Abstract
Resistance to the extended-spectrum cephalosporins can occur in Salmonella species via the production of extended-spectrum and AmpC β-lactamases. We describe human infections with Salmonella enterica serotype Newport phage type 14 strains resistant to ceftazidime (CAZ) and cefoxitin (FOX) related to the handling of pet treats containing dried beef. These strains were isolated from five patients in Calgary, Alberta, Canada, during 2002 and were compared to a strain cultured from a commercial pet treat present at the property of one of the patients. The strains were resistant to FOX, CAZ, cefpodoxime, ampicillin, and chloramphenicol; intermediate resistant to ceftriaxone and cefotaxime; and sensitive to the aminoglycosides, ciprofloxacin, cefepime, and imipenem. Isoelectric focusing, multiplex PCR, and sequencing of the amplicons showed that all strains produced the plasmid-encoded AmpC β-lactamase, CMY-2. Restriction analysis of plasmid DNA following transformation demonstrated that blaCMY-2 was encoded on an approximately 140-kb plasmid. Pulsed-field gel electrophoresis showed the human and pet treat Salmonella strains to be highly related. This study is the first to implicate the transfer of multidrug-resistant Salmonella species through the handling of commercial pet treats containing animal products. In addition to documenting the first cases of human infection caused by CMY-2-producing S. enterica serotype Newport strains in Canada, this study illustrates the necessity of rapid and accurate laboratory-based surveillance in the identification of novel types of antimicrobial resistance.
Nontyphoidal Salmonella species are important food-borne pathogens that cause gastroenteritis, bacteremia, and focal infections in humans and animals (14). It is estimated that Salmonella species are responsible for ∼1.4 million illnesses and 600 deaths annually in the United States (16). Notable recent outbreaks of Salmonella infection have been linked to the consumption of eggs, undercooked ground beef, cheese, dry cereal, ice cream premix, a variety of fresh sprouts, other fresh vegetables, juice, and cantaloupes (14, 30).
Resistance to the extended-spectrum cephalosporins can occur in Salmonella species via the production of plasmid-mediated extended-spectrum β-lactamases that are capable of hydrolyzing the newer oxyimino-cephalosporins and monobactams, but not the 7-α-methoxy-cephalosporins (6, 13, 17, 18, 19, 27, 28). In addition, Salmonella species have developed resistance to the cephalosporins via the acquisition of plasmids containing the AmpC β-lactamase genes derived from the chromosomes of Citrobacter freundii and Morganella morganii (4, 13, 17, 29). These enzymes have the ability to hydrolyze the 7-α-methoxy-cephalosporins, oxyimino-cephalosporins, and monobactams.
This report describes a strain of Salmonella enterica serotype Newport phage type 14 that was resistant to ceftazidime (CAZ) and cefoxitin (FOX). The strain was isolated from a stool specimen of a 1-month-old infant from Calgary, Alberta, Canada. Since Salmonella species resistant to the extended-spectrum cephalosporins and cephamycins are relatively rare and had not been previously reported in human isolates of Salmonella species from Canada, a study was designed to characterize the resistance mechanism(s). The Department of Environmental Health of the Calgary Health Region was contacted to undertake an epidemiological investigation to determine the possible source(s) of this resistant strain.
MATERIALS AND METHODS
Laboratory setting.
The city of Calgary and immediate surrounding areas have a single centralized high-volume laboratory, namely, Calgary Laboratory Services (CLS), that provides services to all acute-care settings (hospitals), doctor's offices, nursing homes, and clinics in the vicinity. Strains from patients with all reportable diseases, including infections with Salmonella species, are forwarded to the Provincial Laboratory for Public Health (Microbiology) [PLPH (M)] in Calgary.
Surveillance.
Salmonellosis is a reportable condition in Alberta, Canada. Clinical microbiology laboratories forward all clinical isolates of Salmonella species to PLPH (M) for serotyping and molecular typing. The strains are also referred to the National Microbiology Laboratory in Winnipeg, Manitoba, Canada, for phage typing. Data on S. enterica serotype Newport strains isolated between 1998 and 2002 in Alberta were provided by PLPH (M).
Case interviews.
Strains of S. enterica serotype Newport phage type (PT) 14 resistant to CAZ and FOX were isolated from cultures of stool specimens from five patients (three from the same household) during 2002. These patients were interviewed to obtain data related to demographics, date of onset of illness, hospitalizations, physician visits, antimicrobial use, similar illnesses among household members or contacts, travel histories, attendance at social gatherings, pet ownership, acquisition of pet treats containing raw animal products, type of water consumed, and restaurants visited during the 3 days prior to the onset of symptoms.
Sources of specimens cultured for Salmonella.
As part of the epidemiological investigation conducted by the Department of Environmental Health of the Calgary Health Region, the following specimens were obtained from the respective households: any water, food, or food products that were considered a possible source of the Salmonella strains; any commercial pet treats known to contain raw animal products; and stool specimens from pets. These were processed at PLPH (M) by previously described methods (25).
Bacterial strains.
The Salmonella isolates were identified biochemically by routine laboratory procedures (10). Serotyping was performed at PLPH (M) by the Kauffman and White scheme (15) with antisera for somatic (O) and flagellar (H) antigens (Difco Laboratories, Detroit, Mich.). The strains were phage typed at the National Microbiology Laboratory by an in-house phage-typing scheme with a panel of nine lytic phages providing 1 to 17 unique lytic patterns by the standard method (3). The phages were isolated, purified, and characterized as described previously (1).
Antimicrobial susceptibility testing.
The MICs of the following drugs were determined by broth microdilution with Pasco gram-negative panels (6952-30; Becton Dickinson, Wheatridge, Colo.): ampicillin (AMP), ampicillin-sulbactam (SAM), FOX, cefpodoxime (CPD), cefotaxime (CTX), ceftriaxone (CRO), CAZ, cefepime (FEP), aztreonam (ATM), gentamicin (GEN), tobramycin (TOB), amikacin (AMK), ciprofloxacin (CIP), chloramphenicol (CHL), and trimethoprim-sulfamethoxazole (SXT). Susceptibility to imipenem (IPM) and nalidixic acid (NAL) was determined by the standard disk diffusion method (20). Disks were obtained from Becton Dickinson Microbiology Systems (Cockeysville, Md.). Throughout this study, the results were interpreted by using National Committee for Clinical Laboratory Standards criteria for broth dilution and disk diffusion (20).
β-Lactamase identification.
Isoelectric focusing, CTX hydrolysis, and inhibitor profile analysis in polyacrylamide gels were performed with sonic extracts, as described previously (24). The genes responsible for the production of the β-lactamases were amplified on a Thermal Cycler 9600 instrument (Applied Biosystems, Norwalk, Conn.) by using the multiplex PCR conditions and primers described previously (21). Gene identification was accomplished by cycle sequencing of the full-length amplified products with Precision Taq (Stratagene) and the primers described previously (13). The following positive control strains were used during the amplification (21): Misc 339 (MOX-1), PVAMC55 (CMY-2), UMJH14 (DHA-1), CDC2085 (ACC), Kleb 225 (ACT-1), and CCF52 (FOX-5).
Plasmid profiles and transformation.
Plasmid DNA was isolated by alkaline lysis (23), with the following modifications. The cells were resuspended by using 100 μl of 0.28 μg of RNase A per ml and 17.8 mg of lysozyme per ml in 1× GTE buffer (50 mM glucose, 25 mM Tris-Cl, 10 mM EDTA [pH 8]), followed by addition of 200 μl of 0.2 mM NaOH-1% sodium dodecyl sulfate in distilled H2O. The DNA was resuspended in 1× TE buffer (10 mM Tris-Cl, 1 mM EDTA [pH 8]) and stored at −20°C. The plasmids were separated by using 0.8% agarose gels (Sigma Inc., St. Louis, Mo.) in TAE buffer (40 mM Tris-acetate, 2 mM EDTA [pH 8.5]) at 30 V for a total of 27 h for the separation of high-molecular-weight plasmids. Bacterial transformations were carried out as follows. Plasmid DNA was isolated by using a mini-plasmid isolation kit (Qiagen Inc., Alameda, Calif.). Approximately 1.0 to 10 ng of plasmid DNA was used to transform electrocompetent Escherichia coli DH101B (Invitrogen) with a Gene Pulser (Bio-Rad Inc., Hercules Calif.) set at 200 Ω/1.25 kV with a time constant of 4 to 5 s in 1-mm electroporation cuvettes (EquiBio Inc., Needham Heights, Mass.). Transformants were selected on Luria-Bertani agar (Difco) containing FOX (5 μg/ml). Plasmid DNA was isolated from the transformants with the midi-plasmid isolation kit (Qiagen Inc.), and PCR with the primers and conditions described previously (21) was used to confirm the presence of blaCMY-2. Approximately 2 μg of plasmid DNA was digested with HpaI (Invitrogen, Burlington, Ontario, Canada), and the restriction fragments were separated on a 0.7% agarose gel with 0.5× Tris-borate-EDTA buffer for 18 h at 2.8 V/cm with buffer circulation. The fragments were sized, and the gels were normalized with the BioNumerics software program (version 2.5; Applied Maths, St-Martens-Latem, Belgium) by using the One Kilobase Extension Ladder (Invitrogen Inc., Carlsbad, Calif.) as a standard.
PFGE.
Extraction of DNA and pulsed-field gel electrophoresis (PFGE) analyses were performed as described previously (12). Briefly, XbaI or BlnI (Invitrogen) was used to digest the genomic DNA, and PFGE was conducted with a CHEF-DRIII apparatus (Bio-Rad Laboratories, Richmond, Va.) with the following running conditions: an initial switch time of 2.2 s and a final switch time of 63.8 s at 6 V/cm and an angle of 120 degrees for 19 h. The gel was stained with ethidium bromide (0.2 μg/ml). A Polaroid negative was captured with an HP Scanjet 7400c camera (Hewlett-Packard, Mississauga, Ontario, Canada) and analyzed with the BioNumerics software program (version 3.0; Applied Maths). PFGE pattern numbers were assigned to each isolate with one band difference in the PFGE profile.
RESULTS
Laboratory setting.
During June 2002, routine cephalosporin susceptibility testing of all stool Salmonella isolates was introduced at CLS. Soon thereafter, the first cephalosporin-resistant Salmonella isolate was encountered at CLS, and the Department of Environmental Health of the Calgary Health Region was contacted to undertake an epidemiological investigation to determine the possible source(s) of this resistant strain.
Surveillance.
The number of Salmonella species, PTs, and antibiotic resistance patterns for Salmonella strains isolated from humans in Alberta, Canada, from January 1998 to December 2002 are illustrated in Table 1. A total of 61 S. enterica serotype Newport strains were isolated: 9 in 1998, 16 in 1999, 8 in 2000, 7 in 2001, and 21 in 2002. Nine strains were resistant to both FOX and CAZ and appeared for the first time in 2002 (Table 1); seven of the nine strains were from the Calgary area (described in this study), while the remaining two were isolated from patients in a small town in rural Alberta near Calgary. The seven strains from Calgary were PT 14, while the remaining two isolates were PT 14 and PT 17, respectively.
TABLE 1.
S. enterica serotype Newport strains isolated from humans in Alberta, Canada, from January 1998 to December 2002
| Yr | No. of Salmonella sp.a | No. of Salmonella serotype Newport isolates | PTsb | No. of strains resistant to:
|
||||
|---|---|---|---|---|---|---|---|---|
| AMP | CAZ | FOX | CHL | SXT | ||||
| 1998 | 1,138 | 9 | 8, 9, 14 | 0 | 0 | 0 | 0 | 0 |
| 1999 | 1,106 | 16 | 8, 9, 13, 14 | 0 | 0 | 0 | 0 | 0 |
| 2000 | 1,065 | 8 | NT, 14 | 0 | 0 | 0 | 0 | 0 |
| 2001 | 1,244 | 7 | 3, 4, 8, 9, 10 | 0 | 0 | 0 | 0 | 0 |
| 2002 | 889 | 21 | 4, 8, 9, 10, 14, 15, 17 | 9 | 9 | 9 | 9 | 1 |
Total number of Salmonella species isolated from humans in Alberta, Canada.
Resistant strains were PT 14 (eight of nine) and PT 17 (one of nine). NT, nontypeable.
Case interviews. (i) Patient A.
The index case was a 1-month-old infant (patient A) who presented in June 2002 with bloody diarrhea and fever and who was admitted to the Alberta Children's Hospital in Calgary. She was treated empirically with CRO. A stool sample for culture taken during admission was positive for S. enterica serotype Newport PT 14 (Table 2, strain Snp1). Follow-up stool specimens taken 7 days later and in August 2002 were also positive for S. enterica serotype Newport PT 14 (Table 2, strains Snp2 and Snp3, respectively).
TABLE 2.
MICs for clinical strains of S. enterica serotype Newport
| Clinical straina | Origin | MIC (μg/ml)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | SAM | FOX | CPD | CAZ | CTX and CROb | ATM | FEP | CHL | SXT | GEN | TOB | AMK | CIP | ||
| Snp1 | Patient A | >16 | >16/8 | >16 | >8 | >16 | 16 | 16 | ≤2 | >16 | ≤1/19 | ≤0.25 | ≤0.25 | ≤1 | ≤0.5 |
| Snp2 | Patient A | >16 | >16/8 | >16 | >8 | >16 | >32 | >16 | 8 | >16 | ≤1/19 | ≤0.25 | ≤0.25 | ≤1 | ≤0.5 |
| Snp3 | Patient A | >16 | >16/8 | >16 | >8 | >16 | >32 | >16 | 8 | >16 | ≤1/19 | ≤0.25 | ≤0.25 | ≤1 | ≤0.5 |
| Snp4 | Patient B | >16 | >16/8 | >16 | >8 | >16 | 16 | 16 | ≤2 | >16 | ≤1/19 | ≤0.25 | ≤0.25 | ≤1 | ≤0.5 |
| Snp5 | Patient C | >16 | >16/8 | >16 | >8 | >16 | 16 | 16 | ≤2 | >16 | ≤1/19 | ≤0.25 | ≤0.25 | ≤1 | ≤0.5 |
| Snp6 | Patient D | >16 | >16/8 | >16 | >8 | >16 | 16 | 16 | ≤2 | >16 | ≤1/19 | ≤0.25 | ≤0.25 | ≤1 | ≤0.5 |
| Snp7 | Patient E | >16 | >16/8 | >16 | >8 | >16 | 16 | 16 | ≤2 | >16 | ≤1/19 | ≤0.25 | ≤0.25 | ≤1 | ≤0.5 |
| Snp8 | Pet treat | >16 | >16/8 | >16 | >8 | >16 | 16 | 16 | ≤2 | >16 | ≤1/19 | ≤0.25 | ≤0.25 | ≤1 | ≤0.5 |
Snp1 to Snp8 designate the strains described in the text.
The MICs of CRO and CTX were identical.
(ii) Patient B.
The older sister (patient B; age, 5 years) of patient A presented in February 2002 to the Emergency Department at the Alberta Children's Hospital with a 24-h history of fever and bloody diarrhea. She received treatment for her symptoms and did not require admission. A stool specimen was obtained, and S. enterica serotype Newport PT 14 (Table 2, strain Snp4) was isolated.
(iii) Patient C.
Two weeks after patient B had presented to the Emergency Department, the father (patient C; age, 35 years) of patients A and B presented to his general practitioner with watery diarrhea and abdominal cramps of 3 days' duration and received treatment for his symptoms. A stool specimen was positive for S. enterica serotype Newport PT 14 (Table 2, strain Snp5). The family had acquired a puppy in January 2002 and fed the animal commercial dog treats consisting of dried steak patties obtained from a local pet store. The family had no other additional exposure or recent travel history.
(iv) Patient D.
A 29-year-old female (patient D) from Calgary presented in July 2002 to her general practitioner with bloody diarrhea, fever, and abdominal cramps of 2 days' duration and received treatment for her symptoms. A stool specimen was positive for S. enterica serotype Newport PT 14 (Table 2, strain Snp6). She had several dogs on her property and also fed them the same commercial dog treats mentioned above. She had no other additional exposure or recent travel history.
(v) Patient E.
A 47-year-old male (patient E) from Calgary presented in August 2002 to his general practitioner with watery diarrhea, fever, and abdominal cramps of 3 days' duration and received treatment for his symptoms. A stool specimen was positive for S. enterica serotype Newport PT 14 (Table 2, strain Snp7). He had several dogs, cats, and horses on his property and also fed them the same commercial dog treats mentioned above. He had recently visited a small rural community near Calgary.
Sources of Salmonella specimens.
Cultures of the environmental specimens and stools of the pets belonging to the households mentioned above were negative for Salmonella species; however, one commercial dog treat obtained from the property of patient D was positive. The brand of pet treat was imported from Texas. The type of dog treat consisted of dried steak patties containing beef, and the culture of this treat generated S. enterica serotype Newport PT 14 (Table 2, strain Snp8).
Antimicrobial susceptibility testing.
Strains Snp1 to Snp8 were susceptible to GEN, TOB, AMK, NAL, CIP, SXT, IPM, and FEP; intermediate to CTX and CRO; and resistant to CHL, AMP, SAM, FOX, CPD, and CAZ (Table 2). Of interest was the posttreatment Salmonella strains of patient A (Snp2 and Snp3), for which FEP MICs were higher than those for the other strains (strains Snp1 and Snp4 to Snp8) (Table 2).
β-Lactamase identification.
Isoelectric focusing revealed that the isolates produced a single β-lactamase with an isoelectric point (pI) of 9.0 and were inhibited by cloxacillin, as determined by the overlay technique. This correlates with a Bush group 1 cephalosporinase (7). Multiplex PCR amplified a 462-bp amplicon by using as a template DNA prepared from every strain tested. The amplicon was consistent with those amplified from strains with the plasmid-encoded types of AmpC β-lactamases originating from the chromosomal gene of C. freundii (21). Sequence analysis of full-length PCR products from every strain revealed 100% identity to blaCMY-2 (5).
Plasmid profiles and transconjugation.
Multiple plasmids were visualized from the Salmonella isolates, ranging from a plasmid of 4 kb to a large-molecular-mass plasmid. The large-molecular-mass plasmid was gel purified from clinical strains Snp1, Snp5, and Snp6 and electroporated into E. coli strain DH101B. The plasmid DNA isolated from these transformants was used as a template for both PCR and fingerprinting analysis. Plasmid DNA restricted with the endonuclease HpaI indicated that the large-molecular-mass plasmid was ∼140 kb. The plasmid DNA used as the template for PCR generated a blaCMY-specific amplicon, indicating that blaCMY-2 was encoded on the ∼140-kb plasmid.
PFGE.
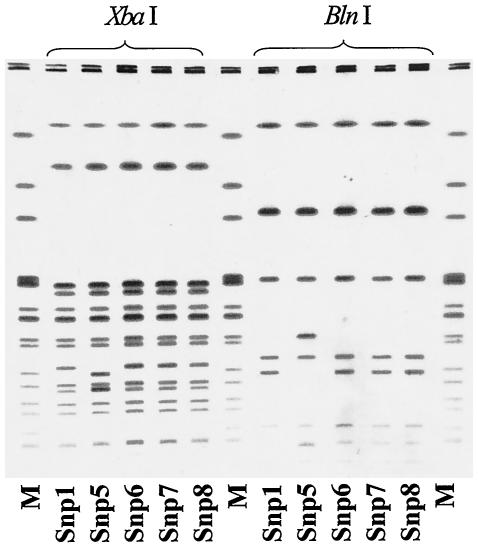
The molecular relatedness between the isolate from the dog treat (isolate Snp8) and the human isolates (isolates Snp1 to Snp7) was determined by PFGE. Figure 1 presents the restriction fragment patterns generated when DNA isolated from strains Snp1, Snp5, Snp6, Snp7, and Snp8 was digested with restriction endonuclease XbaI or BlnI. One cluster pattern that was indistinguishable was observed for strains Snp1, Snp6, and Snp8; and this pattern was highly related to that of strain Snp5. PFGE of DNA from strains Snp2, Snp3, and Snp4 under the same conditions described above also demonstrated the same cluster pattern (data not shown). This PFGE pattern was highly related to those of multidrug-resistant strains of S. enterica serotype Newport causing an outbreak in the United States. These strains were compared by PFGE to two strains of S. enterica Newport PT 14, collected in September 2002, that were susceptible to FOX and CAZ. The PFGE patterns of the susceptible strains were different from the patterns of strains Snp1 to Spn8 (data not shown).
FIG. 1.
PFGE analysis of DNA isolated from human Salmonella strains (strains Snp1, Snp5, Snp6, and Snp7) and a Salmonella strain from a commercial product (strain Snp8). DNA was cleaved with the endonuclease XbaI or BlnI. Snp1, Snp5, Snp6, Snp7, and Snp8 designate the strains described in the text. Lanes M, S. enterica serotype Newport AM01144 (915 to 35.8 kb), used as a size marker. The braces indicate the restriction endonuclease used for each digestion.
DISCUSSION
CMY-2 is the most prevalent and the most geographically widely distributed of the plasmid-encoded AmpC β-lactamases. Members of the family Enterobacteriaceae producing CMY-2 have been isolated since the late 1980s from humans from various countries, including Algeria, France, Spain, Germany, Greece, Romania, India, Pakistan, Turkey, the United Kingdom, and the United States (11, 17, 22). Recently, CMY-2-producing Salmonella species were isolated from food-producing animals in Canada and the United States (2, 26, 29). This is the first study to implicate CMY-2-producing Salmonella species as a cause of human infection in Canada.
CRO-resistant Salmonella species producing AmpC β-lactamases appear to be more widespread in the United States than was previously believed. A study reported by Dunne et al. (9) showed that the rates of CRO resistance in Salmonella species increased from 0.1% in 1996 to 1.9% in 1999. A recent report from the Centers for Disease Control and Prevention indicated that the proportion of laboratory-confirmed cases of S. enterica serotype Newport infection in relation to the number of all Salmonella isolates recovered in the United States was 5% of 34,608 strains in 1997, with this proportion increasing to 10% of 31,607 strains in 2001 (30). Furthermore, the same report revealed that in 1998, 1% of S. enterica serotype Newport isolates produced AmpC β-lactamases, whereas that proportion was 26% in 2001 (30). A case-control study from the Centers for Disease Control and Prevention found that CRO-resistant S. enterica serotype Newport human infections were associated with the consumption of ground beef (30). To extend the concept of linking the presence of these strains in both animals and humans, a study from Iowa described Salmonella strains from both humans and animals that share plasmids encoding genes which express AmpC β-lactamases (29).
This study indicates that the incidence of S. enterica serotype Newport infections is also increasing in Alberta, Canada, and suggests that AmpC-producing strains were introduced into this area in 2002 (Table 1). This study also describes cephalosporin-resistant S. enterica serotype Newport isolates that were recovered from humans as well as a commercial pet treat containing dried beef. These strains were highly related by the use of PFGE criteria and expressed the same plasmid-encoded AmpC β-lactamase, CMY-2. However, when these strains were compared to susceptible S. enterica serotype Newport strains, they were found to be unrelated. The phenotypic and genotypic similarities identified in both the human and the pet treat strains suggest the transfer of multidrug-resistant S. enterica serotype Newport isolates to humans via the handling of a pet treat. The handling of commercial pig ear dog treats has been implicated as a possible source of Salmonella infection in humans (8). To our knowledge, this is the first study to imply that the handling of pet treats containing animal products could be involved in the transmission of multidrug-resistant Salmonella strains to humans. The plasmid-encoded AmpC β-lactamase, CMY-2, has been identified in strains of S. enterica serotype Newport present in cattle, horses, dogs, and pigeons (26). Therefore, the association between animals, humans, and the handling of pet treats containing animal products, which are available in pet shops and retail stores, could play a role in the increasing prevalence of AmpC-producing S. enterica serotype Newport infections observed in the United States and Alberta.
This study highlights the important roles that clinical microbiology laboratories play in surveillance for the early detection of drug resistance among Salmonella isolates in a region or a country. In June 2002, CLS introduced cephalosporin susceptibility testing of all Salmonella stool isolates, and AmpC-producing strains were recognized soon thereafter. An epidemiological investigation was undertaken to identify the potential sources of these strains, and appropriate steps were taken to prevent an outbreak. This investigation led to the recommendation of the removal of the implicated brand of commercial pet treats from pet shops and retail stores, as well as a public advisory issued through newspapers regarding the potential dangers associated with the handling of these treats.
It is important to realize that no guidelines on the detection of AmpC β-lactamases or extended-spectrum β-lactamases in Salmonella species are available from the National Committee for Clinical Laboratory Standards, or elsewhere (20). Therefore, we recommend that all clinically relevant Salmonella species be routinely tested for their susceptibilities to CPD, FOX, CAZ, and CRO or CTX. Strains intermediate or resistant to any of these antibiotics should be referred to a reference laboratory for further characterization of the resistance mechanism(s). These vital steps will ensure the early detection of Salmonella species resistant to the extended-spectrum cephalosporins as well as the possible mechanism(s) by which resistance is spread. The outcome will ensure optimal patient care with the introduction of appropriate infection control procedures in a timely fashion.
Acknowledgments
This study was funded in part by the Antibiotic Resistant Organism Research fund, which is sponsored by a partnership between CLS, the Calgary Health Trust, the University of Calgary, and the Calgary Health Region.
We thank Geoff Soule, Philip Lee, and Kathy Moore for technical expertise; Karen Grimsrud, Judy McDonald, Ellen Blewett, and Dora Lee for providing the data on the prevalence of Salmonella isolates in Alberta; and Kirsty Townsend for reviewing the manuscript.
REFERENCES
- 1.Ahmed, R., C. Bopp, A. Borczyk, and S. Kasatiya. 1987. Phage-typing scheme for Escherichia coli O157:H7. J. Infect. Dis. 155:806-809. [DOI] [PubMed] [Google Scholar]
- 2.Allen, K. J., and C. Pope. 2002. Occurrence and characterization of resistance to extended-spectrum cephalosporins mediated by β-lactamase CMY-2 in Salmonella isolated from food-producing animals in Canada. Can. J. Vet. Res. 66:137-144. [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, E. S., L. R. Ward, M. J. Saxe, and J. D. de Sa. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. (Camb.) 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnaud, G., G. Arlet, C. Verdet, O. Gaillot, P. H. Lagrange, and A. Philippon. 1998. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob. Agents Chemother. 42:2352-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauernfeind, A., I. Stemplinger, R. Jungwirth, and H. Giamarellou. 1996. Characterization of the plasmidic β-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob. Agents Chemother. 40:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, P. A., Y. Yang, D. Sahm, I. Grope, D. Gardovska, and G. Storch. 1998. CTX-M-5, a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob. Agents Chemother. 42:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, C., J. Cunningham, R. Ahmed, D. Woodward, K. Fonseca, S. Isaacs, A. Ellis, C. Anand, K. Ziebel, A. Muckle, P. Sockett, and F. Rodgers. 2001. Characterization of Salmonella associated with pig ear dog treats in Canada. J. Clin. Microbiol. 39:3962-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunne, E. F., P. D. Fey, P. Kludt, R. Reporter, F. Mostashari, P. Shillam, J. Wicklund, C. Miller, B. Holland, K. Stamry, T. J. Barret, J. K. Rasheed, F. C. Tenover, E. M. Ribot, and F. J. Angulo. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamases. JAMA 284:3151-3156. [DOI] [PubMed] [Google Scholar]
- 10.Ewing, W. H. 1986. The genus Salmonella, p. 181-340. In Edwards and Ewings's identification of Enterobacteriaceae, 4th ed. Elsevier Science Publishing Co., Inc., New York, N.Y.
- 11.Fey, P. D., T. H. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. K. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 12.Food-Borne and Diarrheal Diseases Branch, Division of Bacterial and Mycotic Diseases, National Center for Infectious Diseases, Centers for Disease Control and Prevention. 1998. PulseNet—the National Molecular Subtyping Network for Food-Borne Disease Surveillance. One day standard protocol for molecular subtyping of Escherichia coli O157:H7 by pulsed-field gel electrophoresis. Centers for Disease Control and Prevention, Atlanta, Ga.
- 13.Hanson, N. D., E. S. Moland, A. Hossain, S. A. Nevilli, I. B. Gosbell, and K. S. Thomson. 2002. Unusual Salmonella enterica serotype Typhimurium isolate producing CMY-7, SHV-9 and OXA-30 β-lactamases. J. Antimicrob. Chemother. 49:1011-1014. [DOI] [PubMed] [Google Scholar]
- 14.Hohmann, E. L. 2001. Non-typhoidal salmonellosis. Clin. Infect. Dis. 32:263-269. [DOI] [PubMed] [Google Scholar]
- 15.Kauffman, F. 1966. The bacteriology of Enterobacteriaceae. The Williams & Wilkins Co., Baltimore, Md.
- 16.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffon, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miriagou, V., R. Filip, G. Coman, and L. S. Tzouvelekis. 2002. Expanded-spectrum cephalosporin-resistant Salmonella strains in Romania. J. Clin. Microbiol. 40:4334-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morosini, M. I., R. Canton, J. Martinez-Beltran, M. C. Negri, J. C. Perez-Diaz, F. Baguero, and J. Blazquez. 1995. New extended-spectrum TEM-type β-lactamase from Salmonella enterica subsp. enterica isolated in a nosocomial outbreak. Antimicrob. Agents Chemother. 39:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulvey, M. R., G. Soule, D. Boyd, W. Demczuk, and R. Ahmed. 2003. Characterization of the first extended-spectrum β-lactamase-producing Salmonella isolate identified in Canada. J. Clin. Microbiol. 41:460-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial and susceptibility testing; twelfth informational supplement, M-100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.Perez-Perez, F. J., and N. D. Hanson. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philippon, A., C. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitout, J. D. D., K. S. Thomson, N. D. Hanson, A. F. Ehrhardt, P. Coudron, and C. C. Sanders. 1998. Plasmid-mediated resistance to expanded-spectrum cephalosporins among Enterobacter aerogenes strains. Antimicrob. Agents Chemother. 42:596-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitout, J. D. D., K. S. Thomson, N. D. Hanson, A. F. Ehrhardt, E. S. Moland, and C. C. Sanders. 1998. β-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered from South Africa. Antimicrob. Agents Chemother. 42:1350-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poppe, C., and E. D. Mann. 1998. Procedures for the isolation of Salmonella species by the modified semi-solid Rappaport Vassiliadis (MSRV) method, MFLP-75. In D. Warburton (ed.), Compendium of analytical methods, Health Protection Branch, Health Canada. Polyscience Publications, Laval, Québec, Canada.
- 26.Rankin, S. C., H. Aceto, J. Cassidy, J. Holt, S. Young, B. Love, D. Tewari, D. S. Munro, and C. E. Benson. 2002. Molecular characterization of cephalosporin-resistant Salmonella enterica serotype Newport isolates from animals in Pennsylvania. J. Clin. Microbiol. 40:4679-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tassios, P. T., M. Gazouli, E. Tzelepi, H. Milch, N. Kozlova, S. Sidorenko, N. J. Legakis, and L. S. Tzouvelekis. 1999. Spread of a Salmonella typhimurium clone resistant to expanded-spectrum cephalosporins in three European countries. J. Clin. Microbiol. 37:3774-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villa, L., C. Mommina, V. Miriagou, L. S. Tzouvelekis, P. T. Tassios, A. Nastasi, and A. Carattoli. 2002. Multidrug and broad-spectrum cephalosporin resistance among Salmonella enterica serotype Enteritidis clinical isolates in southern Italy. J. Clin. Microbiol. 40:2662-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winokur, P. L., A. Brueggemann, D. L. de Salvo, L. Hoffman, M. D. Apley, E. K. Uhlenhopp, M. A. Phaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zansky, S., B. Wallace, D. Schoonmaker, P. Smith, F. Ramsay, J. Painter, A. Gupta, P. Kalleri, and S. Noviello. 2002. Outbreak of multidrug-resistant Salmonella Newport—United States, January-April 2002. CDC Surveillance Summaries. Morb. Mortal. Wkly. Rep. 51:545-548. [PubMed] [Google Scholar]