Abstract
To date, studies conducted on cone snail venoms have attributed the origins of this complex mixture of neuroactive peptides entirely to gene expression by the secretory cells lining the lumen of the venom duct. However, specialized tissues such as the salivary glands also secrete their contents into the anterior gut and could potentially contribute some venom components injected into target animals; evidence supporting this possibility is reported here. Sequence analysis of a cDNA library created from a salivary gland of Conus pulicarius revealed the expression of two transcripts whose predicted gene products, after post-translational processing, strikingly resemble mature conopeptides belonging to the α-conotoxin family. These two transcripts, like α-conotoxin transcripts, putatively encode mature peptides containing the conserved A-superfamily cysteine pattern (CC-C-C) but, the highly conserved A-superfamily signal sequences were not present. Analysis of A-superfamily members expressed in the venom duct of the same C. pulicarius specimens revealed three putative α-conotoxin sequences; the salivary gland transcripts were not found in the venom duct cDNA library, suggesting that these α-conotoxins are salivary gland-specific. Therefore, expression of conotoxin-like gene products by the salivary gland could potentially add to the complexity of Conus venoms.
Keywords: Conus pulicarius, α-conotoxins, cDNA libraries, salivary gland, alpha4/7, conopeptide, exogenomics
1. Introduction
Venoms of predatory gastropods in the genus Conus (“cone snails”) have been intensively investigated; these venom components have the potential to be useful pharmacological agents that act predominantly on molecular targets in nervous systems (Wang and Chi 2004, Norton and Olivera 2006, Olivera 2006, Olivera and Teichert 2007). Decades of research have documented that these venoms comprise a complex mixture of secreted gene products synthesized by a specialized organ referred to as the venom duct (Olivera and Cruz 2001, Olivera 2002). During envenomation, contraction of the venom bulb causes venom duct’s contents to be forced through the proboscis, which is preloaded with a single harpoon-like tooth; the harpoon, which is hollow, is used to inject venom into the target animal (Schulz et al. 2004).
The venom toxins are mostly small, disulfide-rich peptides, whose transcripts encode precursors with a characteristic organization – at the N-terminus is a signal sequence, followed by an intervening “pro” region, which includes a protease cleavage site, and at the C-terminus, the mature toxin, always in single copy and generally rich in cysteine codons (Woodward et al. 1990, Olivera et al. 1999). Within the mature toxin region, only a few characteristic arrangements of cys residues occur; these are referred to as “Cys patterns.” Each Cys pattern generally specifies a particular disulfide framework, the dominant structural determinants that stabilize the three-dimensional conformation of these unusually small (12–30 AA) peptide toxins. A most remarkable feature of Conus toxin precursors from the venom duct is that toxins with a particular Cys pattern also share a highly conserved signal sequence. The combined presence of these superfamily-specific characters distinguishes particular conopeptide gene superfamilies (Terlau and Olivera 2004).
In Conus and other Conoidean gastropods, other secretory organs, such as the salivary or accessory salivary gland, can potentially secrete gene products into the lumen of the anterior gut (Taylor et al. 1993). Therefore, these glands may also contribute their own gene products to the venom. Although no previous studies have addressed what gene products are produced by the salivary glands of Conus, the salivary secretions of a closely related family of carnivorous snails (i.e., the Turrids) have been postulated to perform one or more functions, which include: i) increasing envenomation efficiency during prey capture, either by proteolytically removing debris, or lubricating the radular tooth, while preparing for hypodermic injection; ii) proteolytic processing of venom components just prior to, or at the site of injection via events resembling the conversion of trypsinogen to trypsin in mamilian systems; and/or iii) enhancing the potency of the venom by also producing bioactive components (Shimek 1975). We have discovered an unusual and unexpected gene product expressed in the salivary gland of the worm-hunting species Conus pulicarius, which may be consistent with the last possibility.
2. Material and Methods
2.1. Specimen Collection; RNA Isolation
Specimens of Conus pulicarius (Fig. 1) were collected at depths ranging from 0.5 to 1.0 m among the intertidal sea grass beds of Pago Bay, Guam, USA. Each specimen was dissected to isolate the venom duct and the salivary gland, which were immediately suspended in 1.0 mL RNAlater solution (Ambion, Austin, TX) and stored at −80 °C until used. The venom duct and salivary gland of a single specimen were each homogenized within their original microcentrifuge tube (1.6 mL) using 1.0 mL of TRIzol reagent and a hand-held Teflon pestle. Total RNA from each tissue was isolated by phase separation and column purification, according to the manufacturer’s recommendations (TRIzol® Plus RNA Purification System; Invitrogen Corp., Carlsbad, CA).
Fig. 1.
A picture of the shell of one of the actual Conus pulicarius specimens collected from Pago Bay, Guam. C. pulicarius is a vermivorous cone snail that inhabits shallow coastal waters throughout the Indo-Pacific.
2.2. Preparation and Sequencing of cDNA Clones
First-strand cDNA was prepared from 2 µg of either salivary gland or venom duct Total RNA, followed by second strand synthesis and 20 additional amplification cycles, which were performed by long-distance polymerase chain reaction (LD-PCR; Creator™ SMART™ cDNA Library Construction Kit; Clontech Laboratories, Inc., Palo Alto, CA) using an MJ Research PTC-200 Peltier Thermal Cycler. The resulting PCR products were purified using the QIAquick® PCR Purification Kit (Qiagen Sciences, Germantown, MD) following the manufacturer’s standard protocol. The resulting venom duct cDNA library was submitted for 454-Sequencing (Courtesy of Roche Colorado Corporation, Boulder, CO); due to the inherent difficulties in resolving the redundancies among transcripts found within this Conus pulicarius venom duct, the results from this work will be presented at a later date. For the salivary gland cDNA pool, a second amplification (5 cycles), using tailed PCR primers, was performed on the PCR products. The resulting PCR products were size-fractionated using the CHROMA SPIN-400 DEPC columns supplied with the cDNA library construction kit. Eluted DNA was cloned into the pNEB206A vector and the resulting products transformed into chemically competent NEB 5-alpha cells, using the USER™ Friendly Cloning Kit (New England Biolabs, Inc., Ipswich, MD). The nucleic acid sequences of the resulting clones were determined using an ABI automated sequencer (Courtesy of the DNA Sequencing and Genomics Core Facility, University of Utah). The sequences of these independent clones were compared to identify PCR errors, and the resulting sequences obtained have been submitted to GenBank.
2.3. PCR-based Screening for Salivary Gland α-Conotoxins
A PCR assay capable of identifying salivary gland α-conopeptides from a Conus pulicarius venom duct cDNA library was based on the PCR primers SG-ALPHA-F (5′S-GGT CTG ACG TGA ACA CGA CAA TG-3′) and SG-ALPHA-R (5'-GCA ACC GTC TGA AGT GGA GGG CG -3'). The sequence used to design primers for amplifying standard α-conopeptide transcripts has been previously published (Santos et al. 2004). Transcripts encoding α-conotoxins were amplified from the cDNA pools by performing Nested-PCR using Platinum Taq DNA Polymerase High Fidelity (Invitrogen) and 40 cycles of amplification (annealing temperature = 55°C; extension temperature = 68°C) in an MJ Research PTC-200 Peltier Thermal Cycler. If a band was visible after loading half (10 µL) of the total reaction onto a 2 % agarose gel, then the resulting PCR products were purified and sequenced using the same procedures as above, except that they were cloned into the pGEM-T Easy vector (Promega, Madison, WI), and submitted for sequencing.
3. Results
3.1. cDNA Clones from Conus pulicarius
Salivary glands were dissected from a diverse set of Conus species for cDNA analysis. cDNA clones from a Conus pulicarius salivary gland library were analyzed as described under Materials and Methods; from this library, a total of 28 cDNAs were sequenced. Using the criteria normally applied for predicting toxin genes in Conus venom ducts (as described in the Introduction), two clones that encoded open reading frames that would be regarded as putative α4/7 toxin precursors if they were expressed in the venom duct were identified. These had clearly defined signal sequences and were small disulfide-rich peptides. Another four predicted larger polypeptides had some toxin-like properties; these will be discussed more thoroughly in a separate manuscript. Two other transcripts encoded polypeptides that had significant sequence similarity to the mitochondrial 16S and ribosomal 12S-subunits of other Conus species.
Among the clones characterized from the salivary gland cDNA library of Conus pulicarius, two had cysteine patterns similar to those found in standard conotoxin gene superfamilies. These two closely-related α-conotoxin-like peptides were found at a high rate of frequency (11/28) in the cDNA library and appeared to be highly expressed in the salivary gland of this species. These are designated PuSG1.1 and PuSG1.2 and are of particular interest since they resemble the most extensively characterized group of conopeptides, the α–conotoxins that belong to the A-superfamily. The two predicted α-like open reading frames are shown in Fig. 2; the two sequences are highly similar and differ by one amino acid in the signal sequence and three in the mature toxin region.
Fig. 2.
Conus pulicarius salivary gland toxin precursors contain α4/7-conotoxin-like Cys patterns. Identical residues between the sequences are enclosed within boxes. The ends of the predicted secretion signals and proteolytic cleavage sites are delineated by arrows. Both of these precursors are predicted to yield mature toxins resembling members of the well-characterized alpha4/7 conopeptide subfamily.
3.2. Comparison of α-conotoxin precursors expressed in the venom duct to the salivary gland precursors
In order to determine whether the transcripts in Fig. 2 were salivary gland specific, or were expressed in the venom duct, a cDNA library from the venom duct was analyzed. Primers corresponding to both the standard α-conotoxin signal sequence (Santos et al. 2004) as well as to the signal sequences of the salivary gland transcripts were employed (see Methods). Three sequences were found that corresponded to α-conotoxin precursors in the venom duct cDNA pools, but there were no sequences homologous to the salivary gland transcripts found using salivary gland signal sequence primers.
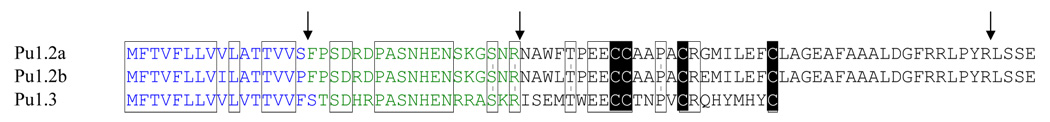
The α-conotoxins expressed in venom ducts that were found in Conus pulicarius are unusual; they have a long N-terminal and C-terminal extensions in the mature toxin region predicted from the cDNA sequences; these are shown in Fig. 3 (lower panel). A comparison of the mature salivary gland α-conotoxins to the three venom duct α-conotoxins is shown in Fig. 3; although all of these sequences fit the spacing between cysteine residues characteristic of the α4/7 subfamily, both sets of sequences can be considered unconventional when compared to standard precursor sequences of the α4/7 subfamily (see Santos et al. 2004). Interestingly, the salivary gland transcripts appear to be specific for the salivary gland, while the other α-conotoxin sequences are specifically expressed in the venom duct.
Fig. 3.
Comparison of the three toxin precursors identified from a Conus pulicarius venom duct from Pago Bay, Guam. Identical residues between the sequences are enclosed within boxes. The ends of the predicted secretion signals and proteolytic cleavage sites are delineated by arrows. The cysteine residues are shaded in black. Each of these precursors is predicted to yield mature toxins resembling members of the well-characterized alpha4/7 conopeptide subfamily.
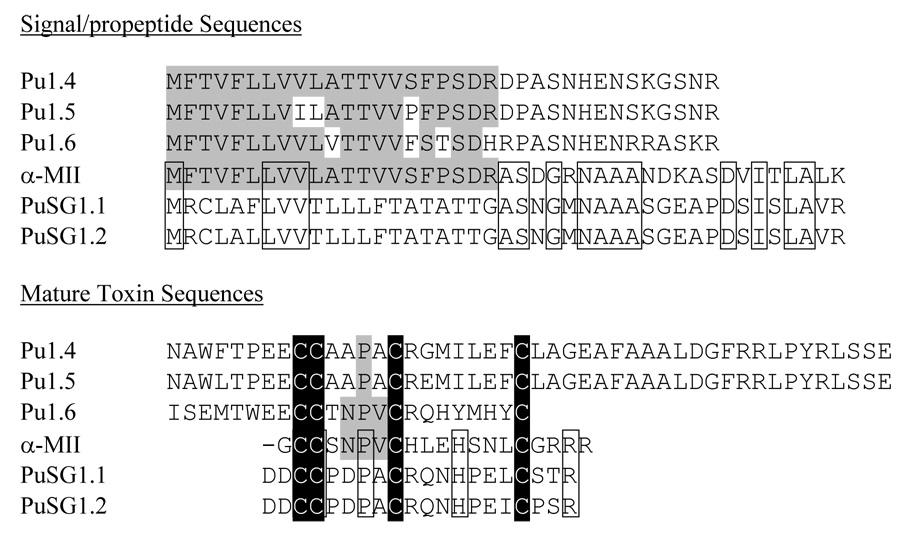
The divergence between C. pulicarius salivary gland transcripts and the venom duct transcripts are detailed in the sequence alignments shown in Fig. 4. The figure also aligns an α4/7 subfamily transcript from a fish-hunting cone snail; this is compared to the two sets of Conus pulicarius sequences. An extremely striking and surprising pattern of similarity emerges from this juxtaposition of precursor sequences.
Fig. 4.
Comparison of the two Conus pulicarius salivary gland toxin precursors with the standard A-superfamily precursors. PuSG1.1 and PuSG1.2 toxin precursors are compared to the sequences of α-MII, and three toxin precursors (Pu1.4, Pu1.5, and Pu1.66) from the venom duct of the same snail. Amino acid residues within the venom duct sequences that are identical to α-MII are shaded in grey; amino acid residues within the salivary gland precursors that are identical to α-MII are delineated by boxes. The cysteine residues are shaded in black.
One of the best characterized α4/7 subfamily conopeptides, α-conotoxin MII from Conus magus, is the prepropeptide sequence compared to the transcripts expressed in the Conus pulicarius venom duct and salivary gland in Fig. 4. The Conus pulicarius venom duct sequences and the α-conotoxin MII sequences share almost identical signal sequences. However, these diverge almost completely in the “pro” region; there is little sequence similarity in this region between the two sets of sequences. What is striking however is that there is a much higher degree of sequence similarity between the “pro” regions of the C. pulicarius salivary gland transcripts with the α-conotoxin MII sequence.
4. Discussion
The characterization of secreted gene products of Conus venom ducts has provided powerful pharmacological tools for studying the nervous system, as well as insights into the biology and evolution of Conus. It has also led to the development of therapeutic drug leads for the treatment of a number of disease states including neuropathic pain (Olivera and Teichert 2007). Comprising ~700 species, which collectively produce an estimated 100,000 bioactive peptides, Conus venoms provide a vast reservoir of potentially useful compounds for neuropharmacological research. The results of this study suggest that Conus venom components may not all be gene products synthesized in the venom duct; other glands in the anterior gut may make a minor contribution. However, some caution in this interpretation should be exercised — although an attractive hypothesis, it remains to be definitely proven. Although it seems likely that the transcripts we have discovered in the salivary gland are indeed processed to yield α-conopeptide like gene products, these could function endogenously within Conus pulicarius. Even if they prove to be nicotinic receptor antagonists (as are all α-conotoxins), they may well be designed to antagonize nicotinic receptors within the body of Conus pulicarius itself (such as targets in the proboscis, as a specific possibility). Nevertheless, the results demonstrate that conotoxin-like genes can be recruited for expression in the salivary glands; a preliminary survey of other Conus salivary glands would suggest that this may be a sporadic occurrence across Conus. Similar types of transcripts were not found in salivary glands from several fish-hunting Conus that were surveyed. Thus, the precise physiological function of the salivary gland transcripts still needs to be defined.
The present study was part of a broader initiative to obtain an overview of genes expressed in Conus salivary glands. Surprisingly, some cDNA clones encoding conopeptide-like gene products were identified. For venom duct cDNAs, in order to conclude that a gene belongs to a particular conopeptide gene superfamily requires both high similarity to the consensus signal sequence of that superfamily, as well as the characteristic arrangement of Cys residues in the mature toxin region corresponding to that superfamily. Although Conus pulicarius salivary gland mRNA transcripts are predicted to encode mature peptides exhibiting the characteristic cysteine pattern of the α-conotoxin family (CC-C-C), the signal sequences of these precursors do not resemble the highly conserved A-superfamily signal sequence; thus, they can not be unequivocally defined as A-superfamily gene transcripts. Nonetheless, the striking similarity within these putative peptides and α-conotoxins demonstrates that “α-like” conopeptide transcripts are specifically expressed in the salivary gland of Conus pulicarius; such transcripts are apparently not expressed in the venom duct.
Although the salivary gland transcripts encode secreted gene products predicted to have the typical α4/7 subfamily cys pattern (-CCX4CX7C--), and the spacing between cysteine residues are the same, little sequence similarity is observed otherwise between salivary gland transcripts and the α-conotoxin transcripts expressed in the venom duct of the same Conus pulicarius specimens. However, when all of these sequences are compared to more classical examples of α-conotoxin prepropeptides (e.g., α-MII; Fig. 4), an interesting conclusion can be made – i.e., the α-conotoxin MII precursor shows greater homology to the Conus pulicarius venom duct sequences when the N-terminal signal sequences are compared, but an even greater similarity to the salivary gland sequences if the “pro” regions are compared.
The unexpected patterns of sequence similarity in Fig. 4 can be rationalized by the following working hypothesis, which would need to be tested by additional experimental data: The original gene that is an ortholog of the α-conotoxin MII precursor (which we might regard as a primordial α4/7 subfamily gene) may be largely conserved in the α-conotoxin MII precursor (which has considerable sequence similarity to many other α-conotoxin precursor sequences in various Conus lineages). In C. pulicarius, two derivatives of the primordial α4/7 gene encode the peptides expressed in the salivary gland and venom duct of the C. pulicarius specimen analyzed, respectively. The venom duct expresses an unusual set of α4/7 subfamily members, with N-terminal and C-terminal extensions (the latter only in the case of the Pu1.4 and Pu1.5 genes). A similar evolutionary path of the A-superfamily has been found in fish-hunting cone snails to generate excitatory peptides (that are not α-conotoxins) in the venom of species such as Conus striatus and Conus magus. In these cases, a large change in the propeptide region occurred to accommodate the longer N-terminal extension. A similar pathway of evolution has apparently occurred with the Pu1.4-6 genes, which are expressed in the venom ducts of Conus pulicarius. However, as in all members of the A-superfamily expressed in venom ducts, the signal sequences are highly conserved.
What is novel is the lack of signal sequence conservation in the two salivary gland transcripts, despite the observed sequence conservation in the propeptide region (and the similarity of Cys patterns in the mature toxin regions). This is not surprising because in most secreted proteins, signal sequences are the least conserved elements. Therefore, we hypothesize that the conservation of conopeptide signal sequences is a phenomenon specific to the venom duct: special mechanisms in this tissue maintain the signal sequence for each superfamily. The underlying rationale for this conservation, and the mechanism that prevents even silent mutations from occurring, needs to be investigated and elucidated. In effect, such a mechanism could, in evolutionary terms, serve as a tissue-specific sequence clamp (somewhat similar to amplified loci in the ribosomal RNA gene cluster, and the ITS sequences between the RNA regions).
What the data in Fig. 4 suggest is that once an α-conotoxin gene is expressed in a tissue other than the venom duct, the constraints on divergence of signal sequences are lifted, and the usual rapid divergence of signal sequences routinely found in genes encoding other secreted gene products occurs. Thus, since PuSG1.1 and PuSG1.2 are genes expressed in the salivary gland rather than in the venom duct, if a mechanism for the conservation of signal sequence is absent, this would result, over time, in divergence of these signal sequences from the standard venom duct A-superfamily signal sequences. In this view, the salivary gland transcripts appear to be more conventional in their pattern of evolution; it is the A-superfamily genes expressed in venom ducts that behave anomalously over evolutionary time, with a highly unconventional pattern of signal sequence conservation over the ~55 million years that the radiation of the Conus lineage occurred.
Acknowledgments
We would like to thank Steve Norby for collecting some of the specimens analyzed in this study and Thomas Flores Jr., Jay Gutierrez, Tino Aguon, and the rest of the members of the Division of Aquatic & Wildlife Resources of Guam for their help in acquiring the collection and transport permits required to conduct these studies on Conus pulicarius from Guam.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Support for this work was provided by grants from the NIHGMS PO1 GM048677 (to BMO) and the NIHGMS Diversity Supplement Fellowship 3 PO1 GM048677 (to JSB)
References
- Norton RS, Olivera BM. Conotoxins down under. Toxicon. 2006;48:780–798. doi: 10.1016/j.toxicon.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Olivera BM. CONUS VENOM PEPTIDES: Reflections from the Biology of Clades and Species. Annual Review of Ecology and Systematics. 2002;33:25–47. [Google Scholar]
- Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Cruz LJ. Conotoxins, in retrospect. Toxicon. 2001;39:7–14. doi: 10.1016/s0041-0101(00)00157-4. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Teichert RW. Diversity of the neurotoxic Conus peptides: a model for concerted pharmacological discovery. Mol Interv. 2007;7:251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Walker C, Cartier GE, Hooper D, Santos AD, Schoenfeld R, Shetty R, Watkins M, Bandyopadhyay P, Hillyard DR. Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Potential evolutionary significance of introns. Ann N Y Acad Sci. 1999;870:223–237. doi: 10.1111/j.1749-6632.1999.tb08883.x. [DOI] [PubMed] [Google Scholar]
- Santos AD, McIntosh JM, Hillyard DR, Cruz LJ, Olivera BM. The A-superfamily of conotoxins: structural and functional divergence. J Biol Chem. 2004;279:17596–17606. doi: 10.1074/jbc.M309654200. [DOI] [PubMed] [Google Scholar]
- Schulz JR, Norton AG, Gilly WF. The projectile tooth of a fish-hunting cone snail: Conus catus injects venom into fish prey using a high-speed ballistic mechanism. Biol Bull. 2004;207:77–79. doi: 10.2307/1543581. [DOI] [PubMed] [Google Scholar]
- Shimek RL. The morphology of the buccal apparatus of Oenopota levidensis (Gastropoda, Turridae) Zoomorphology. 1975;80:59–96. [Google Scholar]
- Taylor JD, Kantor YI, Sysoev AV. Foregut anatomy, feeding mechanisms, relationships and classification of the Conoidea (=Toxoglossa) (Gastropoda) Bulletin of the Natural History Museum. Zoology series. 1993;59:125–170. [Google Scholar]
- Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- Wang CZ, Chi CW. Conus peptides--a rich pharmaceutical treasure. Acta Biochim Biophys Sin (Shanghai) 2004;36:713–723. doi: 10.1093/abbs/36.11.713. [DOI] [PubMed] [Google Scholar]
- Woodward SR, Cruz LJ, Olivera BM, Hillyard DR. Constant and hypervariable regions in conotoxin propeptides. Embo J. 1990;9:1015–1020. doi: 10.1002/j.1460-2075.1990.tb08204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]