Abstract
Inflammatory bowel disease (IBD) is thought to result from commensal flora, aberrant cellular stress, and genetic factors. Here we show that the expression of colonic Ste20-like proline-/alanine-rich kinase (SPAK) that lacks a PAPA box and an F-α helix loop is increased in patients with IBD. The same effects were observed in a mouse model of dextran sodium sulfate-induced colitis and in Caco2-BBE cells treated with the pro-inflammatory cytokine tumor necrosis factor (TNF)-α. The 5′-flanking region of the SPAK gene contains two transcriptional start sites, three transcription factor Sp1-binding sites, and one transcription factor nuclear factor (NF)-κB-binding site, but no TATA elements. The NF-κB-binding site was essential for stimulated SPAK promoter activity by TNF-α, whereas the Sp1-binding sites were important for basal promoter activity. siRNA-induced knockdown of NF-κB, but not of Sp1, reduced TNF-α-induced SPAK expression. Nuclear run-on and mRNA decay assays demonstrated that TNF-α directly increased SPAK mRNA transcription without affecting SPAK mRNA stability. Furthermore, up-regulation of NF-κB expression and demethylation of the CpG islands induced by TNF-α also played roles in the up-regulation of SPAK expression. In conclusion, our data indicate that during inflammatory conditions, TNF-α is a key regulator of SPAK expression. The development of compounds that can either modulate or disrupt the activity of SPAK-mediated pathways is therefore important for the control and attenuation of downstream pathological responses, particularly in IBD.
Inflammatory bowel diseases (IBD), including mainly ulcerative colitis (UC) and Crohn’s disease (CD), are chronic, idiopathic, inflammatory disorders of the gastrointestinal tract and are thought to arise from the interplay of genetic and environmental factors.1 Barrier function loss has especially gained increasing support in IBD pathogenesis for the past decades, particularly as the epithelium represents an interface for genetic and environmental influences. Moreover, epigenetic studies have also confirmed a strong link between barrier function and IBD loci.2,3 Ste20-like kinases, including p21-activated kinase (PAK) and germinal center kinase (GCK), exert various functions, including barrier function.4,5 For example, ste20-like oxidant stress-activated kinase (SOK)6 can induce apoptosis through activating the JNK pathway; lymphocyte-oriented kinase (LOK)7 and ste20-like kinase (SLK)8 can regulate Rac1-mediated actin reorganization during cell adhesion and spreading; and PAK can increase endothelial permeability.9
SPAK is defined as a ste20-like proline-/alanine rich kinase that contains an N-terminal series of proline and alanine repeats (PAPA box) followed by a kinase domain, a nuclear localization signal, a consensus caspase cleavage motif, and a C-terminal regulatory region.10 Our laboratory showed that colonic SPAK exists as a unique isoform that lacks the PAPA box and F-α helix loop in the N-terminus.11 The diversity of domains present in SPAK protein might be associated with a variety of biological roles. For example, SPAK has been shown to play roles in cell differentiation,10,12 cell transformation and proliferation,13 and regulation of chloride transport.14,15,16 More importantly, a linkage has been established between SPAK and inflammation, SPAK, as an upstream kinase to Na+-K+-2Cl− co-transporter 1 (NKCC1), can phosphorylate Thr203, Thr207, and Thr212 amino acids on NKCC1, which play an important role in inflammation.17,18 Furthermore, we have demonstrated that SPAK can activate p38 pathway11 that is well known involving inflammation19,20,21,22 and interestingly can regulate the intestinal barrier function11 as well. However, SPAK expression, activation, and roles in barrier functions of intestinal epithelial cells (IECs), have not been evaluated comprehensively. However, no previous study has evaluated SPAK expression, activity, or signaling in intestinal epithelial cells. Thus, we intend to investigate the expression and role(s) of the SPAK in intestinal inflammation.
Materials and Methods
Mouse Colitis Model
All experiments were performed in female C57BL/6 mice (8 weeks of age, 18 to 22 g) obtained from The Jackson Laboratory (Bar Harbor, ME). Mice were group housed under a controlled temperature (25°C) and photoperiod (12:12-hour light-dark cycle) and allowed unrestricted access to standard mouse chow and tap water. They were allowed to acclimate to these conditions for at least 7 days before inclusion in the experiments. Colitis was induced by the addition of dextran sodium sulfate (DSS) [40,000 Da, 3.5% (w/v); ICN Biochemicals, Aurora, OH] to the drinking water. The mean DSS water consumption was recorded for each group. Groups of mice (n = 6 mice per group) were treated with 3.5% DSS or regular water for the indicated days. Body weights were assessed every day during the treatment period. Histological assessment of colitis was performed by hematoxylin and eosin (H&E) staining and analyzed by microcopy. All animal experiments were approved by The Animal Care Committee of Emory University, Atlanta, and were in accordance with the guide for the Care and Use of Laboratory Animal, published by the US Public Health Service.
Cell Culture
The human intestinal cell line Caco2-BBE at passage 30 to 50 was cultured in high-glucose Dulbecco’s Vogt modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 14 mmol/L NaHCO3 and 10% newborn calf serum. Cells were incubated at 37°C in 5% CO2 and 90% humidity.
Human Material
The diagnosis of IBD was based on clinical, endoscopic, and histological criteria. Clinical data for IBD patients were obtained by medical record review. Infectious colitis was ruled out by stool cultures. The collection of samples was approved by the Institutional Review Board of Emory University. Mucosal biopsy specimens from four UC active patients were obtained during routine endoscopy that was performed after written informed consent was obtained. Control biopsy samples were collected from six volunteers (three females and three males) undergoing colonoscopy for colorectal cancer screening who had no overt pathology including polyps. Biopsy specimens were snap-frozen in optimal cutting temperature immediately after endoscopic resection and stored at −80°C for histological immunostaining or homogenized to extract the protein for Western immunoblotting or RNA for real-time polymerase chain reaction (PCR).
Myeloperoxidase (MPO) Activity
Neutrophil infiltration into colon was quantified by measuring MPO activity. Briefly, a portion of colon was homogenized in 1:20 (w/v) of 50 mmol/L phosphate buffer (pH 6.0) containing 0.5% hexadecyltrimethyl ammonium bromide (Sigma-Aldrich, St. Louis, MO) on ice using a Polytron homogenizer. The homogenate was sonicated for 10 seconds, freeze-thawed three times, and centrifuged at 14,000 rpm for 15 minutes. Supernatant (14 μl) was added to 1 mg/ml o-dianisidine hydrochloride (Sigma-Aldrich) and 0.0005% hydrogen peroxide, and the change in absorbance at 460 nm was measured. One unit of MPO activity was defined as the amount that degraded 1 μmol peroxidase per minute at 25°C. The results were expressed as absorbance per gram of tissue.
Immunohistochemistry
Immunostaining was performed according to the standard protocol. Slices (1 to 2 mm thick) of human and mouse colonic mucosal were frozen in liquid nitrogen and stored at −80°C until use. The specimen were blocked for 10 minutes with 5% normal donkey serum in phosphate-buffered saline (PBS)/Triton and then sequentially incubated with 1:200 diluted primary antibodies against SPAK (Cell Signaling Technology Inc., Danvers, MA) for overnight at 4°C. Subsequently incubated with fluoresceinated secondary antibody plus 100 ng/μl DAPI (4,6-diamidino-2-phenylindole hydrochloride) (Invitrogen) to visualize nuclear for 1 hour at room temperature in the dark. Samples were mounted in p-phenylenediamine/glycerol (1:1) and analyzed by epifluorescence microscopy (Carl Zeiss, Thornwood, NY).
Plasmid Construction
Genomic DNA was extracted from Caco2-BBE cells using the REDExtract-N-Amp tissue PCR kit (Sigma-Aldrich). The 5′-flanking region of the SPAK gene was PCR amplified with ThermalACE DNA polymerase (Invitrogen) in the presence of 1 mol/L betaine (Sigma-Aldrich) using SPAK For and SPAK Rev (Table 1) primers based on the genomic sequence of the SPAK gene (accession no. NT_005403). The expected approximate 1.5-kb-long fragment was gel-purified with the Qiaquick gel extraction kit (Qiagen, Valencia, CA) and cloned into the PCR2.1/TOPO vector (Invitrogen) and confirmed by sequencing. The plasmid was then digested with KpnI and XhoI (New England Biolabs, Ipswich, MA), subcloned into the promoter-less pGL3 firefly luciferase basic vector (Promega, San Luis, CA), and confirmed by sequencing. Truncated constructs were generated by PCR using the full-length sequence as a template along with the following primers (Table 1): SPAK promoter I (−1050, +4) (SPAK1 and SPAK Rev); SPAK promoter II (−398, +4) (SPAK2 and SPAK Rev); SPAK promoter III (−331, +4) (SPAK3 and SPAK Rev); SPAK promoter IV (−149, +4) (SPAK4 and SPAK Rev); and SPAK promoter V (−72 to + 4) (SPAK5 and SPAK Rev). Mutations of Sp1 and nuclear factor (NF)-κB binding sites were introduced by overlapping PCR using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The mutagenic oligonucleotide sequences are shown in Table 1 with the substitutions underlined. In detail, the I Sp1 mutant was generated with primers I Sp1 Mut For and I Sp1 Mut Rev; the II Sp1 mutant with primers II Sp1 Mut For and II Sp1 Mut Rev; the III Sp1 mutant with primers III Sp1 Mut For and III Sp1 Mut Rev. The NF-κB binding domain mutant was generated with primers NF-κB Mut For and NF-κB Mut Rev, respectively. All mutants were confirmed by DNA sequencing.
Table 1.
Primers Used for This Research
| Name | Sequence | Name | Sequence |
|---|---|---|---|
| Primers used for real-time PCR | Oligonucleotides used for EMSA | ||
| SPAK HQF | 5′-TGGAATTAGCAACAGGAGCAGCG-3′ | I Sp1 For | 5′-TTCAACGACCCCGCCCTCGGGGT-3′ |
| SPAK HQR | 5′-TTTCCAAAGTGGGTGGATCATTT-3′ | I Sp1 Rev | 5′-ACCCCGAGGGCGGGGTCGTTGAA-3′ |
| SPAK MQF | 5′-GTAAGGCGAGTTCCTGGGTCG-3′ | I Sp1 MFor | 5′-CACGACCACCTTGCCCTCGGGGT-3′ |
| SPAK MQR | 5′-CCAGTCGCCGTCTTCAGTCTT-3′ | I Sp1 MRev | 5′-ACCCCGAGGGCAAGGTGGTCGTG-3′ |
| GAPDH For | 5′-ACCACAGTCCATGCCATCAC-3′ | II Sp1 For | 5′-CGGAGAGGGGGGGCGGCGGTTCC-3′ |
| GAPDH Rev | 5′-TCCACCACCCTGTTGCTGTA-3′ | II Sp1 Rev | 5′-GGAACCGCCGCCCCCCCTCTCCG-3′ |
| 36B4 For | 5′-TCCAGGCTTTGGGCATCA-3′ | II Sp1 MFor | 5′-CGGAGAGGGGGGTCCGCGGCGGT-3′ |
| 36B4 Rev | 5′-CTTTATCAGCTGCACATCACTCAGA | II Sp1 MRev | 5′-ACCGCCGCGGACCCCCCTCTCCG-3′ |
| Primers used for ChIP | III Sp1 For | 5′-TCCTCCCTTCCCCGCCTCCGCCG-3′ | |
| Sp1 IF | 5′-GTAAATGAACTTCAGGTTCTCTTTG-3′ | III Sp1 Rev | 5′-CGGCGGAGGCGGGGAAGGGAGGA-3′ |
| Sp1 IR | 5′-CGCCCTGCGCCTTGGCCCCAGACGA-3′ | III Sp1 MFor | 5′-CCTCCCTGTCCCGCTCCTCCGCC-3′ |
| Sp1 IIF | 5′-AGCACACACAAAGCGGCCTGACTCC-3′ | III Sp1 MRev | 5′-GGCGGAGGAGCGGGACAGGGAGG-3′ |
| Sp1 IIR | 5′-CCCAGAGCCTAGCGCGCGCTGTTCT-3′ | NF-κB For | 5′-TTTGGTGGGAACTTCCCCGGCCG-3′ |
| Sp1 IIIF | 5′-CTGGCTTCGGCGGGGACGGCGGCGG-3′ | NF-κB Rev | 5′-CGGCCGGGGAAGTTCCCACCAAA-3′ |
| NF-κBF | 5′-GGCGCAGGGCGAGCAGGGAGGGAGG-3′ | NF-kB MFor | 5′-GGTGGGACCGACTTCCTTTCCGG-3′ |
| NF-κBR | 5′-TGTTCTCCGCCTCGGCGAGGGGAAC-3′ | NF-κB MRev | 5′-CCGGAAAGGAAGTCGGTCCCACC-3′ |
| Primers used for other detections | Primers for SPAK promoter constructs | ||
| SPAKR1 | 5′-GTGTTCTTGTAAGCAGCTTCTCAATC-3′ | SPAK For | 5′-CTTCAAGACCAGAGCGAGGTAGC-3′ |
| SPAKR2 | 5′-CACAAGAAGAAGCTTCTCTGTAGTCT-3′ | SPAK Rev | 5′-CCATGATGCTGCGGAGGAGAGCAGGAG-3′ |
| SPAKNorFor | 5′-CTGATTGAGAAGCTGCTTACAAG-3′ | SPAK 1 | 5′-CCTATGGTTTTACCATAGCAGTTTTACAG-3′ |
| SPAKNorRev | 5′-CAAGAAGAAGCTTCTCTGTAGTC-3′ | SPAK 2 | 5′-AGCACACACAAAGCGGCCTGA-3′ |
| 18SFor | 5′-CCCCTCGATGCTCTTAGCTGAGTGT-3′ | SPAK 3 | 5′-AGGGGGCGGCGGAGAGGGGGG-3′ |
| 18SRev | 5′-CGCCGGTCCAAGAATTTCACCTCT-3′ | SPAK 4 | 5′-TCCTAGCTGGCTTCGGCGGGGA-3′ |
| AMPfor | 5′-GGTTTGATTTTGGTGGGAA-3′ | SPAK 5 | 5′-CGAGGCAAAGGAAGTTTCAAGTGG-3′ |
| AMPrev | 5′-TTAAAACTTCCTTTACCTC-3′ | I Sp1 Mut For | 5′-GCGGCCGTTCAACGACCCCGACCTCGGGGTCCCCAG-3′ |
| TQMprobe | 5′FAM-AATAGCGCGCGTTAGGTT-BHQ-3′ | I Sp1 Mut Rev | 5′-CTGGGGACCCCGAGGTCGGGGTCGTTGAACGGCCGC-3′ |
| TQUprobe | 5′-HEX-AATAGTGTGTGTTAGGTT-BHQ-3′ | II Sp1 Mut For | 5′-GCGGCGGAGAGGGGGGACGGCGGTTCCCCTCGCCGA-3′ |
| II Sp1 Mut Rev | 5′TCGGCGAGGGGAACCGCCGTCCCCCCTCTCCGCCGC-3′ | ||
| III Sp1 Mut For | 5′CGGCGGCGGCGGCGGCGGACGGGGAGGGCGTGCGCC-3′ | ||
| III Sp1 Mut Rev | 5′-GGCGCACGCCCTCCCCGTCCGCCGCCGCCGCCGCCG-3′ | ||
| NF-kB Mut For | 5′-ACTCCTTTGGTGGGAACTTCACCGGCCGCCAGCGGA-3′ | ||
| NF-kB Mut Rev | 5′-TCCGCTGGCGGCCGGTGAAGTTCCCACCAAAGGAGT-3′ | ||
Tumor Necrosis Factor (TNF)-α Treatment and Western Blot
Caco2-BBE cells were treated with 40 ng/ml of TNF-α, total protein was collected and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot by standard method with relevant antibodies anti-SPAK (Cell Signaling Technology Inc.), anti-GAPDH (Ambion, Austin, TX), anti-Sp1 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-NF-κB (p65) (Santa Cruz Biotechnology), anti-histone1 (Upstate, Charlottesville, Virginia).
Co-Immunoprecipitation and Western Blot
Caco2-BBE cells were subjected to co-immunoprecipitation with relevant antibodies using Catch and Release v2.0 kit (Upstate Cell Signaling Solutions, Lake Placid, NY), then Western blots were performed based on the standard methods with anti-phosphoserine antibody (Sigma-Aldrich).
Real-Time PCR
Total RNA was extracted using TRIzol (Invitrogen) from mucosa of colon biopsy of IBD patients, DSS-treated mice, and Caco2-BBE cells treated with TNF-α. RNA was reverse-transcribed using the Thermoscript RT-PCR system (Invitrogen) and purified with the RNeasy mini kit (Qiagen, Germantown, MD). Real-time PCR was performed using iQ SYBR Green Supermix kit (Bio-Rad, Hercules, CA) with the iCycler sequence detection system (Bio-Rad). Specific primers were designed using the Primer Express Program (Applied Biosystems, Foster City, CA): human samples, SPAK primers HQF and HQR, internal control GAPDH For and GAPDH Rev (Table 1); mouse sample, SPAK primers MQforward and MQreverse and internal control 36B4For and 36B4Rev (Table 1). For graphical representation of quantitative PCR data, raw cycle threshold values (Ct values) obtained for target samples were deducted from the Ct value obtained for internal control transcript levels, using the ΔΔCt method as follows: ΔΔCT = (Ct,target − Ct,con)treatment − (Ct,target − Ct,con)nontreatment, and the final data were derived from 2−ΔΔCT.
Nuclear Protein Extraction
Caco2-BBE cells were washed once in ice-cold PBS, and centrifuged at 800 rpm for 5 minutes. The resulting pellets were resuspended in 5 ml of cold lysis buffer (10 mmol/L HEPES, 10 mmol/L KC1, 1.5 mmol/L MgC12, and 0.1% Nonidet P-40, pH 7.9 at 4°C) for 10 minutes on ice. For the isolation of nuclei, the lysate was vigorously mixed and centrifuged for 5 minutes at 12,000 × g and 4°C, and the nuclear pellet was washed once with 1 ml of Nonidet P-40-free lysis buffer. For the extraction of nuclear proteins, the nuclear pellet was resuspended in 1 ml of protein extraction buffer [420 mmol/L NaCl, 20 mmol/L HEPES, 1.5 mmol/L MgCl2, 0.2 mmol/L ethylenediaminetetraacetic acid (EDTA), and 25% glycerol, pH 7.9] for 10 minutes at 4°C. After vigorously mixed, the nuclear suspension was centrifuged for 5 minutes at 4°C, the pellet was discarded, and the supernatant was centrifuged (12,000 rpm) again for 5 minutes at 4°C. The protein content in the final supernatant (nuclear protein extract) was measured using the Bradford method (Bio-Rad). Dithiothreitol (0.5 mmol/L), phenylmethyl sulfonyl fluoride (0.5 mmol/L), and leupeptin (10 pg/ml) were added to the lysis and extraction buffers just before use. The diluting buffer contained the same amounts of dithiothreitol and leupeptin but only 0.2 mmol/L phenylmethyl sulfonyl fluoride. Samples were stored at −70°C until use.
Transient Transfection and Luciferase Reporter Gene Assay
Five ng of Renilla (phRL-CMV) and 4 μg of the relevant SPAK promoter constructs were co-transfected into Caco2-BBE cells with Lipofectin (Invitrogen). After 24 hours, cells were stimulated with 40 ng/ml of TNF-α for 16 hours or with 10 ng/ml of IL-1β for 24 hours. Cell were washed and lysed with the provided buffer in a luciferase assay kit (Promega). For the luciferase activity assay, 20 μl of cell extract was mixed with 100 μl of luciferase assay substrate (Promega), and the resulting luminescence was measured for 10 seconds in a luminometer (Luminoskan; Thermo Labsystems, Needham Heights, MA). For normalization of luciferase activity, we used the phRL-CMV control vector containing the CMV immediate-early enhancer/promoter region, which provides strong constitutive expression of Renilla luciferase in a variety of cell types. Each extract was analyzed in duplicate, and three independent experiments were performed for each experiment. The observed luciferase activity reported in the figures is expressed as the percentage of the activity obtained with regard to the most active construct used in the figures.
Electrophoretic Mobility Shift Assays (EMSAs)
Probes for EMSAs were designed using a standard protocol and are shown in Table 1. In detail, I Sp1 For and I Sp1 Rev are complementary oligonucleotides for the first Sp1 binding site, I Sp1MFor and I Sp1MRev are complementary oligonucleotides for the first Sp1 binding site mutant, II Sp1 For and II Sp1 Rev are complementary oligonucleotides for the second Sp1 binding site, II Sp1MFor and II Sp1MRev are complementary oligonucleotides for the second Sp1 binding site mutant, III Sp1 For and III Sp1 Rev as well as III Sp1MFor and III Sp1MRev correspond to the third Sp1 binding site and its mutant, and primers NF-κB For and NF-κB Rev as well as NF-κB MFor and NF-κB MRev correspond to the NF-κB binding site and its mutant. Probes were synthesized with HPLC purification and end labeled with a biotin 3′ end DNA labeling kit (Pierce, Rockford, IL). Briefly, 0.2 U/μl of terminal deoxynucleotidyl transferase (TdT), 0.5 μmol/L biotin-11-UTP, and 100 nmol/L of each single-stranded oligonucleotide were incubated in 1× TdT reaction buffer for 30 minutes at 37°C. EDTA (2.5 μl of 0.2 mol/L) was added to stop the reaction, and 50 μl of chloroform:isoamyl alcohol (24:1) was added to extract the labeled oligonucleotides. For annealing, concentrated complementary oligonucleotides were mixed together at a 1:1 molar ratio and incubated at 95°C for 5 minutes. The heat was then gradually reduced for hours until the oligonucleotides reached room temperature. Standard EMSAs were performed using the LightShift chemiluminescent EMSA kit (Pierce). Briefly, 20 fmol of each biotin end-labeled target oligonucleotide pair were incubated in EMSA binding buffer (100 mmol/L Tris, 500 mmol/L KCl, and 10 mmol/L dithiothreitol, pH 7.5) containing 50 ng/μl of poly(dI-dC) and 5 μg of Caco2-BBE nuclear proteins for 15 minutes at room temperature. For the competition EMSA, we added 50-fold (4 pmol) of unlabeled paired oligonucleotides. For the supershift EMSA, we added 2 μg of related Sp1 antibody (Santa Cruz Biotechnology) or anti-NF-κB p65 antibody (Santa Cruz Biotechnology). Complexes were resolved by electrophoresis on native 5% TBE Criterion Precast Gels (Bio-Rad) in 0.5× TBE buffer at 110 V. Gels were transferred to Biodyne B pre-cut modified nylon membranes (0.45 μmol/L, Pierce) using a Trans-Blot SD semi-dry transfer cell (Bio-Rad). Membranes were then cross-linked using UVC-508 UV cross-linker (Ultra-Lum, Claremont, CA) and visualized using the chemiluminescent nucleic acid detection system (Pierce).
Chromatin Immunoprecipitation (ChIP) Assay
Sp1 and NF-κB (p65) ChIP assays were performed using the ChIP assay kit (Upstate Cell Signaling Solutions) according to the manufacturer’s instructions with some modifications. Briefly, Caco2-BBE cells, treated with 40 ng/μl of TNF-α for 16 hours or with 10 ng/μl of IL-1β for 24 hours, were fixed with 1% formaldehyde for 10 minutes at 37°C to initiate cross-linking, scraped off the plate, washed with ice-cold PBS, and resuspended in 200 μl of sodium dodecyl sulfate lysis buffer for 10 minutes on ice. Cells were then sonicated with three sets of 10-second pulses at 35% power to shear the DNA into 200 1,000-bp fragments. Samples were centrifuged, and the supernatant (used as total DNA input) was diluted in ChIP dilution buffer and precleared with a protein A agarose-salmon sperm DNA slurry to reduce the nonspecific background. Samples were then immunoprecipitated with 2 μg of mouse anti-Sp1 (Upstate Cell Signaling Solutions) or 3 μg of rabbit anti-p65 antibody (Santa Cruz Biotechnology) overnight at 4°C. Complexes were collected in a protein A agarose-salmon sperm DNA slurry for 1 hour at 4°C, washed once each with the provided low-salt, high-salt, and LiCl wash buffers, and then washed twice in Tris-EDTA buffer [10 mmol/L Tris-HCl (pH 8.0) and 1 mmol/L EDTA]. The immunoprecipitated chromatin was eluted from protein A using freshly prepared elution buffer (100 mmol/L NaHCO3 and 1% sodium dodecyl sulfate), and the protein-DNA cross-links were reversed by treatment with NaCl (200 mmol/L) at 65°C for 4 hours. The DNA was purified by incubation with proteinase K at 45°C for 1 hour, followed by phenol-chloroform extraction and ethanol precipitation with glycogen. Sp1 (I, II, and III) and NF-κB binding sites in immunoprecipitates were detected by PCR using the following specific primers (Table 1): Sp1 IF and Sp1 IR, Sp1 IIF and Sp1 IIR, Sp1 IIIF and SPAK Rev, NF-κBF and NF-κBR. The products were resolved on a 1% agarose gel and visualized with ethidium bromide.
Transfection of siRNA
Subconfluent (60%) Caco2-BBE cells on six-well plates (Costar, Corning, NY) were transfected with siRNA (Ambion) against Sp1 and NF-κB (p65) using Lipofectamine 2000 (Invitrogen) in serum-free medium, a nontargeting siRNA was used as the control for nonsequence-specific effects of siRNAs (Ambion). Serum was added after 24 hours, Caco2-BBE cells underwent 40 ng/ml of TNF-α treatment for 16 hours; cells were then collected for Western blot analysis with relevant antibodies.
Nuclear Run-On Assay
Nuclear run-on assay was performed following the protocol of Gnoni and colleagues.23 Briefly; nuclei were isolated from Caco2-BBE cells treated with or without 40 ng/ml of TNF-α for 6 hours. Approximately 5 × 107 cells were pelleted in a conical tube and washed twice with ice-cold PBS. Cells were then lysed on ice for 10 minutes in lysis buffer containing 0.3 mol/L sucrose, 0.4% (v/v) Nonidet P-40, 10 mmol/L Tris-HCl at pH 7.4, 10 mmol/L NaCl, and 3 mmol/L MgCl2. After centrifugation (15 minutes at 500 relative centrifuge force), the nuclear pellet was resuspended and subjected to a repeat (5 minutes) lysis to remove any remaining intact cells. After centrifugation, nuclei were purified by centrifugation through a 2.0 mol/L sucrose cushion. The nuclei were resuspended in 300 μl of transcription buffer (50 mmol/L Tris-HCl, pH 8.0, 150 mmol/L KCl, 5 mmol/L MgCl2, 0.5 mmol/L MnCl2, 1 mmol/L dithiothreitol, 0.1 mmol/L EDTA, 10% glycerol). After pretreatment with 1 μl of 50 μg/ml RNase A and followed by 2.5 μl of 100 U of RNasin, the in vitro elongation reaction was initiated with the addition of ribonucleotides to a final concentration of 0.25 mmol/L each ATP, GTP, CTP, and UTP. The reaction was performed for 25 minutes at 30°C. After incubation with RNase-free DNase, RNA was extracted with phenol-chloroform, precipitated with ammonium acetate and isopropanol, washed with 70% ethanol, and dissolved in water. cDNA was synthesized with the SuperScript III first-strand synthesis system (Invitrogen) and amplified with Platinum TaqDNA polymerase (Invitrogen) using SPAK HQF and SPAK HQR (Table 1) as primers. GAPDH acts as internal control. The products were resolved on a 2% agarose gel and visualized with ethidium bromide.
SPAK mRNA Stability Assay and Northern Blot
For mRNA decay experiments, Caco2-BBE cells pretreated with actinomycin D (5 μg/ml) for 1 hour to arrest transcription were incubated with TNF-α (40 ng/ml) or saline at indicated time points. The decay of SPAK mRNA was examined by real-time PCR with primers SPAK HQF and SPAK HQR (Table 1). Levels of mRNA were standardized against 18S rRNA levels with primers 18SFor and 18SRev (Table 1), taking into account a previous determination of 65 hours for 18S rRNA half life,24 and plotted as the percentage of remaining mRNA compared to message levels at the 0 time point (where there is a 100% maximum mRNA level). The 24-hour time point sample was also used for Northern blot analysis with North2South complete biotin random prime labeling and detection kit (Pierce) with probe generated by PCR with primers SPAKNorFor and SPAKNorRev (Table 1).
Real-Time TaqMan Methylation-Sensitive Polymerase Chain Reaction
Sodium bisulfite treatment of genomic DNA and real-time TaqMan methylation-sensitive polymerase chain reaction was performed according to the protocol described previously.25 In detail, 1 μg of genomic DNA from Caco2-BBE cells in 25 μl of dH2O was denatured by the addition of 2.75 μl of 2 N NaOH at 37°C for 10 minutes, later, 15 μl of freshly prepared 10 mmol/L hydroquinone (no. H9003, Sigma) and 260 μl of 3.6 mol/L sodium bisulfite pH5.0 (no. 243973, Sigma) were added, and the reaction was layered with mineral oil and incubated overnight (16 hours) at 54°C. The Wizard SV Genomic DNA Clean-Up System (Promega) was used to isolate the DNA after sodium bisulfite treatment. The DNA eluted from the column was desulfonated by the addition of 5.5 μl 3 N NaOH and incubated at room temperature for 5 minutes. The DNA was ethanol-precipitated, washed one time with 75% ethanol, and suspended in 50 μl of 10 mmol/L Tris-HCl (pH 8.0). All primers for the TaqMan MSP were synthesized by Integrated DNA Technologies (Coralville, IA). The amplification primers used in the real-time TaqMan methylation-sensitive PCR reaction were designed to avoid CpG dinucleotides in the sense strand in the promoter of the SPAK gene. The SPAK amplification primers are AMPfor, AMPrev, TQMprobe, TQUprobe (Table 1). The methylated DNA-specific probe is 5′ end-labeled with FAM and quenched by the addition of Black Hole Quencher 1 to the 3′ end of the oligonucleotide. The unmethylated DNA-specific probe was 5′ end-labeled with HEX and quenched by the addition of Black Hole Quencher 1 to the 3′ end of the oligonucleotide.
In separate reactions, the methylation status of the SPAK promoter was assessed using 30 ng of sodium bisulfite-treated genomic DNA with iQ supermix kit (Bio-Rad). The PCR cycling parameters using iQ5 Light-Cycler (Bio-Rad) were incubated at 95°C for 3 minutes followed by 95°C for 10 seconds, 52°C for 30 seconds, and 72°C for 30 seconds, repeated 40 times. A relative methylation index (MI) is defined as 2−ΔΔCT, the relative ratio change of unmethylated/methylated CpG island without or with TNF-α treatment: here ΔΔCT = (CT,Unmethylated − CT,Methylated)TNF-α − (CT,Unmethylated − CT,Methylated)Con. MI would be assessed in triplicate.
Results
Human Clonic SPAK Is Up-Regulated in Intestinal Inflammation
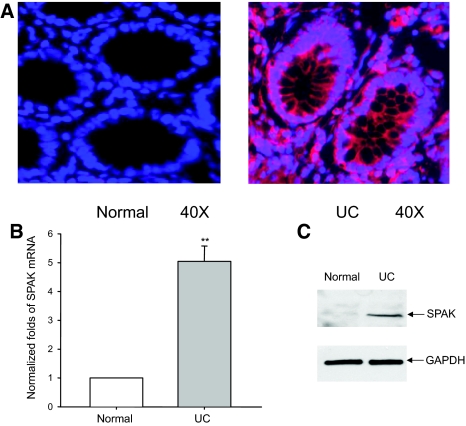
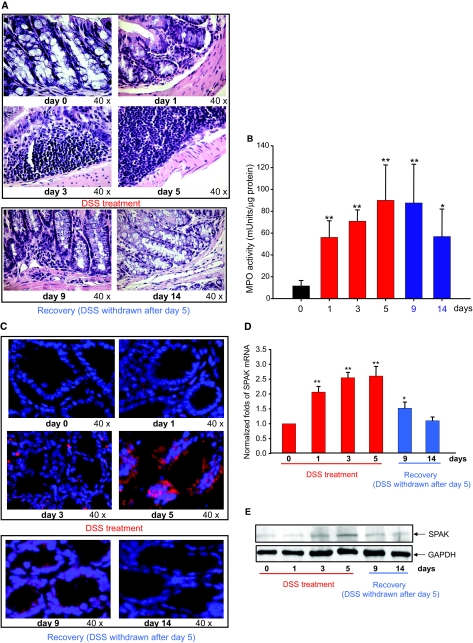
As a preliminary step toward a functional study of SPAK in IBD, we assayed SPAK expression during intestinal inflammation. We examined the relative SPAK expression in colonic biopsies in normal and IBD patients and colon samples from normal mice and mice treated with DSS. Immunofluorescence performed on the biopsy specimens collected from UC patients showed enhanced colonic SPAK expression mainly in epithelial cells in comparison with sections obtained from healthy colon tissue (Figure 1A). Here we do not exclude that SPAK is also overexpressed in other cells such as immune cells during intestinal inflammation. However, our present study is focused on colonic SPAK isoform expression. By real-time PCR and Western blots, colonic mucosa from the same UC patients showed significantly increased SPAK expression levels compared with tissue from normal patients, at both the mRNA and protein levels (Figure 1, B and C), respectively. By interfering with intestinal barrier function and stimulating local inflammation, DSS is often used to induce the mouse model of colitis26 that can mimic IBD with UC characteristics. As shown in Figure 2A, DSS induces cell wall damage, interstitial edema, and a general increase in the numbers of inflammatory cells in the lamina propria. After withdrawing DSS, the inflammatory effects gradually reduce to normal levels. Next, we measured colonic MPO activity as an indicator of the extent of neutrophil infiltration into the mucosa. We found that DSS-induced increases of MPO activity reached to the maximum at day 5 after DSS was replaced with drinking water. MPO level remained significantly higher at day 9; and at day 14, the MPO level had declined significantly (Figure 2B). Interestingly, the SPAK protein (Figures 2C and 1E) and mRNA (Figure 2D) levels in colonic tissue of DSS-treated mice increased in a time-dependent manner and correlated with the extent of colonic injury (Figure 2, C–E), the greatest increases in SPAK expression levels were on days 3 and 5, ∼2.5-fold greater levels of SPAK transcript versus those from untreated mice (Figure 2D), as shown by real-time PCR, immunohistochemistry, and Western blot analyses. Furthermore, when mice were allowed to recover from DSS-induced colitis after 5 days of DSS treatment (9 and 14 days), colonic SPAK mRNA and protein levels decreased throughout time and reached to the levels reported in untreated mice (Figure 2, C–E). Together, these results indicate a strong correlation between the level of SPAK expression and the extent of intestinal inflammation, which in turn indicates that SPAK could be involved in the pathogenesis and/or maintenance of IBD.
Figure 1.
SPAK expression profile in colon tissue from patients with UC. A: Immunostaining of SPAK in normal human colon tissue and UC patient colon tissue from mucosal biopsies. SPAK expression (red); nuclear staining by DAPI (blue); SPAK is primarily expressed in epithelial cells. B: The expression of SPAK mRNA in normal human and UC patient colon tissues from mucosal biopsies were quantified by real-time PCR. **P < 0.01. C: Ten μg of protein from normal human colon and UC patient colon from mucosal biopsies were examined by Western blot with SPAK antibody, colon tissue from UC patients demonstrated a significantly higher level of SPAK expression (top) versus healthy colon, with GAPDH as the internal loading control.
Figure 2.
SPAK expression profile in colon tissue from mice with experimental colitis. A: H&E-stained colon sections of mice treated with DSS at 0, 1, 3, 5, 9 (withdraw after 5 days treatment, 4 days to recover), and 14 (withdraw after 5 days treatment, 9 days to recover) days. B: Determination of MPO enzymatic activity in the colon as an index of neutrophil infiltration into the injured tissue. Results are expressed as MPO mU per μg protein and represent mean ± SEM of three determinations. *P < 0.05, **P < 0.01. C: Immunostaining of SPAK in same mice colon tissue as in H&E-stained sections. SPAK (red); nuclear staining by DAPI (blue). D: Real-time PCR analysis of SPAK mRNA expression in mucosa from colon tissue of DSS-treated mice. *P < 0.05, **P < 0.01. E: Western blot analysis of SPAK expression in mucosa from colon tissue of DSS-treated mice.
TNF-α Increases SPAK Expression
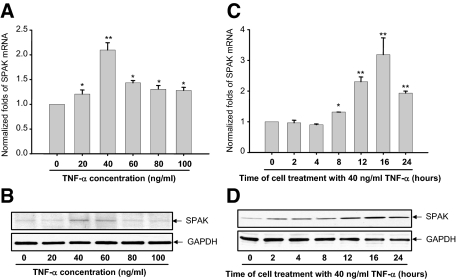
TNF-α is an important pro-inflammatory cytokine and has a key role in the pathogenesis of IBD,27 overexpression of TNF-α is sufficient to generate colitis in a mouse model,28 anti-TNF-α therapies have been successfully proven to attenuate IBD patients29 as well. Here, we show that TNF-α significantly increases SPAK mRNA and protein levels in a dose-dependent and time-dependent manner, with a maximal effect reported with 12- to 16-hour treatment with 40 ng/ml of DSS (Figure 3, A–D). These results suggest a link between intestinal inflammation, TNF-α expression, and SPAK expression.
Figure 3.
Real-time PCR (A) and Western blot (B) demonstrate dose responses of TNF-α on SPAK expression in Caco2-BBE cells. *P < 0.05, **P < 0.01. Real-time PCR (C) and Western blot (D) show the time course of TNF-α on SPAK expression in Caco2-BBE cells. *P < 0.05, **P < 0.01.
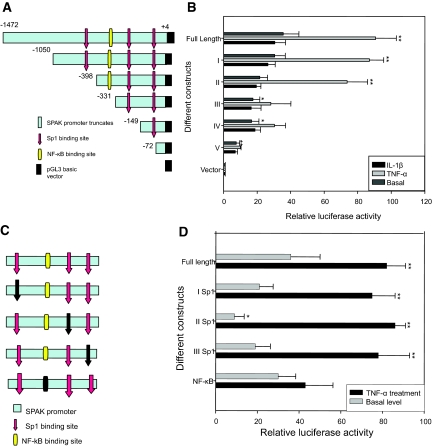
Cloning and Characterization of the Human SPAK Promoter
To understand the molecular mechanisms underlying the regulation of SPAK expression, an ∼1.5-kb fragment of the 5′ flanking region of the SPAK gene was cloned from human genomic DNA using a pair of gene-specific primers: SPAKFor and SPAKRev (Table 1). Sequencing of this 1.5-kb fragment revealed an overlap (100% sequence identity) with the 139-bp portion of the 5′ untranslated region (UTR) of the SPAK mRNA sequence from GenBank (accession no. AY629298) (Supplementary Figure 1, see http://ajp.amjpathol.org). Features of this novel sequence (accession no. DQ537524), including putative cis-acting elements predicted by database analyses (TFSEARCH and TESS), are presented in Supplementary Figure 1, see http://ajp.amjpathol.org. The SPAK promoter appears to be a TATA-less promoter with three Sp1-binding domains at positions −496, −303, and −114, and one NF-κB binding site consensus sequence at position −354 (Figure 4A and Supplementary Figure 1, see http://ajp.amjpathol.org); the two Sp1-binding sites at positions −303 and −114 are embedded in CpG-rich islands. The presence of the basal expression regulator Sp1, the inducible gene expression mediator NF-κB and CpG islands indicate that complex but complementary mechanisms are involved in SPAK regulation.
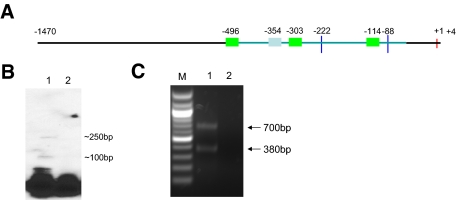
Figure 4.
Characteristics of 5′ flanking region of human SPAK gene. A: Schematic presentation of characteristics of 5′ flanking region of SPAK gene. The red vertical line represents translational initiation site; two blue vertical lines represents the two TSSs; three green bars represent Sp1 binding sites; one gray bar represents NF-κB binding site; the horizontal line represent the 5′ flanking region of SPAK gene, of which the light blue line represents CpG island. The digits indicate the position of corresponding sites. B: Mapping of the TSS by primer extension analysis. Lane 1, Primer extension results with template; lane 2, Primer extension results without template. C: Mapping of the TSS by 5′-RACE. Lane M, 100-bp DNA molecular weight marker; lane 1, gel electrophoresis of PCR products; lane 2, gel electrophoresis of PCR products without template.
To map the transcription start site (TSS) of the human SPAK gene, primer extension was performed using total RNA from Caco2-BBE cells (Figure 4B). Two products were generated using the primer SPAK Rev (Table 1), located upstream of the translational start codon ATG (Figure 4B), indicating the probable existence of two TSSs. To determine the exact location of the TSSs of the human SPAK gene, 5′-rapid amplification of complementary DNA ends (5′-RACE) was performed with total RNA from Caco2-BBE cells and primer SPAKR1(Table 1), and nested PCR was then performed using primer SPAKR2 (Table 1). Two DNA fragments of ∼700 bp and 380 bp were generated (Figure 4C). Sequencing analysis revealed that these two fragments mapped to NCBI reference sequences 88 bp and 222 bp upstream of the ATG codon. Our results indicate these sequences to be the TSSs of the SPAK gene, and the first bases of these sequences are referred to as nucleotides −88 bp and −222 bp in this study.
Sp1-Binding Sites Play a Role in the Basal SPAK Promoter Activities and the NF-κB-Binding Site Is Crucial in TNF-α-Stimulated SPAK Promoter Activities
To identify the core promoter region of SPAK gene, we generated five partial deletion constructs fused with the luciferase reporter gene. Caco2-BBE cells were transiently transfected with these five constructs, then stimulated with or without 40 ng/ml of TNF-α for 16 hours. The transcriptional activities were examined by transient transfection of using the Dual-Luciferase Reporter Assay System (Promega). Construct I displayed ∼35-fold and 83-fold greater basal and TNF-α-stimulated promoter activities, respectively, compared with the control empty pGL3 basic vector (Figure 5, A and B). Construct II, construct III, and construct IV had ∼18-fold higher promoter activity than the pGL3 vector at the basal level, a response that was similar (93%) to that of the full-length SPAK promoter. With TNF-α stimulation, construct II had ∼70-fold greater promoter activity compared with the basic vector pGL3, which was ∼84% of the full-length SPAK promoter activity. However, the promoter activities of constructs III and IV were low under both basal and TNF-α-stimulated conditions (∼40-fold greater than basic vector pGL3), only ∼45% of the full-length SPAK promoter activity. In contrast, construct V had low basal and TNF-α-stimulated promoter activities (Figure 5B). In short, the promoter activity of construct II was similar (∼80-fold increase compared with the basic vector pGL3) to that of construct I (Figure 5B), whereas constructs III, IV, and V did not show significant increases in TNF-α-stimulated promoter activity compared with controls (Figure 5B). Together, these results indicate that elements responsible for the basal and TNF-α-stimulated promoter activities of the SPAK gene are both in the full-length SPAK promoter and the truncate construct II (Figure 5, A and B). However, constructs III and IV, which contain Sp1-binding sites but not NF-κB-binding sites, show reduced TNF-α-stimulated promoter activities and slightly reduced basal activities compared with the full-length SPAK promoter. Finally, construct V, which has neither NF-κB- nor Sp1-binding sites, shows minimal basal activity of the SPAK promoter. To determine the level of specificity of TNF-α-induced response, Caco2 cells transfected with SPAK promoter constructs were treated with 10 ng/ml of IL-1β; luciferase assay did not show significant different promoter activity, which means TNF-α specifically induces SPAK promoter activity. Together, these results indicate that the Sp1-binding sites play a role in basal promoter activities, and the NF-κB-binding site is crucial for TNF-α-stimulated SPAK promoter activities.
Figure 5.
Characterization of SPAK promoter: A: Schematic representation of human SPAK promoter constructs. The full-length SPAK promoter (nt −1472 to +4); construct I (nt −1050 to +4); construct II (nt −398 to +4); construct III (nt −331 to +4); construct IV (nt −149 to +4); and construct V (nt −72 to +4). Numbers are given in relation to the translational start codon (+1) and indicate 5′-ends of the deletion constructs. The location of the identified positive regulatory region is indicated by a light blue box. Positions of the putative Sp1 (red) and NF-κB (yellow) sites are indicated by arrows. B: Promoter activities of the 5′ deleted constructs in untreated or TNF-α- and IL-1β-stimulated Caco2-BBE cells normalized to Renilla Luc activities driven by the phRL-CMV control vector. Activities are expressed as fold inductions over cells transfected with the empty pGL3-basic vector. Each value represents the mean ± SD of at least three independent sets of transfection experiments performed in triplicate. *P < 0.05, **P < 0.01. C: Schematic representation of mutated SPAK promoter constructs: the full-length SPAK promoter; I Sp1 binding site (−496); II Sp1 binding site (−303); III Sp1 binding site (−114); and NF-κB binding site (−354). The digits are given in relation to the translational start codon (+1). The location of the identified positive regulatory region is indicated by a light blue box. Positions of the putative Sp1 sites are indicated by arrows and NF-κB is indicated by a rectangle. The corresponding mutated transcription factor binding site is indicated by a black arrow or black rectangle. D: Effects of mutations of Sp1 or NF-κB binding sites on SPAK promoter activity. The various mutated constructs were transiently transfected into Caco2-BBE cells under the basal (gray bar) or TNF-α-stimulated promoter activities of the full-length wild-type construct were set to 100% (control). Values represent means ± SD of at least three independent sets of transfection experiments performed in triplicate. *P < 0.05, **P < 0.01.
To investigate and confirm the functional role of the relevant binding motifs in regulating SPAK promoter activity, we generated various Sp1- and NF-κB-binding mutants (Figure 5C) with relevant primers (Table 1). With the transcriptional activity assay, we found that all of the Sp1 mutants had significantly reduced basal promoter activity levels compared with the wild-type promoter, whereas the TNF-α-stimulated activity levels of these Sp1 mutants were similar to wild-type promoter activity levels (Figure 5, C and D). In contrast, although the NF-κB mutant basal luciferase activity was similar to that of the wild type, there was a marked reduction in TNF-α-stimulated promoter activity (∼50% reduction) (Figure 5D). Taken together, these results show that the Sp1-binding sites are important in basal SPAK promoter activities, and the NF-κB-binding site has a crucial role in TNF-α-stimulation SPAK promoter activities.
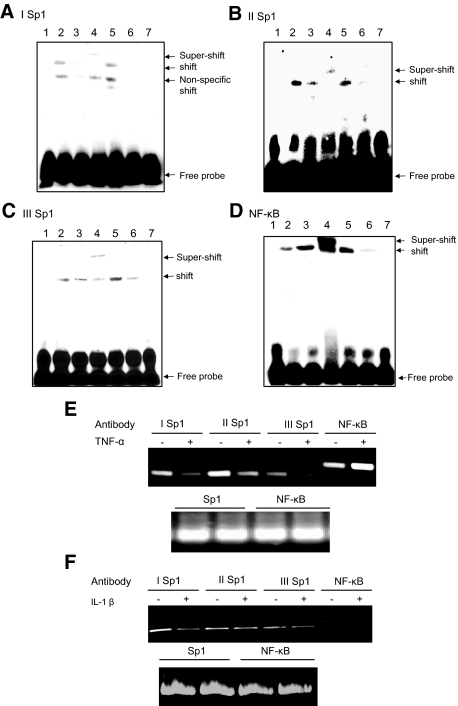
The Transcription Factors Sp1 and NF-κB Are Physically Associated with Their Corresponding Binding Sites in the SPAK Promoter
To study the association between the transcription factors and the corresponding binding sequences, and further confirm the importance of Sp1 and NF-κB for activation of the SPAK gene, we used EMSA to characterize the binding of the transcription factors Sp1 and NF-κB to their respective binding sites: Sp1 −496 (Figure 6A), Sp1 −303 (Figure 6B), Sp1 −114 (Figure 6C), and NF-κB −354 (Figure 6D). EMSA revealed that incubation of the DNA-protein complexes with anti-Sp1 or anti-p65 antibodies shifted the migrating bands in an upward direction, indicating specificity to Sp1 and NF-κB (p65) proteins (Figure 6, A–D; lane 4). Surprisingly, under treatment with TNF-α (40 ng/ml) for 16 hours, we noticed that the binding of Sp1 to the corresponding oligonucleotides was much weaker compared with the untreated control (Figure 6, A–C; lane 3). This indicates that TNF-α might reduce Sp1 expression levels or inhibit the binding of Sp1 to oligonucleotides.30 However, as shown in Figure 6C, TNF-α treatment increases NF-κB (p65) binding to the corresponding oligonucleotides.
Figure 6.
EMSA of I Sp1 (−496) (A), II Sp1 (−303) (B), III Sp1 (−114) (C), NF-κB (−354) (D). Lane 1, biotin-labeled oligonucleotide alone; lane 2, biotin-labeled oligonucleotides incubated with 5 μg of Caco2-BBE nuclear extracts; lane 3, biotin-labeled oligonucleotides incubated with 5 μg of TNF-α-treated Caco2-BBE nuclear extracts; lane 4, biotin-labeled oligonucleotides incubated with 5 μg of Caco2-BBE nuclear extracts in the presence of anti-Sp1 (A–C) or NF-κB (p65) (D) antibodies; lane 5, biotin-labeled oligonucleotides incubated with 5 μg of Caco2-BBE nuclear extracts in the presence of nonspecific IgG; lane 6, biotin-labeled oligonucleotides incubated with 5 μg of Caco2-BBE nuclear extracts in the presence of a 50-fold excess of cold competitor oligonucleotide; lane 7, biotin-labeled binding site-mutated oligonucleotides incubated with 5 μg of Caco2-BBE nuclear extracts. ChIP assay: the antibodies indicated were incubated with cross-linked DNA isolated from Caco2-BBE cells treated with (+) or without (−) TNF-α (E) or IL-1β (F). Sp1 (I, II, and III) and NF-κB promoter elements in the immunoprecipitates were detected by PCR using the specific primers shown in Table 1. The bottom panel uses DNA input as template for internal control.
To confirm the in vivo importance of Sp1- and NF-κB (p65)-binding sites in response to TNF-α in Caco2-BBE cells, we performed ChIP analysis. As shown in Figure 6E, under resting conditions, the transcription factor Sp1 binds to the I Sp1 (lane 1), II Sp1 (lane 3), and III Sp1 (lane 5) binding sites and the transcription factor NF-κB (p65) binds to the NF-κB binding site (lane 7). After TNF-α treatment, the DNA binding activities of I Sp1 (lane 2), II Sp1 (lane 4), and III Sp1 (lane 6) are reduced compared with untreated cells. However, the NF-κB (p65) (lane 6) binding site showed a more-dense band, which indicates that more NF-κB (p65) binds to this site. However, if Caco2-BBE cells are treated with irrelevant cytokine IL-1β, no significant different dense bands are shown either for Sp1 or NF-κB binding sites both at the resting or treated conditions (Figure 6F). Together, these results indicate that Sp1 has an important role in basal SPAK promoter activity and NF-κB has a role in TNF-α-stimulated SPAK promoter activity.
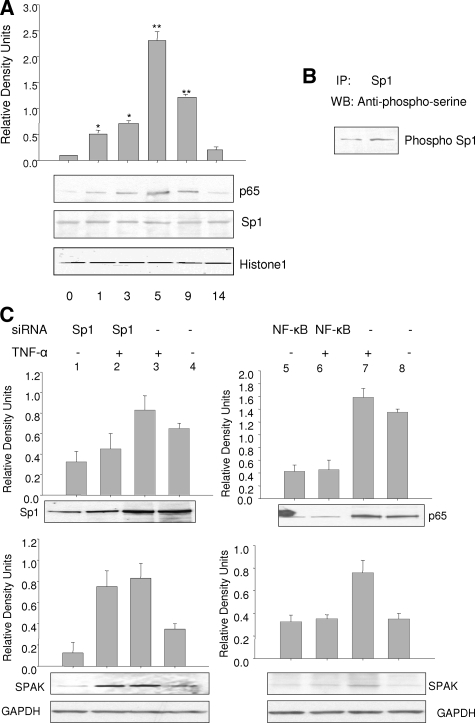
DSS Increases SPAK Expression via Increased NF-κB Expression
To confirm the effects of DSS stimulation on SPAK expression, nuclear protein was extracted from colonic epithelial cells from mice with colitis induced by DSS and subjected to Western blot analysis with relevant antibodies. As shown in Figure 7A, after different durations of DSS treatment, there was no significant change in Sp1 expression level; however, expression of NF-κB (p65) reached a maximum level after 5 days, then returned to basal levels after withdrawal of DSS treatment. These in vivo data show that DSS does not alter overall nuclear levels of Sp1. In contrast, DSS treatment can translocate NF-κB (p65) to the nucleus and up-regulate its expression as well. These results indicate that DSS increases expression of NF-κB-dependent SPAK expression.
Figure 7.
A: Western blots of Sp1 and NF-κB (p65) demonstrating DSS effect on Sp1 and NF-κB protein levels. Histone 1 acts as a control. *P < 0.05, **P < 0.01. B: Co-immunoprecipitation assay of TNF-α increasing phosphorylation of Sp1. C: Reduction of NF-κB not Sp1 expression reduced SPAK protein expression in unstimulated and in TNF-α-stimulated Caco2-BBE cells. Cells were harvested and subjected to Western blot analysis using Sp1, NF-κB (p65), and SPAK antibodies as described in the Materials and Methods. GAPDH acts as a loading control.
Reduction of Sp1 or NF-κB Expression Differentially Regulates SPAK Expression
One approach to study the functional role of a specific protein is to knockdown its expression. As shown in Figure 7C, Caco2-BBE cells transfected with siRNA against Sp1 and NF-κB (p65) showed lower expression levels of the targeted proteins Sp1 (lanes 1 and 2) and NF-κB (p65) (lanes 5 and 6) compared with Caco2-BBE cells transfected with scrambled control Sp1 siRNA (lanes 3 and 4) and NF-κB siRNA (lanes 7 and 8). Basal SPAK protein expression was also significantly reduced in Caco2-BBE cells transfected with Sp1 siRNA (SPAK lane 1 versus lane 4); however, there were no significant reductions in SPAK expression with NF-κB (p65) siRNA (SPAK lane 5 versus lane 8). Thus, not like NF-κB (p65) siRNA, Sp1 siRNA effectively reduces basal SPAK expression in Caco2-BBE cells by reducing expression of its target protein Sp1. We further investigated whether siRNA against Sp1 and/or against NF-κB reduced TNF-α-induced SPAK expression. As shown in Figure 7C, there were significant increases in SPAK expression in TNF-α-stimulated Caco2-BBE cells transfected with Sp1 siRNA but not in cells transfected with NF-κB siRNA (SPAK lanes 6 and 8, respectively), when compared with cells transfected with scrambled Sp1 siRNA (SPAK lane 3) and p65 (SPAK lane 7).
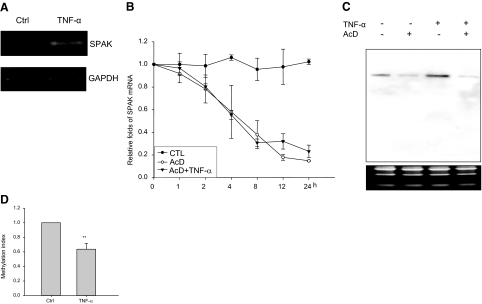
Induction of SPAK Is Primarily at the Transcriptional Level
To determine whether the induction of SPAK gene expression by TNF-α was mediated through transcriptional or posttranscriptional mechanisms, nuclear run-on assays (Figure 8A) were performed on Caco2-BBE cells pretreated with or without TNF-α. Data demonstrate that after stimulation with TNF-α, SPAK mRNA was increased compared to the mRNA transcribed from nuclei without TNF-α treatment. This experiment was repeated three times with the same result. These results indicate that the TNF-α-induced increase in SPAK mRNA is transcriptional.
Figure 8.
A: Nuclear run-on assay indicated the increase of SPAK mRNA transcription under the treatment of TNF-α, with the mRNA transcription of GAPDH as internal control. B: TNF-α does not change SPAK mRNA stability, the percentage of remaining SPAK mRNA is shown at the different time points. Filled circle represents the value of real-time PCR with the samples from Caco2-BBE cells without treatment, open circle represents the value of real-time PCR with the samples from Caco2-BBE treated with actinomycin D, solid triangle represents real-time PCR with the samples from Caco2-BBE treated with actinomycin D and TNF-α. C: Northern blot analysis of total RNA from Caco2-BBE cells. Lane 1, no treatment; lane 2, AcD; lane 3, TNF-α; and lane 4, AcD and TNF-α. The lower RNA electrophoresis shows equal loading of each condition. D: Methylation assay of CpG island in SPAK promoter. *P < 0.05 versus control.
The Increase in SPAK Transcription by TNF-α Is Not Mediated by Alteration of mRNA Stability
Changes in steady-state mRNA levels may be attributable to changes in the degradation rate of a transcript and/or rate of gene transcription. Hence, it was important to investigate the relative contribution of a posttranscriptional mechanism in the modulation of SPAK mRNA levels by TNF-α. To measure SPAK mRNA stability, Caco2-BBE cell lines were treated with 5 ng/ml of AcD to inhibit mRNA synthesis, and SPAK mRNA levels were measured at the indicated time points in the presence and absence of 40 ng/ml of TNF-α by real-time RT-PCR. 18S rRNA was used as an internal control to normalized SPAK mRNA levels. The half-life of SPAK mRNA (t1/2 = ∼240 minutes) in AcD-treated Caco2-BBE cells was almost the same as that of SPAK mRNA in cells treated with AcD plus TNF-α (t1/2 = ∼250 minutes), no significant difference was detected (Figure 8B). In parallel, Northern blots were performed with 20 μg of Caco2-BBE mRNA treated with or without AcD (5 ng/ml) and/or TNF-α (Figure 8C). Results showed that AcD can significantly reduce mRNA levels (Figure 8C, lane 2) compared with untreated Caco2-BBE cells (Figure 8C, lane 1), TNF-α treatment (without AcD treatment) markedly increased levels of SPAK mRNA transcripts. However, if Caco2-BBE cells were treated with AcD 1 hour before TNF-α treatment, TNF-α cannot reverse the AcD-induced inhibition. Together, these findings indicate that the observed changes in SPAK protein level are attributable to increased levels of SPAK mRNA transcripts, not because of changes in mRNA stability.
CpG Island in the SPAK Promoter Was Demethylated during Inflammation
As we have shown previously, there is one CpG island in the 5′ flanking region of the SPAK gene. CpG islands are often located near the promoters of genes that typically lack TATA core promoter elements but might contain multiple GC box motifs that bind Sp1 and related transcription factors.31,32 Analysis of 1031 human genes revealed that approximately half of the potential promoter regions are located in CpG islands.33 Regulation of CpG islands, particularly the methylation of CpG islands, affects the activity of adjacent genes and is critical to the regulation of gene expression. As shown in Figure 8D, the methylation index is reduced in Caco2-BBE cells stimulated by TNF-α treatment. In other words, TNF-α treatment of Caco2-BBE cells may significantly demethylate the CpG island in the SPAK promoter compared with untreated cells, which is consistent with recent results showing that TNF-α can demethylate the 5′-long terminal repeat (LTR) of the HTLV-1 provirus34,35 when it induces its expression.
Discussion
In this study, we demonstrate for the first time that colonic epithelial SPAK expression is increased in IBD patients and in mice with experimentally induced colitis. Importantly, we have also found that the pro-inflammatory cytokine TNF-α increases colonic SPAK expression, which is in agreement with studies that have shown that TNF-α may regulate the expression of other Ste20-like kinases. For example, TNF-α increased Map4k4 expression levels through TNF-α receptor-1 signaling to c-Jun.36 Based on these findings, we speculated that colonic SPAK might play a role in multifactorial IBD. To study the mechanisms by which SPAK expression is regulated during intestinal inflammation, we characterized the SPAK promoter and its regulation by TNF-α.
We first cloned the 5′-flanking region of the colonic SPAK gene from the epithelial cell line Caco2-BBE. Computer analysis indicates that this region contains a CpG island, three potential Sp1 binding sites, one NF-κB binding site, and two TSSs located at −88 bp and −222 bp upstream of the first ATG, but no TATA or CCAAT boxes. Several studies have investigated the incidence of TATA-less human promoters and have yielded apparently conflicting results from 11% up to 76%.37 More than half of these TATA-less promoters have CpG islands and contain more than one weak TSS, which may have different functions.38 The presence of the basal expression regulator Sp1, inducible gene expression mediator NF-κB and CpG islands indicate that SPAK regulation may involve multiple complex and complementary mechanisms. Sp1, one of the first isolated and characterized eukaryotic transcription factors,39 is a sequence-specific and ubiquitously expressed zinc finger element that supports constitutive basal expression of a variety of eukaryotic genes that specially lack a functional TATA box and is believed to have a key role in maintaining expression of housekeeping genes.19,40,41 In addition, there is now emerging evidence that shows the function of Sp1 in modulating specific gene expression.42 The NF-κB family forms a variety of hetero- and homodimers with its subunits to differentially control gene expression elicited by cytokines, bacterial products, viral expression, growth factors, and stress stimuli.43 Increased activation of NF-κB subunit p65 has been shown to be involved in the regulation of inflammatory responses in both CD and UC.44,45 Literature also showed that in CD and UC, NF-κB precursor p105 is processed by proteasomes to active form p50.45 However no studies demonstrated that precursor p100 was cleaved to its active form p52 during IBD; furthermore we did Western blot with p100/p52 antibody (Cell Signaling technology Inc.) to check the p100/p52 status in DSS mouse model of colitis induced during the period of DSS treatment, we only saw the precursor p100 form, not the p52 (data not shown). So NF-κB heterodimer p65/p50 is the form that seems to play a crucial role in the regulation of inflammatory responses.
Here, EMSA and ChIP assay show that TNF-α treatment reduces the binding of Sp1 to its binding site, this unanticipated result could be attributable to NF-κB binding to Sp1 binding site owing to the similar consensus sequence. If this was the case, increased binding of NF-κB under TNF-α treatment would be expected to compete with and prevent the binding of Sp1, thereby reducing Sp1 binding. This possibility is supported by the results of previous studies that demonstrated such an interaction in IL-6 and P-selectin promoters.46,47 The reduced levels of Sp1 binding with TNF-α stimulation could also be attributable to TNF-α-induced Sp1 phosphorylation (Figure 7B) because Sp1 phosphorylation could reduce Sp1 binding48 and dephosphorylation of Sp1 has been suggested to enhance Sp1 DNA binding activity.49
In DSS-treated mice, we found that DSS does not alter overall nuclear levels of Sp1, similar studies showed that TNF-α did not change Sp1 expression30 but increased NF-κB (p65) expression.50 Similarly, DSS treatment can up-regulate the expression of NF-κB (p65) in colon epithelial cells, and more importantly, DSS treatment can translocate p65 protein into nucleus as active form, which suggests that the increased expression of SPAK might be attributable to the increased nuclear protein NF-κB (p65). Our data also showed that knock-down of NF-κB (p65) by siRNA will decrease SPAK expression significantly, which implied the potential of NF-κB as a good target against inflammatory diseases such as IBD.51,52,53,54,55 Hoffmann and Baltimore56 pointed out that almost all genes induced by inflammatory stimuli are transcriptionally up-regulated in an NF-κB-dependent manner in all of the cell types examined so far, including gene-encoding proinflammatory cytokines, IL-1, IL-6, IL-12, TNF-α, IFN-γ, IL-8, chemokine MIP-1, and adhesion molecules ICAM.57,58,59 We showed here that NF-κB is crucial to activate SPAK gene expression, however, we cannot exclude the possibility that NF-κB acts in concert with other transcription factors or co-activators to affect chromatin remodeling, recruitment of the transcription initiation machinery, and/or the elongation process.60,61
As we know from our study, TNF-α treatment can enhance the SPAK mRNA in a time- and dose-dependent manner, but this increase in RNA abundance could be brought about either by an increase in transcription, a decrease in the rate of degradation, or both. To distinguish between these possibilities, the transcription rate should be measured by nuclear run-on assay. By this method, per mRNA levels62 in Drosophila were found to be regulated at a posttranscriptional level without evident transcriptional regulation. In contrast, another nuclear run-on assay63 demonstrated that inhibition of both Erk and p38 kinase pathways simultaneously resulted in a decrease in cytokine gene transcription (IL-6 and TNF-α) to near-control levels. Here our data also show that a short time treatment (6 hours) of TNF-α increases SPAK mRNA transcription.
In the study, we found that TNF-α demethylates the CpG island in the SPAK promoter, which is consistent with a recent study in which TNF-α was shown to demethylate the 5′-long terminal repeat (LTR) of HTLV-1 provirus when TNF-α induces its expression.35 DNA methylation has been found involving in intestinal inflammation. High levels of methylation for ER, MYOD, p16 exon 1, and CSPG2 in UC patients have been reported.64,65,66 To silence the joint gene expression and maintain the homeostasis, CpG methylation is normally combined with the modification of certain molecules. For example, in APRT gene, the modification of Sp1 plays a role in maintenance of housekeeping genes by preventing joint gene silencing by DNA methylation.31,32,67 There are different mechanisms by which TNF-α regulates the expression of target genes. For one, TNF-α increases the expression of secreted phospholipases A2 (PLA2), c-fos, and c-jun by increasing its promoter activity68,69; in contrast, TNF-α induced increases of Clara cell secretory protein (CCSP) mRNA and wild-type p53-activated fragment 1 (WAF1) at posttranscriptional levels70,71; here, mRNA half-life assay indicated that TNF-α up-regulates SPAK expression at the transcriptional level.
It has been shown that with different doses of TNF (40 ng/ml, 50 ng/ml, and 100 ng/ml), with different time treatments of TNF (24 hours and 48 hours) in different colon cell lines (Caco2, HT29), TNF itself cannot cause obvious apoptosis.72,73,74 On the other hand, the combination of TNF with other cytokine such as IFN,75 or with protein synthesis inhibitor cycloheximide,76 or tyrosine kinase inhibitor73 can induce strong apoptosis. Furthermore, we performed Western blots on TNF-α treated Caco2-BBE cells with caspase3 antibody, we could not find the cleavage of caspase3 either (data not shown). However, we agree with the reviewer that DSS can also cause apoptosis.77 Here we cannot rule out the possibility that the increase of SPAK expression is partially due to the DSS-induced apoptosis, as well as inflammation.
In the context of intestinal inflammation such as IBD, recent studies have identified at least two signaling cascades that are activated preferentially by the inflammatory cytokines TNF-α, IL-1β, and IL-6, as well as by a wide variety of cellular stresses such as UV and ionizing radiation, hyperosmolarity, heat stress, and oxidative stress.22,78 It has been reported that ste20 is activated by at least three pathogen-associated molecular patterns, namely lipopolysaccharide, peptidoglycan, and flagellin. These pathogen-associated molecular patterns are produced by invading microbial pathogens and initiate innate immune responses by binding to pattern recognition receptors, pathogen-associated molecular patterns can also activate GCK signals through MLK-2 and -3 to recruit the JNK p38 and their effectors.79
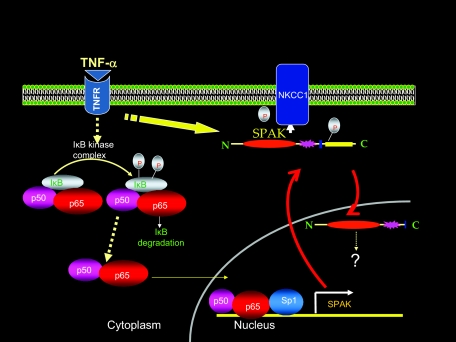
In conclusion, we report that during inflammatory conditions (such as IBDs, mouse model of colitis induced by DSS, and TNF-α-treated Caco2-BBE cells), the expression of SPAK was significantly up-regulated. This expression augmentation is accredited to a combination of multiple factors, including more NF-κB translocation to the nucleus, phosphorylation of Sp1, and demethylation of the CpG in the SPAK promoter. We speculate that the enhanced SPAK exported into the cytoplasm after being phosphorylated and activated, then was cleaved and translocated to the nucleus, leading to different functions such as cell differentiation, cell transformation and proliferation, cytoskeleton rearrangement, cell volume regulation, and intestinal epithelial permeability (Figure 9).11 Regulation of expression of SPAK and other ste20 kinases, and development of compounds that can modulate or disrupt the activity of intracellular pathways, are important for control or attenuation of downstream pathological responses in IBD.
Figure 9.
Proposed model of activation and regulation of SPAK expression.
Footnotes
Address reprint requests to Didier Merlin, Ph.D., Emory University, Department of Medicine, Division of Digestive Diseases, 615 Michael St., Atlanta, GA 30322. E-mail: dmerlin@emory.edu.
Supported by the National Institute of Diabetes and Digestive and Kidney Diseases (grants R24-DK-064399 to the center, RO1-DK-061941, RO1-DK-071594 to D.M., RO1-DK55850 to S.V.S.) and the Crohn’s and Colitis Foundation of America (research fellowship award to Y.Y.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Goyette P, Labbé C, Trinh TT, Xavier RJ, Rioux JD. Molecular pathogenesis of inflammatory bowel disease: genotypes, phenotypes and personalized medicine. Ann Med. 2007;39:177–199. doi: 10.1080/07853890701197615. [DOI] [PubMed] [Google Scholar]
- Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, Griffiths AM, St George-Hyslop PH, Siminovitch KA. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- Stoll M, Corneliussen B, Costello CM, Waetzig GH, Mellgard B, Koch WA, Rosenstiel P, Albrecht M, Croucher PJ, Seegert D, Nikolaus S, Hampe J, Lengauer T, Pierrou S, Foelsch UR, Mathew CG, Lagerstrom-Fermer M, Schreiber S. Genetic variation in DLG5 is associated with inflammatory bowel disease. Nat Genet. 2004;36:476–480. doi: 10.1038/ng1345. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM. Signaling by the germinal center kinase family of protein kinases. J Biol Chem. 1999;274:5259–5262. doi: 10.1074/jbc.274.9.5259. [DOI] [PubMed] [Google Scholar]
- Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- Pombo CM, Bonventre JV, Molnar A, Kyriakis J, Force T. Activation of a human Ste20-like kinase by oxidant stress defines a novel stress response pathway. EMBO J. 1996;15:4537–4546. [PMC free article] [PubMed] [Google Scholar]
- Endo J, Toyama-Sorimachi N, Taya C, Kuramochi-Miyagawa S, Nagata K, Kuida K, Takashi T, Yonekawa H, Yoshizawa Y, Miyasaka N, Karasuyama H. Deficiency of a STE20/PAK family kinase LOK leads to the acceleration of LFA-1 clustering and cell adhesion of activated lymphocytes. FEBS Lett. 2000;468:234–238. doi: 10.1016/s0014-5793(00)01219-9. [DOI] [PubMed] [Google Scholar]
- Wagner S, Flood TA, O'Reilly P, Hume K, Sabourin LA. Stk10, a new member of the polo-like kinase kinase family highly expressed in hematopoietic tissue. J Biol Chem. 2002;277:37685–37692. [Google Scholar]
- Stockton R, Reutershan J, Scott D, Sanders J, Ley K, Schwartz MA. Induction of vascular permeability: beta PIX and GIT1 scaffold the activation of extracellular signal-regulated kinase by PAK. Mol Biol Cell. 2007;18:2346–2355. doi: 10.1091/mbc.E06-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AM, Naselli G, Gonez LJ, Martin RM, Harrison LC, DeAizpurua HJ. SPAK, a STE20/SPS1-related kinase that activates the p38 pathway. Oncogene. 2000;19:4290–4297. doi: 10.1038/sj.onc.1203784. [DOI] [PubMed] [Google Scholar]
- Yan Y, Nguyen H, Dalmasso G, Sitaraman SV, Merlin D. Cloning and characterization of a new intestinal inflammation-associated colonic epithelial Ste20-related protein kinase isoform. Biochim Biophys Acta. 2007;1769:106–116. doi: 10.1016/j.bbaexp.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Li Y, Hu J, Vita R, Sun B, Tabata H, Altman A. SPAK kinase is a substrate and target of PKCtheta in T-cell receptor-induced AP-1 activation pathway. EMBO J. 2004;23:1112–1122. doi: 10.1038/sj.emboj.7600125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechotta K, Lu J, Delpire E. Cation chloride cotransporters interact with the stress-related kinases Ste20-related proline-alanine-rich kinase (SPAK) and oxidative stress response 1 (OSR1). J Biol Chem. 2002;277:50812–50819. doi: 10.1074/jbc.M208108200. [DOI] [PubMed] [Google Scholar]
- Dowd BF, Forbush B. PASK (proline-alanine-rich STE20-related kinase), a regulatory kinase of the Na-K-Cl cotransporter (NKCC1). J Biol Chem. 2003;278:27347–27353. doi: 10.1074/jbc.M301899200. [DOI] [PubMed] [Google Scholar]
- Gagnon KB, England R, Delpire E. Characterization of SPAK and OSR1, regulatory kinases of the Na-K-2Cl cotransporter. Mol Cell Biol. 2006;26:689–698. doi: 10.1128/MCB.26.2.689-698.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper JN, Wasserman SM, Anderson KR, Cai J, Falb D, Gimbrone MA., Jr Expression of the bumetanide-sensitive Na-K-Cl cotransporter BSC2 is differentially regulated by fluid mechanical and inflammatory cytokine stimuli in vascular endothelium. J Clin Invest. 1997;99:2941–2949. doi: 10.1172/JCI119489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Pace AJ, Koller BH. Mice lacking NKCC1 is protected from development of bacteremia and hypothermic sepsis secondary to bacterial pneumonia. J Exp Med. 2007;204:1383–1393. doi: 10.1084/jem.20061205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Supakar PC, Vellanoweth RL, Song CS, Chatterjee B, Roy AK. Functional role of a conformationally flexible homopurine/homopyrimidine domain of the androgen receptor gene promoter interacting with Sp1 and a pyrimidine single strand DNA-binding protein. Mol Endocrinol. 1997;11:3–15. doi: 10.1210/mend.11.1.9868. [DOI] [PubMed] [Google Scholar]
- Badger AM, Cook MN, Lark MW, Newman-Tarr TM, Swift BA, Nelson AH, Barone FC, Kumar S. SB 203580 inhibits p38 mitogen-activated protein kinase, nitric oxide production, and inducible nitric oxide synthase in bovine cartilage-derived chondrocytes. J Immunol. 1998;161:467–473. [PubMed] [Google Scholar]
- Craxton A, Shu G, Graves JD, Saklatvala J, Krebs EG, Clark EA. p38 MAPK is required for CD40-induced gene expression and proliferation in B lymphocytes. J Immunol. 1998;161:3225–3236. [PubMed] [Google Scholar]
- Hollenbach E, Neumann M, Vieth M, Roessner A, Malfertheiner P, Naumann M. Inhibition of p38 MAP kinase- and RICK/NF-kappaB-signaling suppresses inflammatory bowel disease. FASEB J. 2004;18:1550–1552. doi: 10.1096/fj.04-1642fje. [DOI] [PubMed] [Google Scholar]
- Gnoni GV, Geelen MJ, Bijleveld C, Quagliariello E, van den Bergh SG. Short-term stimulation of lipogenesis by triiodothyronine in maintenance cultures of rat hepatocytes. Biochem Biophys Res Commun. 1985;128:525–530. doi: 10.1016/0006-291x(85)90078-6. [DOI] [PubMed] [Google Scholar]
- Nwagwu M, Nana M. Ribonucleic acid synthesis in embryonic chick muscle, rates of synthesis and half-lives of transfer and ribosomal RNA species. J Embryol Exp Morphol. 1980;56:253–267. [PubMed] [Google Scholar]
- Coffee B, Muralidharan K, Highsmith WE, Jr, Lapunzina P, Warren ST. Molecular diagnosis of Beckwith-Wiedemann syndrome using quantitative methylation-sensitive polymerase chain reaction. Genet Med. 2006;8:628–634. doi: 10.1097/01.gim.0000237770.42442.cc. [DOI] [PubMed] [Google Scholar]
- Dieleman LA, Palmen MJ, Akol H, Bloemena E, Peña AS, Meuwissen SG, Van Rees EP. Chronic experimental colitis induced by dextran sulphate sodium (DSS) is characterized by Th1 and Th2 cytokines. Clin Exp Immunol. 1998;114:385–391. doi: 10.1046/j.1365-2249.1998.00728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- Targan SR, Hanauer SB, van Deventer SJ, Mayer L, Present DH, Braakman T, DeWoody KL, Schaible TF, Rutgeerts PJ. A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor alpha for Crohn’s disease. Crohn’s Disease cA2 Study Group. N Engl J Med. 1997;337:1029–1035. doi: 10.1056/NEJM199710093371502. [DOI] [PubMed] [Google Scholar]
- Denson LA, Menon RK, Shaufl A, Bajwa HS, Williams CR, Karpen SJ. TNF-α down-regulates murine hepatic growth hormone receptor expression by inhibiting Sp1 and Sp3 binding. J Clin Invest. 2001;107:1451–1458. doi: 10.1172/JCI10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Tsunoda T, Sese J, Taira H, Mizushima-Sugano J, Hata H, Ota T, Isogai T, Tanaka T, Nakamura Y, Suyama A, Sakaki Y, Morishita S, Okubo K, Sugano S. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 2001;11:677–684. doi: 10.1101/gr.164001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gonzalo M, Carretero M, Rullas J, Lara-Pezzi E, Aramburu J, Berkhout B, Alcamí J, López-Cabrera M. The hepatitis B virus X protein induces HIV-1 replication and transcription in synergy with T-cell activation signals: functional roles of NF-kappaB/NF-AT and SP1-binding sites in the HIV-1 long terminal repeat promoter. J Biol Chem. 2001;276:35435–35443. doi: 10.1074/jbc.M103020200. [DOI] [PubMed] [Google Scholar]
- Ishida T, Hamano A, Koiwa T, Watanabe T. 5′ Long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology. 2006;3:69–75. doi: 10.1186/1742-4690-3-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesz GJ, Guilherme A, Guntur KV, Hubbard AC, Tang X, Chawla A, Czech MP. Tumor necrosis factor alpha (TNFalpha) stimulates Map4k4 expression through TNFalpha receptor 1 signaling to c-Jun and activating transcription factor 2. J Biol Chem. 2007;282:19302–19312. doi: 10.1074/jbc.M700665200. [DOI] [PubMed] [Google Scholar]
- Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATAless core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WC, Geng Y, Johnson LF. In vitro transcription of the TATAA-less mouse thymidylate synthase promoter: multiple transcription start points and evidence for bidirectionality. Gene. 1994;146:183–189. doi: 10.1016/0378-1119(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Dynan WS, Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983;32:669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Briggs MR, Kadanoga JT, Bell SP, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Saffer JD, Jackson SP, Annarella MB. Developmental expression of Sp1 in the mouse. Mol Cell Biol. 1991;11:2189–2199. doi: 10.1128/mcb.11.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Kadonaga JT, Luciw PA, Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science. 1986;232:755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Schreiber S, Nikolaus S, Hampe J. Activation of nuclear factor kappa B inflammatory bowel disease. Gut. 1998;42:477–484. doi: 10.1136/gut.42.4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visekruna A, Joeris T, Seidel D, Kroesen A, Loddenkemper C, Zeitz M, Kaufmann SH, Schmidt-Ullrich R, Steinhoff U. Proteasome-mediated degradation of IkappaBalpha and processing of p105 in Crohn disease and ulcerative colitis. J Clin Invest. 2006;116:3195–3203. doi: 10.1172/JCI28804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancéau J, Kaisho T, Hirano T, Wietzerbin J. Triggering of the human interleukin-6 gene by interferon-gamma and tumor necrosis factor-alpha in monocytic cells involves cooperation between interferon regulatory factor-1, NF kappa B, and Sp1 transcription factors. J Biol Chem. 1995;270:27920–27931. doi: 10.1074/jbc.270.46.27920. [DOI] [PubMed] [Google Scholar]
- Hirano F, Tanaka H, Hirano Y, Hiramoto M, Handa H, Makino I, Scheidereit C. Functional interference of Sp1 and NF-kappaB through the same DNA binding site. Mol Cell Biol. 1998;18:1266–1274. doi: 10.1128/mcb.18.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Ferro TJ. Identification of a hydrogen peroxide-induced PP1-JNK1-Sp1 signaling pathway for gene regulation. Am J Physiol. 2006;291:L983–L992. doi: 10.1152/ajplung.00454.2005. [DOI] [PubMed] [Google Scholar]
- Borellini F, Aquino A, Josephs SF, Glazer RI. Increased expression and DNA-binding activity of transcription factor Sp1 in doxorubicin-resistant HL-60 leukemia cells. Mol Cell Biol. 1990;10:5541–5547. doi: 10.1128/mcb.10.10.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BN, Tirumurugaan KG, Deshpande DA, Amrani Y, Panettieri RA, Walseth TF, Kannan MS. Transcriptional regulation of CD38 expression by tumor necrosis factor-alpha in human airway smooth muscle cells: role of NF-kappaB and sensitivity to glucocorticoids. FASEB J. 2006;20:1000–1002. doi: 10.1096/fj.05-4585fje. [DOI] [PubMed] [Google Scholar]
- Scheinman RI, Cogswell PC, Lofuist AK, Baldwin AS. Role of transcriptional activation of IκB-α in mediation of immunosuppression by glucocorticoids. Science. 1995;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- Auphan N, DiDonato JA, Rosette C, Helmberg A, Karin M. Immunosuppression by glucocorticoids: inhibition of NF-κB activity through induction of IκB synthesis. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- Lenardo MJ, Baltimore D. NF-kappa B: a pleiotropic mediator of inducible and tissue specific gene control. Cell. 1989;58:227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- Lenardo MJ, Fan CM, Maniatis T, Baltimore D. The involvement of NF-kappa B in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- Kopp E, Ghosh S. Inhibition of NF-κB by sodium salicylate and aspirin. Science. 1994;265:956–959. doi: 10.1126/science.8052854. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Baltimore D. Circuitry of nuclear factor κB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- Kunsch C, Rosen C. NF-κB subunit-specific regulation of the interleukin-8 promoter. Mol Cell Biol. 1993;13:6137–6146. doi: 10.1128/mcb.13.10.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Zhang G, Ghosh S. Toll-like receptor-mediated NF-κB activation: a phylogenetically conserved paradigm in innated immunity. J Clin Invest. 2001;107:13–19. doi: 10.1172/JCI11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard KA, Rose DW, Haque ZK, Kurokawa R, McInerney E, Westin S, Thanos D, Rosenfeld MG, Glass CK, Collins T. Transcriptional activation by NF-κB requires multiple coactivators. Mol Cell Biol. 1999;19:6367–6378. doi: 10.1128/mcb.19.9.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr LD, Ransone LJ, Wamsley P, Schmitt MJ, Boyer TG, Zhou Q, Berk AJ, Verma IM. Association between proto-oncoprotein Rel and TATA-binding protein mediates transcriptional activation by NF-kappa B. Nature. 1993;365:412–419. doi: 10.1038/365412a0. [DOI] [PubMed] [Google Scholar]
- So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AB, Monick MM, Hunninghake GW. Both Erk and p38 kinases are necessary for cytokine gene transcription. Am J Respir Cell Mol Biol. 1999;20:751–758. doi: 10.1165/ajrcmb.20.4.3420. [DOI] [PubMed] [Google Scholar]
- Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- Hsieh CJ, Klump B, Holzmann K, Borchard F, Gregor M, Porschen R. Hypermethylation of the p16INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res. 1998;58:3942–3945. [PubMed] [Google Scholar]
- Sato F, Harpaz N, Shibata D, Xu Y, Yin J, Mori Y, Zou TT, Wang S, Desai K, Leytin A, Selaru FM, Abraham JM, Meltzer SJ. Hypermethylation of the p14(ARF) gene in ulcerative colitis-associated colorectal carcinogenesis. Cancer Res. 2002;62:1148–1151. [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Hanazawa S, Takeshita A, Amano S, Semba T, Nirazuka T, Katoh H, Kitano S. Tumor necrosis factor-α induces expression of monocyte chemoattractant JE via fos and jun genes in clonal osteoblastic MC3T3-E1 cells. J Biol Chem. 1993;268:9526–9532. [PubMed] [Google Scholar]
- Beck S, Lambeau G, Scholz-Pedretti K, Gelb MH, Janssen MJ, Edwards SH, Wilton DC, Pfeilschifter J, Kaszkin M. Potentiation of tumor necrosis factor alpha-induced secreted phospholipase A2 (sPLA2)-IIA expression in mesangial cells by an autocrine loop involving sPLA2 and peroxisome proliferator-activated receptor alpha activation. J Biol Chem. 2003;278:29799–29812. doi: 10.1074/jbc.M211763200. [DOI] [PubMed] [Google Scholar]
- Shiohara M, Akashi M, Gombart AF, Yang R, Koeffler HP. Tumor necrosis factor alpha: posttranscriptional stabilization of WAF1 mRNA in p53-deficient human leukemic cells. J Cell Physiol. 1996;166:568–576. doi: 10.1002/(SICI)1097-4652(199603)166:3<568::AID-JCP11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Cowan MJ, Huang X, Yao XL, Shelhamer JH. Tumor necrosis factor alpha stimulation of human Clara cell secretory protein production by human airway epithelial cells. Ann NY Acad Sci. 2000;923:193–201. doi: 10.1111/j.1749-6632.2000.tb05530.x. [DOI] [PubMed] [Google Scholar]
- Schmitz H, Fromm M, Bentzel CJ, Scholz P, Detjen K, Mankertz J, Bode H, Epple HJ, Riecken EO, Schulzke JD. Tumor necrosis factor-alpha (TNFalpha) regulates the epithelial barrier in the human intestinal cell line HT-29/B6. J Cell Sci. 1999;112:137–146. doi: 10.1242/jcs.112.1.137. [DOI] [PubMed] [Google Scholar]
- Yan F, John SK, Wilson G, Jones DS, Washington MK, Polk DB. Kinase suppressor of Ras-1 protects intestinal epithelium from cytokine-mediated apoptosis during inflammation. J Clin Invest. 2004;114:1272–1280. doi: 10.1172/JCI21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley KF, Pantano C, Ciolino A, Mawe GM. IFN-gamma and TNF-alpha decrease serotonin transporter function and expression in Caco2 cells. Am J Physiol. 2007;292:G779–G784. doi: 10.1152/ajpgi.00470.2006. [DOI] [PubMed] [Google Scholar]
- Abreu-Martin MT, Vidrich A, Lynch DH, Targan SR. Divergent induction of apoptosis and IL-8 secretion in HT-29 cells in response to TNFa and ligation of Fas antigen. J Immunol. 1995;155:4147–4154. [PubMed] [Google Scholar]
- Jin S, Ray RM, Johnson LR. TNF-alpha/cycloheximide-induced apoptosis in intestinal epithelial cells requires Rac1-regulated reactive oxygen species. Am J Physiol. 2008;294:G928–G937. doi: 10.1152/ajpgi.00219.2007. [DOI] [PubMed] [Google Scholar]
- Vetuschi A, Latella G, Sferra R, Caprilli R, Gaudio E. Increased proliferation and apoptosis of colonic epithelial cells in dextran sulfate sodium-induced colitis in rats. Dig Dis Sci. 2002;47:1447–1457. doi: 10.1023/a:1015931128583. [DOI] [PubMed] [Google Scholar]
- Waetzig GH, Seegert D, Rosenstiel P, Nikolaus S, Schreiber S. p38 mitogen-activated protein kinase is activated and linked to TNF-α signaling in inflammatory bowel disease. J Immunol. 2002;168:5342–5351. doi: 10.4049/jimmunol.168.10.5342. [DOI] [PubMed] [Google Scholar]
- Zhong J, Kyriakis JM. Dissection of a signaling pathway by which pathogen-associated molecular patterns recruit the JNK and p38 MAPKs and trigger cytokine release. J Biol Chem. 2007;282:24246–24254. doi: 10.1074/jbc.M703422200. [DOI] [PubMed] [Google Scholar]