogen-, and HLA-wide screening. Vaccine. 2007;25:2823–2831. doi: 10.1016/j.vaccine.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 57.Peters B., et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nichol K.L., et al. Effectiveness of live, attenuated intranasal influenza virus vaccine in healthy, working adults: a randomized controlled trial. JAMA. 1999;282:137–144. doi: 10.1001/jama.282.2.137. [DOI] [PubMed] [Google Scholar]
- 59.Belshe R.B., et al. Efficacy of vaccination with live attenuated, cold-adapted, trivalent, intranasal influenza virus vaccine against a variant (A/Sydney) not contained in the vaccine. J. Pediatr. 2000;136:168–175. doi: 10.1016/S0022-3476(00)70097-7. [DOI] [PubMed] [Google Scholar]
- 60.Suguitan A.L., Jr., et al. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 2006;3:e360. doi: 10.1371/journal.pmed.0030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roederer M., Koup R.A. Optimized determination of T cell epitope responses. J. Immunol. Methods. 2003;274:221–228. doi: 10.1016/S0022-1759(02)00423-4. [DOI] [PubMed] [Google Scholar]
- 62.Seder R.A., Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. . Nat. Immunol. 2003;4:835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- 63.Butz E.A., Bevan M.J. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. . Immunity. 1998;8:167–175. doi: 10.1016/S1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blattman J.N., et al. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J. Exp. Med. 2002;195:657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rowland-Jones S.L., et al. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Invest. 1998;102:1758–1765. doi: 10.1172/JCI4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dong T., et al. An HLA-B35-restricted epitope modified at an anchor residue results in an antagonist peptide. Eur. J. Immunol. 1996;26:335–339. doi: 10.1002/eji.1830260210. [DOI] [PubMed] [Google Scholar]
- 67.Chakrabarti S., Brechling K., Moss B. Vaccinia virus expression vector: coexpression of beta-galactosidase provides visual screening of recombinant virus plaques. Mol. Cell. Biol. 1985;5:3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith G.L., Murphy B.R., Moss B. Construction and characterization of an infectious vaccinia virus recombinant that expresses the influenza hemagglutinin gene and induces resistance to influenza virus infection in hamsters. Proc. Natl. Acad. Sci. U. S. A. 1983;80:7155–7159. doi: 10.1073/pnas.80.23.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mackett M., Smith G.L., Moss B. General method for production and selection of infectious vaccinia virus recombinants expressing foreign genes. J. Virol. 1984;49:857–864. doi: 10.1128/jvi.49.3.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Linnemann T., Jung G., Walden P. Detection and quantification of CD4(+) T cells with specificity for a new major histocompatibility complex class II-restricted influenza A virus matrix protein epitope in peripheral blood of influenza patients. J. Virol. 2000;74:8740–8743. doi: 10.1128/JVI.74.18.8740-8743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McMichael A.J., Gotch F., Cullen P., Askonas B., Webster R.G. The human cytotoxic T cell response to influenza A vaccination. Clin. Exp. Immunol. 1981;43:276–284. [PMC free article] [PubMed] [Google Scholar]
- 72.Adler S., Reay P., Roy P., Klenk H.D. Induction of T cell response by bluetongue virus core-like particles expressing a T cell epitope of the M1 protein of influenza A virus. Med. Microbiol. Immunol. 1998;187:91–96. doi: 10.1007/s004300050078. [DOI] [PubMed] [Google Scholar]
- 73.DiBrino M., et al. Identification of the peptide binding motif for HLA-B44, one of the most common HLA-B alleles in the Caucasian population. Biochemistry. 1995;34:10130–10138. doi: 10.1021/bi00032a005. [DOI] [PubMed] [Google Scholar]
- 74.Brett S.J., et al. Human T cell recognition of influenza A nucleoprotein. Specificity and genetic restriction of immunodominant T helper cell epitopes. J. Immunol. 1991;147:984–991. [PubMed] [Google Scholar]
- 75.DiBrino M., et al. HLA-A1 and HLA-A3 T cell epitopes derived from influenza virus proteins predicted from peptide binding motifs. J. Immunol. 1993;151:5930–5935. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.