Abstract
Transforming growth factor-β (TGF-β) plays a key role in scleroderma pathogenesis. The transcription factor early growth response-1 (Egr-1) mediates the stimulation of collagen transcription elicited by TGF-β and is necessary for the development of pulmonary fibrosis in mice. Here, we report that TGF-β causes a time- and dose-dependent increase in Egr-1 protein and mRNA levels and enhanced transcription of the Egr-1 gene via serum response elements in normal fibroblasts. The ability of TGF-β to stimulate Egr-1 was preserved in Smad3-null mice and in explanted Smad3-null fibroblasts. The response was blocked by a specific mitogen-activated protein kinase kinase 1 (MEK1) inhibitor but not by an ALK5 kinase inhibitor. Furthermore, MEK1 was phosphorylated by TGF-β, which was sufficient to drive Egr-1 transactivation. Stimulation by TGF-β enhanced the transcriptional activity of Elk-1 via the MEK-extracellular signal-regulated kinase 1/2 pathway. Bleomycin-induced scleroderma in the mouse was accompanied by increased Egr-1 accumulation in lesional fibroblasts. Furthermore, biopsies of lesional skin and lung from patients with scleroderma showed increased Egr-1 levels, which were highest in early diffuse disease. Moreover, both Egr-1 mRNA and protein were elevated in explanted scleroderma skin fibroblasts in vitro. Together, these findings define a Smad-independent TGF-β signal transduction mechanism that underlies the stimulation of Egr-1, demonstrate for the first time sustained Egr-1 up-regulation in fibrotic lesions and suggests that Egr-1 has a role in the induction and progression of fibrosis.
Scleroderma or systemic sclerosis (SSc), a potentially fatal disease of unknown cause, is characterized by fibrosis with collagenous scar tissue formation in the skin and lungs.1 The pathogenesis of fibrosis remains incompletely understood, and effective treatments are lacking.2 As the primary collagen-producing cells activated by paracrine and autocrine transforming growth factor-β (TGF-β), fibroblasts are the key cellular effectors of fibrosis.3 Fibroblast responses elicited by TGF-β involve both Smad-dependent and -independent signal transduction. In the Smad pathway, activated ALK5 phosphorylates Smad2/3, inducing interaction with Smad4, translocation into the nucleus, and binding to consensus Smad-binding DNA elements of target genes, such as COL1A2.4 In addition, TGF-β also activates non-Smad pathways in a cell- and context-dependent manner.5
The early growth response-1 (Egr-1) transcription factor was originally identified as an early-immediate gene product induced by environmental stress.6 In addition to Egr-1, the early response gene family also includes Egr-2, Egr-3, and Egr-4, along with their endogenous inhibitors NAB1 and NAB2. The Egr-1 gene codes for an 82-kDa zinc finger protein that binds to GC-rich regulatory DNA elements found in the promoter region of many genes. Although negligible in normal cells, Egr-1 expression is rapidly and transiently induced during acute injury by growth factors, cytokines, oxidant stress, hormones, neurotransmitters, UV light, and mechanical forces.7,8 As a transcriptional master regulator that integrates converging signaling pathways, Egr-1 has a central role in orchestrating adaptive cellular response to injury.9
Because of its significance in cardiovascular pathobiology, the regulation of Egr-1 in the vascular system has been investigated extensively. In vascular smooth muscle cells, TGF-β was shown to induce Egr-1 via the extracellular signal-regulated kinase 1/2 (ERK1/2) mitogen-activated protein kinase (MAPK) pathway.10 In contrast, the regulation of Egr-1 expression in the context of fibrogenesis has not been characterized to date, despite an emerging recognition of the importance of Egr-1 in the pathogenesis of fibrosis. We had previously demonstrated that TGF-β stimulation of normal fibroblasts resulted in enhanced Egr-1 DNA-binding activity specific for a GC-rich sequence within the promoter region of COL1A2.11 Furthermore, Egr-1 was shown to be both necessary and sufficient to stimulate type I collagen production in vitro, suggesting a fundamental role for Egr-1 in mediating TGF-β-dependent profibrotic responses. In accordance with these observations, Egr-1-null mice were shown to be protected from pulmonary fibrosis induced by TGF-β.12
In the present study, we investigated the mechanism of Egr-1 stimulation by TGF-β. The results indicate that TGF-β caused rapid and transient up-regulation of Egr-1 via a Smad-independent pathway involving mitogen-activated protein kinase kinase 1 (MEK1)-ERK1/2- Elk-1. The expression of Egr-1 was markedly up-regulated in fibrotic lesions in a mouse model of scleroderma, and elevated Egr-1 was noted in skin and lung biopsies from patients with active scleroderma. Because Egr-1 is known to mediate stimulation of collagen by TGF-β and to induce the production of fibrogenic signals and their receptors, our findings suggest that sustained up-regulation of Egr-1 in scleroderma might contribute to amplification or persistence of the TGF-β-driven fibrotic process. Accordingly, targeting Egr-1 regulation and biological activity represents a novel therapeutic strategy to block TGF-β-dependent pathological fibrogenesis.
Materials and Methods
Cell Culture and Reagents
Cultures of normal human dermal fibroblasts were established by explant from neonatal foreskin or forearm skin biopsies of healthy adults and studied at early (<8) passage. Dermal fibroblasts were obtained by biopsy of the affected dorsal forearm and/or from clinically unaffected skin from four patients with early diffuse cutaneous SSc13 and studied in parallel with fibroblasts from three healthy donors. All patients fulfilled the American College of Rheumatology criteria for SSc. Cultures of murine dermal fibroblasts were established from newborn female Smad3-null mice and their wild-type littermates.14 Murine NIH3T3 fibroblasts were from the American Type Culture Collection (Manassas, VA). All fibroblasts were maintained in modified Eagle’s minimal essential medium or Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 1% vitamins, and 2 mmol/L l-glutamine and studied at early confluence. Culture media and tissue culture reagents were from Biowhittaker (Walkersville, MD). For experiments, fresh serum-free media containing 0.1% bovine serum albumin was added to cultures for 24 hours before TGF-β. In selected experiments, fibroblasts were preincubated in media containing the kinase inhibitors SB431542 (Sigma-Aldrich, St. Louis, MO) or U0126 (Cell Signaling, Beverly, MA) for 30 minutes, followed by 12.5 ng/ml TGF-β1 (PeproTech, Rocky Hill, NJ) for up to 48 hours. Expression plasmids for constitutively active MEK1, Gal4-dbd-Elk-1, and its control vector and the reporter plasmid 5xGAL4-luciferase were all from Stratagene (La Jolla, CA).
Northern Analysis and PCR
Total RNA was isolated from confluent fibroblasts using TRIzol Reagent (Life Technologies, Grand Island, NY). For Northern analysis, total RNA was examined using [α-32P]dCTP-labeled cDNA probes for human Egr-1, glyceraldehyde-3-phosphate dehydrogenase, and 18S. After extensive washing of the nitrocellulose membranes, cDNA-mRNA hybrids were visualized by autoradiography. Signal intensities were quantitated by densitometry, and results were normalized with levels for glyceraldehyde-3-phosphate dehydrogenase mRNA or 18S RNA in each sample. For PCR analysis, total RNA (1 μg) from cultured fibroblasts or lesional skin tissue from mice was reverse-transcribed to cDNA using AMV Reverse Transcriptase System (Promega, Madison, WI) and subjected to amplification. PCR products were separated by electrophoresis in 1.5% agarose gels. For real-time quantitative PCR analysis, total RNA was reverse-transcribed, and the products (50 ng) were amplified with the oligonucleotide primers shown in Table 1 using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) on the ABI 7500 Prism Sequence Detection System machine.
Table 1.
Sequences Used for RT-PCR and Real-Time Quantitative PCR
| Gene | cDNA sequence |
|---|---|
| RT-PCR | |
| Egr-1 | Forward: 5′-GACAGCAACCTTTTCTCC CAGG-3′ |
| Reverse: 5′-GTTAGGTCCTCACTTGGG GGAA-3′ | |
| Egr-2 | Forward: 5′-CCGCCAAGGCCGTAGACA AAA-3′ |
| Reverse: 5′-GGGTCAATGGAGAACTT GCCC-3′ | |
| Egr-3 | Forward: 5′-GAGAAGCTGCCGGTGACC ATGA-3′ |
| Reverse: 5′-AGTTGGAAGGGGAGTCGAA GGC-3′ | |
| Nab1 | Forward: 5′-TGCTGACAAGAAGAGA TGAG-3′ |
| Reverse: 5′-TCCTGGTTTCCACAGA CTAC-3′ | |
| Quantitative PCR | |
| Egr-1 | Forward: 5′-TGCGGCAGAAGGACAAGA AAGC-3′ |
| Reverse: 5′-TGAGGAAGGGAAGCTGCT GACC-3′ | |
| Egr-2 | Forward: 5′-CCGCCAAGGCCGTAGACA AAAT-3′ |
| Reverse: 5′-GGGTCAATGGAGAACTTG CCCA-3′ | |
| Egr-3 | Forward: 5′-GAGAAGCTGCCGGTGACC ATGA-3′ |
| Reverse: 5′-AGTTGGAAGGGGAGTCGA AGGC-3′ | |
| Nab2 | Forward: 5′-TGACAGCCAGAAGGAAG AGGA-3′ |
| Reverse: 5′-AGGTGCTCTCTCTCGGGCT ACTT-3′ | |
| Actin | Forward: 5′-AATGTCGCGGAGGACTT TGAT-3′ |
| Reverse: 5′-AGGATGGCAAGGGACTT CCTG-3′ | |
| Mouse Egr-1 | Forward: 5′-TTTGCCTCCGTTCCA CCTGC-3′ |
| Reverse: 5′-TGCCAACTTGATGGTCAT GCGC-3′ |
Western Analysis
At the end of the experiments, fibroblasts were harvested, whole-cell lysates were prepared, and equal amounts of proteins (20 to 50 μg/lane) were subjected to electrophoresis in 8% SDS polyacrylamide gels. Proteins were transferred to Immobilon-P membranes (Millipore, Billerica, MA), and membranes were probed sequentially with primary antibodies specific for Egr-1 (C19), MEK1 (C18), phospho-MEK1(Ser298), and Actin (C2) (all from Santa Cruz Biotechnology, Santa Cruz, CA); Smad3 (Zymed Laboratories, South San Francisco, CA); or phospho-ERK1/2 (Thr202 Tyr204), ERK1/2, or phospho-Smad2 (Ser465,467) (all from Cell Signaling), as indicated. Membranes were then incubated with appropriate secondary antibodies and subjected to enhanced chemiluminescence detection using ECL Reagent (Amersham Pharmacia, Piscataway, NJ).
Transient Transfection Assays
Plasmids harboring 1.2 kb of the mouse Egr-1 gene promoter or 5′-truncated fragments of the promoter15 fused to luciferase gene were used for transient transfection assays. At early confluence, fibroblasts were transfected using SuperFect Transfection kit (Qiagen, Valencia, CA). Twenty-four hours later, fresh serum-free media containing 0.1% bovine serum albumin and indicated kinase inhibitors (10 μmol/L) were added, followed 30 minutes later by TGF-β1. At the end of the experiments cultures were harvested, and cell lysates were assayed for their luciferase activities using the Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase pRL-TK (Promega) was cotransfected along with the reporter constructs, and luciferase activity was used to monitor transfection efficiency. Experiments were performed in triplicate, and the results were normalized for protein concentrations. To assess Elk-1 transactivation, NIH3T3 fibroblasts were cotransfected with Gal4-dbd-Elk-1 along with 5xGAL4-luciferase reporter plasmids, and after incubation with TGF-β, luciferase activities were quantified as above.
Immunofluorescence
The regulation of endogenous Egr-1 expression was further examined by immunocytochemistry and fluorescence confocal microscopy.16 Briefly, foreskin fibroblasts (104 cells/well) incubated in serum-free media with 0.1% bovine serum albumin were exposed to 1 ng/ml TGF-β1 for 30 minutes. Cultures were then fixed with 100% methanol and incubated with rat anti-human Egr-1 antibodies (C19) at a 1:100 dilution, followed by fluorescein isothiocyanate-labeled secondary antibodies. Nuclei were identified using 4,6-diamidino-2-phenylindone. Subcellular distribution of immunofluorescence was then evaluated under a Zeiss LSHS10 laser scanning confocal microscope.
Egr-1 Expression in Mouse Skin in Vivo
Six-week-old female Smad3-null mice and wild-type Back Swiss ×129 SJ littermates (Taconic, Germantown, NY) weighing 15 to 20 g were administered 250 ng TGF-β1 s.c. or PBS s.c. One or 24 hours later, mice were euthanized, and the injected skin was processed for immunohistochemical analysis.17 Four-micrometer sections from lesional skin were deparaffinized, rehydrated, and immersed in Tris-buffered saline-0.1%Tween 20 buffer followed by target retrieval solution (DAKO, Carpinteria, CA). After incubation of the slides with rabbit anti-mouse Egr-1 polyclonal antibodies (C-19; Santa Cruz Biotechnology) at a dilution of 1:300, donkey anti-rabbit Ig as secondary antibodies (Promega) at 1:1000 dilution were applied. Bound antibodies were detected using DAKO Envision + System according to the manufacturer’s instructions. After counterstaining with hematoxylin, sections were mounted with Permount (Fisher Scientific, Pittsburgh, PA) and viewed under an BH-2 microscope (Olympus, Tokyo, Japan). Skin samples from Egr-1-null C57/BL6 mice18 were stained in parallel as negative controls. An additional negative control consisted of substituting nonspecific rabbit IgG (Invitrogen, Carlsbad, CA) for the primary antibody. To quantify Egr-1 expression in lesional skin, at least 30 fibroblastic cells (identified based on their characteristic spindle-shaped morphology) from multiple microscopic fields were scored as clearly immunopositive or negative for Egr-1 by three independent examiners blinded to the treatment, and the ratio of positive cells to total cells was calculated. In other experiments, 6-week-old BALB/c mice weighing 20 g were given s.c. bleomycin or PBS daily for up to 28 days.17 At the end of the experiments, mice were sacrificed, and lesional skin was processed for quantitative PCR or for immunohistochemistry as above. Each experimental group contained at least five mice, and three separate experiments were performed with similar results. All animal studies were performed using protocols and guidelines approved by the Northwestern University Animal Care and Use Committee.
Egr-1 Expression in SSc Lesional Tissue
Skin tissue samples were obtained by biopsy from the affected forearm from 23 patients with SSc (16 patients with disease duration <3 years, early stage; and seven patients with disease duration ≥3 years, late stage) and five healthy adult controls. The protocols for tissue collection were approved by the Institutional Review Boards for Human Studies at Northwestern University, the University of Pittsburgh, and Kanazawa University. Some of the SSc tissues were processed following exactly the same procedure as described under Egr-1 Expression in Mouse Skin in Vivo, where the slides were counterstained with hematoxylin. For other tissues, methyl green was used for counterstain. Paraffin-embedded or frozen tissue sections were subjected to immunostaining using primary anti-Egr-1 antibody (C19; Santa Cruz Biotechnology) at 1:300 dilution, followed by incubation with biotinylated anti-rabbit IgG secondary antibodies (Santa Cruz sc-2089) at 1:100 dilution. Slides were then reacted with avidin-biotin peroxidase (Vectastain ABC kit; Vector Laboratories, Burlingame, CA) and peroxidase activity was detected with diaminobenzidine substrate (brown color). To quantify Egr-1 in lesional dermis, Egr-1-positive fibroblasts were counted by blinded examiners, and the ratio of positive cells to total cells was calculated. Tissue sections from prostate cancer were used as positive control, and substitution of primary antibody with nonspecific rabbit IgG (Invitrogen) served as negative control. Lung tissue was obtained from four patients with SSc-associated pulmonary fibrosis who were undergoing lung transplant surgery, and from unused normal donor lungs from three patients. Tissues were processed as above, the slides were counterstained with hematoxylin. To quantify Egr-1 expression, the numbers of cells in several microscopic fields were scored as clearly immunopositive or negative for Egr-1 by two independent examiners, and the ratio of positive cells to total cells was calculated. Tissue sections from a colon cancer were used as a positive control, and nonspecific rabbit IgG was used as a negative control.
Statistical Analysis
Statistical significance was determined using the unpaired Student’s t-test. Values of P ≤ 0.05 were considered significant.
Results
TGF-β Stimulates Egr-1 Expression in Normal Fibroblasts
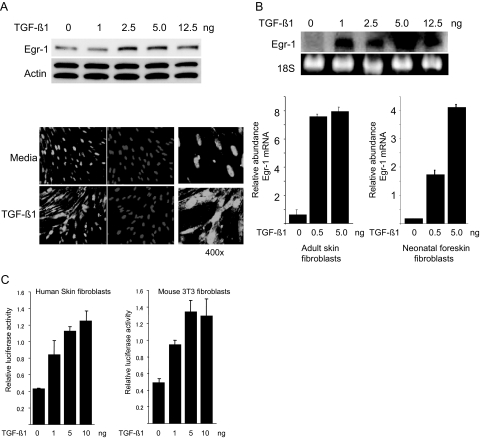
To characterize Egr-1 regulation by TGF-β, quiescent foreskin fibroblasts were stimulated with increasing concentrations of TGF-β1. The results of Western and Northern analysis showed rapid (30 minutes) and dose-dependent increase in Egr-1 protein (Figure 1A, top) and mRNA (Figure 1B, top) expression. Immunofluorescence confocal microscopy indicated little detectable Egr-1 in the absence of TGF-β and marked accumulation in both cytoplasm and nucleus in TGF-β-treated fibroblasts (Figure 1A, bottom). Real-time quantitative PCR analysis confirmed the dose-dependent stimulation of Egr-1 mRNA in neonatal and adult dermal fibroblasts (Figure 1B, bottom).
Figure 1.
TGF-β stimulates Egr-1 expression in fibroblasts. Serum-starved neonatal human foreskin fibroblasts (A–C) or adult skin fibroblasts (B) or mouse NIH3T3 fibroblasts (C) at confluence were incubated with indicated concentrations of TGF-β1. A: After 120 minutes incubation, whole-cell lysates were prepared and subjected to Western analysis (top). Representative immunoblots are shown. Fibroblasts incubated with 1 ng/ml TGF-β for 120 minutes were immunostained with antibodies to Egr-1 or with 4,6-diamidino-2-phenylindone and viewed by laser scanning confocal microscopy (bottom). Representative images are shown. Magnification: ×100 (left and middle panels); ×400 (right panels). B: After 30-minute incubation, total RNA was isolated and examined by Northern analysis (top) or by real-time quantitative PCR (bottom; bars indicate means ± SD from triplicate determinations). Values are the mean and SD and normalized with Actin. Representative results are shown. C: Fibroblasts were transiently transfected with Egr-1 promoter-luc constructs. After incubation of the cultures with increasing concentrations of TGF-β1 for 24 hours, cell lysates were assayed for their luciferase activities. The results are the means ± SD of triplicate determinations.
To characterize the level of regulation of Egr-1, quiescent fibroblasts were transiently transfected with a plasmid containing the proximal 1.2 kb of the human Egr-1 gene promoter linked to the luciferase reporter and then incubated in media with increasing concentrations of TGF-β1. Twenty-four hours later, the cultures were harvested, and cell lysates were assayed for their luciferase activities. The results showed that TGF-β induced a dose-dependent increase in Egr-1 promoter activity in transfected primary foreskin fibroblasts (Figure 1C, left) and in NIH3T3 fibroblasts (Figure 1C, right). In both human and mouse fibroblasts, TGF-β at a concentration of 10 ng/ml induced a ∼threefold stimulation of Egr-1 promoter activity.
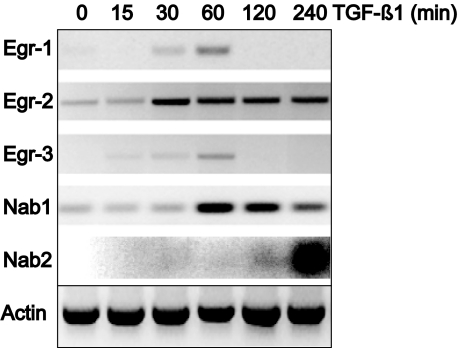
In addition to Egr-1, the family of early growth response transcription factors also includes Egr-2, Egr-3, and Egr-4. Because little is known about the function or regulation of the Egr moieties in the context of fibrosis, foreskin fibroblasts were incubated with TGF-β for indicated periods, and mRNA expression was evaluated by RT PCR analysis. The results showed that TGF-β1 caused rapid stimulation of Egr-3 and Egr-1 mRNA in these cells (Figure 2). Marked increase was seen at 60 minutes, followed by decline after 120 minutes. The expression of Egr-2 mRNA was also stimulated, but in contrast to Egr-1, the response was relatively sustained, with elevated mRNA expression persisting at 4 hours. We also examined TGF-β regulation of Nab1 and Nab2, which are inhibitory members of the early growth response family that bind to Egr-1 and regulate its activity.19 The results of PCR analysis showed that Nab1 was constitutively expressed in fibroblasts, as it is in most cell types, and was further increased by TGF-β. In contrast, Nab2 mRNA was undetectable in the absence of stimulation but was strongly induced by TGF-β; however, compared with the rapid stimulation of Egr-1 and Egr-3, the Nab2 response was delayed, peaking at 4 hours (Figure 2). Thus, endogenous inhibitors of Egr-1 are induced by the same environmental signals that stimulate Egr-1 and, at least in the case of Nab2, show delayed response compatible with a negative feedback function. The stimulation of mRNA of each of the Egr moieties by TGF-β was confirmed by real-time quantitative PCR (data not shown).
Figure 2.
TGF-β regulation of the expression of Egr moieties. Confluent foreskin fibroblasts were incubated with TGF-β1 (12.5 ng) for indicated periods. Total RNA was isolated and subjected to RT-PCR analysis. The results are representative of at least two independent experiments.
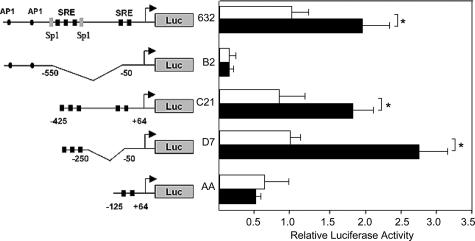
Delineation of the Egr-1 Promoter TGF-β Response Region
As illustrated schematically (Figure 3, left), the proximal promoter of the Egr-1 gene harbors multiple potential regulatory elements. These include binding sites for AP-1, GC-rich elements, and five serum-responsive elements (SREs). Transient transfection assays were performed to localize the functional TGF-β response regions of the Egr-1 gene. For this purpose, mouse NIH3T3 fibroblasts were transfected with the indicated deletion or truncation mutants of the mouse Egr-1 promoter linked to the luciferase reporter gene. Incubation of transfected fibroblasts with TGF-β resulted in ∼twofold stimulation of luciferase activity driven by the full-length Egr-1 promoter (Figure 3). The activity of luciferase fused to a promoter mutant lacking all five SRE sites (B2) showed a substantial decrease in basal activity and complete lack of stimulation by TGF-β.
Figure 3.
Delineation of TGF-β response region of the Egr-1 gene promoter. NIH3T3 fibroblasts transiently transfected with indicated 5′ deletion or truncation mutants of the mouse Egr-1 gene promoter linked to the luciferase reporter were incubated with TGF-β1 for 24 hours. Cell lysates were assayed for their luciferase activities. The results, normalized with Renilla luciferase activities, are the means ± SD of triplicate determinations from at least two independent experiments. □, untreated fibroblasts; ▪, TGF-β-treated fibroblasts. *P < 0.005.
Deletion of neither the region containing the distal AP-1 and Sp1 binding sites (C21) nor the two proximal SRE sites (D7) significantly affected either basal promoter activity or the TGF-β response. In contrast, truncation of the three distal SRE sites (AA) resulted in complete abrogation of TGF-β response, indicating that the distal SRE sites mediate TGF-β-induced Egr-1 promoter transactivation.
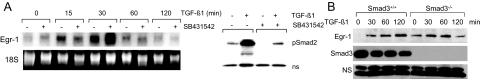
Role of Smad3 in Egr-1 Stimulation
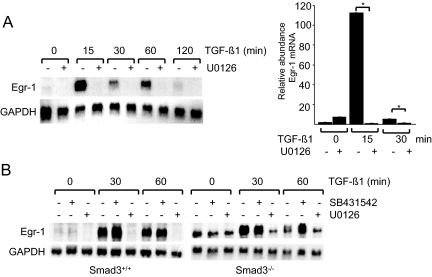
Downstream targets for activated ALK5, Smad2, and Smad3 are the primary signal transducers for most TGF-β responses in fibroblasts.20 To evaluate their role as potential mediators of Egr-1 stimulation, SB431542, a potent and specific inhibitor of ALK5-dependent TGF-β signaling, was used.21 Confluent serum-starved human fibroblasts were preincubated with 10 μmol/L SB431542 for 30 minutes, followed by TGF-β1 for indicated periods. The results of Northern analysis showed that SB431542 pretreatment failed to prevent rapid and transient stimulation of Egr-1 mRNA induced by TGF-β (Figure 4A). Under these experimental conditions, SB431542 significantly prevented the phosphorylation of Smad2 induced by TGF-β (Figure 4B, right).
Figure 4.
Smad3-independent TGF-β stimulation of Egr-1. A: Confluent serum-starved foreskin fibroblasts were pretreated with 10 μmol/L SB431542 for 30 minutes, followed by TGF-β1 for indicated periods. At the end of the incubation, total RNA was isolated and examined by Northern analysis. The results are representative of two independent experiments. B: Time-dependent Egr-1 stimulation in mouse skin fibroblasts. Confluent serum-starved fibroblasts explanted from Smad3-null mice and wild-type littermates in parallel were incubated with TGF-β1 for indicated periods. Whole-cell lysates were examined by Western analysis. NS, nonspecific band. Representative immunoblots are shown.
Complementary experiments used primary dermal fibroblasts from Smad3-null mice. We have shown previously that Smad3-null fibroblasts displayed impaired TGF-β responses in vitro, with substantially reduced stimulation of collagen.14 Confluent fibroblasts from Smad3-null mice and their wild-type littermates were incubated in parallel with TGF-β1 (12.5 ng) for up to 120 minutes. The results of Western analysis showed that Egr-1 was rapidly induced by TGF-β1 in wild-type murine fibroblasts in a manner comparable with that observed in normal human fibroblasts (Figure 4B). Smad3-null fibroblasts showed virtually identical early induction of Egr-1. Interestingly, basal levels of Egr-1 were found to be modestly elevated. In agreement with results from Western analysis, examination of Egr-1 mRNA levels showed that TGF-β1 caused comparable stimulation in both wild-type fibroblasts and Smad3-null fibroblasts (data not shown; Figure 5B). Taken together, the results from experiments using ALK5 inhibition and Smad3-deficient mouse fibroblasts indicated that stimulation of Egr-1 by TGF-β occurred largely independent of the ALK5/Smad3 signaling axis.
Figure 5.
MEK1-dependent stimulation of Egr-1 mRNA. Confluent serum-starved human (A) or mouse (B) skin fibroblasts were preincubated with protein kinase inhibitors (10 μmol/L) for 30 minutes, followed by TGF-β1 for indicated periods. A: Cultures were harvested, and total RNA was subjected to Northern analysis (left) or real-time quantitative PCR analysis (right). For real-time PCR, values are the mean and SD and normalized with Actin. *P < 0.05. B: RNA from Smad3-null dermal fibroblasts or wild-type control fibroblasts was subjected to Northern analysis. Representative autoradiograms are shown.
MAPK Pathway Mediates Egr-1 Induction
Because stimulation of Egr-1 appeared to be a Smad3-independent TGF-β response, we proceeded to examine the functional role of non-Smad TGF-β signaling pathways. For this purpose, confluent foreskin fibroblasts were pretreated with the selective MEK1/2 kinase inhibitor U0126 for 30 minutes, followed by incubation with TGF-β1 for up to 120 minutes. Results from Northern analysis showed that the rapid stimulation of Egr-1 mRNA was completely abolished by pretreatment of the cultures with U0126 (Figure 5A, left). Under the experimental conditions used here, U0126 had no demonstrable cellular toxicity. Consistent results were obtained in separate experiments using real-time quantitative PCR analysis (Figure 5A, right). Furthermore, in contrast to the ALK5 inhibitor SB431542 (Figure 5B) or inhibitors of p38 (data not shown), pretreatment of the cultures with U0126 abrogated the stimulation of Egr-1 in both wild-type and Smad3-null dermal fibroblasts (Figure 5B). These results suggested that MEK1/2, which is upstream of, and directly activates, ERK1/2, and which is induced by a variety of growth factors and cytokines, might serve as a mediator for TGF-β stimulation of Egr-1.
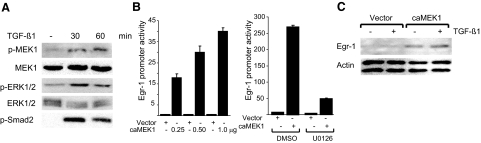
To examine the effect of TGF-β on MEK1/2, serum-starved fibroblasts were incubated with TGF-β for indicated periods and subjected to Western analysis. The results showed that TGF-β stimulation induced rapid phosphorylation of cellular MEK1/2 (Figure 6A). Phosphorylation of ERK1/2 and Smad2 also occurred in parallel with unaltered ERK1/2. To examine the functional contribution of MEK1 in Egr-1 stimulation, NIH3T3 fibroblasts were cotransfected with expression vector for a constitutively active MEK1 mutant, along with Egr-1 promoter-luc reporter, and incubated in the presence or absence of TGF-β for a further 24 hours. The results of transient transfection assays showed that constitutively active MEK1 was by itself capable of causing dose-dependent stimulation of Egr-1 promoter activity (Figure 6B, left) and endogenous Egr-1 protein accumulation (Figure 6C). No stimulation occurred in NIH3T3 fibroblasts transfected with empty vector. Pretreatment of the cultures with the MEK1 kinase inhibitor U0126 significantly attenuated the stimulation of Egr-1 promoter activity induced by MEK1 (Figure 6B, right). Taken together, these results demonstrate that MEK1 is activated by TGF-β1 in fibroblasts and plays a major functional role in mediating the stimulation of Egr-1 promoter activity and Egr-1 protein accumulation in these cells.
Figure 6.
MEK1 is activated by TGF-β and induces Egr-1. A: Foreskin fibroblasts were incubated with TGF-β for indicated periods and whole-cell lysates were subjected to Western analysis. Representative autoradiograms are shown. B: NIH3T3 fibroblasts were cotransfected with indicated concentrations of constitutively active MEK1 (caMEK1) or empty vector along with Egr-1-luc (left). After incubation for 24 hours, cultures were harvested, and cell lysates were assayed for their luciferase activities. Transfected NIH3T3 fibroblasts were incubated in media containing U0126 for 24 hours (right). Results, normalized with Renilla luciferase, are expressed as means ± SD of triplicate determinations from two independent experiments. C: Whole-cell lysates prepared from transfected NIH3T3 fibroblasts incubated with TGF-β for 24 h were examined by Western analysis. Representative immunoblots are shown.
TGF-β Stimulates Elk-1 Transcriptional Activity
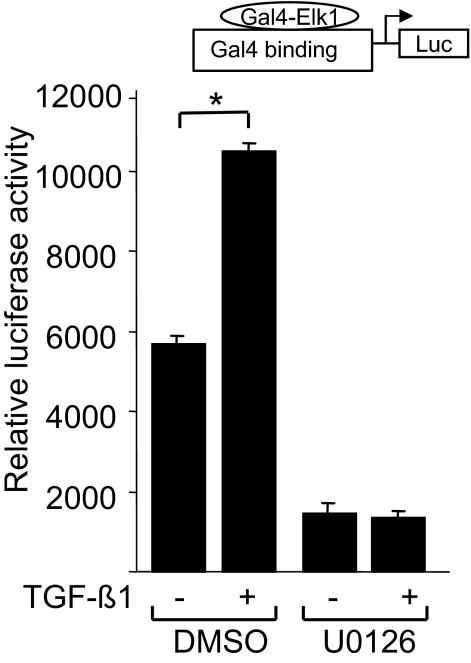
We next turned our attention to downstream effectors of the MAPK-mediated TGF-β response. Deletion analysis identified the distal SRE sites of the Egr-1 promoter as the elements responsible for stimulation by TGF-β. Serum response elements are known to bind to multicomponent complexes that contain the Ets-like transcription factor Elk-1.22 Elk-1 is a direct ERK1/2 substrate in the MAPK signaling cascade, and its phosphorylation has been shown to enhance its binding to SRE sequences. In fibroblasts, TGF-β stimulation induced activation of the MEK-ERK1/2 pathway (Figure 6A). Furthermore, in HaCaT cells, TGF-β has been shown to increase Elk-1 transcriptional activity.23 Because the distal SRE sites appear to serve as TGF-β response elements in the Egr-1 promoter and because Elk-1 is known to mediate the induction of Egr-1 by inflammation-associated stimuli, we examined the functional role of Elk-1 in mediating the TGF-β response using heterologous Gal4-Elk-1 transactivation assays. For this purpose, NIH3T3 fibroblasts were cotransfected with a reporter construct containing five tandem repeats of the yeast Gal4 DNA binding element linked to the luciferase reporter along with pGAL4-DBD-Elk-1, which expresses a chimeric protein that contains the GAL4 DNA binding domain fused to the transactivation domain of Elk-1. Transfected cells were incubated with TGF-β overnight, and cell lysates were assayed for their luciferase activities. The results showed that TGF-β induced a ∼twofold increase in Elk-1-dependent transcription (Figure 7). No induction was observed in fibroblasts transfected with a plasmid that expresses only the pGAL4-binding domain (data not shown). Pretreatment of the cultures with U0126 completely abrogated the TGF-β response. We conclude therefore that TGF-β stimulation leads to Elk-1 activation via the MEK-ERK1/2 pathway.
Figure 7.
TGF-β stimulates Elk-1-dependent transcription. Subconfluent NIH3T3 fibroblasts were cotransfected with the expression vector pGal4-dbd-Elk-1 along with the reporter construct pGal4-DNA-luc. Transfected fibroblasts were preincubated in the presence or absence of 10 μmol/L U0126 for 30 minutes before incubation with TGF-β1 for 24 hours. Cell lysates were assayed for their luciferase activities. The results, shown as the means ± SD of triplicate determinations, are representative of three independent experiments. *P < 0.01.
TGF-β Induces Egr-1 Expression in Vivo
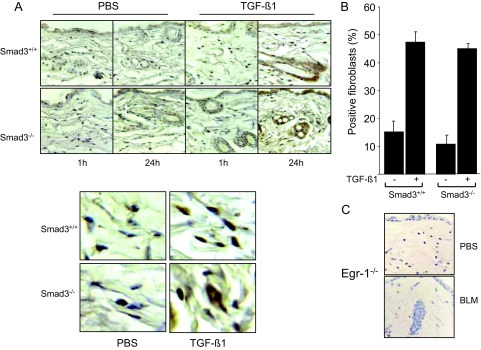
To examine the in vivo regulation of Egr-1 in skin fibroblasts, young Smad3-null mice and wild-type littermates received a single s.c. injection of 250 ng of TGF-β1 or PBS in parallel, and 1 or 24 hours later, Egr-1 expression in the lesional dermis was examined. The results of immunohistochemistry indicated that by 24 hours, TGF-β1 induced a marked increase in Egr-1 in fibroblastic cells (Figure 8A). Quantitation of the results from multiple determinations indicated a reproducible two- to threefold increase in skin Egr-1 expression in both Smad3-null mice and wild-type littermates (Figure 8B). Parallel examination of lesional skin from Egr-1-null mice showed no detectable Egr-1 expression, indicating the specificity of the immunostaining (Figure 8C). Therefore, TGF-β induces Egr-1 expression in a Smad3-independent manner in vivo and as in vitro.
Figure 8.
TGF-β induces Egr-1 expression in vivo. Six-week-old female Smad3-null mice and wild-type littermate mice received a single s.c. injection of 250 ng TGF-β1 or PBS. Lesional skin was harvested 1 or 24 hours later and examined by immunohistochemistry using antibodies against Egr-1. A: Representative photomicrographs; bottom panels, higher magnification. B: The proportion of Egr-1-positive fibroblasts in the lesional dermis (at 24 hours) was determined. The results represent the means ± SE. C: Negative control. Skin tissue from Egr-1-null mice was processed in parallel.
Bleomycin-Induced Mouse Scleroderma Is Associated with Egr-1 Up-Regulation
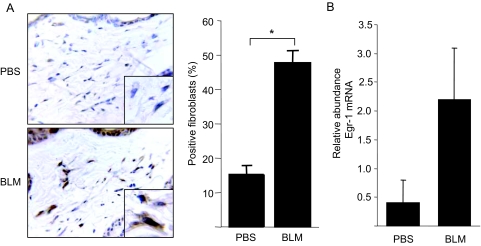
By inducing fibroblast activation and extracellular matrix accumulation, TGF-β plays a key role in fibrosis. In light of the present results showing that Egr-1 is stimulated by TGF-β and is involved in mediating TGF-β responses downstream of the activated TGF-β receptors, we proceeded to evaluate Egr-1 expression in a murine model of skin fibrosis. In agreement with our previous findings,14 s.c. administration of bleomycin in BALB/c mice resulted in scleroderma-like dermal fibrosis and local TGF-β signaling (data not shown). Immunohistochemical examination showed an almost threefold increase in Egr-1 expression in lesional dermis at day 21 compared with PBS-treated control mice (Figure 9A). Immunostaining was localized mostly within the nucleus of fibroblastic cells. The levels of Egr-1 mRNA in lesional skin were increased in parallel with Egr-1 protein (Figure 9B).
Figure 9.
Bleomycin-induced skin fibrosis associated with Egr-1 expression in vivo. Six-week-old female BALB/C mice received daily s.c. injections of bleomycin (BLM) or PBS. A: At day 21, lesional skin was harvested. Immunohistochemistry using antibodies against Egr-1. Hematoxylin counterstain. Representative images are shown. Magnification: ×400; ×1000 (inset, left). The proportion of immunopositive fibroblasts in the lesional dermis was determined (right). Results represent means ± SD. B: Total RNA from lesional skin was analyzed by real-time quantitative PCR. Results are the means ± SD of duplicate determinations from three mice/group. *P < 0.005.
Elevated Egr-1 Expression in Lesional SSc Skin and Lungs
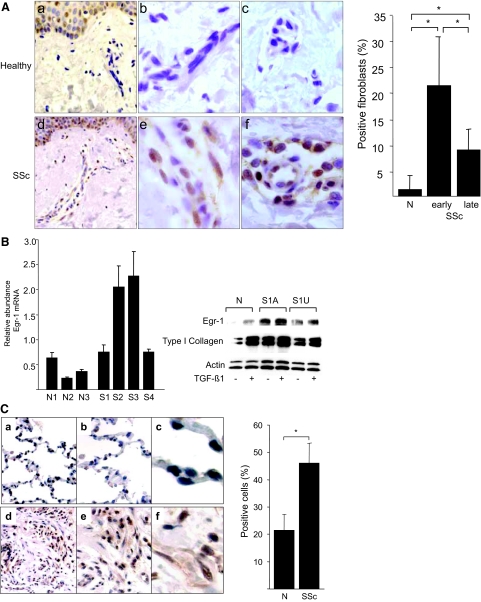
Although Egr-1 is normally low or undetectable in most tissues, rapid and transient Egr-1 accumulation takes place in response to injury, ischemia, and stress. We have shown previously that Egr-1 can directly transactivate COL1A2 transcription and mediates stimulation of collagen production and other fibrotic responses elicited by TGF-β.11 Because TGF-β is known to play a pivotal role in the pathogenesis of SSc and its activity is up-regulated in lesional tissues, aberrant Egr-1 regulation in response to chronic TGF-β signaling in lesional fibroblasts might contribute to the development or persistence of fibrosis. To further explore this notion, the expression of Egr-1 in SSc lesional tissue was examined by immunohistochemistry. For this purpose, tissue samples from 23 patients with SSc and 5 matched healthy controls were studied in parallel. The clinical characteristics of the patients are presented in Table 2. The results showed that in contrast to skin tissue from healthy individuals that had little or no detectable Egr-1, a significant proportion of fibroblastic cells in SSc lesional dermis showed positive Egr-1 immunostaining (Figure 10A). Almost all SSc skin biopsies demonstrated intracellular Egr-1 that was detectable in the papillary and the reticular dermis and at the dermis-adipose layer interface; the epidermis in both SSc samples and healthy controls showed Egr-1 expression, which was most pronounced in the basal layer. Immunostaining was specific, because substitution of the primary antibody resulted in the absence of brown staining. Quantitative analysis showed that SSc patients with relatively early-stage disease (<3 years duration) had a ∼20-fold increase in the proportion of Egr-1-positive fibroblasts in the lesional dermis and that patients with late-stage disease (>3 years) had a ∼10-fold elevation compared with healthy controls (Figure 10A, right). Moreover, a trend toward a negative correlation between the proportion of Egr-1-positive fibroblasts and disease duration was noted (r = −0.538, P = 0.0072).
Table 2.
Clinical Characteristics of SSc Patients Studied
| Sample | Age | Sex | Disease duration (years) | SSc subset |
|---|---|---|---|---|
| Skin tissue immunostaining | ||||
| SSc1 | 59 | F | 0.40 | dcSSc |
| SSc2 | 44 | F | 1.80 | dcSSc |
| SSc3 | 21 | F | 6.00 | dcSSc |
| SSc4 | 42 | F | 0.58 | dcSSc |
| SSc5 | 44 | F | 0.40 | lcSSc |
| SSc6 | 49 | F | 2.00 | lcSSc |
| SSc7 | 54 | M | 3.00 | dcSSc |
| SSc8 | 64 | M | 0.17 | dcSSc |
| SSc9 | 30 | F | 2.5 | dcSSc |
| SSc10 | 24 | F | 9.00 | dcSSc |
| SSc11 | 73 | F | 0.20 | dcSSc |
| SSc12 | 43 | F | 10.00 | dcSSc |
| SSc13 | 31 | F | 15.00 | lcSSc |
| SSc14 | 57 | F | 10.00 | dcSSc |
| SSc15 | 54 | F | 12.00 | dcSSc |
| SSc16 | 25 | F | 1.00 | dcSSc |
| SSc17 | 16 | F | 7.00 | dcSSc |
| SSc18 | 55 | F | 0.30 | lcSSc |
| SSc19 | 67 | M | 0.25 | dcSSc |
| SSc20 | 57 | F | 3.00 | dcSSc |
| SSc21 | 30 | F | 2.50 | dcSSc |
| SSc22 | 43 | M | 0.08 | dcSSc |
| SSc23 | 63 | F | 0.90 | lcSSc |
| Lung tissue immunostaining | ||||
| SSc25 | 57 | M | N/A | dcSSc |
| SSc29 | 60 | F | N/A | dcSSc |
| SSc30 | 58 | F | N/A | lcSSc |
| SSc31 | 35 | M | N/A | SSc sine scleroderma |
| Explanted skin fibroblasts | ||||
| S10 | 36 | F | 0.91 | lcSSc |
| S11 | 46 | F | 1.00 | dcSSc |
| S12 | 54 | M | 1.00 | dcSSc |
| S34 | 38 | F | 1.00 | dcSSc |
| SU9 | N/A | N/A | <5.00 | dcSSc |
lcSSc, limited cutaneous SSc; dcSSc, diffuse cutaneous SSc.
Figure 10.
Elevated Egr-1 accumulation in SSc. A: Lesional skin tissue from patients with SSc (n = 23) and the healthy controls (n = 5) were processed for immunohistochemistry with antibodies against Egr-1. Representative images (left). a and d: Dermis of healthy controls and SSc, respectively (original magnification, ×400). b and e: Fibroblasts in the dermis of healthy controls and SSc tissue, respectively(original magnification, ×1000). c and f: Perivascular and vascular staining of healthy controls and SSc patients, respectively. The proportion of Egr-1-positive fibroblastic cells was calculated (right). B: SSc dermal fibroblasts explanted from lesional skin (S1–4) and control fibroblasts from healthy individuals (N1–3) were harvested, and total RNA was subjected to quantitative PCR analysis (left). The results, expressed relative to the levels of actin mRNA, are the means of triplicate determinations, and representative of two independent experiments. Whole-cell lysates from confluent fibroblasts explanted from affected (A) and unaffected (U) skin from a patient with SSc were incubated with or without TGF-β1 and subjected to Western analysis (right). Results with a representative SSc cell line (S1) are shown. Similar results were obtained with five additional SSc fibroblast lines. C: Lung tissues from patients with SSc-associated pulmonary fibrosis undergoing lung transplantation surgery (n = 4) and normal donor lungs (n = 3) were examined by immunohistochemistry as above. a–c: Normal lungs. d–f: SSc-associated pulmonary fibrosis. Magnification: ×100 (a and d); ×400 (b and e); ×1000 (c and f). Brown staining indicates Egr-1-positive cells. Nuclei are counterstained with hematoxylin (blue). The proportion of Egr-1-positive cells was determined, and results are shown as means ± SD (right). *P < 0.001.
Next, Egr-1 expression was examined in SSc dermal fibroblasts explanted from the lesional skin. For this purpose, fibroblasts from four patients with early and untreated SSc, and three matched healthy controls were studied at low passage (<5). Confluent cultures were incubated in serum-free Dulbecco’s modified Eagle’s medium for 48 hours, and total RNA or whole-cell lysates were examined by real-time quantitative PCR analysis or Western analysis, respectively. Quantitative PCR analysis showed elevated Egr-1 mRNA levels in all four SSc fibroblast lines, with a ∼fivefold (mean) increase compared with healthy control fibroblasts studied in parallel (Figure 10B, left). Western analysis showed that Egr-1 protein was elevated in unstimulated SSc fibroblasts explanted from both clinically involved and uninvolved skin, compared with healthy control fibroblasts (Figure 10B, right). Examination of lesional fibroblasts from an additional five patients indicated a two- to fourfold elevation in Egr-1 protein (data not shown).
Fibrosis in SSc commonly affects the lungs, and the process is associated with fibroblast activation and local TGF-β signaling.24 To examine Egr-1 expression in the lung in SSc, samples from four patients with SSc-associated pulmonary fibrosis and normal donor lungs were examined by immunohistochemistry. The results showed low Egr-1 expression in normal donor lung tissue, detected primarily in alveolar macrophages and in occasional type I and type II pneumocytes (Figure 10C, left). In contrast, lung tissues from all four SSc patients showed strong Egr-1 expression at fibrotic loci. In these regions, Egr-1 immunostaining was detected in epithelial cells and mesenchymal cells. Quantitation confirmed a marked increase in the numbers of Egr-1-positive cells in SSc lungs (Figure 10C, right).
Discussion
Stimulation of collagen transcription by TGF-β involves both canonical Smad signaling and intracellular pathways that operate through cross-talk with the Smad axis or in a Smad-independent manner via ERK, c-Jun N-terminal kinase, p38, cyclin-dependent kinase 2/4, Rho, focal adhesion kinase, c-Abl, and phosphatidylinositol 3-kinase.25,26,27,28 We had previously identified Egr-1 as a novel TGF-β mediator with a significant role in collagen regulation.10 Whereas induction of Egr-1 by acute injury has been investigated extensively, its regulation by TGF-β in the context of fibrogenesis and its role in fibrosis remain poorly understood. In this study, we therefore explored the molecular basis of Egr-1 stimulation by TGF-β in fibroblasts. We find that the response involves Smad-independent transcriptional regulation via the MEK1-ERK1/2-Elk-1 signaling cascade. The expression of Egr-1 was up-regulated in skin and lung tissues and in explanted dermal fibroblasts from patients with active scleroderma and in fibrotic tissue from mice with bleomycin-induced scleroderma. These findings suggest that unchecked TGF-β activity associated with pathological fibrogenesis causes MAPK-dependent up-regulation and persistent local expression of Egr-1 that in turn might contribute to the progression of fibrosis.
Egr-1, a prototypic member of the early-immediate growth response gene family, couples acute perturbations in the extracellular milieu to short-lived changes in the target gene expression.29 In immortalized vascular endothelial cells, ectopic Egr-1 expression resulted in up-regulation of >300 genes, including transcriptional regulators, signaling proteins, cell cycle regulators, cytokines, and extracellular matrix components,30 whereas in normal skin fibroblasts, Egr-1 altered the expression of >100 genes involved in inflammation, apoptosis, cell cycle, cell growth, cell migration, and extracellular matrix (S. Bhattacharyya, S. Lin, and J. Varga, unpublished data). Normal tissue has low Egr-1, but expression is rapidly and transiently induced in acute injury. In turn, Egr-1 up-regulates tumor necrosis factor-α, TGF-β, platelet-derived growth factor, CD44, and intercellular adhesion molecule-1, indicating a role in the cellular stress response program, and the tumor suppressors PTEN and p53, consistent with a role in cell proliferation, apoptosis, and tumor suppression.31 Additional biological roles of Egr-1 include regulation of synaptic plasticity and reproduction.
Emerging evidence implicates Egr-1 in physiological wound healing on the one hand and pathological tissue repair on the other. The list of Egr-1 targets relevant to chronic tissue injury and pathological fibrosis includes plasminogen activator inhibitor-1,32,33 platelet-derived growth factor,34,35 Snail,36 fibronectin,37,38 TGF-β,39 type 2 TGF-β receptor,40 basic fibroblast growth factor,41 matrix metalloproteinase-2,42,43 tissue inhibitor of metalloproteinases-1,44 and type I collagens.10 The importance of Egr-1 in fibrosis is highlighted by its elevated expression and colocalization with connective tissue growth factor in lung tissue from smokers42 and by protection of Egr-1-null mice from lung fibrosis induced by hypoxia,45 by TGF-β12 or IL-13.46 Our present results demonstrate that Egr-1 is induced by TGF-β in skin and lung fibroblasts and NIH3T3 cells. The response is characterized by time- and dose-dependent Egr-1 protein accumulation, mRNA expression, and promoter activation. Stimulation by TGF-β also caused rapid induction of other members of the early growth response gene family. In contrast to Egr-1, its endogenous inhibitor Nab2 followed more delayed kinetics, showing maximal induction at 4 hours. The discrepant time course for the induction of Egr-1 and Nab2, also reported in vascular smooth muscle cells, is consistent with the putative role of Nab2 as a negative feedback on Egr-1 activity.47
In contrast to other TGF-β-inducible profibrotic genes, Egr-1 regulation log TGF-β appears to be largely Smad-independent and parallels Egr-1 induction by inflammatory signals, such as platelet-derived growth factor, lipopolysaccharide, oxidative stress, ischemia reperfusion, cigarette smoke, and mechanical stretch.33,34,37,42 The proximal 1 kb of the Egr-1 gene promoter, sufficient to direct tissue-specific Egr-1 expression in vivo,48 harbors five CarG elements, also known as serum response elements (SREs), in addition to binding sites for Sp1, NF-κB, and Smad2/3.6 The present results identify the distal SRE sites as a principal TGF-β response elements for Egr-1. The SRE sites are recognized by serum response factor and ternary complex factors, such as Elk-1 and related Ets family oncogenes.22 Phosphorylation of cellular Elk-1 by ERK results in enhanced DNA binding activity and SRE-mediated transcription. To induce transcription, the ternary complex factors must bind both to serum response factor and to SRE sequences, and induction of Egr-1 typically depends on cooperative interaction between serum response factor and one or more ternary complex factors. Interestingly, the Egr-1 promoter also contains an Egr-1 binding site, enabling Egr-1 to down-regulate its transcription. Thus, Egr-1 expression is tightly regulated via the positively acting transcription factors serum response factor and Elk-1 and negatively acting factors, including Egr-1, thereby establishing a negative feedback loop restricting the duration of Egr-1 expression.
To better understand Egr-1 regulation in the context of fibrosis, we examined Egr-1 expression in bleomycin-induced murine scleroderma.49 We noted that whereas Egr-1 protein and mRNA were essentially undetectable in BS-treated control mice, levels were elevated in lesional skin from bleomycin-treated mice, with expression localized principally to fibroblastic cells. Although the pathogenesis of skin fibrosis in this mouse model of scleroderma is not fully understood, transient local inflammation and TGF-β production and activity are considered to be important, and fibrosis is attenuated in Smad3-null mice.14 Therefore, elevated Egr-1 expression in lesional skin may be a reflection of local TGF-β activity or persistent fibroblast stimulation. In addition to TGF-β, cytokines such as IL-13 are also known to contribute to bleomycin-induced fibrogenesis, and IL-13 is a potent inducer for Egr-1 gene expression.50 Chronic tissue hypoxia and ischemia reperfusion are prominent features of scleroderma that are also capable of stimulating Egr-1, and thus may account for the sustained expression of Egr-1 in fibrotic lesions.51,52 Additionally, bleomycin itself may directly regulate Egr-1 expression.52
Little or no Egr-1 expression was detected in normal adult skin or nonfibrotic lung tissue. Normally, Egr-1 expression and activity are tightly regulated by endogenous inhibitors, such as NAB2, which is induced by the same signals that also regulate Egr-1, and by Egr-1 itself which can repress its own transcription. In scleroderma lesional tissues, Egr-1 was consistently detected, with highest levels in patients with early and progressive disease, supporting a functional role for Egr-1 in the development and progression of fibrosis. Transcriptional profiling of lesional skin tissue from patients with early-stage scleroderma using cDNA microarrays showed elevated expression of Egr-1 mRNA (F. Tan, unpublished information).53 Whereas lungs from patients without fibrosis showed only low levels of Egr-1, detected in alveolar macrophages and occasional lining cells, fibrotic lesions from scleroderma lungs consistently showed strong Egr-1 staining in stromal cells.
Scleroderma skin fibroblasts displayed elevated Egr-1 mRNA and protein in vitro, although heterogeneity from one cell line to another was observed. A previous study similarly demonstrated overexpression of Egr-1 associated with increased levels of TGF-β and connective tissue growth factor in lung fibroblasts from patients with chronic obstructive pulmonary disease.43 It has been proposed that the scleroderma fibroblast phenotype reflects constitutive autocrine stimulation by TGF-β.54,55 Thus, an autocrine TGF-β stimulatory loop might underlie the persistent expression of Egr-1 in explanted scleroderma fibroblasts. Moreover, Egr-1, by virtue of its capacity to directly stimulate TGF-β production and expression of type 2 TGF-β receptor, might itself further contribute to autocrine TGF-β signaling. However, the strong TGF-β activation signature detected in transcriptional analysis of SSc skin biopsies was largely extinguished in cultured fibroblasts explanted from the same lesions.53,56 Currently, we are exploring the mechanisms for constitutive Egr-1 up-regulation in scleroderma fibroblasts.
Although most tissues demonstrate low basal Egr-1 expression, Egr-1 is readily inducible by acute injury and stress as an early and transient response. The same stimuli that induce Egr-1 expression normally also induce its inhibitors, such as Nab2 and Egr-1, and this negative feedback on Egr-1 activity ensures its rapid extinction. Although short-lived Egr-1 expression and activity orchestrate adaptive tissue response to acute injury, persistent Egr-1 expression results in unchecked target gene activation that contributes to tissue damage. For instance, up-regulated Egr-1 expression in emphysema might be involved in aberrant tissue remodeling,57 persistent Egr-1 expression in vascular smooth muscle cells contributes to atherosclerotic injury,58 and in bowel mucosal cells, it plays a role in chronic inflammation in inflammatory bowel disease.59 Furthermore, in contrast to acute Egr-1 induction that stimulates angiogenesis, sustained Egr-1 expression blocks it.41,60 We have recently found that Egr-1-null mice are partially protected from fibrosis in the bleomycin-induced model of scleroderma (unpublished data). Therefore, persistent Egr-1 expression or activity in lesional tissue might contribute to progression of TGF-β-induced fibrosis and could also suppress local vascular repair contributing to vascular rarefaction characteristic of scleroderma.61
In summary, the present results show that TGF-β is a potent inducer of Egr-1 transcription and Egr-1 accumulation. Stimulation involves a Smad-independent MAPK signaling pathway comparable with Egr-1 regulation associated with acute cellular stress. The upstream components of this pathway that link the activated TGF-β receptors to MEK1 are currently unknown. It is relevant to note in this regard that induction of Egr-1 by oxidative stress was shown to involve the nonreceptor tyrosine kinase c-Abl.62 Because c-Abl can also be activated by TGF-β (W. Ishida, S. Bhattacharyya, and J. Varga, unpublished data), this tyrosine kinase therefore might be responsible for coupling TGF-β to the MEK-ERK-Elk-1 pathway for inducing Egr-1. We are currently exploring this mechanism. The expression of Egr-1, normally tightly regulated, was elevated in bleomycin-induced murine scleroderma, in explanted SSc dermal fibroblasts, and in fibrotic skin or lung tissue from patients with SSc. The inducibility of Egr-1 by TGF-β and by IL-13; its capacity to stimulate production of extracellular matrix genes (collagen and fibronectin), enzymes (matrix metalloproteinases and plasminogen activator inhibitor-1), and fibrogenic cytokines (TGF-β and platelet-derived growth factor); and its persistent expression in fibrotic lesions all point to its potential importance in fibrosis. Blocking Egr-1 expression or biological activity appears to be a potential strategy to control pathological fibrogenesis. Significantly, several drugs in current clinical use have potent inhibitory effects on Egr-1 expression or activity. These drugs include mycophenolate mofetil,63 cyclosporine,64 simvastatin,65 and insulin-sensitizing peroxisome proliferator-activated receptor-γ ligands, such as rosiglitazone (M. Wu and J. Varga, unpublished data).65 Because Egr-1-null mice have no obvious phenotype, it might be feasible to use such drugs to therapeutically target abnormal Egr-1 expression or function in fibrosis with little ill effect.
Acknowledgments
We are grateful to David M. Cohen (Oregon Health and Science University), Eileen D. Adamson (Burnham Institute), and Warren G. Tourtellotte and William Schnaper (Northwestern University) for providing valuable reagents and suggestions and to members of the laboratory for valuable technical assistance and discussions.
Footnotes
Address reprint requests to John Varga, Division of Rheumatology, Northwestern University Feinberg School of Medicine, 240 E. Huron St., Chicago, IL 60611. E-mail: j-varga@northwestern.edu.
Supported by grants from the National Institutes of Health (AR-04239), the Department of Defense, and the Scleroderma Foundation.
S.B. and S.-J.C. contributed equally to this work.
References
- Jimenez SA, Derk CT. Following the molecular pathways toward an understanding of the pathogenesis of systemic sclerosis. Ann Intern Med. 2004;140:37–45. [PubMed] [Google Scholar]
- Varga J, Abraham D. Systemic sclerosis: a prototypic multisystem fibrotic disorder. J Clin Invest. 2007;117:557–567. doi: 10.1172/JCI31139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauviel A. Transforming growth factor-beta: a key mediator of fibrosis. Methods Mol Med. 2005;117:69–80. doi: 10.1385/1-59259-940-0:069. [DOI] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. Smad transcription factors (review). Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. Non-Smad TGF-beta signals (review). J Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Eg-1 (review). J Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- Gashler A, Sukhatme VP. Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- Khachigian LM. Early growth response-1 in cardiovascular pathobiology. Circ Res. 2006;98:186–191. doi: 10.1161/01.RES.0000200177.53882.c3. [DOI] [PubMed] [Google Scholar]
- Ngiam N, Post M, Kavanagh BP. Early growth response factor-1 in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1089–L1091. doi: 10.1152/ajplung.00265.2007. [DOI] [PubMed] [Google Scholar]
- Fu M, Zhang J, Lin Y, Zhu X, Zhao L, Ahmad M, Ehrengruber MU, Chen YE. Early stimulation and late inhibition of peroxisome proliferator-activated receptor gamma (PPARγ) gene expression by transforming growth factor beta in human aortic smooth muscle cells: role of early growth-response factor-1 (Egr-1), activator protein 1 (AP1) and Smads. Biochem J. 2003;370:1019–1025. doi: 10.1042/BJ20021503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SJ, Ning H, Ishida W, Sodin-Semrl S, Takagawa S, Mori Y, Varga J. The early-immediate gene EGR-1 is induced by transforming growth factor-beta and mediates stimulation of collagen gene expression. J Biol Chem. 2006;281:21183–21197. doi: 10.1074/jbc.M603270200. [DOI] [PubMed] [Google Scholar]
- Lee CG, Cho SJ, Kang MJ, Chapoval SP, Lee PJ, Noble PW, Yehualaeshet T, Lu B, Flavell RA, Milbrandt J, Homer RJ, Elias JA. Early growth response gene 1-mediated apoptosis is essential for transforming growth factor beta1-induced pulmonary fibrosis. J Exp Med. 2004;200:377–389. doi: 10.1084/jem.20040104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Chen SJ, Varga J. Modulation of endogenous Smad expression in normal skin fibroblasts by transforming growth factor-beta. Exp Cell Res. 2000;258:374–383. doi: 10.1006/excr.2000.4930. [DOI] [PubMed] [Google Scholar]
- Lakos G, Takagawa S, Chen SJ, Ferreira AM, Han G, Masuda K, Wang XJ, DiPietro LA, Varga J. Targeted disruption of TGF-beta/Smad3 signaling modulates skin fibrosis in a mouse model of scleroderma. Am J Pathol. 2004;165:203–217. doi: 10.1016/s0002-9440(10)63289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhatme VP, Cao XM, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB, Curran T. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- Mori Y, Chen SJ, Varga J. Expression and regulation of intracellular SMAD signaling in scleroderma skin fibroblasts. Arthritis Rheum. 2003;48:1964–1978. doi: 10.1002/art.11157. [DOI] [PubMed] [Google Scholar]
- Lakos G, Takagawa S, Varga J. Animal models of scleroderma. Methods Mol Med. 2004;102:377–393. doi: 10.1385/1-59259-805-6:377. [DOI] [PubMed] [Google Scholar]
- Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S-J, Yuan W, Mori Y, Levenson A, Trojanowska M, Varga J. Stimulation of type I collagen transcription in human skin fibroblasts by TGF-β: involvement of Smad 3. J Invest Dermatol. 1999;112:49–57. doi: 10.1046/j.1523-1747.1999.00477.x. [DOI] [PubMed] [Google Scholar]
- Mori Y, Ishida W, Bhattacharyya S, Li Y, Platanias LC, Varga J. Selective inhibition of activin receptor-like kinase 5 signaling blocks profibrotic transforming growth factor beta responses in skin fibroblasts. Arthritis Rheum. 2004;50:4008–4021. doi: 10.1002/art.20658. [DOI] [PubMed] [Google Scholar]
- Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Hu PP, Shen X, Huang D, Liu Y, Counter C, Wang XF. The MEK pathway is required for stimulation of p21(WAF1/CIP1) by transforming growth factor-beta. J Biol Chem. 1999;274:35381–35387. doi: 10.1074/jbc.274.50.35381. [DOI] [PubMed] [Google Scholar]
- Broekelmann TJ, Limper AH, Colby TV, McDonald JA. Transforming growth factor beta 1 is present at sites of extracellular matrix gene expression in human pulmonary fibrosis. Proc Natl Acad Sci USA. 1991;88:6642–6646. doi: 10.1073/pnas.88.15.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CE, Wilkes MC, Edens M, Kottom TJ, Murphy SJ, Limper AH, Leof EB. Imatinib mesylate inhibits the profibrogenic activity of TGF-beta and prevents bleomycin-mediated lung fibrosis. J Clin Invest. 2004;114:1308–1316. doi: 10.1172/JCI19603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida T, Wu MH, Pierce A, Poncelet AC, Varga J, Schnaper HW. MAP-kinase activity necessary for TGFbeta1-stimulated mesangial cell type I collagen expression requires adhesion-dependent phosphorylation of FAK tyrosine 397. J Cell Sci. 2007;120:4230–4240. doi: 10.1242/jcs.03492. [DOI] [PubMed] [Google Scholar]
- Hayashida T, Decaestecker M, Schnaper HW. Cross-talk between ERK MAP kinase and Smad signaling pathways enhances TGF-beta-dependent responses in human mesangial cells. FASEB J. 2003;17:1576–1588. doi: 10.1096/fj.03-0037fje. [DOI] [PubMed] [Google Scholar]
- Sato M, Shegogue D, Gore EA, Smith EA, McDermott PJ, Trojanowska M. Role of p38 MAPK in transforming growth factor beta stimulation of collagen production by scleroderma and healthy dermal fibroblasts. J Invest Dermatol. 2002;118:704–711. doi: 10.1046/j.1523-1747.2002.01719.x. [DOI] [PubMed] [Google Scholar]
- Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13:115–124. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Zhu X, Zhang J, Liang J, Lin Y, Zhao L, Ehrengruber MU, Chen YE. Egr-1 target genes in human endothelial cells identified by microarray analysis. Gene. 2003;315:33–41. doi: 10.1016/s0378-1119(03)00730-3. [DOI] [PubMed] [Google Scholar]
- Krones-Herzig A, Adamson E, Mercola D. Early growth response 1 protein, an upstream gatekeeper of the p53 tumor suppressor, controls replicative senescence. Proc Natl Acad Sci USA. 2003;100:3233–3238. doi: 10.1073/pnas.2628034100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Hyman MC, Lawrence DA, Pinsky DJ. Molecular regulation of the PAI-1 gene by hypoxia: contributions of Egr-1, HIF-1alpha, and C/EBPalpha. FASEB J. 2007;21:935–949. doi: 10.1096/fj.06-6285com. [DOI] [PubMed] [Google Scholar]
- Hasan RN, Schafer AI. Hemin up-regulates Egr-1 expression in vascular smooth muscle cells via ROS ERK-1/2 Elk-1 and NF-{kappa}B. Circ Res. 2008;102:42–50. doi: 10.1161/CIRCRESAHA.107.155143. [DOI] [PubMed] [Google Scholar]
- Hjoberg J, Le L, Imrich A, Subramaniam V, Mathew SI, Vallone J, Haley KJ, Green FH, Shore SA, Silverman ES. Induction of early growth-response factor 1 by platelet-derived growth factor in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2004;286:L817–L825. doi: 10.1152/ajplung.00190.2003. [DOI] [PubMed] [Google Scholar]
- Midgley VC, Khachigian LM. Fibroblast growth factor-2 induction of platelet-derived growth factor-C chain transcription in vascular smooth muscle cells is ERK-dependent but not JNK-dependent and mediated by Egr-1. J Biol Chem. 2004;279:40289–40295. doi: 10.1074/jbc.M406063200. [DOI] [PubMed] [Google Scholar]
- Grotegut S, von Schweinitz D, Christofori G, Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated up-regulation of Snail. EMBO J. 2006;25:3534–3545. doi: 10.1038/sj.emboj.7601213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggioli C, Deckert M, Robert G, Abbe P, Batoz M, Ehrengruber MU, Ortonne JP, Ballotti R, Tartare-Deckert S. HGF induces fibronectin matrix synthesis in melanoma cells through MAP kinase-dependent signaling pathway and induction of Egr-1. Oncogene. 2005;24:1423–1433. doi: 10.1038/sj.onc.1208318. [DOI] [PubMed] [Google Scholar]
- Liu C, Yao J, Mercola D, Adamson E. The transcription factor EGR-1 directly transactivates the fibronectin gene and enhances attachment of human glioblastoma cell line U251. J Biol Chem. 2000;275:20315–20323. doi: 10.1074/jbc.M909046199. [DOI] [PubMed] [Google Scholar]
- Liu C, Adamson E, Mercola D. Transcription factor EGR-1 suppresses the growth and transformation of human HT-1080 fibrosarcoma cells by induction of transforming growth factor beta 1. Proc Natl Acad Sci USA. 1996;93:11831–11836. doi: 10.1073/pnas.93.21.11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder PJ, Bernadt CT, Kim JH, Rizzino A. Stimulation of the murine type II transforming growth factor-beta receptor promoter by the transcription factor Egr-1. Mol Reprod Dev. 2002;63:282–290. doi: 10.1002/mrd.10165. [DOI] [PubMed] [Google Scholar]
- Fahmy RG, Dass CR, Sun LQ, Chesterman CN, Khachigian LM. Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nat Med. 2003;9:1026–1032. doi: 10.1038/nm905. [DOI] [PubMed] [Google Scholar]
- Li CJ, Ning W, Matthay MA, Feghali-Bostwick CA, Choi AM. MAPK pathway mediates EGR-1-HSP70-dependent cigarette smoke-induced chemokine production. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1297–L1303. doi: 10.1152/ajplung.00194.2006. [DOI] [PubMed] [Google Scholar]
- Ning W, Li CJ, Kaminski N, Feghali-Bostwick CA, Alber SM, Di YP, Otterbein SL, Song R, Hayashi S, Zhou Z, Pinsky DJ, Watkins SC, Pilewski JM, Sciurba FC, Peters DG, Hogg JC, Choi AM. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci USA. 2004;101:14895–14900. doi: 10.1073/pnas.0401168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aicher WK, Alexander D, Haas C, Kuchen S, Pagenstecher A, Gay S, Peter HH, Eibel H. Transcription factor early growth response 1 activity up-regulates expression of tissue inhibitor of metalloproteinases 1 in human synovial fibroblasts. Arthritis Rheum. 2003;48:348–359. doi: 10.1002/art.10774. [DOI] [PubMed] [Google Scholar]
- Yan SF, Zou YS, Gao Y, Zhai C, Mackman N, Lee SL, Milbrandt J, Pinsky D, Kisiel W, Stern D. Tissue factor transcription driven by Egr-1 is a critical mechanism of murine pulmonary fibrin deposition in hypoxia. Proc Natl Acad Sci USA. 1998;95:8298–8303. doi: 10.1073/pnas.95.14.8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SJ, Kang MJ, Homer RJ, Kang HR, Zhang X, Lee PJ, Elias JA, Lee CG. Role of early growth response-1 (Egr-1) in interleukin-13-induced inflammation and remodeling. J Biol Chem. 2006;281:8161–8168. doi: 10.1074/jbc.M506770200. [DOI] [PubMed] [Google Scholar]
- Miano JM, Berk BC. NAB2: a transcriptional brake for activated gene expression in the vessel wall? Am J Pathol. 1999;155:1009–1012. doi: 10.1016/S0002-9440(10)65200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JC, Liu L, Cooley BC, DiChiara MR, Topper JN, Aird WC. The Egr-1 promoter contains information for constitutive and inducible expression in transgenic mice. FASEB J. 2000;14:1870–1872. doi: 10.1096/fj.99-1072fje. [DOI] [PubMed] [Google Scholar]
- Clark SH. Animal models in scleroderma. Curr Rheumatol Rep. 2005;7:150–155. doi: 10.1007/s11926-005-0068-x. [DOI] [PubMed] [Google Scholar]
- Ingram JL, Antao-Menezes A, Mangum JB, Lyght O, Lee PJ, Elias JA, Bonner JC. Opposing actions of Stat1 and Stat6 on IL-13-induced up-regulation of early growth response-1 and platelet-derived growth factor ligands in pulmonary fibroblasts. J Immunol. 2006;177:4141–4148. doi: 10.4049/jimmunol.177.6.4141. [DOI] [PubMed] [Google Scholar]
- Yan SF, Fujita T, Lu J, Okada K, Shan Zou Y, Mackman N, Pinsky DJ, Stern DM. Egr-1, a master switch coordinating up-regulation of divergent gene families underlying ischemic stress. Nat Med. 2000;6:1355–1361. doi: 10.1038/82168. ; erratum in Nat Med 2001, 7:509. [DOI] [PubMed] [Google Scholar]
- Day RM, Yang Y, Suzuki YJ, Stevens J, Pathi R, Perlmutter A, Fanburg BL, Lanzillo JJ. Bleomycin up-regulates gene expression of angiotensin-converting enzyme via mitogen-activated protein kinase and early growth response 1 transcription factor. Am J Respir Cell Mol Biol. 2001;25:613–619. doi: 10.1165/ajrcmb.25.5.4521. [DOI] [PubMed] [Google Scholar]
- Tan FK, Hildebrand BA, Lester MS, Stivers DN, Pounds S, Zhou X, Wallis DD, Milewicz DM, Reveille JD, Mayes MD, Jin L, Arnett FC., Jr Classification analysis of the transcriptosome of nonlesional cultured dermal fibroblasts from systemic sclerosis patients with early disease. Arthritis Rheum. 2005;52:865–876. doi: 10.1002/art.20871. [DOI] [PubMed] [Google Scholar]
- Ihn H. Autocrine TGF-beta signaling in the pathogenesis of systemic sclerosis. J Dermatol Sci. 2008;49:103–113. doi: 10.1016/j.jdermsci.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Trojanowska M, Varga J. Molecular pathways as novel therapeutic targets in systemic sclerosis. Curr Opin Rheumatol. 2007;19:568–573. doi: 10.1097/BOR.0b013e3282e6f495. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Finlay DR, Murray JI, Troyanskaya OG, Chi JT, Pergamenschikov A, McCalmont TH, Brown PO, Botstein D, Connolly MK. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci USA. 2003;100:12319–12324. doi: 10.1073/pnas.1635114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yan SD, Zhu A, Zou YS, Williams M, Godman GC, Thomashow BM, Ginsburg ME, Stern DM, Yan SF. Expression of Egr-1 in late stage emphysema. Am J Pathol. 2000;157:1311–1320. doi: 10.1016/S0002-9440(10)64646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey TA, Fu C, Du B, Eksinar S, Kent KC, Bush H, Jr, Kreiger K, Rosengart T, Cybulsky MI, Silverman ES, Collins T. High-level expression of Egr-1 and Egr-1-inducible genes in mouse and human atherosclerosis. J Clin Invest. 2000;105:653–662. doi: 10.1172/JCI8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaramaiah K, Yoshimatsu K, Scherl E, Das KM, Glazier KD, Golijanin D, Soslow RA, Tanabe T, Naraba H, Dannenberg AJ. Microsomal prostaglandin E synthase-1 is overexpressed in inflammatory bowel disease: evidence for involvement of the transcription factor Egr-1. J Biol Chem. 2004;279:12647–12658. doi: 10.1074/jbc.M312972200. [DOI] [PubMed] [Google Scholar]
- Lucerna M, Pomyje J, Mechtcheriakova D, Kadl A, Gruber F, Bilban M, Sobanov Y, Schabbauer G, Breuss J, Wagner O, Bischoff M, Clauss M, Binder BR, Hofer E. Sustained expression of early growth response protein-1 blocks angiogenesis and tumor growth. Cancer Res. 2006;66:6708–6713. doi: 10.1158/0008-5472.CAN-05-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JN, Nash RA, McLeod DO, Fiorentino DF, Shulman HM, Connolly MK, Molitor JA, Henstorf G, Lafyatis R, Pritchard DK, Adams LD, Furst DE, Schwartz SM. Capillary regeneration in scleroderma: stem cell therapy reverses phenotype? PLoS ONE. 2008;16:e1452. doi: 10.1371/journal.pone.0001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JR, Kawai H, Tsai KK, Chuang EY, Yuan ZM. c-Abl regulates early growth response protein (EGR1) in response to oxidative stress. Oncogene. 2005;24:8085–8092. doi: 10.1038/sj.onc.1208953. [DOI] [PubMed] [Google Scholar]
- Farivar AS, MacKinnon-Patterson B, Barnes AD, Mulligan MS. The effect of anti-inflammatory properties of mycophenolate mofetil on the development of lung reperfusion injury. J Heart Lung Transplant. 2005;24:2235–2242. doi: 10.1016/j.healun.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Farivar AS, Mackinnin-Patterson BC, Barnes AD, McCourtie AS, Mulligan MS. Cyclosporine modulates the response to hypoxia-reoxygenation in pulmonary artery endothelial cells. Ann Thorac Surg. 2005;79:1010–1016. doi: 10.1016/j.athoracsur.2004.08.078. [DOI] [PubMed] [Google Scholar]
- Bea F, Blessing E, Shelley MI, Shultz JM, Rosenfeld ME. Simvastatin inhibits expression of tissue factor in advanced atherosclerotic lesions of apolipoprotein E deficient mice independently of lipid lowering: potential role of simvastatin-mediated inhibition of Egr-1 expression and activation. Atherosclerosis. 2003;167:187–194. doi: 10.1016/s0021-9150(02)00387-8. [DOI] [PubMed] [Google Scholar]
- Okada M, Wen F, Yan SF. Peroxisome proliferator-activated receptor-gamma (PPAR-gamma) activation suppresses ischemic induction of Egr-1 and its inflammatory gene targets. FASEB J. 2002;16:1861–1868. doi: 10.1096/fj.02-0503com. [DOI] [PubMed] [Google Scholar]