Abstract
Saccharomyces cerevisiae is dimorphic and switches from a yeast form to a pseudohyphal (PH) form when starved for nitrogen. PH cells are elongated, bud in a unipolar manner, and invade the agar substrate. We assessed the requirements for actin in mediating the dramatic morphogenetic events that accompany the transition to PH growth. Twelve “alanine scan” alleles of the single yeast actin gene (ACT1) were tested for effects on filamentation, unipolar budding, agar invasion, and cell elongation. Some act1 mutations affect all phenotypes, whereas others affect only one or two aspects of PH growth. Tests of intragenic complementation among specific act1 mutations support the phenotypic evidence for multiple actin functions in filamentous growth. We present evidence that interaction between actin and the actin-binding protein fimbrin is important for PH growth and suggest that association of different actin-binding proteins with actin mediates the multiple functions of actin in filamentous growth. Furthermore, characterization of cytoskeletal structure in wild type and act1/act1 mutants indicates that PH cell morphogenesis requires the maintenance of a highly polarized actin cytoskeleton. Collectively, this work demonstrates that actin plays a central role in fungal dimorphism.
INTRODUCTION
Genetic and biochemical studies in the budding yeast Saccharomyces cerevisiae have provided important insights into the cellular functions of actin and actin-binding proteins. More than 80 sequenced mutations affecting the single yeast actin gene (ACT1) have been identified (Ayscough and Drubin, 1996). Collectively, these mutations affect many aspects of cell growth, including secretion (Novick and Botstein, 1985), endocytosis (Riezman et al., 1996), bipolar bud site selection (Yang et al., 1997), and organellar trafficking (Drubin et al., 1993; Simon et al., 1995). Numerous yeast actin-binding proteins have also been identified using genetic, biochemical, and two-hybrid approaches. Mutations in genes encoding many such proteins perturb actin cytoskeletal structure and/or function (for examples and reviews, see Drubin et al., 1988, 1990; Adams and Botstein, 1989; Adams et al., 1989; Liu and Bretscher, 1989; Amatruda et al., 1990; Welch et al., 1994; Amberg et al., 1995, 1997; Brower et al., 1995; Ayscough and Drubin, 1996; Riezman et al., 1996; Botstein et al., 1997).
Most observations regarding the yeast actin cytoskeleton have been made on yeast form (YF)1 cells grown in the presence of ample nutrients. Under these conditions, diploid S. cerevisiae grow by budding to produce oval-shaped daughters that separate from the mother cell at cytokinesis and remain on the surface of the agar substrate. However, S. cerevisiae is dimorphic. When starved for nitrogen in the presence of a fermentable carbon source, S. cerevisiae switches to a pseudohyphal (PH), or filamentous, mode of growth (Gimeno et al., 1992). PH cells are much longer and thinner than YF cells, remain attached to mothers after cytokinesis, and invade the agar substrate. These growth characteristics, coupled with a unipolar budding pattern, with daughters predominantly budding away from mothers, lead to growth of filaments of long cells away from the body of the colony.
Differences in the polarity of the actin cytoskeleton between YF and PH cells suggest a specialized role for the actin cytoskeleton in PH growth. The yeast actin cytoskeleton comprises two major structures, patches and cables. Actin patches are punctate “dots” of filamentous actin present at the cell cortex, whereas cables are oriented along the mother–bud axis and have been described as cytoplasmic (Adams and Pringle, 1984). Patches are highly mobile (Doyle and Botstein, 1996; Waddle et al., 1996) and are localized to areas of new cell growth throughout the cell cycle. In PH cells, actin patches are polarized to the distal tip of the daughter cell throughout bud growth and then repolarize to the site of septation shortly before cytokinesis (Kron et al., 1994). This behavior contrasts with that of patches in YF cells, where there is a distinct period between bud emergence and cytokinesis when patches are distributed around the entire cortex of the emerging bud (the isotropic growth phase) (Lew and Reed, 1993). These differences indicate that actin cytoskeletal structure and dynamics are regulated differently in YF and PH cells.
Genetic analysis also indicates that the actin cytoskeleton has an important function in PH growth. In a general screen for mutants defective for filamentation, Mösch and Fink (1997) identified mutations in several genes encoding cytoskeletal proteins. These include the genes encoding the actin-binding proteins Tpm1p (tropomyosin), Srv2p (cyclase-associated protein), and the formin homology domain protein Bni1p. Deletion of SLA2, which encodes a protein with homology to the focal adhesion protein talin, was also shown to block filamentation (Yang et al., 1997).
Here we have investigated the functional requirements for actin in mediating the dramatic changes in agar invasion, bud site selection pattern, and cell shape that accompany the transition from growth in the YF to PH growth. This work indicates that actin has multiple functions in filamentous growth and provides new insight into actin structure–function relationships. In addition, image analysis of wild type and act1/act1 mutants suggests an important role for cortical actin patch polarization in the morphogenesis of the PH cell.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Microbiological Methods
Yeast strains used in this study are isogenic and derived from the Σ1278b strain background (Table 1). Standard media and methods of sporulation, tetrad dissection, and scoring of mating type were as previously described (Guthrie and Fink, 1991). Synthetic low-ammonia dextrose (SLAD) medium has also been described (Gimeno et al., 1992). Plasmids used in this study have been previously described: pIL30 (Laloux et al., 1994), pRS314 and pRS316 (Sikorski and Hieter, 1989), and AAB117 and AAB289 (Brower et al., 1995). Transformation was performed by the lithium acetate transformation method (Ito et al., 1983; Gietz et al., 1992).
Table 1.
Yeast strains
| Strain | Genotype | Plasmids |
|---|---|---|
| BCY116 | MATa/MATα his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | |
| BCY333 | MATa/MATα his3::hisG/HIS3 ura3–52/ura3–52 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | |
| BCY144 | MATa/MATα act1Δ::LEU2/ACT1 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | |
| BCY118 | MATa/MATα act1Δ::LEU2/ACT1 his3::hisG/HIS3 ura3–52/ura3–52 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pRS316 |
| BCY156 | MATa/MATα act1–109::HIS3/ACT1 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY157 | MATa/MATα act1–110::HIS3/ACT1 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY168 | MATa/MATα act1–131::HIS3/ACT1 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY199 | MATa/MATα act1–124::HIS3/act1–124::HIS3 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY201 | MATa/MATα act1–113::HIS3/act1–113::HIS3 his3::hisG/his3::hisG ura 3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY202 | MATa/MATα act1–112::HIS3/act1–112::HIS3 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY204 | MATa/MATα act1–111::HIS3/act1–111::HIS3 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY209 | MATa/MATα ACT1::HIS3/ACT1::HIS3 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/ leu2::hisG | pIL30 |
| BCY210 | MATa/MATα act1–120::HIS3/act1–120::HIS3 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY212 | MATa/MATα act1–117::HIS3/act1-117::HIS3 his3::hisG/his3::HisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY231 | MATa/MATα act1–129::HIS3/act1–129:HIS3 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY300 | MATa/MATα act1–104::HIS3/act1–104::HIS3 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY405 | MATa/MATα act1–132::HIS3/act1–132::HIS3 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY310 | MATa/MATα act1–111::HIS3/act1–120::HIS3 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY302 | MATa/MATα act1–111::HIS3/act1–112::HIS3 his3::hisG/his3::hisG ura3–52/URA3 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30 |
| BCY261 | MATa/MATα sac6Δ::LEU2/SAC6 his3::hisG/HIS3 ura3–52/ura3–52 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | |
| BCY372 | MATa/MATα sac6Δ::LEU2/sac6Δ::LEU2 his3::hisG/HIS3 ura3–52/ura3–52 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pRS316 |
| BCY411 | MATa/MATα act1–120::HIS3/act1–120::HIS3 his3::hisG/his3::hisG ura3–52/ura3–52 trp1::hisG/trp1::hisG leu2::hisG/leu2::hisG | pIL30, AAB289, pRS314 |
| BCY412 | MATa/MATα act1–120::HIS3/act1–120::HIS3 his3::hisG/his3::hisG ura3–52/ura3–52 trp1::hisG/trp1::hisG leu2::hisG/leu2::hisG | pIL30, AAB117, pRS314 |
| BCY432 | MATa/MATα act1–111::HIS3/act1–111::HIS3 his3::hisG/his3::hisG ura3–52/ura3–52 trp1::hisG/TRP1 leu2::hisG/leu2::hisG | pIL30, AAB289 |
| BCY433 | MATa/MATα act1–124::HIS3/act1–124::HIS3 his3::hisG/his3::hisG ura3–52/ura3–52 leu2::hisG/leu2::hisG | pIL30, AAB289 |
Genetic Manipulations and Strain Constructions
Actin mutant strains were constructed by single-step gene replacement using donor DNA fragments containing the desired act1 allele with HIS3 integrated just downstream of the act1 coding sequence (act1-x::HIS3) and an act1Δ::LEU2/ACT1 heterozygous diploid as recipient. To generate the act1Δ::LEU2/ACT1 heterozygote for eventual gene replacement, an ACT1 disruption construct was made by first subcloning the 3.8-kb EcoRI fragment containing ACT1 from pKFW29 (Wertman et al., 1992) into the EcoRI site of pUC19 to generate BCB109. BCB109 was digested with BclI and XhoI and ligated in the presence of an XhoI–BglII fragment from YEp13 that contains LEU2. The resultant act1Δ::LEU2 construct in pUC19 (BCB207) deletes all but the first three amino acids of the ACT1 coding region and contains 1246 bp upstream and 1118 bp downstream of ACT1. BCY116 was transformed with the ApaI–SmaI fragment of BCB207, and Leu+ transformants were identified. Genomic DNA from 27 Leu+ transformants was tested by PCR for linkage of a LEU2-specific oligonucleotide (LEU2-P3: 5′-GATTTCTTGACCAACGTGGTC) with an oligonucleotide (ACT1–8: 5′-TTCAAACTGCTGTTGATCGG) specific to a genomic region just outside that contained in BCB207 and oriented toward LEU2-P3. Twelve transformants gave the 2.0-kb product expected for a disruptant. Three of these putative heterozygotes segregated two viable Leu−:2 lethal spores, confirming heterozygosity for act1Δ::LEU2. One such heterozygote was kept as BCY144.
To introduce the act1 alleles by gene replacement, BCY144 was transformed independently with each of the EcoRI-digested plasmid derivatives of pKFW46 carrying the relevant act1 alleles with HIS3 integrated downstream (Wertman et al., 1992). His+ transformants were selected and screened for those that were Leu−. Two independent His+ Leu− transformants were retained as candidate act1-x::HIS3/ACT1 heterozygotes, where x represents any given act1 allele. As HIS3 is tightly linked to each of these alleles, they can be followed in crosses by monitoring histidine prototrophy. Heterozygotes for each of the alleles were sporulated, and the tetrads were dissected. Nine alleles were viable as haploids at 30°C. Strains heterozygous for three of the alleles (act1–109, act1–110, and act1–131) segregated two viable His−:2 lethal spores, indicating that these act1 alleles are recessive lethal in Σ1278b, as they are in S288c (Wertman et al., 1992). Presence of the desired act1 mutation was confirmed by restriction analysis of an act1 PCR product amplified from the genome. This assignment was possible because each of the act1 alleles analyzed in this study creates or destroys a restriction site in ACT1 (Wertman et al., 1992). To confirm the presence of the act1 mutation in each of these strains, act1 was amplified by PCR with Taq polymerase using oligonucleotides act1–4 (5′-CTTTTCCTTAAAAATACTTTATTA) and ACT1–10 (5′-GTACTAACATCGATTGCTTC) under standard conditions. The expected 1275-bp product was digested with enzymes diagnostic for each mutation as previously described (Wertman et al., 1992). Restriction fragments of the size expected for the appropriate allele were seen in all cases. For all viable alleles, act1 was amplified from two independent His+ haploids segregated from the original heterozygous diploids. For the recessive lethal alleles act1–109, act1–110, and act1–131, the ACT1 alleles were amplified from the heterozygous diploid, and both wild-type and mutant restriction patterns were observed.
act1/act1 homozygotes were constructed by first transforming act1-x::HIS3/ACT1 heterozygous diploids to Leu+ with the LEU2 CEN plasmid pIL30 (Laloux et al., 1994). Sporulation, dissection, and mating of appropriate haploids were then used to generate prototrophic diploids homozygous for each of the viable act1 alleles (Table 1). For analysis of the recessive lethal alleles, the original act1-x::HIS3/ACT1 heterozygotes were transformed to Leu+ with pIL30 and used for subsequent analysis.
To generate a sac6Δ::LEU2/sac6Δ::LEU2 diploid, a 2.9-kb genomic fragment carrying the sac6Δ::LEU2 allele of AAY1047 (Adams et al., 1995) was generated by PCR with oligonucleotides SAC6-1 (5′-GCAGTTAAAGGTGCTTTGTC) and SAC6-2 (5′-AAAGTTCACAGGATATATGG) and used to transform BCY333 to Leu+. Candidate sac6Δ::LEU2/SAC6 heterozygotes were tested by PCR to identify linkage of oligonucleotides LEU2-P3 and SAC6-3 (5′-GCTCAAGGACGCCCCATTTG); one transformant that gave the appropriate product was sporulated and dissected; sac6Δ::LEU2 was confirmed to segregate 2:2. Mating of appropriate haploids from this dissection and subsequent transformation with pRS316 generated the sac6Δ::LEU2/sac6Δ::LEU2 homozygote BCY372.
Assessment of Filamentation, Cell Elongation, and Invasion Phenotypes
Strains to be analyzed were streaked for well-isolated single colonies on SLAD plates, with no more than four strains per plate. Plates were incubated for 4 d at 30°C, and well-isolated colonies were scored for filamentation. Strains with no filaments extending beyond the perimeter of the colony were scored as (−) for filamentation; those with 1–10% of colonies showing some filaments were scored as (+). (++) indicates that 80–100% of colonies had filaments, yet these filaments were disorganized relative to wild type. (+++) = wild-type filamentation.
Cell elongation was quantified by growing colonies as above, scraping cells from the agar with a toothpick into 10 μl of water, and scoring morphology on a hemacytometer. Cells with length/width ratios ≥2 were scored as long cells.
To score invasion, the same plates used to score filamentation and cell elongation were washed with a stream of water. Noninvasive cells were removed by gently rubbing the agar surface. Those strains with <10% of washed colonies having residual cells in the agar, and only in the center of the colony, were scored as (−). (+) denotes strains with 10–50% of washed colonies having residual cells. Typically, only cells in the center of the colony had invaded in this class. Mutants that showed 50–100% of colonies with invaded cells were scored either as (++) or (+++) depending on whether part (++) or all (+++) of the colony diameter contained invaded cells.
Preparation of PH Cells for Rhodamine–Phalloidin and Calcoflour Staining
Strains to be assayed were grown overnight at 30°C in 5 ml of YNB. Cells were washed once in 5 ml of water, spread on the surface of a 10.5-cm SLAD plate, and incubated 24–48 h at 30°C. For rhodamine–phalloidin staining studies, prestarved cells were collected in water and serially diluted in 1.25% alginate (wt/vol) (Sigma, St. Louis, MO; catalog number A-2158) in water. Suspensions of cells (0.5 ml) were spread thinly over the surface of a SLAD agar plate, alginate was allowed to solidify, and plates were incubated 48 h at 30°C. For fixation, alginate was removed with forceps from plates with well-separated single colonies and placed in 6 ml of 0.8 M EGTA/3.7% formaldehyde in a 15-ml polypropylene tube (chelation of calcium by EGTA causes alginate to liquefy). After 1 h with intermittent shaking, cells were washed three times in 1× PBS. Rhodamine–phalloidin staining was performed as described previously (Kron et al., 1994). For calcoflour staining, prestarved cells were grown in SLAD liquid overlay media, fixed, and stained as previously described (Kron et al., 1994).
Microscopic Methods
Cells fixed and stained as above were mounted in 90% glycerol/1× PBS and viewed using epiflourescence on a Nikon (Garden City, NY) E6 inverted microscope, controlled by the Applied Precision (Issaquah, WA) DeltaVision wide-field microscope system. Fluorescent images were obtained at 100× magnification (numerical aperture, 1.4) and captured with a Princeton Instruments (Trenton, NJ) cooled charged-couple device camera. Stacks of rhodamine and DAPI fluorescence images were captured at 0.2-μm intervals in the z-plane (1- to 3-s image capture time), and out-of-focus fluorescence was removed by iterative deconvolution (Agard et al., 1989; Scalettar et al., 1996). Three-dimensional representations of these two-dimensional stacks were then generated, and rotated views through the stack were obtained and used to generate stereo images.
RESULTS
To determine the functional requirements for actin during PH growth, we have taken advantage of an extant set of “charged-to-alanine scan” alleles of ACT1 (Wertman et al., 1992). These mutations provide a number of distinct advantages for dissecting the roles of actin in PH growth. First, the residues affected by these mutations are known and can be mapped to the crystal structure of actin. Collectively these mutations affect a large portion of the solvent-exposed surface of the actin monomer. Second, the effects of these mutations on a number of actin-dependent processes have been well documented (for examples, see Read et al., 1992; Drubin et al., 1993; Smith et al., 1995). Third, the effects of these alanine scan alleles on the binding of many actin-interacting proteins have been determined (Holtzman et al., 1994; Honts et al., 1994; Amberg et al., 1995, 1997). This detailed information on the functions affected by these alleles provides an invaluable guide in assessing the possible roles of actin and actin-binding proteins in PH growth.
Actin Mutants Exhibit Filamentation Defects
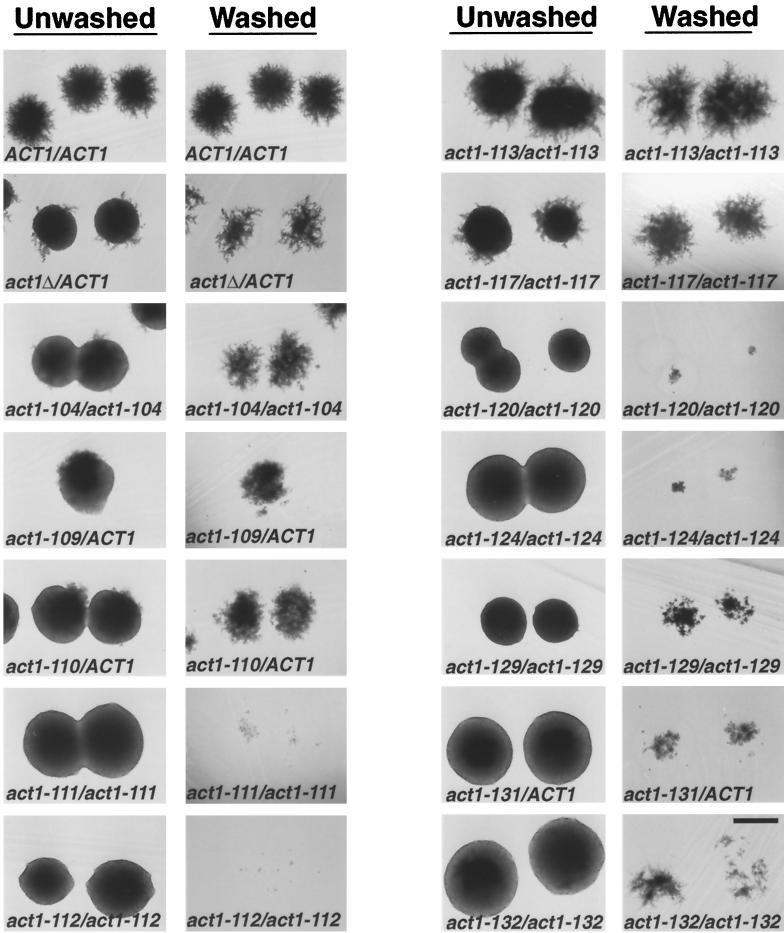
Twelve alanine scan act1 alleles were introduced into a strain (a Σ1278b derivative) capable of PH growth (Gimeno et al., 1992) (see MATERIALS AND METHODS for details). These constructions were necessary because the act1 alleles were originally studied in S288c (Wertman et al., 1992), a background that does not exhibit PH growth (Liu et al., 1996). Nine of these alleles are viable as haploids at 30°C, whereas three alleles (act1–109, act1–110, and act1–131) are recessive lethal in Σ1278b, as they are in S288c (Wertman et al., 1992).
As haploids do not make florid pseudohyphae (Gimeno, et al., 1992; Mösch and Fink, 1997), diploids homozygous for each of the viable alleles were constructed. Growth of all act1/ACT1 heterozygotes and viable act1/act1 homozygotes was assessed on YPD agar plates at 30 and 36°C to determine the recessive and dominant effects of these mutations on YF growth and viability in the Σ1278b strain background (Table 2). Strains homozygous for seven of the nine recessive viable alleles (act1–111, act1–112, act1–113, act1–120, act1–124, act1–129, and act1–132) exhibit temperature-sensitive growth; only two of these alleles (act1–111 and act1–132) cause slow growth at 30°C. Diploids homozygous for the recessive viable alleles act1–104 and act1–117 grow as well as wild type at both 30 and 36°C. All alleles show the same general trends of temperature sensitivity and dominance as previously reported for these mutations in S288c with the exception of act1–112, which has a dominant effect on growth in Σ1278b but was reported to be recessive in S288c (Wertman et al., 1992).
Table 2.
Summary of growth phenotypes of actin mutants on YPD
| Allele | Growth of homozygote at 30°C | Growth of homozygote at 36°C | Dominance |
|---|---|---|---|
| WT | +++ | ++ | |
| 104 | +++ | ++ | |
| 109 | ND | ND | Recessive |
| 110 | ND | ND | Dominant |
| 111 | ++ | − | Recessive |
| 112 | +++ | +/− | Dominant |
| 113 | +++ | +/− | Recessive |
| 117 | +++ | ++ | |
| 120 | +++ | − | Recessive |
| 124 | +++ | +/− | Recessive |
| 129 | +++ | +/− | Recessive |
| 131 | ND | ND | Dominant |
| 132 | + | − | Recessive |
WT, wild-type; ND, not determined, as these mutations are recessive lethal. Growth phenotypes were determined by spotting serial dilutions of a saturated culture and assessing plating efficiency and size of colonies after 3 d growth on YPD agar plates. Dominance refers specifically to effect of heterozygous mutation on growth on YPD at 36°C.
To determine the effects of actin mutations on PH growth, strains homozygous for the recessive viable act1 alleles and strains heterozygous for act1Δ::LEU2 or the recessive lethal act1 alleles were streaked to SLAD media to induce PH growth, and the morphology of the resulting colonies was examined after 4 d growth at 30°C (Figure 1, unwashed colonies). Of the nine viable alleles analyzed, all but act1–113 cause filamentation defects when homozygous. The severity of the filamentation phenotype is allele specific. For example, act1–112/act1–112 mutants make no filaments, whereas act1–117/act1–117 strains make many clumpy and disorganized filaments. Each of the recessive lethal alleles (act1–109, act1–110, and act1–131) has a dominant effect on PH growth, with act1–131 showing the most severe phenotype. A decrease in actin dosage also has a dominant effect on PH growth, as the act1Δ::LEU2/ACT1 heterozygote shows reduced filamentation. However, the phenotype of the act1Δ::LEU2/ ACT1 heterozygote is less severe than that of strains heterozygous for act1–109, act1–110, and act1–131.
Figure 1.
Filamentation and invasion phenotypes of actin mutants. Strains of indicated genotype were streaked for single colonies on SLAD agar and incubated for 4 d at 30°C. Complete genotypes of strains are listed in Table 1. Colonies were photographed before (unwashed column) and after (washed column) washing the surface of the agar to remove noninvaded cells. Bar, 500 μm.
Allele-specific Effects of Actin Mutations on Invasion, Cell Elongation, and Unipolar Bud Site Selection
The actin alleles were tested for their effects on other features of filamentous growth: 1) invasion—PH cells can invade the agar substrate, whereas YF cells do not; 2) cell elongation—PH cells are much longer and thinner than their YF counterparts; and 3) unipolar bud site selection—PH daughters tend to bud primarily from the pole opposite the birth end of the mother (the distal end). In contrast, YF diploids bud in a bipolar manner—the first and second daughters tend to bud from the distal end of the mother cell, and subsequent daughters bud with equal likelihood from either the proximal or distal poles. The subsequent section describes the analysis, the results of which are summarized in Table 3.
Table 3.
Summary of effects of actin mutations on filamentous growth-specific properties.
| Relevant Genotype | Filamentation | Invasion | Cell Elongation | Budding Pattern |
|---|---|---|---|---|
| ACT1/ACT1 | +++ | +++ | +++ | Unipolar |
| act1–104/act1–104 | + | +++ | ++ | Unipolar |
| act1–109/ACT1 | + | +++ | ++ | Random |
| act1–110/ACT1 | + | +++ | + | Random |
| act1–111/act1–111 | − | − | +++ | ND |
| act1–112/act1–112 | − | − | − | Random |
| act1–113/act1–113 | +++ | +++ | +++ | Unipolar |
| act1–117/act1–117 | ++ | +++ | ++ | Bipolar |
| act1–120/act1–120 | − | − | − | Random |
| act1–124/act1–124 | − | + | + | Random |
| act1–129/act1–129 | − | ++ | + | Random |
| act1–131/ACT1 | − | + | + | Random |
| act1–132/act1–132 | − | + | +++ | ND |
Filamentation and invasion were scored as described in MATERIALS AND METHODS. Cell elongation scores were determined by comparing percent long cell values of mutant and wild type shown in Figure 2A. Mutants whose percent long cell value (plus the SE) was between 24 and 34% (the wild type value ± 1 SE) were scored as +++, those between 14 and 24% as ++, those between 4 and 14% as +, and those <4% as −.
Agar Invasion.
Colonies grown on SLAD plates were compared before and after washing (Figure 1). Five alleles (act1–104, act1–109, act1–110, act1–113, and act1–117) have little or no effect on agar invasion. However, the remaining seven alleles show defects ranging from a slight reduction to virtually complete elimination of agar invasion. For example, act1–129/act1–129 homozygotes invade slightly less well than ACT1 strains, whereas act1–112/act1–112 homozygotes show an extreme invasion defect.
Cell Elongation.
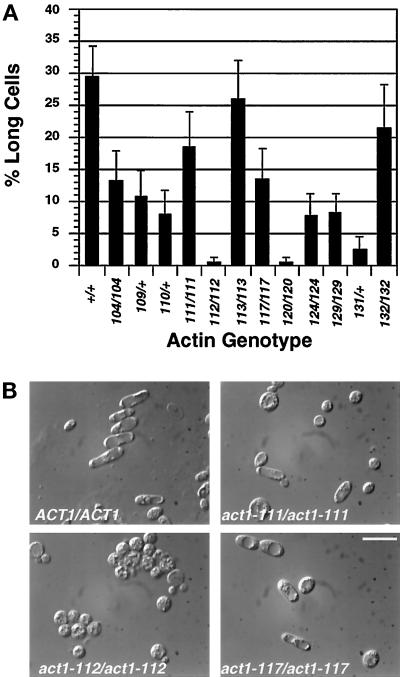
The percentage of long cells made under conditions that induce PH growth was determined for both the wild type and mutant strains harboring actin alanine scan alleles shown in Figure 1. The different alleles have distinct effects on elongation. Most mutants make from 30 to 80% of wild-type numbers of long cells, but act1–112/act1–112 and act1–120/act1–120 strains make virtually no long cells (Figure 2A).
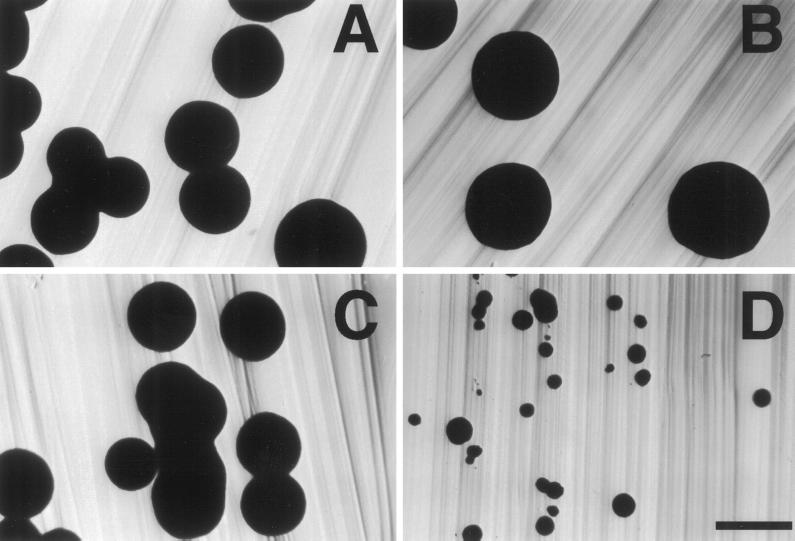
Figure 2.
Effect of act1 mutations on cell elongation. (A) Percentage of long cells in each of the actin mutants. Complete genotypes of strains are as indicated in Figure 1. Long cell counts were performed as described in MATERIALS AND METHODS. Bars show the SE of two independent experiments. (B) Examples of different cell elongation phenotypes. Images shown are differential interference contrast images of cells scraped from the agar as above. Bar, 10 μm.
Interestingly, the effect of a given mutation on cell elongation does not correlate with its effect on invasion. For example, both act1–111 and act1–112 block invasion (Figure 1), but act1–111/act1–111 strains make substantial numbers of long cells, whereas act1–112/act1–112 homozygotes do not (Figure 2, A and B). Thus, cell elongation and invasion appear to require different functions of actin that are differentially affected by certain mutations.
Unipolar Budding
Many mutations affecting actin (as well as mutations affecting other cytoskeletal proteins) cause random budding in YF diploids but have no effect on axial budding in haploids (Drubin et al., 1993; Sivadon et al., 1995; Haarer et al., 1996; Zahner et al., 1996; Yang et al., 1997). We examined whether this subset of actin mutations affects the unipolar budding pattern that diploids exhibit during PH growth. The budding patterns of 10 of the 12 act1/act1 mutants grown under PH growth-inducing conditions were assessed (act1–111/act1–111 and act1–132/act1–132 homozygotes could not be scored because they exhibit abnormal chitin deposition and high background staining with calcoflour). Seven alleles cause random budding under conditions that induce PH growth, whereas three (act1–104, act1–113, and act1–117) do not (Table 3).
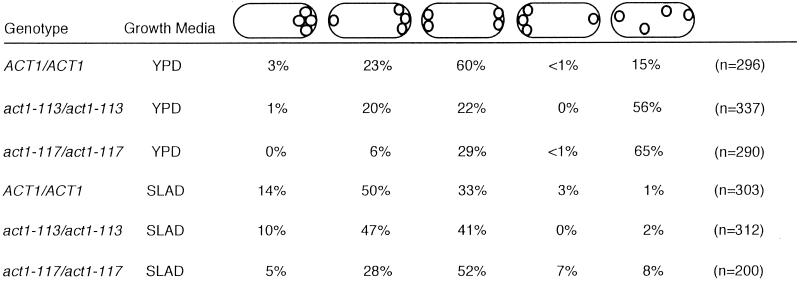
act1–113 and act1–117 are distinct because their effect on budding pattern is determined by nutritional conditions. act1–113/act1–113 and act1–117/act1–117 YF diploids were shown to bud in a random pattern in the S288c strain background (Yang et al., 1997). The same alleles do not cause random budding in the Σ1278b strain background under conditions that induce PH growth. Quantitative analysis of bud site selection patterns of ACT1/ACT1, act1–113/act1–113, and act1–117/act1–117 strains grown under PH growth-inducing and noninducing conditions shows that this difference reflects an effect of growth condition, not strain background (Table 4).When grown in rich medium, act1–113/act1–113 and act1–117/act1–117 diploids in the Σ1278b strain background show a random budding pattern similar to that seen for these alleles in S288c (Yang et al., 1997). However, when grown under conditions that induce PH growth, act1–117/act1–117 diploids exhibit a budding pattern that most closely resembles the bipolar budding pattern of wild-type YF cells. act1–113/act1–113 diploids bud in a unipolar manner similar to that of wild-type PH cells under such conditions.
Table 4.
Effect of growth condition on bud-site selection in ACT1/ACT1, act1–113/act1–113 and act1–117/act1–117 diploids
Exponential cultures grown in either SLAD/LA or YPD were fixed and stained with calcoflour as described in MATERIALS AND METHODS. Cells with four bud scars (or three scars and an emerging bud) were scored. The percentages of cells exhibiting the bud patterns schematized above are shown (numbers do not total to 100% because of rounding). Cells are shown with the birth end on the left.
Intragenic Complementation between act1 Alleles
Different act1 alleles with similar PH phenotypes might disrupt filamentation by affecting either the same or different aspects of actin function. If two mutations affect different functions required for PH growth, then they might complement each other for filamentations defects. Mutations affecting the same functions would not. To distinguish these possibilities, all heteroallelic combinations of the nine viable act1 alleles were constructed and tested for ability to complement each other for filamentation defects. The filamentation phenotypes of each of these strains were compared with those of strains homozygous and heterozygous for each allele (Table 5).
Table 5.
Filamentation phenotypes of act1 allelic combinations
| Allele | WT | Δ | 104 | 111 | 112 | 113 | 117 | 120 | 124 | 129 | 132 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | +++ | ++ | +++ | +++ | − | +++ | +++ | +++ | ++ | + | + |
| 104 | + | + | − | ++ | + | +++ | + | − | − | ||
| 111 | − | −a | +++ | ++ | +++ | − | − | − | |||
| 112 | − | − | − | − | − | − | − | ||||
| 113 | +++ | +++ | +++ | ++ | + | + | |||||
| 117 | ++ | +++ | + | − | + | ||||||
| 120 | − | + | + | − | |||||||
| 124 | − | − | − | ||||||||
| 129 | − | − | |||||||||
| 132 | − |
Filamentation of heteroallelic act1/act1 diploids was scored after 4 d growth at 30°C on SLAD agar plates as described in MATERIALS AND METHODS. WT, wild type.
Very poor growth of heteroallelic strain at 30°C (see Figure 4).
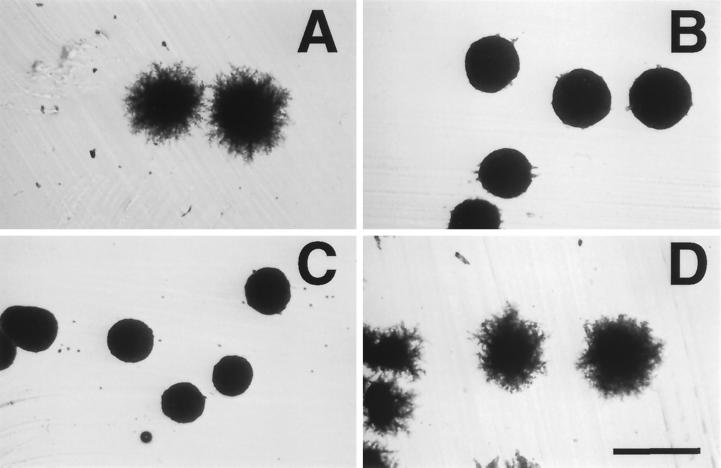
Both positive and negative interactions were observed in our complementation analysis. act1–120, which completely eliminates all aspects of PH growth as a homozygote, is able to complement the filamentation defects of five other act1 alleles (act1–104, act1–111, act1–117, act1–124, and act1–129). An example of this effect is shown in Figure 3. Although both act1–111/act1–111 and act1–120/act1–120 homozygotes fail to make filaments, act1–111/act1–120 heterozygotes exhibit filamentation indistinguishable from that of wild type. One striking negative interaction was also observed. As shown in Figure 4, act1–111/act1–112 strains grow much more poorly at 30°C than either act1–111/act1–111 or act1–112/act1–112 homozygotes.
Figure 3.
act1–111 and act1–120 exhibit intragenic complementation for filamentation defects. Shown is the growth on SLAD agar after four days at 30°C. (A) BCY209 (ACT1/ACT1); (B) BCY204 (act1–111/act1–111); (C) BCY210 (act1–120/act1–120); (D) BCY310 (act1–111/act1–120). Bar, 500 μm.
Figure 4.
act1–111 and act1–112 exhibit a negative growth interaction. Shown are colonies after growth for 3 d at 30°C on YNB. (A) BCY209 (ACT1/ACT1); (B) BCY204 (act1–111/act1–111); (C) BCY202 (act1–112/act1–112); (D) BCY302 (act1–111/act1–112). Bar, 1 mm.
Dominant Effects on PH Growth
The complementation analysis also revealed that some act1 alleles that are recessive with regard to YF growth and viability (Table 2) have dominant effects on PH growth (Table 5). Four alleles that reduce filamentation as homozygotes (act1–112, act1–124, act1–129, and act1–132) also reduce filamentation in act1/ACT1 heterozygotes, whereas another set (act1–104, act1–111, act1–117, and act1–120) are completely recessive. We considered the possibility that the dominance of these alleles is due to a reduction in actin dosage, as act1Δ/ACT1 heterozygotes show a reduction in filamentation (Figure 1 and Table 5). However, strains heterozygous for act1–112, act1–129, and act1–132 show stronger filamentation defects than the act1Δ/ACT1 control (Table 5). This result shows that the phenotype of these heterozygotes is due to a dominant effect of the mutant actin encoded by these alleles, rather than a simple reduction in actin levels.
The Role of the Fimbrin–Actin Interaction in PH Growth
Although act1–120/act1–120 homozygotes are defective for all aspects of PH growth investigated, this allele complements the PH growth defects of many other actin mutants (Table 5). Thus, act1–120 appears to encode a mutant actin that retains substantial function. Genetic and biochemical studies in yeast have established that act1–120 perturbs binding of the actin filament-bundling protein fimbrin (Sac6p) both in vitro and in vivo (Holtzman et al., 1994; Honts et al., 1994, Sandrock et al., 1997, Doyle and Botstein, unpublished observations). This is consistent with structural data showing that the residues affected by act1–120 (E99 and E100) are in a region of subdomain 1 of actin that forms extensive contacts with the amino-terminal domain of fimbrin (Hanein et al., 1997). Specific mutations affecting either of the two actin-binding domains of fimbrin can suppress the temperature-sensitive lethal phenotype of act1–120 (Brower et al., 1995; Sandrock et al., 1997).
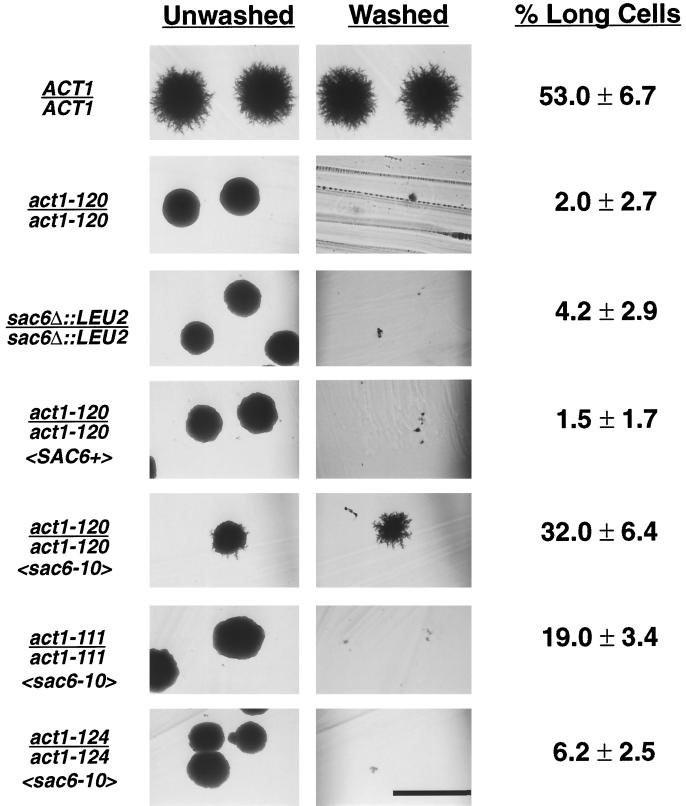
We propose that the major effect of act1–120 on PH growth is through its effect on fimbrin binding. This model can be directly tested by evaluating two important predictions: 1) sac6/sac6 and act1–120/act1–120 mutants will show similar PH growth defects; and 2) sac6 alleles that restore the interaction between fimbrin and the mutant actin encoded by act1–120 will suppress some or all of the PH growth defects of act1–120/act1–120 mutants. Both of these predictions are borne out. As shown in Figure 5, sac6Δ::LEU2/ sac6Δ::LEU2 diploids, like act1–120/act1–120 mutants, make no filaments, are invasion defective, and are severely reduced for cell elongation at 30°C. Expression of a mutant fimbrin encoded by sac6–10 rescues both the cell elongation and invasion phenotypes of these mutants. sac6–10 alters a conserved tryptophan in the first actin binding domain of fimbrin (W252C) and was shown to suppress the temperature-sensitive phenotype of act1–120, presumably by restoring the fimbrin–actin interaction (Brower et al., 1995; Sandrock et al., 1997). Importantly, sac6–10 does not suppress the PH growth defects of two actin mutations (act1–111 and act1–124) that affect residues in subdomains 4 and 2, respectively, and are not predicted to interfere with fimbrin binding to actin. These data indicate that the invasion and cell elongation phenotypes of act1–120/act1–120 mutants are due primarily to a defect in fimbrin binding.
Figure 5.
Expression of a mutant fimbrin (Sac6p) with an altered actin-binding domain suppresses the invasion and cell elongation defects of an act1–120/act1–120 diploid. Strains BCY209 (ACT1/ACT1), BCY210 (act1–120/act1–120), BCY372 (sac6/sac6), BCY412 (act1–120/act1–120 〈SAC6+〉), BCY411 (act1–120/act1–120 〈sac6–10〉), BCY432 (act1–111/act1–111 〈sac6–10〉), and BCY433 (act1–124/act1–124 〈sac6–10〉), where genotypes in angle brackets refer to relevant genotypes of plasmids AAB117 (SAC6 URA3 CEN) and AAB289 (sac6–10 URA3 CEN) (Brower et al., 1995), were streaked to SLAD agar and incubated at 30°C for 4 d. The resultant colonies were photographed before and after washing the agar surface. Long cells were quantified as described in MATERIALS AND METHODS and are reported with SE. Bar, 500 μm.
Characterization of the Actin Cytoskeleton in PH Cells in Wild Type and act1 Mutants
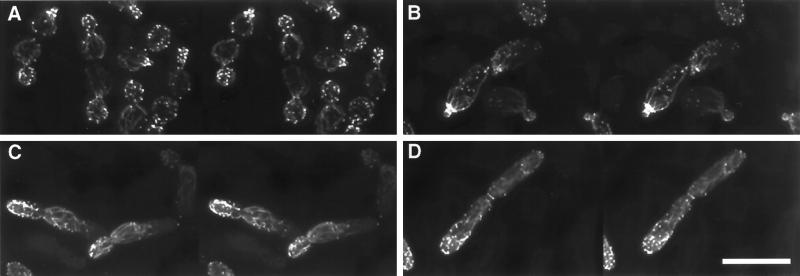
As a basis for understanding the similarities and differences in the YF and PH actin cytoskeletons and how this may account for the differences in cell shape and other growth properties in these distinct cell types, the actin cytoskeleton was imaged in both YF and PH cells at various stages of the cell cycle (Figure 6). The major structural features of the PH and YF actin cytoskeletons are similar in that a prominent ring of filamentous actin is seen at the site of bud emergence, cortical patches are found almost exclusively in the emerging bud, and cables are oriented toward the site of bud emergence in both mother and daughter cells. However, there are notable differences in cytoskeletal structure between these cell types that bear mention. As observed previously (Kron et al., 1994), we find that the polarity of patch localization is enhanced in PH cells relative to YF cells, as patches tend to remain at the distal pole throughout bud emergence in PH cells until a new ring of filamentous actin is formed at the eventual site of cytokinesis. In addition, actin cables are more pronounced in PH cells than in YF cells.
Figure 6.
Comparison of the YF and PH actin cytoskeletons. Stereo images of the filamentous actin cytoskeleton (stained with rhodamine–phalloidin) in strain BCY209 grown either in YPD (A) or in SLAD alginate (B–D). (A) YF cells: all stages of cell cycle. (B) PH cell, early bud emergence; note prominent ring of actin in mother cell at site of bud emergence and at the neck of the very small emerging bud. (C) PH cell, midbud emergence; image shows the pronounced actin cables and patch polarization to the distal end of the emerging bud. (D) PH cell shortly before cytokinesis; a ring of actin has formed at the septation site between mother and daughter, whereas some patches remain near the distal end of the daughter cell. Bar, 10 μm.
As was recently shown for YF cells in the S288c background (Botstein et al., 1997), we find that actin cables are cortical in both YF and PH cells in the Σ1278b genetic background. Cables appear to run just under the plane in which patches reside and to follow the overall contour of the cell. Thus, it appears generally true that actin cables do not run directly through the cytoplasm as has long been thought. It seems likely that the cortical nature of actin cables was not noticed previously because the generation and manipulation of high resolution stereo images of the yeast actin cytoskeleton has only recently become possible.
Actin patch dynamics were also examined in wild-type PH cells using a previously described GFP-SAC6 fusion (Doyle and Botstein, 1996). Expression of this fusion protein rescues all PH growth defects associated with deletion of SAC6, providing additional evidence to that previously presented that this fusion protein is fully functional (Doyle and Botstein, 1996). Analysis of actin patch dynamics showed that actin patches are mobile in PH cells, as they are in YF cells (Doyle and Botstein, 1996; Waddle et al., 1996). Patch dynamics did not appear to differ significantly between YF and PH cells.
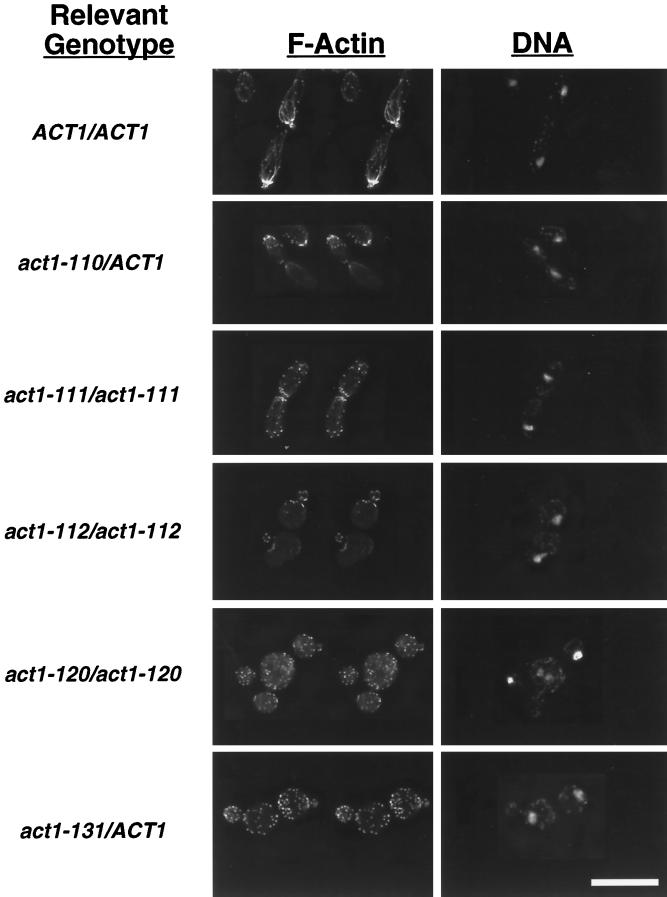
Rhodamine–phalloidin staining of all act1/act1 mutants (with the exception of act1–129, which encodes an actin that does not bind phalloidin; Drubin et al., 1993) grown under PH growth-inducing conditions revealed numerous defects in actin cytoskeletal structure. Cytoskeletal structure in a sample of mutants is shown in Figure 7. (Rotations of these mutants and of the wild-type PH cells in Figure 6 further illustrate many of the features of the actin cytoskeleton discussed here and are available on the internet version of this manuscript.) In all mutants with effects on PH growth, actin cables are either absent or thin and disorganized. Perturbation of actin patch polarization and morphology was also observed, with the most pronounced effects in those mutants displaying the most severe cell elongation defects. For example, in act1–120/act1–120 homozygotes and act1–131/ACT1 heterozygotes, which both show strong defects in cell elongation (Figure 2), actin patches are depolarized and located in both mother and daughter cells. act1–112/act1–112 homozygotes, which are also severely affected in cell elongation, show defects in patch polarization, an overall reduction in patch number and staining intensity, and a large proportion of cells with thick or clumpy patches (Figure 7). Multibudded and multinucleate cells were also apparent in many mutants under PH growth-inducing conditions (e.g., –act1–120/act1–120 and act1–112/act1–112 in Figure 7). The abnormal patch structures and nuclear segregation and budding defects we observed under these conditions closely resemble those previously described for these and other act1 mutants in YF cells (Drubin et al., 1993).
Figure 7.
The actin cytoskeleton in actin mutants grown under conditions that induce PH growth. Shown are wild-type and actin mutant strains grown in SLAD alginate and stained with rhodamine–phalloidin (to detect F-actin) and DAPI (to detect DNA) as described in MATERIALS AND METHODS. F-actin images are in stereo. Strains shown are BCY209 (ACT1/ACT1), BCY157 (act1–110/ACT1), BCY204 (act1–111/act1–111), BCY202 (act1–112/act1–112), BCY210 (act1–120/act1–120), and BCY168 (act1–131/ACT1). Bar, 10 μm.
For those mutants capable of making long cells, actin patch polarization is more normal under PH growth-inducing conditions than for those mutants with the most severe cell elongation defects. For example, act1–111/act1–111 homozygotes and act1–110/ACT1 heterozygotes both show relatively normal patch morphology and polarization in long cells (Figure 7), whereas actin cables are rarely seen in these mutants. These data underscore the importance of patch polarization to the distal end of the emerging bud for cell elongation during PH growth (see DISCUSSION).
DISCUSSION
Actin Has Multiple Functions in Filamentous Growth
We have shown that act1 mutations disrupt the PH growth-specific properties of agar invasion, cell elongation, and unipolar bud site selection. Some act1 mutations show allele-specific effects on invasion, cell elongation, and unipolar budding (Table 3). In addition, certain act1 mutations exhibit specific intragenic complementation with respect to PH growth defects (Table 5). These data suggest that there are multiple requirements for actin function in PH growth.
Several observations indicate that the effects of these mutations on PH growth are due to specific effects on functions of actin required for PH growth and not general effects on cell growth and physiology. First, two alleles that have no effect on viability at any temperature (act1–104 and act1–117) both exhibit pronounced effects on filamentation. Second, filamentation defects are seen at the “permissive” temperature (30°C) for the temperature-sensitive act1 alleles and the sac6 mutant analyzed in this study. Third, only two alleles analyzed in this study (act1–111 and act1–132) show obvious growth defects at the temperature at which these studies were conducted (30°C) (Table 2), and these mutations have less severe effects on PH growth than other alleles with no overt growth phenotypes at this temperature (e.g., –act1–112 and act1–120) (Table 3).
The Utility of Filamentous Growth for Studying Actin Function
Filamentous growth provides a uniquely sensitive assay for actin cytoskeletal function during a developmental switch. The sensitivity of this system allowed us to identify new phenotypic consequences for mutations affecting actin. We found dominant effects on filamentation for several actin mutations previously classified as recessive based on their effects on YF growth. Effects on PH growth were also found for certain act1 alleles that have no (or subtle) effects on YF growth and viability. In addition, using the filamentation phenotype, we have been able to identify genetic interactions between actin alleles. Similar efforts to define act1 intragenic complementation by assessing growth at restrictive temperature have led to ambiguous results.
New Insights into Actin Structure and Function Revealed by Analysis of PH Growth Phenotypes
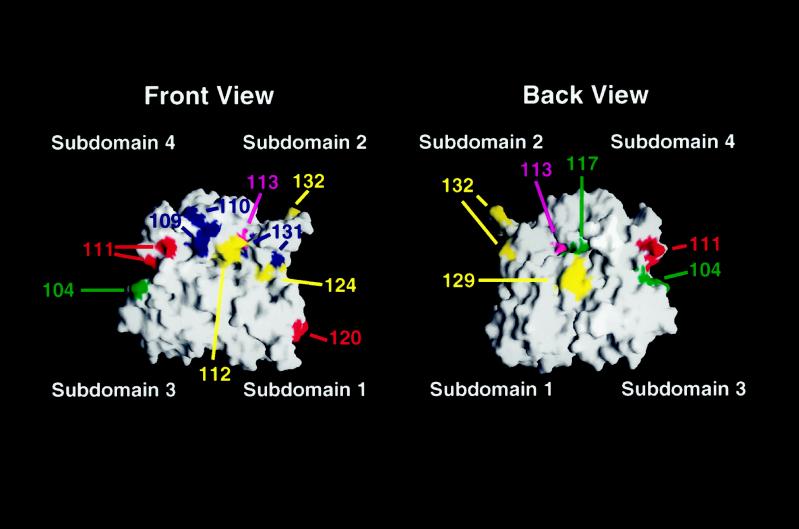
Many actin mutations are likely to cause defects in PH growth by blocking the interaction between actin and other proteins. Indeed, we have shown that act1–120, which is known to disrupt binding of fimbrin to actin, can be suppressed for its invasion and cell elongation defects by a compensatory mutation in the actin-binding domain of fimbrin (Figure 5). Other act1 alleles with effects on PH growth are known or presumed to affect the binding of proteins required for filamentation, suggesting that the phenotypes of these act1 mutants may be also be due, in part, to an effect on the binding of these proteins. For example, act1–110 alters residues (E237 and K238) that are part of the binding site for the actin filament-stabilizing protein tropomyosin (Milligan et al., 1990; Saeki et al., 1996), which is encoded by TPM1 and TPM2 in yeast (Drees et al., 1995). tpm1/tpm1 mutants, like act1–110/ACT1 heterozygotes, are defective for filamentation (Mösch and Fink, 1997). act1–104 is particularly interesting, in that this allele has no known YF phenotype and is not known to affect the binding of any actin-binding protein, yet it has an obvious effect on filamentation. act1–104 alters residues (K315 and E316) predicted to lie on the surface of both the actin monomer (Figure 8) and the actin filament. Thus, these amino acids are well positioned to be a binding site for an as yet unidentified protein with a specific role in PH growth.
Figure 8.
Modeling of the mutations analyzed in this study onto the solvent-exposed surface of an actin monomer. Shown is the calculated surface of the actin monomer based on the coordinates of the rabbit muscle actin filament structure (Lorenz et al., 1993). Actin subdomains are indicated. Red, locations of residues altered by viable, temperature-sensitive alleles with recessive effects on PH growth; yellow, location of residues altered by viable, temperature-sensitive alleles with dominant effects on PH growth; blue, positions of residues affected by recessive lethal alleles with dominant effects on PH growth; green, locations of residues affected by mutations that have no effect on viability but disrupt PH growth; magenta, positions of residues altered by mutations with no effect on PH growth. Surfaces were generated using GRASP (Nicholls et al., 1991) on a Silicon Graphics Iris computer.
Several mutations may act by altering the interaction of actin with itself. Four alleles that show effects on PH growth (act1–111, act1–112, act1–129, and act1–132) disrupt actin–actin interactions in the two-hybrid assay (Amberg et al., 1995) and alter residues predicted to stabilize actin–actin contacts (Holmes et al. 1990; Lorenz et al., 1993). Actin monomers defective for one such interaction could disrupt filament assembly in a dominant manner by their incorporation into a growing filament and blocking additional contacts necessary for filament stability and/or elongation. Interestingly, three alleles that affect actin–actin contacts (act1–112, act1–129, and act1–132) do indeed show dominant effects on PH growth.
The synthetic growth defect of act1–111/act1–112 (Figure 4) may also be explained by effects on actin filament assembly and stability. Insertion of a hydrophobic loop from one actin monomer into a hydrophobic hole defined in part by the cleft between subdomains 2 and 4 in an adjacent actin monomer has been suggested to stabilize interactions within the actin filament (Chen et al., 1993; Lorenz et al., 1993; Kuang and Rubenstein, 1997). act1–111 affects amino acids (D222, E224, and E226) that form a pocket in which the hydrophobic loop resides in monomeric actin and may perturb the ability of the hydrophobic loop to swing out of the pocket to interact with the hydrophobic cleft (Amberg et al., 1995). act1–112 alters residues (K213, E214, and K215) that form one-half of the cleft into which the hydrophobic loop is inserted. Thus, the negative interaction between these alleles could reflect a synergistic effect of perturbing both structural features required for filament stability.
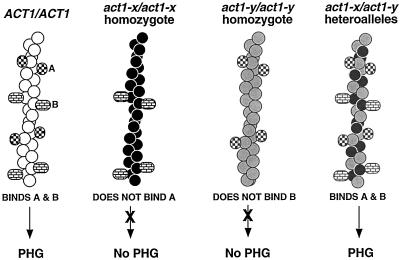
The demonstration that some act1 mutations exert their effects by altering the binding of specific actin-binding proteins suggests a mechanism for the intragenic complementation we have observed. Such alleles might allow formation of heteropolymeric actin filaments that can bind ligands that neither homopolymer can bind. For example, as shown in Figure 9, two actin alleles (X and Y) might affect binding of distinct proteins (A and B) that are both required for filamentation. If allele X blocks binding of A, and allele Y blocks binding of B, act1-x/act1-x and act1-y/act1-y homozygotes would be defective for PH growth, but because of distinct molecular effects. However, actin filaments comprising X and Y monomers would be competent to bind both A and B, and so act1-x/act1-y strains would exhibit filamentation. Intragenic complementation between act1 alleles based on such a mechanism has been predicted (Ayscough and Drubin, 1996). However, to our knowledge the data presented here represent the first experimental demonstration of this effect.
Figure 9.
Model for mechanism of intragenic complementation between act1 alleles (after Ayscough and Drubin, 1996). Actin monomers encoded by ACT1, act1-x, and act1-y are represented by white, black, and gray circles, respectively. The ability of hypothetical actin-binding proteins A and B to interact with actin filaments comprising wild-type actin, each mutant actin singly, or a combination of mutant actins is schematized. The PH growth phenotype of strains containing such actin filaments is also indicated.
This model predicts that mutations exhibiting intragenic complementation should affect binding of distinct subsets of ligands and will likely reside in different regions of the actin monomer and/or filament. Indeed, act1–120 is the only mutant analyzed in this study localized to subdomain 1 of actin (Figure 8), and it is able to complement mutations in subdomains 2, 3, and 4 that are quite distant (in both the monomer and filament) from those mutations it complements for PH growth defects (act1–104 [subdomain 3], act1–111 [subdomain 4], act1–117 [subdomain 4], act1–124 [subdomain 2], and act1–129 [subdomain 3]). In addition, in the two-hybrid assay, the actin encoded by act1–120 binds many ligands whose binding was disrupted by act1 mutations that act1–120 can complement (Amberg et al., 1995).
Actin Function in Cell Elongation, Unipolar Budding, and Agar Invasion
Our results suggest that cytoskeletal integrity is essential for filamentation. What might be the specific requirements for actin in each of the PH growth-specific traits we analyzed?
Cell Elongation.
The PH and YF actin cytoskeletons are different in structure. In YF cells, there is a distinct period during bud emergence in which actin patches are distributed over the cortex of the emerging bud. In PH cells, actin patches remain polarized at the tip of the cell throughout bud emergence (Kron et al., 1994). Our imaging of the PH actin cytoskeleton in wild type and act1 mutants confirms and extends this observation to suggest that actin patch polarization plays an important role in the elongation of the PH cell. In act1/act1 mutants with the most pronounced effects on cell elongation (act1–112/act1–112 and act1–120/act1–120 homozygotes and act1–131/ACT1 heterozygotes), substantial perturbation of actin patch polarization during bud emergence is observed (Figure 7). In contrast, actin patches are polarized to the distal end of many emerging buds in those actin mutants capable of making substantial numbers of long cells. These results are consistent with the proposal that actin patches play an important role in directing secretion of new membrane and cell wall material in the emerging bud (Novick and Botstein, 1985; Mulholland et al., 1994). Thus, the extended polarization of actin patches to the distal end of PH cells throughout bud emergence would promote elongation of the PH cell.
Unipolar Budding.
The demonstration that actin cytoskeletal mutants are unable to exhibit bipolar budding in YF cells led to the suggestion that the actin cytoskeleton is required to maintain, and perhaps direct, the placement of bipolar bud site selection cues (Zahner et al., 1996; Yang et al., 1997). These presently unidentified signals, present at both the proximal and distal ends of the cell, would then serve to direct new bud formation at either end of the mother cell. The fact that nearly all actin mutants we characterized exhibit random budding under conditions that induce PH growth demonstrates that, like bipolar bud site selection in YF diploids, unipolar bud site selection in PH cells requires actin cytoskeletal function. However, act1–113/act1–113 and act1–117/act1–117 strains are suppressed for their YF random budding phenotype when grown under conditions that induce PH growth (Table 4). Thus, despite their similar dependency for actin function, these results demonstrate that the YF and PH bud site selection programs are regulated somewhat distinctly in response to nutrient conditions.
Invasion.
The mechanism by which PH cells invade agar is not known. However, it has been suggested that these cells might secrete enzymes that hydrolyze agar and thus facilitate growth through the substrate (Gimeno et al., 1992). Indeed, many invasive fungi, including Candida albicans, have been shown to secrete proteases and other enzymes believed to facilitate their invasion (Macdonald and Odds, 1983). Given the well-established role for actin in secretion, it is tempting to speculate that actin is required for the secretion of such enzymes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alison Adams for plasmids used in this work, Ken Holmes for the actin filament coordinates, and James Berger for assistance using GRASP. We are grateful to members of the Fink and Botstein laboratories for helpful discussions and Todd Milne for critical reading of the manuscript. Many thanks to Katja Schwartz for providing help with some of the microscopy in Figure 6. B.M.C. is a Chiron Fellow of the Life Sciences Research Foundation. G.R.F. is an American Cancer Society Professor of Genetics. The DeltaVision microscope system is supported by National Institutes of Health Research Resources grant RR11939-01. This work was supported by grants from the National Institutes of Health (GM46406-07 to D.B. and GM35010 to G.R.F.).
Footnotes
REFERENCES
- Adams AE, Botstein D. Dominant suppressors of yeast actin mutations that are reciprocally suppressed. Genetics. 1989;121:675–683. doi: 10.1093/genetics/121.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AE, Botstein D, Drubin DG. A yeast actin-binding protein is encoded by SAC6, a gene found by suppression of an actin mutation. Science. 1989;243:231–233. doi: 10.1126/science.2643162. [DOI] [PubMed] [Google Scholar]
- Adams AE, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AE, Shen W, Lin CS, Leavitt J, Matsudaira P. Isoform-specific complementation of the yeast sac6 null mutation by human fimbrin. Mol Cell Biol. 1995;15:69–75. doi: 10.1128/mcb.15.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Amatruda JF, Cannon JF, Tatchell K, Hug C, Cooper JA. Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature. 1990;344:352–354. doi: 10.1038/344352a0. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Nat Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Drubin DG. ACTIN: general principles from studies in yeast. Annu Rev Cell Dev Biol. 1996;12:129–160. doi: 10.1146/annurev.cellbio.12.1.129. [DOI] [PubMed] [Google Scholar]
- Botstein D, Amberg D, Mulholland J, Huffaker T, Adams A, Drubin D, Stearns T. The yeast cytoskeleton. In: Pringle JR, Broach JR, Jones EW, editors. The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1997. pp. 1–90. [Google Scholar]
- Brower SM, Honts JE, Adams AE. Genetic analysis of the fimbrin-actin binding interaction in Saccharomyces cerevisiae. Genetics. 1995;140:91–101. doi: 10.1093/genetics/140.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cook RK, Rubenstein PA. Yeast actin with a mutation in the “hydrophobic plug” between subdomains 3 and 4 (L266D) displays a cold-sensitive polymerization defect. J Cell Biol. 1993;123:1185–1195. doi: 10.1083/jcb.123.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B, Brown C, Barrell BG, Bretscher A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J Cell Biol. 1995;128:383–392. doi: 10.1083/jcb.128.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Miller KG, Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Mulholland J, Zhu ZM, Botstein D. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-I. Nature. 1990;343:288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. San Diego: Academic Press; 1991. [Google Scholar]
- Haarer BK, Corbett A, Kweon Y, Petzold AS, Silver P, Brown SS. SEC3 mutations are synthetically lethal with profilin mutations and cause defects in diploid-specific bud-site selection. Genetics. 1996;144:495–510. doi: 10.1093/genetics/144.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D, Matsudaira P, DeRosier DJ. Evidence for a conformational change in actin induced by fimbrin (N375) binding. J Cell Biol. 1997;139:387–396. doi: 10.1083/jcb.139.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Holtzman DA, Wertman KF, Drubin DG. Mapping actin surfaces required for functional interactions in vivo. J Cell Biol. 1994;126:423–432. doi: 10.1083/jcb.126.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honts JE, Sandrock TS, Brower SM, O’Dell JL, Adams AE. Actin mutations that show suppression with fimbrin mutations identify a likely fimbrin-binding site on actin. J Cell Biol. 1994;126:413–422. doi: 10.1083/jcb.126.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron SJ, Styles CA, Fink GR. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang B, Rubenstein PA. Beryllium fluoride and phalloidin restore polymerizability of a mutant yeast actin (V266G, L267G) with severely decreased hydrophobicity in a subdomain 3/4 loop. J Biol Chem. 1997;272:1237–1247. doi: 10.1074/jbc.272.2.1237. [DOI] [PubMed] [Google Scholar]
- Laloux I, Jacobs E, Dubois E. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 1994;22:999–1005. doi: 10.1093/nar/22.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HP, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Lorenz M, Popp D, Holmes KC. Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J Mol Biol. 1993;234:826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- Macdonald F, Odds FC. Virulence for mice of a proteinase-secreting strain of Candida albicans and a proteinase deficient mutant. J Gen Microbiol. 1983;129:431–438. doi: 10.1099/00221287-129-2-431. [DOI] [PubMed] [Google Scholar]
- Milligan RA, Whittaker M, Safer D. Molecular structure of F-actin and location of surface binding sites. Nature. 1990;348:217–221. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- Mösch HU, Fink GR. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Read EB, Okamura HH, Drubin DG. Actin- and tubulin-dependent functions during Saccharomyces cerevisiae mating projection formation. Mol Biol Cell. 1992;3:429–444. doi: 10.1091/mbc.3.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H, Munn A, Geli MI, Hicke L. Actin-, myosin- and ubiquitin-dependent endocytosis. Experientia. 1996;52:1033–1041. doi: 10.1007/BF01952099. [DOI] [PubMed] [Google Scholar]
- Saeki K, Sutoh K, Wakabayashi T. Tropomyosin-binding site(s) on the Dictyostelium actin surface as identified by site-directed mutagenesis. Biochemistry. 1996;35:14465–14472. doi: 10.1021/bi961292c. [DOI] [PubMed] [Google Scholar]
- Sandrock TM, O’Dell JL, Adams AEM. Allele-specific suppression by formation of new protein-protein interactions in yeast. Genetics. 1997;147:1635–1642. doi: 10.1093/genetics/147.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalettar BA, Swedlow JR, Sedat JW, Agard DA. Dispersion, aberration and deconvolution in multi-wavelength fluorescence images. J Microsc. 1996;182:50–60. doi: 10.1046/j.1365-2818.1996.122402.x. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon VR, Swayne TC, Pon LA. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems, identification of a motor activity on the mitochondrial surface. J Cell Biol. 1995;130:345–354. doi: 10.1083/jcb.130.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivadon P, Bauer F, Aigle M, Crouzet M. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. Mol Gen Genet. 1995;246:485–495. doi: 10.1007/BF00290452. [DOI] [PubMed] [Google Scholar]
- Smith MG, Simon VR, O’Sullivan H, Pon LA. Organelle-cytoskeletal interactions: actin mutations inhibit meiosis-dependent mitochondrial rearrangement in the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1381–1396. doi: 10.1091/mbc.6.10.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddle JA, Karpova TS, Waterston RH, Cooper JA. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Holtzman DA, Drubin DG. The yeast actin cytoskeleton. Curr Opin Cell Biol. 1994;6:110–119. doi: 10.1016/0955-0674(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Wertman KF, Drubin DG, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Ayscough KR, Drubin DG. A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection. J Cell Biol. 1997;136:111–123. doi: 10.1083/jcb.136.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.