Abstract
Post-translational isoprenylation of proteins is carried out by three related enzymes: farnesyltransferase, geranylgeranyl transferase-I, and Rab geranylgeranyl transferase (RabGGTase). Despite the fact that the last one is responsible for the largest number of individual protein prenylation events in the cell, no structural information is available on its interaction with substrates and products. Here, we present structural and biophysical analyses of RabGGTase in complex with phosphoisoprenoids as well as with the prenylated peptides that mimic the C terminus of Rab7 GTPase. The data demonstrate that, unlike other protein prenyl transferases, both RabGGTase and its substrate RabGTPases completely ‘outsource' their specificity for each other to an accessory subunit, the Rab escort protein (REP). REP mediates the placement of the C terminus of RabGTPase into the active site of RabGGTase through a series protein–protein interactions of decreasing strength and selectivity. This arrangement enables RabGGTase to prenylate any cysteine-containing sequence. On the basis of our structural and thermodynamic data, we propose that RabGGTase has evolved from a GGTase-I-like molecule that ‘learned' to interact with a recycling factor (GDI) that, in turn, eventually gave rise to REP.
Keywords: protein prenylation, RabGTPases, Rab geranylgeranyl transferase, Rab escort protein
Introduction
Over the past 15 years, it has become increasingly clear that post-translational modification with isoprenoids is a widespread phenomenon, affecting up to 2% of proteins in eukaryotic cells (Glomset and Farnsworth, 1994; Gelb, 1997; Gelb et al, 1998). In all cases that have been studied, such a modification has been shown to be crucial for protein function by modulating protein–lipid or protein–protein interactions. Most of the prenylated proteins are GTPases that have key functions in signal-transduction pathways. This includes the vast majority of the Ras superfamily members and subunits of heterotrimeric G proteins. Interest in protein prenylation has increased dramatically following the observation of the importance of this modification for sustaining the transformed phenotype in many tumours (Gelb et al, 1998; Rowinsky et al, 1999; Lackner et al, 2005).
In protein prenylation, either a farnesyl (F) or a geranylgeranyl (GG) moiety is donated by a soluble isoprenoid pyrophosphate (FPP or GGPP) and attached to one or two C-terminal cysteine residues of the target protein through a thioether linkage. This type of reaction is catalysed by three different protein prenyl transferases: protein farnesyltransferase (FTase), protein geranylgeranyl transferase-I (GGTase-I), and Rab geranylgeranyl transferase (RabGGTase or GGTase-II) (for reviews, see Casey and Seabra, 1996; Leung et al, 2006b). The closely related FTase and GGTase-I transfer prenyl groups to proteins that contain a C-terminal CAAX motif (C is cysteine, A is usually an aliphatic amino acid, and X is a variety of amino acids). The X residue of this motif is the most decisive element for differential recognition by FTase versus GGTase-I.
All three enzymes are heterodimers that are composed of α- and β-subunits (Lane and Beese, 2006). The lipid-binding site is located in a deep hydrophobic cavity of the β-subunit and features an essential Zn2+-binding site at its entrance. Lipid and protein substrate discrimination between FTase and GGTase-I results from size and shape differences in the lipid- and peptide-binding sites. RabGGTase (GGTase-II) stands quite apart from the above-mentioned enzymes both functionally and structurally. Its α-subunit is significantly larger than its counterparts in FTase and GGTase-I due to two additional domains, an immunoglobulin (Ig)-like domain and a leucine-rich repeat (LRR) domain, which are inserted between helices 11 and 12 and at the C terminus, respectively (Zhang et al, 2000) (Figure 1A). The function of these domains is unclear, because they are not involved in interactions within the catalytic ternary complex and are even absent from RabGGTases of low eukaryotes (Dursina et al, 2002; Pylypenko et al, 2003). They may be involved in interactions with intracellular membranes or other proteins (Rasteiro and Pereira-Leal, 2007). RabGGTase transfers the geranylgeranyl moiety onto, in most cases, two C-terminal cysteines of Rab proteins in a broad context of amino acids (Leung et al, 2006a). In contrast to the other two protein prenyltransferases, RabGGTase does not recognize its protein substrate directly but exerts its function in concert with another protein, termed REP (Rab escort protein) (Seabra et al, 1992a, 1992b). REP forms a complex with a newly synthesized Rab protein and presents it to RabGGTase (Andres et al, 1993). On prenylation, dissociation of the Rab–REP complex from RabGGTase is induced by the binding of a new GGPP molecule to the latter, and REP escorts the prenylated Rab to its target membrane (Alexandrov et al, 1994; Thoma et al, 2001a). The recently determined structures of the RabGGTase–REP-1 and Rab7GG–REP-1 complexes provide insights into the mechanism of REP-mediated Rab prenylation (Pylypenko et al, 2003; Rak et al, 2004). REP binds the Rab protein by forming two binding interfaces: one that is between the Rab-binding platform (RBP) and effector loops of the GTPase and another that is between the C-terminal-binding region (CBR) and the CBR interacting motif (CIM), which consists of two hydrophobic residues near the C terminus of RabGTPases (Figure 1A) (Alory and Balch, 2003; Rak et al, 2004). Similar interactions are observed in the structure of the related recycling factor, GDI, crystallized in complex with the yeast Rab GTPase YPT1 (Rak et al, 2003; Pylypenko et al, 2006). We previously proposed that the CBR may have a function in coordinating the C terminus of Rab proteins, orienting it towards the active site of RabGGTase that is docked to domain II of the REP molecule (Rak et al, 2004).
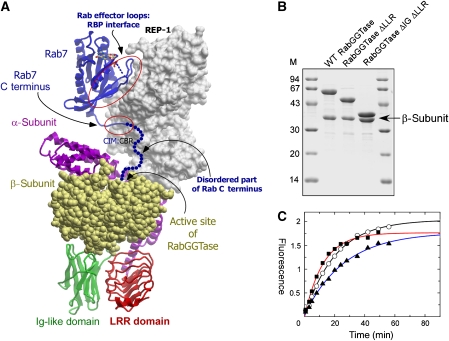
Figure 1.
Engineering of mammalian RabGGTase. (A) Model of the mammalian RabGGTase catalytic ternary complex. The complex was obtained by superimposition of high-resolution structures of Rab7–REP-1 and REP-1–RabGGTase complexes (PDB codes 1VGO and 1LTX). REP-1 is displayed in surface representation and coloured in grey. Rab7 is displayed in ribbon representation and coloured in blue. The disordered part of the C terminus is drawn as a string of blue dots. The α-subunit of RabGGTase is displayed in ribbon representation, and the LRR and Ig-like domains are coloured red and green, respectively, whereas the β-subunit is shown in CPK representation and coloured in yellow. The contact areas between the Rab effector loops and Rab-binding platform (RBP) as well as between the C-terminal interacting motif (CIM) of Rab and the C-terminal binding region (CBR) of REP are circled. Unless otherwise indicated, these and other molecular graphics were generated with ICM Browser Pro. (B) SDS–PAGE analysis of recombinantly produced wild-type and deletion mutants of RabGGTase. (C) In vitro prenylation of Rab7 with wild-type and mutants of RabGGTase using NBD-FPP as a substrate. Incorporation of NBD-F into Rab7 was measured as described in the Materials and methods section and plotted against time. Open circles: RabGGTaseΔLLRΔIg; filled squares: RabGGTaseΔIg; filled triangles: wild-type enzyme.
Despite this progress, RabGGTase remains the least studied prenyltransferase, and the exact mechanism of substrate selection and product release remains unclear. For instance, no structural information is available for RabGGTase in complex with its substrates or products. In addition to being essential for understanding the RabGGTase mechanism, this information is particularly relevant, because knock down or chemical inhibition of RabGGTase was shown to induce apoptosis in cancer cell lines (Lackner et al, 2005; Roelofs et al, 2006).
The lack of structural information is due largely to the difficulties in obtaining high-quality crystals of RabGGTase. In this study, we describe the design of a RabGGTase variant that overcomes this problem. Using the engineered enzyme, we were able to solve the structure of RabGGTase in the apo form and in complex with phosphoisoprenoids and a series of prenylated peptides that simulate both intermediates and the final product of the Rab prenylation reaction. Further, we demonstrate that CIM–CBR interactions have a central function in Rab prenylation and are functionally analogous to CAAX-peptide:active site interactions of FTase and GGTase-I.
Results and discussion
Design and functional analysis of RabGGTase mutants
As mentioned above, structural analysis of RabGGTase is hampered by difficulties in obtaining well-diffracting crystals. Although RabGGTase has previously been crystallized in the apo form as well as in complex with REP-1 and a farnesyl moiety; in both cases, the search for well-diffracting crystals required considerable effort (Zhang et al, 2000; Pylypenko et al, 2003) (NH Thomae and K Alexandrov, unpublished results).
In contrast, both GGTase-I and FTase could be reliably crystallized, and a large number of structures in complex with substrate analogues, reaction products, and inhibitors have been solved (Lane and Beese, 2006). Because we had proposed earlier that the LRR and Ig domains of mammalian RabGGTase are not involved in its catalytic activity (Dursina et al, 2002), we attempted to create mutant variants of the α-subunit gene that lacked sequences for either the LRR or Ig domain, or both. Genes for mutant α- and wild-type β-subunits were co-expressed in Escherichia coli as described (Kalinin et al, 2001) and analysed for expression and solubility. Soluble recombinant proteins were observed in the case of the α-subunit that lacked either the LRR domain or both the LRR and Ig domains (not shown). In both cases, the α/β-heterodimer remained stable throughout purification, indicating that the engineering had not affected the structural integrity of the α-subunit (Figure 1B). Both mutants were able to prenylate Rab7 with an efficiency that was comparable with that of the wild-type enzyme (Figure 1C), as determined by a fluorescent in vitro Rab prenylation assay (Wu et al, 2007a). This directly confirms our earlier proposal that the IGG and LRR domains of mammalian RabGGTase are not involved in the prenylation reaction and must therefore presumably perform an as yet unidentified function (Dursina et al, 2002).
Crystallization and structure solution of RabGGTaseΔIGΔLRR mutant
The overall shape of the RabGGTaseΔIgΔLRR mutant is expected to be similar to that of FTase or GGTase-I, which could make it more amenable to crystallization than the parental enzyme. Indeed, the enzyme crystallized readily and its structure was determined to 1.8-Å resolution by molecular replacement using wild-type RabGGTase as a search model (Table I). Figure 2A shows a ribbon representation of the engineered molecule, which bears the typical hallmarks of a prenyltransferase: a crescent-shaped α-subunit that is formed by seven successive pairs of helical hairpins that are wrapped around the β-subunit. The β-subunit folds into an α–α barrel that consists of six core and six peripheral helices. As expected, the overall structure of the engineered enzyme changed very little compared with the wild-type protein, with an overall Cα r.m.s.d. of 0.85 Å for 609 residues. Most of the changes comprise shifts in the position of α-helices in the α-subunit, whereas the β-subunit is essentially unchanged, with a Cα r.m.s.d. of 0.35 Å. Electron density is missing for the 11 flexible N-terminal amino acids and for the loop region between helices 9 and 10. The engineered link between helices 11 and 10, on the other hand, is visible in the electron density, indicating that the chosen length and sequence were appropriate to create a defined structure. As in the native enzyme, the mutant harbours a catalytic Zn2+ ion at the top of the active site funnel.
Table 1.
Data collection and refinement statistics
| Data collection | RabGGTase Apo | RabGGTase–GGPP | RabGGTase–FPP |
|---|---|---|---|
| Wavelength (Å)a | 0.9800 | 1.0007 | 0.9998 |
| Resolution (highest shell, Å) | 29.2–1.8 (1.9–1.8) | 29.4–1.9 (2.0–1.9) | 29.4–1.9 (2.0–1.9) |
| Space group | P212121 | P212121 | P212121 |
| Cell constants (Å; deg) | a=66.7, b=90.6, c=114.1; α=β=γ=90 | a=67.3, b=90.6, c=114.5; α=β=γ=90 | a=66.9, b=90.9, c=114.2; α=β=γ=90 |
| VM | 2.5 | 2.5 | 2.5 |
| Total measurements | 294 558 | 257 863 | 313 284 |
| Unique reflections | 64 333 | 55 563 | 54 566 |
| Average redundancy | 4.6 (3.4) | 4.6 (4.5) | 5.7 (5.8) |
| I/σ | 16.4 (4.7) | 24.6 (5.4) | 22.5 (4.8) |
| Completeness (%) | 99.3 (99.7) | 99.4 (99.4) | 98.2 (98.1) |
| Rsymb | 6.3 (27.5) | 4.4 (30.8) | 4.6 (40.2) |
| Wilson B-factor (Å2) | 30 | 47 | 34 |
| Refinement | |||
| Resolution (highest shell, Å) | 1.8 (1.85–1.8) | 1.9 (1.95–1.9) | 1.9 (1.95–1.9) |
| Rc | 14.9 (16.3) | 18.0 (22.3) | 15.9 (18.6) |
| Rfreed | 19.4 (23.4) | 22.0 (28.8) | 21.6 (25.9) |
| r.m.s.d. bonds (Å)/angles (deg) | 0.009/1.250 | 0.010/1.375 | 0.010/1.409 |
| B-factor deviation bonds/angles (Å2) | |||
| Main chain | 1.157/1.860 | 0.936/1.497 | 1.119/1.820 |
| Side chain | 2.411/3.825 | 2.064/2.999 | 2.336/3.580 |
| Residues in Ramachandran core (%)e | 93.4 | 92.8 | 92.2 |
| Protein atoms Solvent atoms Ligand atoms | |||
| Average B-factor (Å2) | 18 | 37 | 25 |
| PDB accession code | 3DSS | 3DST | 3DSU |
| Data collection | RabGGTase–Ser-Cys- Ser-Cys(GG) | RabGGTase–Ser-Cys(GG)- Ser-Cys | RabGGTase–Ser-Cys(GG)-Ser-Cys(GG) |
| Wavelength (Å)a | 0.9999 | 0.9825 | 0.9841 |
| Resolution (highest shell, Å) | 29.4–2.1 (2.2–2.1) | 29.4–2.15 (2.25–2.15) | 29.4–2.1 (2.2–2.1) |
| Space group | P212121 | P212121 | P212121 |
| Cell constants (Å; deg) | a=67.0, b=90.6, c=114.4; α=β=γ=90 | a=67.1, b=90.9, c=114.3; α=β=γ=90 | a=67.4, b=91.2, c=114.6; α=β=γ=90 |
| VM | 2.5 | 2.5 | 2.5 |
| Total measurements | 763 680 | 153 685 | 302 144 |
| Unique reflections | 41 339 | 38 414 | 41 826 |
| Average redundancy | 18.5 (16.5) | 4.0 (4.1) | 7.2 (7.4) |
| I/σ | 31.0 (11.1) | 15.2 (5.2) | 23.4 (6.9) |
| Completeness (%) | 99.9 (99.7) | 99.1 (99.8) | 99.8 (100) |
| Rsymb | 8.5 (37.9) | 7.2 (37.3) | 5.7 (39.1) |
| Wilson B-factor (Å2) | 30 | 39 | 39 |
| Refinement | |||
| Resolution (highest shell, Å) | 2.1 (2.15–2.1) | 2.15 (2.2–2.15) | 2.1 (2.15–2.1) |
| Rc | 14.9 (14.0) | 17.0 (17.7) | 16.8 (17.0) |
| Rfreed | 21.1 (22.4) | 23.2 (27.4) | 23.0 (26.0) |
| r.m.s.d. bonds (Å)/angles (deg) | 0.011/1.456 | 0.012/1.446 | 0.011/1.285 |
| B-factor deviation bonds/angles (Å2) | |||
| Main chain | 1.100/1.968 | 1.391/2.440 | 1.166/1.867 |
| Side chain | 2.394/3.701 | 2.759/4.269 | 2.397/3.576 |
| Residues in Ramachandran core (%)e | 91.7 | 92.4 | 92.8 |
| Protein atoms Solvent atoms Ligand atoms | |||
| Average B-factor (Å2) | 26 | 36 | 30 |
| PDB accession code | 3DSV | 3DSW | 3DSX |
| aAll data were collected at beamline X10SA of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland). | |||
| bRsym=∑∑I(h)j−<I(h)>/∑∑I(h)j where I(h)j is the measured diffraction intensity and the summation includes all observations. | |||
| cR is the R-factor=(∑∣FO∣−∑∣FC∣)/∑∣FO∣. | |||
| dRfree is the R-factor calculated using 5% of the data that were excluded from the refinement. | |||
| eRamachandran core refers to the most favoured regions in the ϕ/ψ-Ramachandran plot. | |||
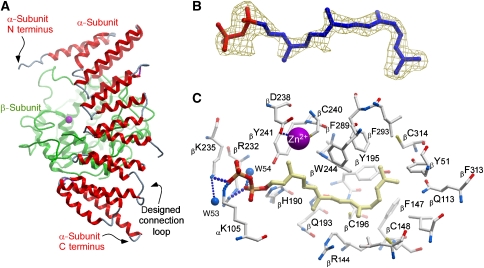
Figure 2.
Structure of the RabGGTaseΔLLRΔIg variant in the apo form and in complex with GGPP. (A) Structure of the apo form in ribbon representation. The β-subunit is coloured green, whereas the α-subunit is coloured by secondary structure. (B) Density map of the active centre of the RabGGTaseΔLLRΔIg–GGPP complex contoured 2.5 σ The electron density (Fo−Fc map) was calculated prior to the incorporation of the ligand into the model. (C) Ball and stick representation of the active site of RabGGTaseΔLLRΔIg with a bound GGPP molecule. The hydrogen bonds are shown as strings of small blue balls sized according to distance. The Zn2+ ion is shown as a magenta ball.
Structure of RabGGTase in complex with its substrate GGPP
The availability of a RabGGTase that displays robust crystallization behaviour and near-native biochemical properties prompted us to attempt to determine the structure of RabGGTase with its native substrate, GGPP. Such a structure would complete the picture of substrate–enzyme complexes for the protein prenyltransferase family and give mechanistic insights into the lipid substrate specificity of RabGGTase. Co-crystallization of the RabGGTase–GGPP complex was achieved using a 1:2 molar mixture of the enzyme and ligand, and the structure of the complex was refined to 1.9-Å resolution (Table I). Initial difference density electron maps (FRabGGTase–GGPP−Fc) showed strong positive density in the hydrophobic cavity of the α–α barrel of the β-subunit (Figure 2B). Similar to structures of other prenyltransferases, the isoprenoid part of the molecule is held in the hydrophobic-binding cleft that is formed by the conserved aromatic residues βY51, βW52, βF147, βY195, βY241, βW243, βW244, βF289, βF293, αF143, and αY107 (Figure 2C). The diphosphate moiety of the GGPP molecule binds in a positively charged cleft that is near the subunit interface and is close to the catalytic zinc ion (Figure 2B and C). The resolution of the structure allows for the unambiguous determination of residues that form hydrogen bonds with the diphosphate moiety of the GGPP molecule. The β-phosphate forms hydrogen bonds with βK235 and αK105, whereas the α-phosphate forms a hydrogen bond with βR232 and a water molecule (Figure 2C). Binding of GGPP to RabGGTase leads to several minor changes in the structure of the active site, which mostly involve rotation or displacement of side chains of the hydrophobic residues βF293, βY241, βW244, βY195, and βC241 (not shown).
Comparison of the RabGGTase:GGPP structure with that of the GGTase-I:GGPP complex
Perhaps the most interesting aspect of the RabGGTase:GGPP complex structure is its comparison with the respective complex of GGTase-I (Taylor et al, 2003). Carbons 1–7 of GGPP adopt different conformations and are located much closer to the Zn2+ ion in RabGGTase than in GGTase-I (Figure 3A). Carbon atoms 8–15 superimpose very well, whereas the distal portions of the isoprenoids are positioned differently. The binding sites can be divided into two halves based on their conservation. The region of the binding site that harbours phosphate groups and carbons 1–12 of the isoprenoid chain are invariant and adopt similar positions in both enzymes. However, the bottom of the binding cavity appears to be less conserved. Although some substitutions do not lead to significant changes in the shape of the binding site or its hydrophobicity, such as GGT-IβF53 to RabGGTaseβW52 or GGT-IβF52 to RabGGTaseβY51, several other substitutions change both properties of the active site: GGT-IβL320 to RabGGTaseβF289, GGT-IβY323 to RabGGTaseβL292, GGT-IβY126 to RabGGTaseβL99, and GGT-IβN345 to RabGGTaseβC314. These changes are responsible for the bend in the last five carbon atoms of GGPP that are associated with RabGGTase. Compared with GGTase-I, the lipid-binding site of RabGGTase is expanded at the bottom, primarily due to the substitutions of GGT-IβY323 and GGT-IβY126 (Figure 3A). This makes RabGGTase more likely to be tolerant to the substitution of the distal part of the isoprenoid chain than CAAX prenyltransferases. This idea finds support in a recent report demonstrating that of the three protein prenyltransferases, only RabGGTase can utilize biotin-geranyl pyrophosphate as a substrate (Nguyen et al, 2007).
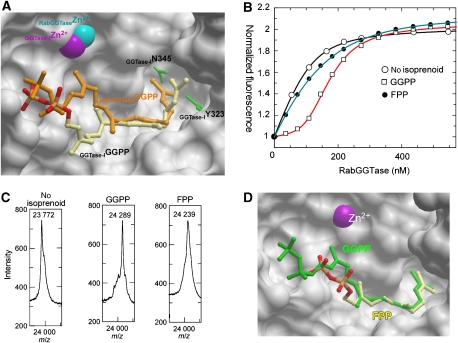
Figure 3.
Analysis of substrate specificity of RabGGTase. (A) Superimposition of structures of the RabGGTase–GGPP and GGTase-I–GGPP (1N4Q) complexes. The active site of RabGGTase is displayed in surface representation, whereas selected residues of the active site of GGTase-I are shown as green ball and stick. The ball-and-stick representation of GGPP bound to GGTase-I is displayed in atomic colours, whereas the GGPP bound to RabGGTase is coloured in orange. Zn2+ ions are displayed as CPK spheres. (B) Fluorescent titration of 200 nM mant-FPP with RabGGTase in the presence of 100 nM of different phosphoisoprenoids. The λex/em was set to 340/426 nm. The data were fitted numerically as described in the Materials and methods section, and the obtained affinities are summarized in Table II. (C) MALDI-TOF analysis of the Rab7 prenylation reaction supplemented with GGPP and FPP. In each experiment, 20 μM of Rab7, REP-1, and RabGGTase were mixed with 50 μM of the respective phosphoisoprenoid and incubated at room temperature for 30 min. (D) Superimposition of RabGGTase in complex with GGPP and FPP, displayed as in (A). GGPP is coloured in green and FPP in atomic colours.
Analysis of the interaction of RabGGTase with farnesyl pyrophosphate
Analysis of structures of prenyltransferases in complex with lipid substrates shows that the phosphate groups are well exposed to the solvent and that the active sites of prenyltransferases could accommodate isoprenoids that have longer or shorter chains. We previously observed incorporation of radioactive FPP into Rab proteins by RabGGTase (Thoma et al, 2000). We decided to revisit this issue and to obtain quantitative and structural information on the interaction of FPP with RabGGTase. To this end, we employed a previously established assay in which binding of isoprenoids to prenyltransferases is monitored by the influence of the former on the interaction of prenyltransferase with a fluorescent isoprenoid analogue (Thoma et al, 2000; Dursina et al, 2006). These experiments led to an estimate of the FPP affinity for RabGGTase (Kd=ca. 90 nM, i.e. >100 times weaker than the native substrate GGPP; Figure 3B and Table II).
Table 2.
Dissociation constants of phosphoisoprenoids:RabGGTase complexes
| GGPP | FPP | |
|---|---|---|
| Kd co-titration (nM) | 0.83±0.22 | 94±14 |
| Kd displacement titration (nM) | 1.8±0.71 | 75±10 |
| Incorporation into Rab7 | + | + |
To obtain a direct measure of farnesyl incorporation, we used mass spectrometry to monitor RabGGTase-catalysed Rab7 prenylation in the presence of FPP. As can be seen in Figure 3C, the farnesyl group was transferred by RabGGTase onto Rab7 efficiently, resulting in the formation of a doubly farnesylated product. Although FPP functions as an efficient lipid donor for Rab prenylation in vitro, farnesylated Rab proteins have not been found in vivo. This is likely to be due to the fact that RabGGTase binds with 100-fold greater preference to GGPP over FPP, providing a thermodynamically driven selection of the appropriate lipid substrate.
Structure solution of RabGGTase in complex with FPP
To gain further insight into the mode of isoprenoid binding to RabGGTase and its ability to utilize substrates of different lengths, we co-crystallized the enzyme with FPP using the procedure that was described for the native substrate. The structure of the complexes was determined to 1.85-Å resolution. Comparison of the structures demonstrated that the 12 carbons of the farnesyl moiety adopt very similar conformations and positions to those of GGPP (Figure 3D). As a result, C1 of FPP is located 5 Å away from the position that is occupied by C1 of the native GGPP. The β-phosphate of FPP forms a strong hydrogen bond with βY241 that is not involved in hydrogen bonding with the native substrate (Figure 2C). On the basis of the crystal structures of the FPP:FTase and GGPP:GGTase-I complexes, a ‘molecular ruler' mechanism was proposed, which postulated that the appropriate prenyl pyrophosphate is selected on the basis of the prenyl-binding site depth (Liang et al, 2002). In a proper isoprenoid:enzyme match, the C1 of FPP or GGPP is placed in the vicinity of the catalytic Zn2+ ion. In keeping with this idea, in the RabGGTase:FPP complex, the C1 atom of FPP is too far away from the Zn2+ ion to participate in the reaction with the Zn2+-coordinated cysteine thiol. However, in contrast to FTase and GGTase-I, RabGGTase transfers GGPP and FPP at a similar rate (Thoma et al, 2000) (data not shown). This suggests that FPP can slide in the active site to allow this interaction to take place. This must occur relatively easily to account for the prenylation reaction, and the rate of sliding must be rapid compared with the actual lipid transfer reaction. These observations are also in keeping with a recent report that showed that RabGGTase has the broadest lipid substrate specificity among prenyltransferases (Nguyen et al, 2007).
Structure of RabGGTase in complex with mono- and diprenylated Rab7 C-terminal peptides
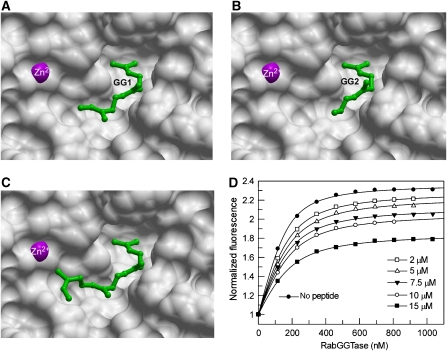
The ability to perform two sequential geranylgeranyl transfer reactions on a single peptide substrate is a salient feature of RabGGTase. One of the longstanding questions in the field is the location of the conjugated isoprenoid during the second transfer reaction. Comparison of the structures of GGTase-I and RabGGTase led to speculation that the conjugated isoprenoid would be located in the cavity that is near the reaction centre (Taylor et al, 2003). To test this experimentally, we attempted crystallization of the enzyme that was bound to the reaction product. To this end, we used solid-phase synthesis to generate three peptides that mimicked the mono- and diprenylated C terminus of Rab7–Ser-Cys-Ser-Cys(GG), Ser-Cys(GG)-Ser-Cys, and Ser-Cys(GG)-Ser-Cys-(GG). Crystals of the complexes with RabGGTase were obtained by co-crystallization. These diffracted to 1.9, 2.1 and 2.0 Å, respectively. In all three cases, the lipid-binding site was occupied by a geranylgeranyl moiety (Figure 4). In the case of RabGGTase–Ser-Cys(GG)-Ser-Cys, the isoprenoid chain was traceable only to C11 and to C6 in the case of Ser-Cys-Ser-Cys(GG). In all three cases, the electron density for the peptide chain was very poor, indicating that it is flexible and does not form highly stabilizing interactions with the active site of the enzyme. Remarkably, the density for the second isoprenoid chain in the RabGGTase–Ser-Cys(GG)-Ser-Cys-(GG) complex could not be detected. However, in this case, the lipid that was bound to the active centre could be traced in the electron density completely, suggesting that the presence of the second lipid leads to a reduction in the flexibility of the peptide. Comparison of the obtained complexes with the structures of FTase and GGTase that were bound to prenylated peptides and phosphoisoprenoids indicates that in contrast to CAAX prenyltransferases, RabGGTase does not possess a defined cysteine-geranyl-binding site that is analogous to the exit groove of CAAX transferases (Long et al, 2002; Taylor et al, 2003). The original study of GGTase-I led to the proposal that a tunnel that is located next to the lipid-binding site of RabGGTase might stabilize the monoprenylated product during the processive reaction (Taylor et al, 2003). On the basis of the presented structures, this scenario appears to be unlikely, because no additional electron density could be detected in any of the presented structures. Further, despite considerable effort, we were not able to obtain the structure of the ternary RabGGTase:GGPP:prenylated peptide complex. The presence of the phosphoisoprenoid in the crystallization set-ups led to the formation of crystals that contained only GGPP that was bound to the active site. This provides an indirect indication that the affinity of the prenylated intermediate or product for RabGGTase is low. To obtain the estimates of the affinities, we performed co-titration experiments in which the interaction of a fluorescent analogue of GGPP (mant-FPP) for RabGGTase was monitored in the absence or presence of the prenylated peptides (Owen et al, 1999; Thoma et al, 2000). The changes in mant fluorescence were fitted to a competitive binding model, allowing for extraction of the affinities of both mant-FPP and peptides for RabGGTase (Figure 4D) (Wu et al, 2007b). It transpired from these data that Ser-Cys-Ser-Cys(GG), Ser-Cys(GG)-Ser-Cys, and Ser-Cys(GG)-Ser-Cys-(GG) bound to RabGGTase with comparable low micromolar affinities (Figure 4D). This indicates that the only strong interaction between the prenylated peptide and RabGGTase is mediated by the lipid-binding site and that a second lipid does not positively contribute to the affinity of the interaction.
Figure 4.
Structure analysis of RabGGTase in complex with Ser-Cys-Ser-Cys(GG), Ser-Cys(GG)-Ser-Cys, and Ser-Cys(GG)-Ser-Cys-(GG) peptides. (A) The active site of RabGGTase in complex with Ser-Cys-Ser-Cys(GG) shown in surface representation. The visible part of the geranylgeranyl moiety is shown in green ball and sticks. The C-terminal isoprenoid is denoted as GG1, whereas the second geranylgeranyl is denoted as GG2. (B) RabGGTase in complex with Ser-Cys(GG)-Ser-Cys, displayed as in (A). (C) RabGGTase in complex with Ser-Cys(GG)-Ser-Cys-(GG), displayed as in (A). (D) Fluorescence titration of mant-FPP with increasing concentrations of RabGGTase in the presence or absence of SC(G)SC(G) peptide. The final concentration of the peptide is indicated in the graph. The filled circles represent a titration in the absence of peptide. To obtain the affinities for enzyme:peptide and enzyme:isoprenoid interactions, the data were fitted to a competitive model as described elsewhere (Wu et al, 2007b) and led to a Kd value of 1.4±0.3 μM.
The CIM motif is essential for Rab prenylation in vitro
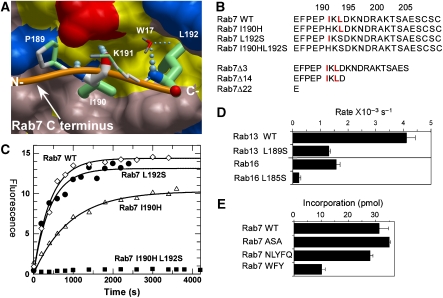
The data that are presented here shed light on the mechanisms of lipid substrate recruitment and events that follow the transfer of the first isoprenoid lipid onto the C terminus of the Rab protein. However, we know very little about the events that lead to the positioning of the first substrate cysteine in the proximity of the catalytic Zn2+ and C1 of GGPP. Clearly, this is triggered by sequential assembly of the Rab:REP–RabGGTase complex; yet it is not known how RabGTPase places its C terminus in the active site of RabGGTase. On the basis of the structure of the Rab7:REP complex, we previously proposed that CIM:CBR interactions may be important for this process. To complete the picture of the RabGGTase functional cycle, we analysed the role of the Rab C terminus and, in particular, the CBR in the formation of the catalytic ternary Rab–REP–RabGGTase complex and in the prenylation reaction. Because the CBR is involved in hydrophobic interactions of Rab C-terminal sequences with the hydrophobic patch on REP, we designed Rab7 mutants in which I190 and L192 of the CBR were mutated to polar amino acids (Figure 5A and B). The resulting mutants were purified and subjected to in vitro prenylation with NBD-FPP (Dursina et al, 2006; Wu et al, 2006). As can be seen in Figure 5C, the individual mutations led to a reduction in the rate and total yield of the prenylation reaction. The Rab7 L192S mutant displayed more significant inhibition of prenylation than Rab7 I190H, consistent with the structure of the CBR:CIM interface, in which L192 forms a larger contact area and protrudes deeply into the hydrophobic cavity on the REP-1 surface, whereas I190 contacts the outer edge of the CBR (Figure 5A). The mutations display an additive effect, as the double mutant Rab7 I190HL192S showed no detectable prenylation, indicating that the CIM motif is indispensable for Rab prenylation (Figure 6). Mutation of the CIM regions of Rab13 and Rab16 led to a reduction in the efficiency of their prenylation, confirming the general role of the CIM:CBR interaction in Rab prenylation (Figure 5D).
Figure 5.
Analysis of the CBR–CIM interaction. (A) Structural basis of the REP-1 CBR interaction with the CIM of Rab7. The hydrophobic CBR patch of REP-1 is displayed in surface representation, and hydrophobic residues are coloured yellow, and polar and charged residues are coloured in pink, blue (+), and red (−). The main chain atoms involved in hydrogen bonding are displayed in atomic colours. The C terminus of Rab7 is displayed as an orange worm, and the residues RabP189-RabL192 are displayed in stick format. N- and C- denote the termini. Atoms involved in hydrophobic interactions are coloured in green. The boldness of the stick representation is an indication of the degree of interaction with REP. The order of contact area for these residues is L192>I190>P189>K191. W17 denotes a water molecule involved in the hydrogen bonding network. (B) C-terminal mutants of Rab7 used in this study. The CIM motif is highlighted in red. Mutation of I190 to histidine was chosen based on the alignment with Rab27A, which does not display a canonical CIM motif. (C) In vitro prenylation of wild-type Rab7 and its CIM mutants with NBD-FPP. The amount of prenylated substrate was determined by fluorescent scanning of SDS–PAGE-resolved samples. (D) Changes in in vitro prenylation rates of wild-type and CIM mutants of Rab13 and Rab16. (E) Effect of extension of the Rab7 C terminus on the ability to serve as substrates of RabGGTase.
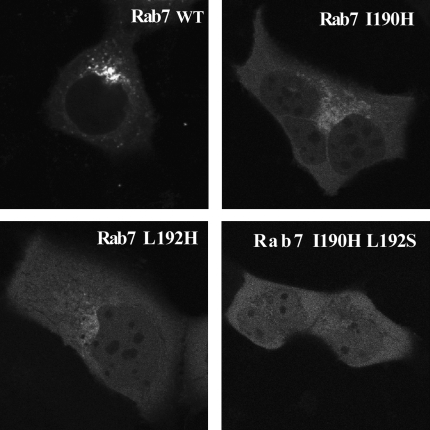
Figure 6.
Analysis of CIM mutants of Rab7 in vivo. Fluorescently tagged recombinant CIM mutants of Rab7 GTPase were microinjected into A431 cells and imaged after 22 h.
The CIM motif modulates the assembly of Rab prenylation machinery
To understand the consequences of the above described CIM mutations at the molecular level, we undertook a quantitative analysis of the role of the CIM in the interactions of the Rab7:REP-1 binary complex and the Rab7:REP-1:RabGGTase ternary complex. Because the affinity of the Rab7:REP interaction is very high (Kd∼1 nM), it can be accurately measured only when a titration experiment is performed at low concentrations of the reactants, thus requiring an assay that has a large fluorescence change and good quantum yield (Alexandrov et al, 1998; Wu et al, 2006). Hence, we employed a semisynthetic Rab7 protein that was C-terminally modified with the NBD group, prepared as described (Rak et al, 2004). Addition of REP-1 protein to a solution of Rab7C(NBD)SC resulted in a four-fold increase in NBD fluorescence, whereas association of RabGGTase with the Rab7-NBD:REP-1 binary complex led to two-fold fluorescence quenching (not shown). We used these signal changes to monitor the interaction of a 50 nM solution of Rab7-NBD and the Rab7-NBD:REP-1 complex with increasing concentrations of REP-1 and RabGGTase, respectively (Supplementary Figure S1). Fitting the data to quadratic equations led to Kd values of 2.8±0.8 nM for Rab7-NBD–REP-1 and 61±7 for Rab7-NBD–REP-1:RabGGTase, in close agreement with the values that have been determined using alternative methods (Alexandrov et al, 1998, 1999). To obtain the Kd values for the interaction of unlabelled wild-type Rab7 and its CIM mutants with REP and RabGGTase, we performed co-titration experiments in which a mixture of Rab7-NBD and Rab7 WT/mutant, or Rab7-NBD–REP-1 and Rab7 WT/mutant:REP-1 were titrated with increasing concentrations of REP-1 or RabGGTase.
As can be seen in Table III, mutation of the CIM motif results in ca. 30- to 70-fold reduction in Rab7–REP-1 affinity, suggesting that the interactions of the CIM with the CBR make a significant contribution to the overall affinity of the binary complex. To further verify our findings, we prepared mutants of Rab7 that retained the CIM motif but were truncated by 3 or 14 residues, and a mutant that was truncated by 22 amino acids, in which CIM was deleted (Figure 5B). Analysis of Rab7Δ3 and Rab7Δ14 interactions with REP reveals that deletion of the prenylation motif (Rab7Δ3) and even the entire downstream residues after the CIM motif (Rab7Δ14) has limited influence on the affinity of the interaction. However, deletion of the 22 residues that include the CIM motif resulted in a 50-fold decrease in the affinity of the binary complex. This observation indicates that, in accordance with the structural data, the CIM motif is the only site in the Rab7 C terminus that contributes significantly to the Rab7:REP interaction. The Kd value for Rab7Δ22 is quite close to that of the double mutant Rab7I 190HL192S, suggesting that I190 and L192S have a central function in the coordination of the Rab C terminus on the surface of the REP molecule.
Table 3.
Dissociation constants of REP-1 in binary complexes with Rab7 variants and of RabbGGTase in ternary complexes with REP-1–Rab7 variants
| Complex | Kd (REP-1) (nM) | Kd (RabGGTase) (nM) |
|---|---|---|
| Rab7C(NBD)SC | 2.8±0.8 | 61±6.9 |
| Rab7C(NBD)SCC | 3.2±0.9 | ND |
| Rab7WT | 7.5±2.7 | 130±9.3 |
| Rab7I 190H | 228±59 | ND |
| Rab7 L192S | 341±74 | ND |
| Rab7I 190HL192S | 547±46 | ND |
| Rab7Δ3 | 16.1±1.0 | 191±22 |
| Rab7Δ14 | 15.8±2.1 | 321±11 |
| Rab7Δ22 | 381±37 | 491±31 |
| Rab7-5A | 21.5±1.1 | 188±45 |
| Rab7_WFY | ND | 107±16 |
| ND, not determined. | ||
Effect of CIM mutations on the processing of Rab7 in vivo
To examine the effect of CIM mutations on prenylation in vivo, we microinjected purified recombinant, fluorescently tagged Rab7 into cultured A431 cells. When unprenylated, wild-type Citrine-Rab7 was microinjected, the protein was initially cytosolic and diffused into the nucleus. Within 2 h, the majority of the wild-type Rab7 became prenylated by the endogenous cellular prenylation machinery and localized to the membranes of perinuclear vesicular structures, and no protein remained in the cytosol or nucleus (Figure 6). When Cherry-Rab7 I190H, Cherry-Rab7 L192S, or Cherry-Rab7 I190H L192S were injected, the rate of localization to membranes was significantly reduced, in keeping with the idea that these mutants were processed inefficiently by the endogenous prenylation machinery. At 2 h after microinjection, the mutants remained entirely cytosolic (not shown). After 22 h, Cherry-Rab7 I190H and Cherry Rab7 L192S showed partial localization to the perinuclear region (Figure 6). The double mutant Cherry-Rab7 I190H L192S showed no localization even after 22 h, indicating that it failed to undergo prenylation. This further demonstrates the significance of the CIM motif for the proper processing of Rab GTPases in vivo.
Effects of sequences downstream of CIM on Rab prenylation
As can be inferred from Table III, the affinity of the Rab7 moiety in the Rab:REP:RabGGTase ternary complex displays a decrease with progressive truncation of C-terminal residues. Replacement of (199)KTSAES(204) of Rab7 by a poly-alanine sequence (Rab7-5A) and addition of aromatic residues at the C terminus (Rab7_WFY) result in a minor reduction and enhancement in ternary complex affinity, respectively (not shown and Figure 5E). These observations demonstrate that the post-CIM portion of the C terminus makes a limited but detectable contribution to the binding energy of the Rab:REP complex interaction with RabGGTase. This suggests that the disordered Rab C terminus associates weakly with RabGGTase. Interestingly, the Rab7_WFY mutant increases the affinity of the ternary complex, whereas its prenylation is reduced (Figure 5E). This suggests that the aromatic WFY residues bind to the lipid-binding site of RabGGTase, increasing the affinity of the complex but at the same time preventing proper positioning of the second cysteine for catalysis. A similar effect was previously described for dansyl groups that were attached to the C terminus of Rab7 (Alexandrov et al, 1999). Remarkably, extension of the C terminus by four or five arbitrarily chosen residues does not decrease prenylation efficiency, suggesting that the C terminus can insert into the active site in a bent conformation and that substrate recognition is truly sequence-independent (Figure 5E).
Functional model of Rab prenylation
The structural and functional data that were obtained in this study allow us to formulate a complete mechanistic model of Rab prenylation. The assembly of the catalytic ternary Rab–REP–RabGGTase complex is triggered by the recognition of the Rab GTPase domain by the RBP of REP. This results in a low- to intermediate-affinity complex, which is further tightened by the interaction of the CIM with the CBR. The resulting complex forms a high-affinity ternary complex with RabGGTase through the interactions between the α-subunit of RabGGTase and domain II of REP. The affinity of the complex is further increased by the weak and largely unspecific interactions of the C terminus with the active site of RabGGTase. This sequential mode of complex assembly engages progressively smaller binding interfaces (Rab–RBP>CIM–CBR>C terminus:RabGGTase) and weaker interactions to recruit and modify a range of peptide substrates that have no common feature other than the presence of cysteines near or at the C terminus. From this perspective, one can view the CIM as being analogous to the AAX motif of the CAAX box in the case of FTase and GGTase-I, working from a remote location. In other words, both the enzyme (RabGGTase) and the substrate (RabGTPase) ‘outsource' their specificity to an accessory factor (REP). One of the consequences of such an arrangement is the ability of the active site to accommodate a broad variety of peptide sequences. The fact that the extension of the Rab7 C terminus by as many as five amino acids does not influence the efficiency of prenylation clearly illustrates this fact (Figure 5E). The need to accommodate very diverse peptide substrates in the active centre puts pressure on the lipid-binding site to provide more room for isoprenoids that might need to adopt different conformations and positions depending on the peptide substrate. This idea finds support in the observation that RabGGTase can transfer both farnesyl and geranylgeranyl groups with comparable rates and can accept very bulky lipid substrates (Nguyen et al, 2007). The lack of a well-defined substrate-binding mode also explains the variation in the observed sequence of isoprenoid addition in the Rab digeranylgeranylation reaction, which can be explained by the influence of the surrounding amino acids on the positioning of reactive cysteines to Zn2+ for catalysis (Shen and Seabra, 1996; Thoma et al, 2001b). The relaxed substrate-binding sites allow the enzyme to operate stochastically, whereby the reaction occurs when one or the other of the cysteine side chains happens to be in the catalytic centre.
Although such an arrangement allows the enzyme to accommodate a broad diversity of C-terminal sequences that occur in RabGTPases, the lack of precise positioning and high affinity of the enzyme:substrate interaction may negatively impact the rate of RabGGTase-mediated catalysis. Although other factors, such as differences in the location of the prenyl group C1 relative to the catalytic Zn2+, substitutions in the binding site, and dielectric effects, can also influence the rate of catalysis, RabGGTase remains the slowest prenyltransferase (FTase: kchem=12–17 s−1; GGTase-I: kchem=0.5 s−1; RabGGTase: k1=0.16 s−1, k2=0.04 s−1).
Following the conjugation of the first lipid, a new molecule of GGPP binds to the active site and dislodges the weakly bound conjugated isoprenoid. The monoprenylated intermediate does not have a defined position in the active site and randomly samples the surface of the transferase. The fact that the rate of the second transfer step is four times slower than the first one, despite the higher affinity of the ternary complex, indicates that the conjugated lipid provides steric hindrance for the second cysteine (Thoma et al, 2001b).
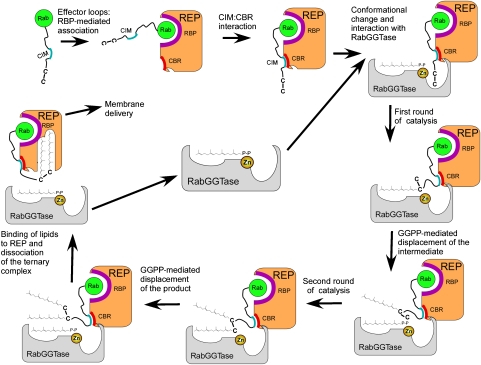
Following the prenylation of the second cysteine moiety, the doubly prenylated peptide is displaced by the tightly binding GGPP molecule (Thoma et al, 2001a). As a result, the bulky prenylated C terminus is dislodged form the active site of RabGGTase and associates with the lipid-binding site of REP, inducing a conformational change in its domain II and triggering the release of RabGGTase (Figure 7) (Thoma et al, 2001a).
Figure 7.
Mechanistic model of RabGGTase-mediated protein prenylation.
Evolution of protein prenylation
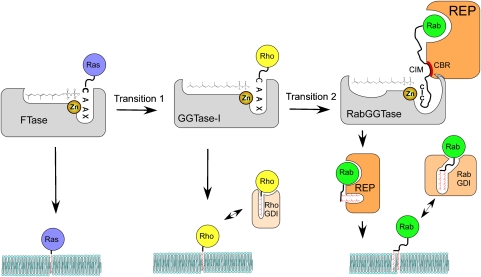
The presented data complete our knowledge of the basic mechanisms of protein prenylation and, in combination with the available phylogenetic data, provide the starting point for an analysis of its evolution (Figure 8). Although the phylogenetic analysis cannot resolve the exact order of appearance of prenyltransferses due to the presence of all three enzymes in the last common eukaryotic ancestor, FTase and GGTase-I are believed to precede RabGGTase (Rasteiro and Pereira-Leal, 2007). Therefore, it appears likely that the primordial protein prenyltransferase was similar to FTase. Emergence of GGPP biosynthesis in early eukaryotes was accompanied by the extension of the lipid-binding site of the prenyltransferase—a step that can be easily recapitulated by mutations in contemporary FTase (Terry et al, 2006). The resulting GGTase-I-like enzyme generated a more hydrophobic product that was unloaded onto the membrane, presumably directly by the enzyme, but could not then be re-solubilized as easily as farnesylated proteins. This promoted the emergence of GDI-like molecules that facilitated the extraction of GTPases from the membranes by binding both the protein and lipid parts of the molecule. To function well as a GDI, the molecule needs to bind relatively weakly to the GTPase domain, and the overall binding affinity should increase significantly following the interaction with the prenylated C terminus (Wu et al, 2007b). Such factors appear to have emerged independently in evolution on several occasions, because RhoGDIs and RabGDIs are not structurally related. Proliferation of intracellular membrane-bound compartments and signalling cascades in eukaryotes resulted in a further increase in the number and specialization of small GTPases. Emergence of the RabGGTase family was probably initiated by the evolutionarily selected modification of GGTase-I to allow binding to a RabGDI-like molecule, which in turn interacted with Rabs. The indirect anchoring of Rab to GGTase through the GDI-like molecule then allowed substantial mutation of the peptide-binding site, because loss of interaction free energy with this site did not lead to loss of the ability to form the active ternary complex. This also allowed development of double prenylation of the proteins, which would be difficult to achieve mechanistically based on the GGTase-I mechanism, which involves highly specific peptide recognition that presumably would exclude the major rearrangement that is needed for the second prenylation step. Further development of the GDI-like molecule to a REP-like molecule then occurred together with loss of peptide specificity of the GGTase, made possible by the indirect cysteine-positioning mechanism than led to contemporary REP and GGTase molecules. The pressure to accommodate ever more diverse C-terminal sequences forced expansion of the peptide and subsequently lipid-binding sites of RabGGTase. This idea finds indirect conformation in the observation that yeast have only 11 Ypt/Rab proteins that have only two types of prenylation motifs (CC or CXC). Correspondingly, yeast RabGGTase only accepts a much narrower range of isoprenoid analogues than its mammalian counterpart (UT Nguyen and K Alexandrov, unpublished results).
Figure 8.
Evolution of protein prenylation. Transition 1 represents expansion of the lipid-binding site of the ancestral FTase. Transition 2 represents invasion of the ancestral GGTase-I by the bifunctional REP/GDI molecule.
Materials and methods
Expression constructs, protein expression and purification
Expression of rat REP-1 in SF21 cells and subsequent purification was performed as described (Armstrong et al, 1995; Alexandrov et al, 1999). Rab7 was expressed in E. coli and purified as described (Alexandrov et al, 1999). Construction of the expression vectors for RabGGTase mutants is described in the Supplementary data. Expression in E. coli and purification of wild type and mutants of rat RabGGTase was performed as described earlier (Kalinin et al, 2001).
Synthesis of Ser-Cys-Ser-Cys(GG), Ser-Cys(GG)-Ser-Cys and Ser-Cys(GG)-Ser-Cys-(GG) peptides
The geranylgeranylated tetrapeptides were synthesized using solid-phase peptide chemistry employing 2-chlorotrityl chloride resin as the solid support. Fmoc-Cys(GG)-OH was synthesized by treating L-cysteine hydrochloride with geranylgeranyl chloride in 4 N ammonia in methanol and finally the amine group was protected by Fmoc-OSu in the presence of triethylamine in methylene chloride. Fmoc-Cys(GG)-OH and Fmoc-Cys(trt)-OH were attached to the resin using N,N-diisopropyethylamine (DIPEA) in methylene chloride. Fmoc deprotections were achieved using a standard protocol of 50% piperidine in DMF. Sequence elongation at the N terminus was performed by coupling the appropriate Fmoc-protected amino acids (Fmoc-Cys(GG)-OH, Fmoc-Cys(Trt)-OH and Fmoc-Ser(Trt)-OH) under standard conditions employing N-hydroxybenzotriazole, O-(6-chlorobenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate and DIPEA as coupling reagents. The peptides were cleaved from the resin using 1% TFA/methylene chloride with 70–75% overall yield.
Fluorescence measurements
Fluorescence measurements were performed in 1 ml quartz cuvettes (Hellma) with continuous stirring and thermostated at 25°C on a Spex Fluoromax-3 spectrofluorometer (Jobin Yvon Inc., Edison, NJ). The detailed description of titration procedure is described in the Supplementary data.
MALDI-TOF analysis
MALDI-TOF spectra were recorded on a Voyager-DE Pro Biospectrometry workstation from Applied Biosystems (Weiterstadt, Germany). Protein samples were desalted using gel filtration spin columns (DyeEx 2.0 Spin Kit; Qiagen, Hilden, Germany) and mixed with an equal volume of matrix (saturated sinapinic acid solution in 0.3% TFA/acetonitrile (2:1 v/v)). The mixture was spotted onto a MALDI sample plate and air-dried. For recording spectra, the laser intensity was manually adjusted during the measurements to obtain optimal signal-to-noise ratios. Calibrations were carried out using a protein mixture of defined molecular mass (Sigma). Data evaluation was performed using the supplied Voyager software package.
In vitro prenylation assays
SDS–PAGE-based assay. Rab (4 μM), REP (4 μM) and wild-type (4 μM) or mutant RabGGTase were incubated in prenylation buffer (50 mM HEPES, pH 7.2, 50 mM NaCl, 5 mM DTE, 2 mM MgCl2, 10 μM GDP) at 25°C, 40 μM NBD-FPP was added to initiate the reaction. In control reactions, REP was omitted. At defined time intervals, 10-μl samples were withdrawn and quenched by the addition of 10 μl 2 × SDS–PAGE sample buffer. The samples were boiled at 95°C for 3 min and were separated by 15% SDS–PAGE. Fluorescent bands corresponding to the NBD-farnesylated protein were visualized in the gel using a fluorescent image reader (FLA-5000, Fuji; excitation laser: 473 nm, cutoff filter: 510 nm) followed by staining with Coomassie blue and scanning. The fluorescence intensities of the bands were quantitatively analysed using AIDA densitometry software. The data were fitted to a single exponential equation using GraFit 5.0 (Micromath Software).
Solution-based prenylation assay. REP-1 and RabGGTase (1 μM each) in a quartz cuvette was mixed with 4 μM NBD-FPP in prenylation buffer (50 mM HEPES, pH 7.2, 50 mM NaCl, 5 mM DTE, 2 mM MgCl2, 10 μM GDP). Rab7 wild-type or mutants were added to a final concentration of 1 μM to start the reaction. The reaction was monitored by the change in NBD fluorescence as described earlier (Wu et al, 2006) with excitation and emission at 479 and 520 nm, respectively. The traces were fitted to a double exponential equation using GraFit 5.0.
Crystallization of RabGGTaseαDLLRDIG, RabGGTaseαDLLRDIG:GGPP, RabGGTaseαDLLRDIG:FPP and RabGGTaseαDLLRDIG:prenylated peptide complexes; data collection and structure determination. Initial crystallization conditions were determined at room temperature using the Classics and PEG Suites from Qiagen in 200 nl sitting drops set up against a 50 μl reservoir with a Mosquito nanolitre dispensing robot (Molecular Dimensions Limited). Promising conditions were optimized with respect to precipitant composition, pH, temperature and protein concentration, and were transferred to hanging drops prepared by mixing 1 μl of protein solution with 1 μl of precipitant mixture. In all cases, crystals for data collection were obtained at 12°C with a reservoir consisting of 14% (w/v) PEG 3350, 0.2 M CaAc2, 0.1 M HEPES pH 7.2. The protein was used at 14 mg/ml in a buffer containing 25 mM HEPES pH 7.2, 40 mM NaCl, 5 mM β-mercaptoethanol. The crystals were cubic and reached a size of approximately 60 μm × 60 μm × 60 μm in 1 day. Crystals of RabGGTase mutants complexed with GGPP, FPP or prenylated peptides were obtained by a co-crystallization approach and were prepared as follows: 1 μl of these compounds at a concentration of 2 mM in methanol were applied to a glass coverslip. After evaporation of the organic solvent, 1 μl of protein solution (14 mg/ml protein in 40 mM NaCl, 5 mM β-mercaptoethanol, 25 mM HEPES pH 7.2) and 1 μl of reservoir containing 14% (w/v) PEG3350, 0.2 M Ca acetate, 0.1 M HEPES pH 7.2 were added. In the case of diprenylated peptide, 2.5% DMSO was added to the reservoir solution to improve its solubility. The drop was equilibrated in a hanging drop set up at 12°C against 500 μl reservoir. The crystals had a rectangular shape and grew to a size of approximately 30 μm × 30 μm × 100 μm in 1 day.
Prior to flash-cooling in liquid nitrogen, the crystals were briefly washed in 20% (w/v) PEG 3350, 5% (v/v) glycerol, 0.2 M CaAc2, 0.1 M HEPES pH 7.2 supplemented with 2 mM ligand where appropriate in the case of complex crystals. Diffraction data were collected at 100 K at station X10SA of the Swiss Light Source (SLS, Villigen, Switzerland). All data were processed with XDS (Kabsch, 1993). The crystals belong to the orthorhombic space group P212121 and contain one RabGGTaseαΔLLRΔIG complex in the asymmetric unit.
Initial phases were determined by molecular replacement with PHASER (McCoy et al, 2005) of the CCP4 suite, using coordinates of RabGGTase in the RabGGTase:REP-1 complex (PDB code 1LTX) from which the IG and LRR domains had been deleted (Pylypenko et al, 2003). The model was then corrected by alternating rounds of refinement in REFMAC5 (Murshudov et al, 1999) and manual adjustment in COOT (Emsley and Cowtan, 2004). Restraint libraries were generated with PRODRG (Schuttelkopf and van Aalten, 2004). Full data collection and refinement statistics are give in Table I.
Microinjection studies. A431 cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 1% penicillin/streptomycin, 1.5 g/l additional sodium bicarbonate and were cultured at 37°C in 5% CO2. Cells were injected using an Eppendorf micromanipulator and transjector, on a Zeiss inverted microscope enclosed in an Okolab temperature-controlled cage. Cells were injected and photographed in imaging medium at 37°C in 5% CO2 and were otherwise incubated in growth medium as for culturing. Cells were injected with 50 μM of recombinant Citrine-Rab7 wild type as a control, Cherry-Rab7 I190H, Cherry-Rab7 L192S or Cherry-Rab7 I190H L192S. Upon microinjection, cells were imaged for 24 h using fluorescent-inverted microscope.
Supplementary Material
Supplementary Information
Acknowledgments
We acknowledge G Holtermann for invaluable technical assistance. We thank the X-ray communities of MPI Dortmund and MPI Heidelberg for their help with data collection and the staff of beamline X10SA at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) for generous access to their facilities. This study was supported in part by grant DFG AL 484/7-2 to KA and grant SFB 642 of the DFG to KA, RSG and HW as well as a grant of the Volkswagen Foundation to the same authors. DD was supported by the postdoctoral felowship of Alexander von Humboldt-Foundation.
References
- Alexandrov K, Horiuchi H, Steele-Mortimer O, Seabra MC, Zerial M (1994) Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated rab proteins to their target membranes. EMBO J 13: 5262–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov K, Simon I, Iakovenko A, Holz B, Goody RS, Scheidig AJ (1998) Moderate discrimination of REP-1 between Rab7 × GDP and Rab7 × GTP arises from a difference of an order of magnitude in dissociation rates. FEBS Lett 425: 460–464 [DOI] [PubMed] [Google Scholar]
- Alexandrov K, Simon I, Yurchenko V, Iakovenko A, Rostkova E, Scheidig AJ, Goody RS (1999) Characterization of the ternary complex between Rab7, REP-1 and Rab geranylgeranyl transferase. Eur J Biochem 265: 160–170 [DOI] [PubMed] [Google Scholar]
- Alory C, Balch WE (2003) Molecular evolution of the Rab-escort-protein/guanine-nucleotide-dissociation-inhibitor superfamily. Mol Biol Cell 14: 3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres DA, Seabra MC, Brown MS, Armstrong SA, Smeland TE, Cremers FP, Goldstein JL (1993) cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 73: 1091–1099 [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Brown MS, Goldstein JL, Seabra MC (1995) Preparation of recombinant Rab geranylgeranyltransferase and Rab escort proteins. Methods Enzymol 257: 30–41 [DOI] [PubMed] [Google Scholar]
- Casey PJ, Seabra MC (1996) Protein prenyltransferases. J Biol Chem 271: 5289–5292 [DOI] [PubMed] [Google Scholar]
- Dursina B, Reents R, Delon C, Wu Y, Kulharia M, Thutewohl M, Veligodsky A, Kalinin A, Evstifeev V, Ciobanu D, Szedlacsek SE, Waldmann H, Goody RS, Alexandrov K (2006) Identification and specificity profiling of protein prenyltransferase inhibitors using new fluorescent phosphoisoprenoids. J Am Chem Soc 128: 2822–2835 [DOI] [PubMed] [Google Scholar]
- Dursina B, Thoma NH, Sidorovitch V, Niculae A, Iakovenko A, Rak A, Albert S, Ceacareanu AC, Kolling R, Herrmann C, Goody RS, Alexandrov K (2002) Interaction of yeast rab geranylgeranyl transferase with its protein and lipid substrates. Biochemistry 41: 6805–6816 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Gelb MH (1997) Protein prenylation, et cetera: signal transduction in two dimensions. Science 275: 1750–1751 [DOI] [PubMed] [Google Scholar]
- Gelb MH, Scholten JD, Sebolt-Leopold JS (1998) Protein prenylation: from discovery to prospects for cancer treatment. Curr Opin Chem Biol 2: 40–48 [DOI] [PubMed] [Google Scholar]
- Glomset JA, Farnsworth CC (1994) Role of protein modification reactions in programming interactions between ras-related GTPases and cell membranes. Annu Rev Cell Biol 10: 181–205 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kalinin A, Thoma NH, Iakovenko A, Heinemann I, Rostkova E, Constantinescu AT, Alexandrov K (2001) Expression of mammalian geranylgeranyltransferase type-ii in Escherichia coli and its application for in vitro prenylation of Rab proteins. Protein Expr Purif 22: 84–91 [DOI] [PubMed] [Google Scholar]
- Lackner MR, Kindt RM, Carroll PM, Brown K, Cancilla MR, Chen C, de Silva H, Franke Y, Guan B, Heuer T, Hung T, Keegan K, Lee JM, Manne V, O'Brien C, Parry D, Perez-Villar JJ, Reddy RK, Xiao H, Zhan H et al. (2005) Chemical genetics identifies Rab geranylgeranyl transferase as an apoptotic target of farnesyl transferase inhibitors. Cancer Cell 7: 325–336 [DOI] [PubMed] [Google Scholar]
- Lane KT, Beese LS (2006) Thematic review series: lipid posttranslational modifications. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J Lipid Res 47: 681–699 [DOI] [PubMed] [Google Scholar]
- Leung KF, Baron R, Ali BR, Magee AI, Seabra MC (2006a) Rab GTPases containing a CAAX motif are processed post-geranylgeranylation by proteolysis and methylation. J Biol Chem 282: 1487–1497 [DOI] [PubMed] [Google Scholar]
- Leung KF, Baron R, Seabra MC (2006b) Thematic review series: lipid posttranslational modifications. Geranylgeranylation of Rab GTPases. J Lipid Res 47: 467–475 [DOI] [PubMed] [Google Scholar]
- Liang PH, Ko TP, Wang AH (2002) Structure, mechanism and function of prenyltransferases. Eur J Biochem 269: 3339–3354 [DOI] [PubMed] [Google Scholar]
- Long SB, Casey PJ, Beese LS (2002) Reaction path of protein farnesyltransferase at atomic resolution. Nature 419: 645–650 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61: 458–464 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr 55: 247–255 [DOI] [PubMed] [Google Scholar]
- Nguyen UT, Cramer J, Gomis J, Reents R, Gutierrez-Rodriguez M, Goody RS, Alexandrov K, Waldmann H (2007) Exploiting the substrate tolerance of farnesyltransferase for site-selective protein derivatization. Chembiochem 8: 408–423 [DOI] [PubMed] [Google Scholar]
- Owen DJ, Alexandrov K, Rostkova E, Scheidig AJ, Goody RS, Waldmann H (1999) Chemo-enzymatic synthesis of fluorescent Rab 7 proteins: tools to study vesicular trafficking in cells. Angew Chem Int Ed Engl 38: 509–512 [DOI] [PubMed] [Google Scholar]
- Pylypenko O, Rak A, Durek T, Kushnir S, Dursina BE, Thomae NH, Constantinescu AT, Brunsveld L, Watzke A, Waldmann H, Goody RS, Alexandrov K (2006) Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling. EMBO J 25: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylypenko O, Rak A, Reents R, Niculae A, Sidorovitch V, Cioaca MD, Bessolitsyna E, Thoma NH, Waldmann H, Schlichting I, Goody RS, Alexandrov K (2003) Structure of rab escort protein-1 in complex with rab geranylgeranyltransferase. Mol Cell 11: 483–494 [DOI] [PubMed] [Google Scholar]
- Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, Waldmann H, Goody RS, Alexandrov K (2003) Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science 302: 646–650 [DOI] [PubMed] [Google Scholar]
- Rak A, Pylypenko O, Niculae A, Pyatkov K, Goody RS, Alexandrov K (2004) Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease. Cell 117: 749–760 [DOI] [PubMed] [Google Scholar]
- Rasteiro R, Pereira-Leal JB (2007) Multiple domain insertions and losses in the evolution of the Rab prenylation complex. BMC Evol Biol 7: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs AJ, Hulley PA, Meijer A, Ebetino FH, Russell RG, Shipman CM (2006) Selective inhibition of Rab prenylation by a phosphonocarboxylate analogue of risedronate induces apoptosis, but not S-phase arrest, in human myeloma cells. Int J Cancer 119: 1254–1261 [DOI] [PubMed] [Google Scholar]
- Rowinsky EK, Windle JJ, Von Hoff DD (1999) Ras protein farnesyltransferase: a strategic target for anticancer therapeutic development. J Clin Oncol 17: 3631–3652 [DOI] [PubMed] [Google Scholar]
- Schuttelkopf AW, van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D Biol Crystallogr 60: 1355–1363 [DOI] [PubMed] [Google Scholar]
- Seabra MC, Brown MS, Slaughter CA, Sudhof TC, Goldstein JL (1992a) Purification of component A of Rab geranylgeranyl transferase: possible identity with the choroideremia gene product. Cell 70: 1049–1057 [DOI] [PubMed] [Google Scholar]
- Seabra MC, Goldstein JL, Sudhof TC, Brown MS (1992b) Rab geranylgeranyl transferase. A multisubunit enzyme that prenylates GTP-binding proteins terminating in Cys-X-Cys or Cys- Cys. J Biol Chem 267: 14497–14503 [PubMed] [Google Scholar]
- Shen F, Seabra MC (1996) Mechanism of digeranylgeranylation of Rab proteins. Formation of a complex between monogeranylgeranyl-Rab and Rab escort protein. J Biol Chem 271: 3692–3698 [DOI] [PubMed] [Google Scholar]
- Taylor JS, Reid TS, Terry KL, Casey PJ, Beese LS (2003) Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J 22: 5963–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry KL, Casey PJ, Beese LS (2006) Conversion of protein farnesyltransferase to a geranylgeranyltransferase. Biochemistry 45: 9746–9755 [DOI] [PubMed] [Google Scholar]
- Thoma NH, Iakovenko A, Kalinin A, Waldmann H, Goody RS, Alexandrov K (2001a) Allosteric regulation of substrate binding and product release in geranylgeranyltransferase type II. Biochemistry 40: 268–274 [DOI] [PubMed] [Google Scholar]
- Thoma NH, Iakovenko A, Owen D, Scheidig AS, Waldmann H, Goody RS, Alexandrov K (2000) Phosphoisoprenoid binding specificity of geranylgeranyltransferase type ii. Biochemistry 39: 12043–12052 [DOI] [PubMed] [Google Scholar]
- Thoma NH, Niculae A, Goody RS, Alexandrov K (2001b) Double prenylation by RabGGTase can proceed without dissociation of the mono-prenylated intermediate. J Biol Chem 276: 48631–48636 [DOI] [PubMed] [Google Scholar]
- Wu YW, Alexandrov K, Brunsveld L (2007a) Synthesis of a fluorescent analogue of geranylgeranyl pyrophosphate and its use in a high-throughput fluorometric assay for Rab geranylgeranyltransferase. Nat Protoc 2: 2704–2711 [DOI] [PubMed] [Google Scholar]
- Wu YW, Tan KT, Waldmann H, Goody RS, Alexandrov K (2007b) Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc Natl Acad Sci USA 104: 12294–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YW, Waldmann H, Reents R, Ebetino FH, Goody RS, Alexandrov K (2006) A protein fluorescence amplifier: continuous fluorometric assay for rab geranylgeranyltransferase. Chembiochem 7: 1859–1861 [DOI] [PubMed] [Google Scholar]
- Zhang H, Seabra CM, Deisenhofer J (2000) Crystal structure of Rab geranylgeranyltransferase at 2.0 Å resolution. Structure 8: 241–251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information