Abstract
Hepatitis delta virus (HDV) contains two RNA species (HDV-S and HDV-L), which encode the small and large forms of hepatitis delta antigens (S- and L-HDAg), respectively. HDV-L RNA is a result of an RNA editing event occurring at an amber/W site of HDV-S RNA. RNA editing must be regulated to prevent premature and excessive accumulation of HDV-L RNA in the viral life cycle. In this study, we used an RNA transfection procedure to study the replication abilities of HDV-L and HDV-S RNA. While HDV-S led to robust RNA replication, HDV-L could not replicate even after 6 days following transfection. The failure of HDV-L to replicate was not due to insufficient amounts of S-HDAg, as identical results were obtained in a cell line that stably overexpresses S-HDAg. Also, it was not due to possible inhibition by L-HDAg, as HDV-S RNA replication was not affected when both HDV-L and HDV-S RNA were cotransfected. Further, when L-HDAg expression from HDV-L RNA was abolished by site-directed mutagenesis, the mutant HDV-L RNA also failed to replicate. Unexpectedly, when the kinetics of RNA replication was examined daily, HDV-L was found to replicate at a low level at the early time points (1 to 2 days posttransfection) but then lose this capability at later time points. Sequence analysis of the replicated HDV-L RNA at day 1 posttransfection showed that it had undergone multiple nucleotide changes, particularly in the region near the putative promoter region of HDV RNA replication. In contrast, very few mutations were found in HDV-S RNA. These results suggest that the editing at the amber/W site triggers a series of additional mutations which rapidly reduce the replication efficiency of the resultant HDV genome and thus help regulate the amount of HDV-L RNA in infected cells. They also explain why L-HDAg is not produced early in HDV infection, despite the fact that HDV-L RNA is present in the virion.
Hepatitis delta virus (HDV) is a highly unusual pathogen that can cause severe chronic and acute hepatitis in humans. HDV is dependent on hepatitis B surface antigen (HBsAg) to provide envelope proteins for virus assembly, and thus it always coexists with hepatitis B virus in natural infections (18). Inside the envelope of the HDV virion is a 1.7-kb circular RNA genome which is complexed with two HDV-encoded capsid proteins, the large (214 amino acids) and small (195 amino acids) forms of hepatitis delta antigen (L-HDAg and S-HDAg, respectively) in variable ratios (for review, see reference 13). Inside the infected cells, the viral RNA replicates by a rolling circle mechanism into a 1.7-kb antigenomic RNA and a 0.8-kb mRNA species which encodes HDAg (5). The antigenomic RNA is subsequently replicated into the genomic RNA. The replication of genomic and antigenomic RNA and transcription of the mRNA appear to be under independent regulation (15). Initially, the 0.8-kb mRNA encodes S-HDAg which, in addition to its role as a viral capsid protein, is required for viral RNA replication (9). As replication proceeds, an RNA editing event occurs at the amber termination codon of the S-HDAg open reading frame (ORF), turning it into a tryptophan codon, thus allowing the ORF to extend for an additional 19 amino acids, generating L-HDAg (10, 16). This editing event is carried out by a cellular double-stranded RNA adenosine deaminase (ADAR) (16).
The amber/W editing of HDV RNA to enable production of L-HDAg plays a key role in the HDV life cycle, as L-HDAg triggers virus assembly (3). In addition, L-HDAg has been described to inhibit viral RNA replication, thus playing a modulating role in viral replication (4). However, recent studies have demonstrated that L-HDAg could inhibit only genomic RNA synthesis, but not antigenomic RNA synthesis (which is the first replicating event from the incoming viral genome) (14), and only during the very early stage of viral replication (11). Thus, the ability of L-HDAg to regulate HDV RNA synthesis may not play a significant role in the natural HDV life cycle. Nevertheless, both the HDV RNA encoding S-HDAg and that encoding L-HDAg (named HDV-S and HDV-L RNA, respectively) are incorporated into the virion (21). Thus, it is expected that L-HDAg will be synthesized from the very beginning of the viral replication cycle; such a scenario would disrupt the normal sequential events of the viral replication cycle. However, from the studies of natural history of experimental HDV infections in woodchucks, this is not the case (22). Furthermore, since the editing event is likely to be irreversible, it might be expected that the bulk of HDV genomes in cells would eventually become edited and, in turn, packaged into HDV particles. This is also not the case. Thus, there must be mechanisms to limit the excessive accumulation and multiplication of HDV RNA encoding L-HDAg.
Earlier studies have shown that HDV-L RNA, in contrast to HDV-S RNA, was not infectious when it was transfected into cells expressing S-HDAg (6) or when it was cotransfected with S-HDAg into cells (19). Its inability to replicate will provide a mechanism to limit the amount of the edited RNA in the cells and the amount of L-HDAg production early in viral replication. In this study, we further explored the replication potential of this type of RNA in the natural settings of viral RNA replication. Our studies provide a novel insight into the molecular mechanism for HDV to regulate the production of L-HDAg.
MATERIALS AND METHODS
Cell culture.
The human hepatoma cell line HuH7 and TSδ3 cells (7), which stably overexpress S-HDAg, were cultured in Dulbecco's modified Eagle's medium supplemented with 10 and 5% fetal bovine serum, respectively, penicillin, and streptomycin and incubated at 37°C in 5% CO2. For transfection studies, cell cultures were seeded overnight either in six-well plates or 60-mm-diameter petri dishes. Following transfection, cells were incubated overnight and the medium was then changed. The cell cultures were incubated for up to an additional 14 days.
Plasmids and cloning.
Plasmids pBSδ1.2G and pBSδ1.2G(2xS) (templates for in vitro transcription of the 1.2X genome-length wild-type and L-HDAg− genomic HDV RNAs, respectively) and pBSδ1.2AG and pBSδ1.2AG(2xS) (templates for in vitro transcription of the 1.2X genome-length wild-type and L-HDAg-deficient antigenomic HDV RNAs, respectively) have been described elsewhere (12). Plasmid pX9-1/II was used for in vitro transcription of S-HDAg mRNA, and pTMδSalB and pBSδHX were used for the generation of 32P-labeled probes to detect genomic and antigenomic HDV RNA (15), respectively. For construction of plasmid pBSδ1.2G(L), which was the template for in vitro transcription of 1.2X genome-length genomic HDV RNA encoding L-HDAg, the UAG stop codon of the S-HDAg ORF in plasmid pBSδ1.2G was converted to UGG, mimicking the natural editing event. This modification was made with a QuickChange (Stratagene) site-directed mutagenesis kit according to the manufacturer's directions, using the primer pair 5′-GGAAACCAGGGATTTCCATGGGATATAC TCTTCCCAGCC-3′ and 5′-GGCTGGGAAGAGTATATCCCATGGAAATCCCTGGTTTCC-3′ and plasmid pBSδ1.2G as template. Plasmid pBSδ1.2AG(L), used for in vitro synthesis of an HDV RNA of opposite polarity to pBSδ1.2G(L), was constructed by subcloning the full-length XbaI HDV cDNA fragment from pBSδ1.2G(L) into the XbaI site of plasmid pBSδAG-Basic (12). Plasmids pBSδ1.2G(mt-L) and pBSδ1.2AG(mt-L) were used for in vitro transcription of the 1.2X genome-length genomic and antigenomic strand, respectively, of an RNA (HDV mt-L) that contains a frameshift mutation in the ORF of L-HDAg. The frameshift mutation was generated by inserting two adenosine residues immediately following the start codon AUG of the L-HDAg ORF. Specifically, the construction of plasmid pBSδ1.2G(mt-L) was made by PCR-based site-directed mutagenesis, using the primer pair 5′-AGCCGGTCCGAAAGAAGGAAAGACCG-3′ and 5′-TTCATCTTCGACTAGAGGCGACGGTCCTC-3′ and plasmid pBSδ1.2G as template and cycled as follows: 95°C for 5 min, then 28 cycles of 94°C 60 s, 60°C for 60 s, and 72°C for 12 min, followed by a final 15 min at 72°C. Plasmid pBSδ1.2AG(mt-L) was constructed by subcloning as previously described. All the mutant constructs were sequenced for at least 300 nucleotides (nt) across the mutation sites to confirm the mutations and to ensure that no other mutations were introduced.
In vitro transcription and RNA transfection.
1.2X genome-length HDV RNAs were transcribed from plasmids pBSδ1.2G, pBSδ1.2AG, pBSδ1.2G(2xS), pBSδ1.2AG(2xS), pBSδ1.2G(L), pBSδ1.2AG(L), pBS1.2G(mt-L), and pBS1.2AG(mt-L) using T7 MEGAscript kits (Ambion) after linearization with the restriction enzyme NotI. Capped mRNA for HDAg was transcribed from plasmid pX9-I/II, which encodes a chimeric S-HDAg of genotype I and II (15), after linearization with the restriction enzyme HindIII, using a T7 m-Message m-Machine kit (Ambion). The method for generation of HDV-specific 32P-labeled riboprobes has been described elsewhere (12).
For all the RNA transfection experiments, a 1.2X genomic-length HDV RNA of either the genomic or antigenomic strand transcribed from one of the above plasmids and mRNA transcribed from plasmid pX9-I/II were used together for transfection. RNA transfection was performed using DMRIE-C reagent (Gibco BRL) according to the manufacturer's directions.
Northern blot hybridization analysis.
RNA was extracted from intact cells using Tri reagent (Molecular Research Center, Inc.) according to the manufacturer's protocol. RNA samples were separated by electrophoresis through morpholinepropanesulfonic acid-formaldehyde-containing 1.2% agarose gels, transferred to membrane, hybridized, and washed as described previously (12). Detection of genomic and antigenomic HDV RNA was performed using in vitro-transcribed 32P-labeled probes generated from pTMδSalB and pBSδHX, respectively. The washed membrane was exposed to Biomax MR or MS X-ray films (Kodak). Quantitation of signals was performed by phosphorimagery using ImageQuant version 1.11 software (Molecular Dynamics). Detection of ChoA mRNA was performed as described previously (12).
RT-PCR and restriction digestion analysis.
Reverse transcription-PCR (RT-PCR) was performed using total RNA from HuH7 cells transfected with HDV RNA. Briefly, 0.5 μg of total cellular RNA was mixed with 10 pmol of the oligonucleotide primers HDV 1224-1204,125-98, or 520-497, which correspond to nt 1224 to 1204, 125 to 98, or 520 to 497, respectively, of the woodchuck HDV sequence according to the numbering of Wang et al. (20), and the volume was adjusted to 10 μl. After 20 min at 70°C, the RT reaction was carried out using Superscript III (Invitrogen) according to the manufacturer's protocol. A portion (1 μl) of the each resultant cDNA mixture was added to the PCR that included primer pairs HDV(A) 1224-1204 and HDV(A) 850-870, HDV(B) 1204-1224 and HDV(B) 125-98, or HDV(C) 16-41 and HDV(C) 520-497 and cycled as follows: 95°C for 2 min, then 28 cycles of 94°C 30 s, 65°C for 30 s, and 72°C for 30 s, followed by a final 5 min at 72°C. For restriction digestion analysis, the PCR products (HDV nt 850 to 1224) were purified using a column (Qiagen PCR purification) and digested with NcoI (Roche), which specifically cleaves HDV cDNA containing the amber/W mutation. Digested and undigested PCR products were analyzed by electrophoresis on 2% agarose gels. The undigested PCR product is 380 bp in length, whereas the NcoI digestion of the PCR product containing the amber/W mutation yielded two bands of 200 and 180 bp. Nonedited HDV cDNA was not cleaved by NcoI. For sequencing analysis, PCR products were cloned into pCR2.1-TOPO by TA-cloning (Invitrogen) as described in the manual. Automated sequencing was performed by Laragen, Inc. (Los Angeles, Calif.).
Immunoblot analysis of HDAg.
Immunoblot analysis of HDAg was performed as described in an earlier study (12). Where indicated, the amount of total protein was assayed using a noninterfering protein assay kit (Geno Technology Inc.) prior to electrophoresis.
RESULTS
Replication potential of HDV RNA encoding L-HDAg.
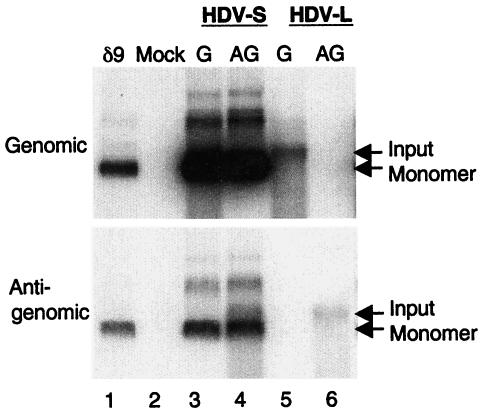
To assess the replication potential of HDV RNA encoding L-HDAg, we used site-directed mutagenesis to construct a mutant HDV genome in which the UAG (amber) stop codon of the wild-type HDV RNA (encoding S-HDAg) was changed to UGG. This modification is identical to that occurring during natural amber/W editing (10, 17) and thus produces an HDV genome that encodes L-HDAg. The 1.2X genome-length genomic- or antigenomic-sense HDV RNA encoding either L- or S-HDAg (termed HDV-L or HDV-S RNA, respectively) was transfected together with a capped in vitro-transcribed mRNA species encoding S-HDAg according to the previously established RNA transfection protocol (15). Six days posttransfection, genomic or antigenomic HDV RNA was examined by Northern blot hybridization (Fig. 1). Following transfection with either genomic or antigenomic HDV-S RNA, large amounts of monomeric and dimeric HDV RNA of both polarities were detected (Fig. 1, lanes 3 and 4), indicating that active HDV RNA replication had occurred. In contrast, when HDV-L RNA was used, no RNA replication was observed (Fig. 1, lanes 5 and 6), irrespective of the polarity of the RNA used for transfection. In these experiments, a small amount of the input HDV RNA species was still visible at day 6 posttransfection, indicating that RNA had been successfully transfected. No HDV RNA was detected in mock-transfected HuH7 cells (lane 2).
FIG. 1.
RNA replication of HDV-S and HDV-L RNA in HuH7 cells, as shown in a Northern blot analysis of genomic and antigenomic HDV RNA in HuH7 cells on day 6 after transfection with genomic (G) or antigenomic (AG) HDV RNA that contained either an S-HDAg ORF (HDV-S) or an L-HDAg ORF (HDV-L). δ9 is the HDV monomer RNA marker.
Since HDV-L RNA could not produce S-HDAg, the only source of S-HDAg in this experiment was the mRNA used for transfection. To rule out the possibility that the failure of HDV-L RNA to replicate was due to an insufficient amount of S-HDAg, we repeated the experiment in TSδ3 cells, a cell line that constitutively expresses S-HDAg (7) (Fig. 2). Similar to the experiments in HuH7 cells, transfection of either genomic or antigenomic HDV-S RNA yielded large amounts of HDV RNA products (lanes 2 and 6), whereas HDV-L did not lead to RNA replication (lanes 4 and 8). Thus, even in the presence of large amounts of S-HDAg, the HDV genome encoding L-HDAg was unable to replicate. This result is consistent with an earlier study by Glenn and White (6).
FIG. 2.
RNA replication of HDV-S and HDV-L RNA in TSδ3 cells, as shown in a Northern blot analysis of genomic and antigenomic HDV RNA in TSδ3 cells. The conditions were the same as those for Fig. 1.
HDV RNA encoding L-HDAg is replication competent at early time points posttransfection.
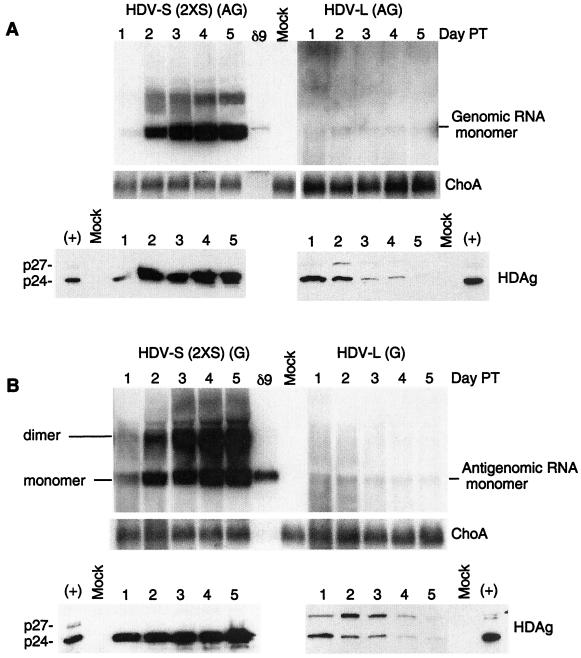
Amber/W editing has been shown to occur on the antigenomic strand of HDV RNA (2), while the template for the synthesis of HDAg mRNA is the genomic strand. Thus, at least a single round of HDV RNA replication has to take place after the editing event before L-HDAg mRNA transcription and subsequent L-HDAg synthesis can occur. We, therefore, performed a kinetic analysis to determine if the L-HDAg-encoding HDV RNA is replication competent at earlier time points posttransfection (Fig. 3). We first examined HuH7 cells cotransfected with an mRNA encoding S-HDAg and 1.9-kb antigenomic-sense HDV-L or HDV-S RNA. The HDV-S RNA used in this experiment (termed 2XS RNA) contains an extra in-frame stop codon immediately downstream of the amber/W editing site and is, thus, unable to synthesize L-HDAg. The use of such an RNA eliminated the possible complication from L-HDAg produced from the wild-type HDV-S RNA after its editing. Transfected cells were examined daily from days 1 to 5 posttransfection for HDV RNA and HDAg synthesis. In cells transfected with the antigenomic HDV-S (2XS) RNA, genomic-sense monomeric HDV RNA was first detected at day 1 posttransfection, which rapidly increased until it reached a steady-state level at day 3 or 4 posttransfection (Fig. 3A, left upper panel). In contrast, no significant HDV RNA synthesis could be detected when HDV-L RNA was used. However, upon closer examination, a trace amount of the genomic HDV RNA could be detected on days 1 to 2 posttransfection, but its amount did not increase; instead, it appeared to decrease slightly thereafter (Fig. 3A, upper right panel). As a control for RNA loading, the amount of cellular ChoA mRNA was constant within each transfection (Fig. 3A, middle panel). This result suggested that HDV-L RNA may replicate at an early time point after transfection.
FIG. 3.
Kinetics of HDV-S and HDV-L RNA replication in HuH7 cells. (A, upper panel) Antigenomic HDV RNA was used for transfection, and Northern blot analysis of the genomic strand was performed. The HDV-S (2XS) RNA contained an additional stop codon downstream of the amber/W editing site, so that L-HDAg could not be synthesized even after the editing event. Total cellular RNA was collected daily after transfection. ChoA mRNA served as a loading control. Autoradiographic exposure time was approximately 5 times longer for HDV-L RNA than for HDV-S RNA. (Lower panel) Immunoblotting analysis of HDAg. The cellular lysates from the same samples as in the upper panel were used for immunoblotting analysis using a mixture of three monoclonal antibodies specific for HDAg (8). (B) The same experiment as in panel A, except that genomic RNA was used for transfection. Northern blot analysis was done to detect antigenomic RNA. The lower panel is the immunoblotting analysis of HDAg of the same samples.
Since the RNA signal was weak, we also examined the production of HDAg after HDV-L RNA transfection. Immunoblot analysis of HDAg revealed abundant amounts of S-HDAg at days 2 to 5 posttransfection with the HDV-S (2XS) RNA (Fig. 3A, lower left panel). As expected, no L-HDAg was observed at any time point. In contrast, in HDV-L-transfected cells, a trace amount of L-HDAg was detected at day 1 posttransfection, which increased slightly by day 2 and then declined sharply at later time points (Fig. 3A, lower right panel). The amount of L-HDAg was less than that of S-HDAg, the latter of which was produced from the cotransfected S-HDAg mRNA. Since L-HDAg can be synthesized only from a newly transcribed mRNA using the replicated genomic RNA as template, these results indicated that at least some HDV RNA replication had occurred for a brief period of time and then stopped replicating. As expected, the amount of S-HDAg also declined at later time points as a result of mRNA degradation.
Similar results were obtained in the converse experiment, in which HuH7 cells were transfected with genomic-sense HDV-L RNA (Fig. 3B). These results were substantiated by both Northern blotting of HDV RNA and immunoblotting of HDAg. However, the amount of L-HDAg, relative to S-HDAg, was more abundant than that observed in the antigenomic RNA transfection. It is likely that the input transfected genomic HDV RNA can directly serve as a template for L-HDAg mRNA synthesis without RNA replication; this result further supports our previous finding that HDV RNA replication and mRNA transcription are independent events (15). Both S- and L-HDAg decreased with time, indicating that HDV-L RNA did not continue to replicate and S-HDAg mRNA eventually was degraded. These results combined suggest that the HDV RNA genome encoding L-HDAg is initially replication competent but rapidly loses this ability soon after transfection.
Sequence analysis of the replicated HDV RNA in HDV-L RNA-transfected cells revealed multiple additional mutations immediately following transfection.
The ability of HDV-L RNA to replicate initially, followed by rapid loss of its replication ability, suggested that HDV-L RNA might have undergone some rapid changes during its initial replication. In contrast, the HDV-S RNA was likely very stable, since its replication capability did not change throughout the replication cycle. This possibility has been suggested previously in an in vitro editing study in which the amber/W editing was suggested to trigger multiple random mutations to inactivate the edited HDV RNA as a means to regulate the extent of HDV RNA editing (17). To examine this possibility, we performed RT-PCR cloning of HDV RNA in the transfected cells on day 1 posttransfection, and multiple individual clones of each PCR product were sequenced. When genomic RNA was used for transfection, antigenomic RNA was used for sequencing, and vice versa. Thus, only the replicated RNA was sequenced. We sequenced three separate regions of HDV RNA, covering almost the entire HDV RNA. Interestingly, on day 1 after genomic or antigenomic HDV-L RNA transfection, almost every PCR clone (clones B) covering nt 1224 to 98, which includes the putative promoter region for HDV RNA replication (1), had accumulated two to three mutations (Table 1). A total of 14 mutations out of 7 PCR clones (558 nt each) sequenced were detected in this region. Other regions of HDV RNA (represented by clones A and C) had relatively fewer mutations. The same results were obtained using either genomic or antigenomic RNA transfection. Interestingly, most of the mutations on the genomic strand involved U-C pairs, corresponding to A-G mutations on the antigenomic strand, which are typical of the mutations mediated by ADARs, the enzymes involved in HDV RNA editing (16). Very small amounts of PCR products could be obtained from the day 6 RNA samples, consistent with the finding that very little HDV-L RNA was detected later in the replication cycle.
TABLE 1.
Summary of mutations detected in HDV RNA 1 day after transfection
| HDV RNA transfected | Individual clonea | Position | Nucleotide change |
|---|---|---|---|
| HDV-L | A-1 | 899 | U to C |
| A-2 | 923 | A to G | |
| B-1 | 1366 | U to C | |
| 1558 | U to C | ||
| 1573 | U to C | ||
| B-2 | 1543 | U to C | |
| B-3 | 1439 | U to C | |
| 1471 | U to C | ||
| B-4 | 1562 | U to C | |
| 1577 | U to C | ||
| B-5 | 1505 | U to C | |
| 1527 | U to C | ||
| B-6 | 1456 | G to A | |
| 229 | U to C | ||
| B-7 | 1462 | U to C | |
| 1492 | U to C | ||
| C-1 | 89 | A to G | |
| 119 | U to C | ||
| C-2 | 182 | A to G | |
| C-3 | 229 | U to C | |
| HDV-S | A-1 | 948 | U to C |
| A-2 | 969 | A to G | |
| A-3 | 1107 | U to C | |
| B-1 | 1619 | U to C | |
| B-2 | 1404 | U to C | |
| 1654 | G to A | ||
| B-3 | 1408 | U to C | |
| C-1 | 167 | A to G | |
| C-2 | 186 | A to G |
PCR products were obtained from three different regions of HDV RNA (clones A, nt 859 to 1224; clones B, nt 1224 to 98; clones C, nt 16 to 520) on day 1 posttransfection. The PCR products were cloned into pCR2.1-TOPO plasmid. Seven individual clones each from HDV-L-transfected cells and 10 clones each from HDV-S-transfected cells were sequenced. The mutations detected in each clone are listed.
In contrast, when HDV-S genomic or antigenomic RNA was used for transfection, the replicated RNA contained very few mutations. A total of four mutations were detected among 10 B clones (558 nt each) sequenced. Thus, the mutation frequency of B clones in HDV-S RNA was nearly 4 times lower than that of HDV-L. The numbers of mutations among clones A and C in HDV-S RNA, however, were similar to those in HDV-L RNA. Therefore, the mutations occurring during HDV RNA replication are most likely related to the amber/W mutation introduced into the HDV-L RNA and are mainly limited to the region that includes the putative promoter for HDV RNA replication.
The loss of replication ability of HDV-L RNA is not due to the production of L-HDAg.
Since HDV-L RNA produces L-HDAg, it is conceivable that the rapid loss of replication ability of HDV-L RNA was due to the production of L-HDAg, which could potentially inhibit HDV RNA replication if it were overexpressed during transfection (11, 14). To rule out this possibility, we cotransfected HDV-L RNA and HDV-S (2XS) RNA, together with S-HDAg mRNA, into HuH7 cells. The possible effect of HDV-L RNA on the replication of HDV-S (2XS) RNA was then evaluated at various time points posttransfection. The results showed that HDV-S (2XS) RNA led to robust RNA replication, which was detectable even on days 1 and 2 (Fig. 4A, lanes c and d). In contrast, HDV-L RNA led to only a very low level of RNA replication on days 1 and 2 (lanes e and f), which became undetectable by day 6 (lane j), consistent with the previous results. When both RNAs were cotransfected, the level of RNA replication was only slightly less than that of HDV-S RNA transfected alone (compare lanes i and k). The same results were obtained using either genomic or antigenomic RNA transfection. These results indicated that HDV-L RNA did not interfere with HDV-S RNA replication. Thus, the inability of HDV-L RNA to replicate could not be due to the putative suppressor function of L-HDAg.
FIG. 4.
Replication of HDV-L and HDV-S RNAs in mixed transfection. (A) HuH7 cells were transfected with HDV-S (2XS) or HDV-L RNA or both by the standard RNA transfection procedures (15). On days 1, 2, or 6, RNA samples were collected and used for Northern blotting analysis. When the genomic (G) strand was used for transfection, the antigenomic (AG) strand was examined (G to AG) and vice versa (AG to G). (B) The protein samples from day 2 or day 6 were used for immunoblotting analysis of HDAg. G or AG denotes the RNA strand used for transfection. S or L denotes HDV-S (2XS) or HDV-L RNA used for transfection.
To confirm this result, we performed immunoblotting analysis of HDAg species (Fig. 4B). In cells transfected with HDV-S (2XS) RNA, only the S-HDAg species was detected. (This RNA could not undergo RNA editing and could not produce L-HDAg even if the RNA editing did occur.) In cells transfected with HDV-L RNA, both S- and L-HDAg were initially detected (day 2). S-HDAg was made from the incoming cotransfected mRNA, whereas L-HDAg could only have come from the replicated HDV-L RNA when antigenomic RNA was used for transfection. Neither of these two HDAg species was detectable on day 6, indicating that HDV-L RNA had stopped replicating. In cells transfected with both HDV-S (2XS) and HDV-L RNAs, both S- and L-HDAg were detected on day 2, but L-HDAg was barely detectable on day 6 while S-HDAg continued to be detectable. Similar results were obtained using either genomic or antigenomic RNA transfection. These results further established that HDV-L RNA could replicate only briefly early in viral replication cycle, whereas HDV-S RNA could replicate even in the presence of HDV-L RNA.
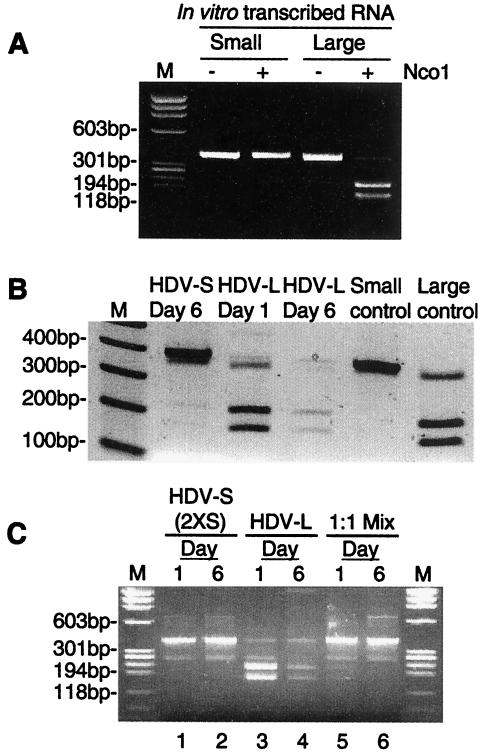
To further establish that only HDV-S, but not HDV-L, RNA can replicate during mixed transfection, we performed RT-PCR analysis of the replicated HDV RNA in cells transfected with both RNAs. This analysis was based on the finding that the cDNA of HDV RNA containing an L-HDAg-coding sequence can be cut with the restriction enzyme NcoI, whereas that of RNA encoding S-HDAg cannot (21), thus enabling the distinction between these two RNA species. To establish the validity of this approach, we first used in vitro-transcribed RNAs encoding either S- or L-HDAg for RT-PCR, and the cDNA product was digested with NcoI. The results showed that cDNA of L-HDAg-encoding HDV RNA could be digested into two fragments (about 80% digestion efficiency), whereas the corresponding S-HDAg-encoding RNA could not (Fig. 5A). Similar studies were then applied to the genomic-sense RNA derived from cells transfected with antigenomic HDV-S or HDV-L RNA. The results clearly showed that HDV-L RNA initially replicated (day 1) and then faded away at later time points (day 6) (Fig. 5B). Significantly, a small amount of the NcoI-digested fragments was detected on day 6 following HDV-S RNA transfection. This was the result of RNA editing in vivo, which generated HDV-L RNA.
FIG. 5.
RT-PCR analysis of the replicated HDV RNA. (A) Optimization of the RT-PCR conditions. In vitro-transcribed mRNAs encoding S-HDAg (small) or L-HDAg (large) were used for RT-PCR using the primers described in Materials and Methods. A portion of the PCR products was digested with NcoI (denoted by +). The products were analyzed by agarose gel electrophoresis. (B) HuH7 cells were transfected with antigenomic HDV-S or HDV-L RNA. On day 1 or 6, RNA samples were collected for RT-PCR analysis as described for panel A. All of the PCR products were digested with NcoI before gel electrophoresis. The HDAg mRNAs from panel A were included as controls. (C) HuH7 cells transfected with HDV-S (2XS), HDV-L, or a 1:1 mixture of these two RNAs were processed for RT-PCR and NcoI digestion as described for panel B. Samples on days 1 and 6 posttransfection were analyzed.
We next applied this analysis to the RNA samples from cells transfected with both HDV-S and HDV-L RNAs (at a 1:1 ratio) of the antigenomic strand. In this experiment, we used HDV-S (2XS) RNA, which could not undergo RNA editing because of the extra nucleotide insertion near the amber/W editing site. Thus, when HDV-S (2XS) RNA was transfected alone, all the cDNA was resistant to NcoI digestion (Fig. 5C, lanes 1 and 2). However, in the cells cotransfected with HDV-S (2XS) and HDV-L RNA, a small amount of the NcoI-digested fragments was detected on day 1, but not on day 6 (lanes 5 and 6). Almost all of the cDNA products on day 6 were resistant to digestion with NcoI. Since HDV-S (2XS) RNA could not be edited, all the NcoI-cleavable cDNA on day 1 must have been derived from the replicated HDV-L RNA. These results also indicated that only HDV-S RNA was detected on day 6. These results combined clearly showed that HDV-L RNA could replicate weakly at early times following RNA transfection but rapidly lost its ability to replicate. In contrast, HDV-S RNA replication was not affected by the presence of HDV-L.
Finally, to establish that the failure of HDV-L RNA to replicate was not due to the inhibition by L-HDAg, we constructed a frameshift mutation near the initiation codon of the HDAg ORF in HDV-L RNA, so that L-HDAg could not be synthesized. This RNA (HDV mt-L) was transfected together with S-HDAg mRNA into HuH7 cells. The results showed that HDV-S RNA by itself led to robust RNA replication, which was detectable on day 6 (Fig. 6A). Neither HDV-L RNA nor HDV mt-L yielded the replicated RNA of the complementary strand on day 6. The same results were obtained using antigenomic RNA transfection (data not shown). In this particular experiment, the replicated RNA from HDV-S or HDV-L could not be detected at day 2 posttransfection, but it could be detected from HDV-S on day 6.
FIG. 6.
Replication of the HDV-S, HDV-L, and HDV mt-L RNAs in HuH7 cells. (A) HuH7 cells were transfected with HDV-S, HDV-L, or HDV mt-L genomic-strand RNAs, respectively (15). On days 2 or 6, RNA samples were collected and used for Northern blotting analysis. (B) The protein samples from day 2 or day 6 were used for immunoblotting analysis of HDAg. Wild-type (WT) HDV RNA from a different strain was used as a positive control. G or AG denotes the RNA strand used for transfection. p27 denotes L-HDAg, and p24 denotes S-HDAg.
These results were confirmed by immunoblotting analysis of HDAg species. In cells transfected with genomic or antigenomic HDV-S RNA, only the S-HDAg species was detected on day 2, which became robust on day 6 (Fig. 6B). In cells transfected with HDV-L RNA, both S- and L-HDAg were initially detected (day 2) (Fig. 6B), consistent with the previous results (Fig. 3B). In cells transfected with HDV mt-L RNA, no L-HDAg could be detected on day 2 (Fig. 6B). Neither S-HDAg nor L-HDAg was detectable on day 6 in the cells transfected with HDV-L or HDV mt-L RNA (Fig. 6B).
These results established that the loss of the replication ability of HDV-L RNA was not due to the production of L-HDAg but was due to the intrinsic properties of HDV-L RNA.
DISCUSSION
Our studies showed that HDV RNA encoding L-HDAg (HDV-L RNA) could not replicate even when an abundant amount of S-HDAg was present. Its inability to replicate is not due to the production of L-HDAg from this RNA, as the HDV RNA encoding S-HDAg (HDV-S RNA) was able to replicate when it was present together with HDV-L RNA. This conclusion is somewhat surprising in view of the earlier finding that L-HDAg inhibits HDV RNA replication (4). However, more recent studies have shown that L-HDAg may not play a prominent role in regulating HDV RNA replication or maintaining the proper balance between HDV-S and HDV-L RNAs in the cells (11, 14). Furthermore, the HDV-L RNA that cannot produce L-HDAg also could not replicate. Therefore, the inability of HDV-L RNA to replicate is most likely an intrinsic property of this RNA. This property explains why HDV-L RNA does not accumulate excessively in the HDV replicating cells despite the high efficiency of RNA editing, and why L-HDAg is not produced excessively during the early stages of viral replication, as HDV-L RNA present in the virion will not be amplified. The surprising finding is that this type of RNA actually replicates initially but then rapidly loses its replication ability as viral replication proceeds. Our studies indicated that HDV-L RNA rapidly undergoes mutations at multiple sites, particularly in the region upstream of the putative HDV replication promoter. The most likely interpretation is that these mutations inactivate the replication ability of HDV-L RNA. The finding that most of the mutations are clustered near the putative promoter for HDV RNA replication makes this interpretation even more plausible. The increased mutations following the amber/W editing have previously been suggested as a possible mechanism for limiting the extent of RNA editing (17). In that particular study, it was noted that the edited RNA often contains additional mutations; however, the kinetics of mutations and the replication ability of these RNAs were not followed in that study. Our findings here give strong support to this interpretation. It is possible that the parental HDV-L RNA, with the amber/W editing but without the additional mutations, is less efficient in replication. The additional mutations may push this RNA to the brink of total inactivation.
This study answered a puzzling question in the early events of HDV replication: why L-HDAg is not produced early despite the presence of HDV-L RNA in the virion. It should be noted that this RNA could be used for the production of L-HDAg (Fig. 3). Thus, unless there is a mechanism to limit the amount of this RNA, L-HDAg will be produced in abundance from the very beginning of virus infection. Our study showed that the in vitro-transcribed HDV-L RNA can replicate briefly after transfection; however, in natural infection, most of the HDV-L RNA incorporated in the virion will likely have already been mutated in the cells, rendering it unable to replicate even at the very outset of infection. This possibility further limits the production of L-HDAg at the beginning of the HDV life cycle. The limitation on the amount of L-HDAg avoids the possible inhibition of HDV RNA replication (at least the synthesis of the genomic strand) early in the viral replication cycle (11, 14) and prevents premature virus assembly. Such a mechanism will also limit the amount of the edited RNA late in the infection. The edited RNA (HDV-L) is able to undergo only a limited number of rounds of replication, while the unedited RNA (HDV-S) continues to replicate, thus reaching a balance between the two RNA species. An interesting possibility is that the transcription of HDAg-encoding mRNA from the genomic HDV-L RNA template may not be inhibited by the mutations observed, thus allowing L-HDAg to be synthesized despite the inhibition of replication of the edited RNA.
The mechanism of mutations associated with the amber/W editing will be of considerable interest. Most of the mutations are limited to the region upstream of the putative promoter region for HDV RNA replication, which has been mapped to around nt 1608 to 1669 (1). These mutations are not contiguous with the amber/W editing site (nt 1012). How the editing affect the fidelity of RNA synthesis at distant sites is a very interesting question. An intriguing possibility is that the editing causes structural changes in the RNA, particularly in the promoter region, such that the fidelity of RNA replication by RNA polymerases is altered. Another interesting question is whether these mutations are triggered by the ADARs, which are responsible for the HDV RNA editing (16). The predominance of the U→C mutations (corresponding to A→G mutations typical of the ADAR specificity on the antigenomic strand) (16, 17) suggested that this is likely the case. Is HDAg itself involved in the regulation of mutations? Our preliminary data showed that in TSδ3 cells, which constitutively express an S-HDAg (7), HDV-L RNA failed to replicate to any significant level, even at the early stage of viral replication (data not shown). These findings suggest that the amount of S-HDAg may regulate the degree of HDV-L RNA inactivation and, consequently, the apparent extent of RNA editing. A previous study indeed suggested that HDAg might suppress HDV RNA editing (17).
In summary, our studies here suggested a mechanism for regulating the extent of RNA editing in HDV replication and ensuring the successful initiation of replication and ordered appearance of S- and L-HDAg during natural virus infection. The mechanism of regulation of these mutations will further contribute to the understanding of HDV RNA editing.
Acknowledgments
We thank Daphne Shimoda for editorial assistance.
This work was partially supported by National Institutes of Health grant no. AI 47348 and the Howard Hughes Medical Institute.
REFERENCES
- 1.Beard, M. R., T. B. Macnaughton, and E. J. Gowans. 1996. Identification and characterization of a hepatitis delta virus RNA transcriptional promoter. J. Virol. 70:4986-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casey, J. L., and J. L. Gerin. 1995. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J. Virol. 69:7593-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, F.-L., P.-J. Chen, S.-J. Tu, C.-J. Wang, and D.-S. Chen. 1991. The large form of hepatitis δ antigen is crucial for assembly of hepatitis δ virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao, M., S.-Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, P.-J., G. Kalpana, J. Goldberg, W. Mason, B. Werner, J. L. Gerin, and J. Taylor. 1986. Structure and replication of the genome of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 83:8774-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glenn, J. S., and J. M. White. 1991. trans-dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 65:2357-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang, S. B., K. S. Jeng, and M. M. C. Lai. 1995. Studies of functional roles of hepatitis delta antigen in delta virus RNA replication, p. 95-109. In G. Dinter-Gottleib (ed.), The unique hepatitis delta virus. R. G. Landes Company, Austin, Tex.
- 8.Hwang, S. B., and M. M. C. Lai. 1993. A unique conformation at the carboxyl terminus of the small hepatitis delta antigen revealed by a specific monoclonal antibody. Virology 193:924-931. [DOI] [PubMed] [Google Scholar]
- 9.Kuo, M. Y.-P., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo, G., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macnaughton, T. B., and M. M. C. Lai. 2002. Large hepatitis delta antigen is not a suppressor of hepatitis delta virus RNA synthesis once RNA replication is established. J. Virol. 76:9910-9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macnaughton, T. B., S. T. Shi, L. E. Modahl, and M. M. C. Lai. 2002. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 76:3920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modahl, L. E., and M. M. C. Lai. 2000. Hepatitis delta virus: the molecular basis of laboratory diagnosis. Crit. Rev. Clin. Lab. Sci. 37:45-92. [DOI] [PubMed] [Google Scholar]
- 14.Modahl, L. E., and M. M. C. Lai. 2000. The large delta antigen of hepatitis delta virus potently inhibits genomic but not antigenomic RNA synthesis: a mechanism enabling initiation of viral replication. J. Virol. 74:7375-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modahl, L. E., and M. M. C. Lai. 1998. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J. Virol. 72:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polson, A. G., B. L. Bass, and J. L. Casey. 1996. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature (London) 380:454-456. [DOI] [PubMed] [Google Scholar]
- 17.Polson, A. G., H. L. Ley, B. L. Bass, and J. L. Casey. 1998. Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol. Cell. Biol. 18:1919-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizzetto, M., B. Hoyer, M. G. Canese, J. W.-K. Shih, R. H. Purcell, and J. L. Gerin. 1980. Delta agent: association of δ antigen with hepatitis B surface antigen and RNA in serum of δ-infected chimpanzees. Proc. Natl. Acad. Sci. USA 77:6124-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheu, G.-T. 2002. Initiation of hepatitis delta virus (HDV) replication: HDV RNA encoding the large delta antigen cannot replicate. J. Gen. Virol. 83:2507-2513. [DOI] [PubMed] [Google Scholar]
- 20.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta viral genome. Nature (London) 323:508-514. [DOI] [PubMed] [Google Scholar]
- 21.Xia, Y.-P., M.-F. Chang, D. Wei, S. Govindarajan, and M. M. C. Lai. 1990. Heterogeneity of hepatitis delta antigen. Virology 178:331-336. [DOI] [PubMed] [Google Scholar]
- 22.Yang, A. L., P. Karayiannis, H. C. Thomas, and J. Monjardino. 1995. Editing efficiency of hepatitis delta virus RNA is related to the course of infection in woodchucks. J. Gen. Virol. 76:3071-3078. [DOI] [PubMed] [Google Scholar]