Abstract
Denaturing gradient gel electrophoresis (DGGE) of a 16S ribosomal DNA PCR product was used to differentiate 32 mycoplasma species of veterinary significance. Twenty-seven (85%) species could be differentiated by DGGE. This method could enable the rapid identification of many mycoplasma species for which there is no specific PCR available and which are currently identified by using culture and serological tests.
Mycoplasmas belong to the class Mollicutes and are among the smallest free-living microorganisms capable of autoreplication. Mycoplasmas are highly fastidious bacteria, difficult to culture and slow growing. Many species are important veterinary pathogens causing respiratory infection, mastitis, conjunctivitis, arthritis, and occasionally abortion; they include Mycoplasma mycoides subsp. mycoides small colony (SC), the causative agent of the Office International des Epizooties list A disease contagious bovine pleuropneumonia (12). Identification of mycoplasmas as the causative agents of disease is often hindered by the lack of rapid diagnostic tests together with similarities in the clinical diseases that they cause. Conventional methods of diagnosis are based on culture and serological tests, such as the complement fixation test (9), enzyme-linked immunosorbent assay (2), and immunoblotting (15), and can be time-consuming, insensitive, and nonspecific.
Recently, PCR has been employed for the laboratory diagnosis of some veterinary mycoplasmas, including species of the closely related M. mycoides cluster (3), Mycoplasma gallisepticum (5), Mycoplasma hyopneumoniae (19), and Mycoplasma bovis (1). Differentiation of mycoplasma by PCR based on specific primers has been limited, as there is little interspecific variation in 16S ribosomal DNA (rDNA). The identification of alternative genes suitable for PCR has been hampered by the lack of sequenced animal mycoplasma genomes. To date, there has not been a single generic test that enables mycoplasma identification to the species level.
Denaturing gradient gel electrophoresis (DGGE) can theoretically detect single base mutations in DNA (4, 7). The method is based on the prevention of migration of DNA fragments following strand separation caused by chemical denaturants. DGGE has been used extensively for diversity analysis in microbial ecology (10) but has not been widely used for the identification and differentiation of pathogenic bacteria except for the detection and identification of Listeria species (N. Schmolker, F. van Ommen Kloeke, and G. Geesey, Bio-Rad Mutation Analysis, technical note 2403) and for the molecular typing of Staphylococcus aureus (6) and Campylobacter species (16). This study used PCR-DGGE of the V3 region of the 16S rDNA gene to differentiate 32 species of mycoplasma commonly associated with animal disease.
The mycoplasma strains used in this study are listed in Table 1. All strains were cultured as described previously (13). Mycoplasma DNA was extracted from a 1-ml aliquot of stationary-phase culture as described previously (17). Amplification of the V3 region of the 16S RNA gene was performed according to the method of Muyzer et al. (11) with universal bacterial primers GC-341F (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG) and 534R (5′-ATT ACC GCG GCT GCT GG) (11). For PCR, 1 μl of lysate was added as a template to 49 μl of a reaction mixture containing 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 0.2 mM (each) deoxynucleoside triphosphates, and 0.5 U of Taqgold (Applied Biosystems). The cycling conditions were: denaturation at 94°C for 5 min followed by 30 cycles of 95°C for 1 min, 55°C for 45 s, and 72°C for 1 min and a final extension step of 72°C for 10 min, and samples were kept at 4°C until analysis. DGGE was performed with the Ingeny PhorU 2 × 2 apparatus (GRI Molecular Biology, Essex, United Kingdom). Samples were loaded onto 10% polyacrylamide-bis (37.5:1) gels with denaturing gradients from 30 to 60% (where 100% is 7 M urea and 40% [vol/vol] deionized formamide) in 1× Tris-acetate-EDTA electrophoresis buffer (Severn Biotech Ltd., Worcestershire, United Kingdom). Electrophoresis was performed at 100 V and 60°C for 18 h. Gels were then stained with SBYR Gold (Cambridge BioScience, Cambridgeshire, United Kingdom) in 1× Tris-acetate-EDTA for 30 min at room temperature and visualized under UV illumination.
TABLE 1.
Mycoplasma species used in this study
| Species | Strain | Host |
|---|---|---|
| M. cloacale | NCTC 10199 | Avian |
| M. gallinarum | NCTC 10120 | Avian |
| M. gallisepticum | NCTC 10115 | Avian |
| M. gallopavonis | NCTC 10186 | Avian |
| M. glycophilum | NCTC 10194 | Avian |
| M. iners | NCTC 10165 | Avian |
| M. iowae | NCTC 10185 | Avian |
| M. lipofaciens | NCTC 10191 | Avian |
| M. meleagridis | NCTC 10153 | Avian |
| M. synoviae | NCTC 10124 | Avian |
| M. agalactiae | NCTC 10123 | Ovine/caprine |
| M. arginini | NCTC 10129 | Ovine/bovine |
| M. capricolum subsp. capricolum | NCTC 10154 | Caprine |
| M. capricolum subsp. capripneumoniae | NCTC 10192 | Caprine |
| M. conjunctivae | NCTC 10147 | Ovine |
| M. cottewii | NCTC 11732 | Caprine |
| M. mycoides subsp. mycoides LC | F30 | Ovine/caprine |
| M. mycoides subsp. capri | NCTC 10137 | Caprine |
| Mycoplasma ovine/caprine serogroup 11 | 2D | Ovine/caprine |
| M. ovipneumoniae | NCTC 10151 | Ovine |
| M. putrefaciens | NCTC 10155 | Ovine/caprine |
| M. flocculare | NCTC 10143 | Porcine |
| M. hyopneumoniae | NCTC 10110 | Porcine |
| M. hyorhinis | NCTC 10130 | Porcine |
| M. hyosynoviae | NCTC 10167 | Porcine |
| M. bovigenitalium | NCTC 10122 | Bovine |
| M. bovirhinis | NCTC 10118 | Bovine |
| M. bovis | NCTC 10131 | Bovine |
| M. bovoculi | NCTC 10141 | Bovine |
| M. canis | PG14 | Bovine/canine |
| M. californicum | NCTC 10146 | Bovine/canine |
| M. dispar | NCTC 10125 | Bovine |
PCR products of approximately 340 bp were obtained for all 32 Mycoplasma species tested. These products were subjected to DGGE in groups according to host animal. In the majority of cases only one band was seen, indicating that there was no intraspecific variation in the amplified sequence. The presence of multiple bands indicated that more than one 16S rDNA operon was present and that there were some sequence differences between the copies.
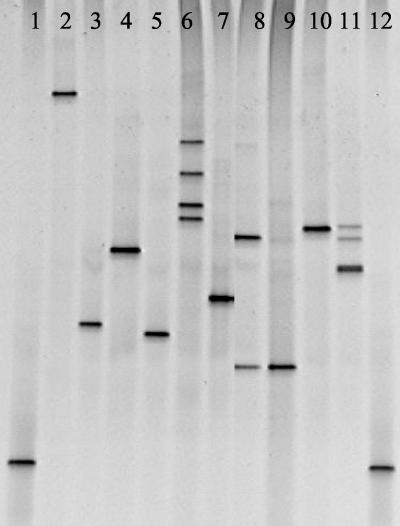
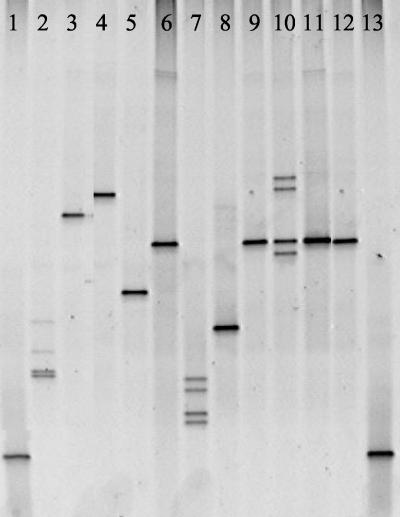
The use of DGGE to separate PCR products according to differences in primary sequence enabled differentiation of all 10 avian mycoplasmas species analyzed (Fig. 1). This indicated that the sequence was different for each isolate. However, DGGE could differentiate only 7 of the 10 small-ruminant mycoplasmas tested, which was due to identical DGGE profiles being obtained for the closely related members of the M. mycoides cluster Mycoplasma capricolum subsp. capripneumoniae, Mycoplasma mycoides subsp. capri, M. mycoides subsp. mycoides large colony, and Mycoplasma putrefaciens (Fig. 2). Interestingly, M. putrefaciens also gave a profile identical to that of the M. mycoides cluster members, providing further evidence that M. putrefaciens should be included in the M. mycoides cluster (21). Although DGGE was unable to distinguish many of the M. mycoides cluster members when PCR of the 16S gene was used, it is possible that DGGE based on alternative targets, such as the 16S-23S intergenic spacer region, might enable differentiation.
FIG. 1.
DGGE of avian mycoplasmas. Lanes: 1 and 12, Escherichia coli marker; 2, M. gallisepticum; 3, M. gallinarum; 4, M. iners; 5, M. gallopavonis; 6, M. lipofaciens; 7, M. cloacale; 8, M. iowae; 9, M. glycophilum; 10, M. synoviae; 11, M. meleagridis.
FIG. 2.
DGGE of ovine and caprine mycoplasmas. Lanes: 1 and 13, E. coli marker; 2, M. agalactiae; 3, M. ovipneumoniae; 4, M. ovine/caprine serogroup 11; 5, M. cottewii; 6, M. capricolum subsp. capripneumoniae; 7, M. arginini; 8, M. conjunctivae; 9, M. mycoides subsp. capri; 10, M. capricolum subsp. capricolum; 11, M. mycoides subsp. mycoides large colony; 12, M. putrefaciens.
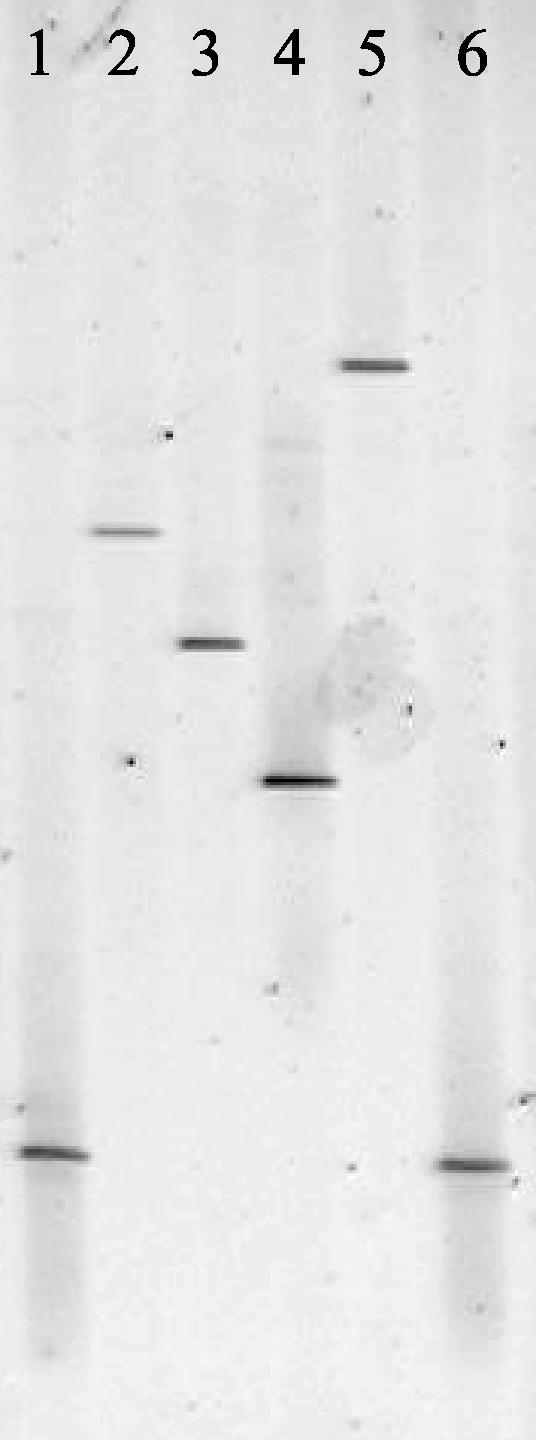
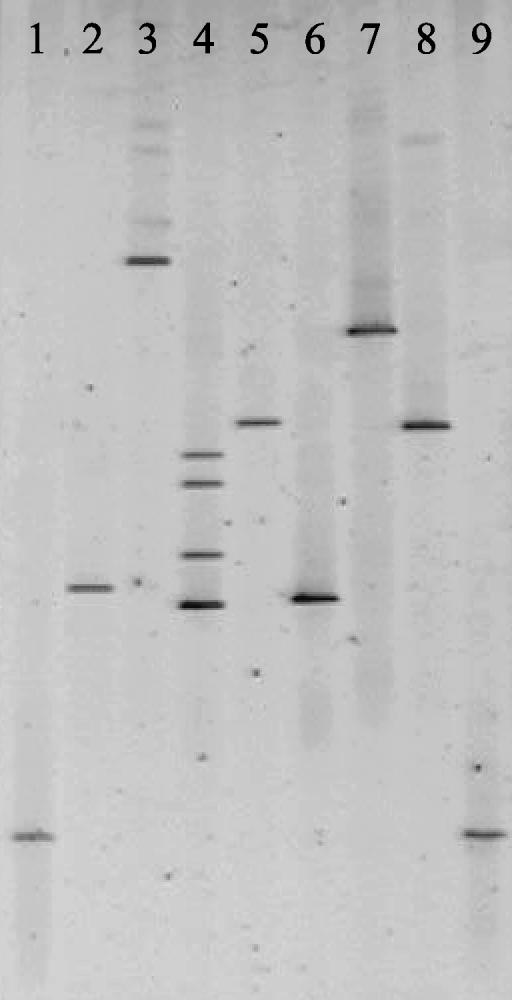
All four porcine mycoplasmas gave different DGGE profiles (Fig. 3). DGGE also enabled the differentiation of five bovine mycoplasmas (Fig. 4). However, Mycoplasma californicum and Mycoplasma dispar could not be distinguished, and the banding patterns of M. bovis and Mycoplasma canis were similar. Interestingly, the migration of the DGGE band obtained for the bovine pathogen Mycoplasma bovigenitalium was identical to the small-ruminant mycoplasma Mycoplasma ovine/caprine serogroup 11 (Fig. 5). These results are not surprising, as it has recently been proposed that Mycoplasma ovine/caprine serogroup 11 and M. bovigenitalium should be redesignated as the same species based on their close phenotypic and genotypic similarity (14).
FIG. 3.
DGGE of porcine mycoplasmas. Lanes: 1 and 6, E. coli marker; 2, M. flocculare; 3, M. hyopneumoniae; 4, M hyosynoviae; 5, M. hyorhinis.
FIG. 4.
DGGE of bovine mycoplasmas. Lanes: 1 and 9, E. coli marker; 2, M. bovis; 3, M. bovoculi; 4, M. bovirhinis; 5, M. californicum; 6, M. canis; 7, M. bovigenitalium; 8, M. dispar.
FIG. 5.
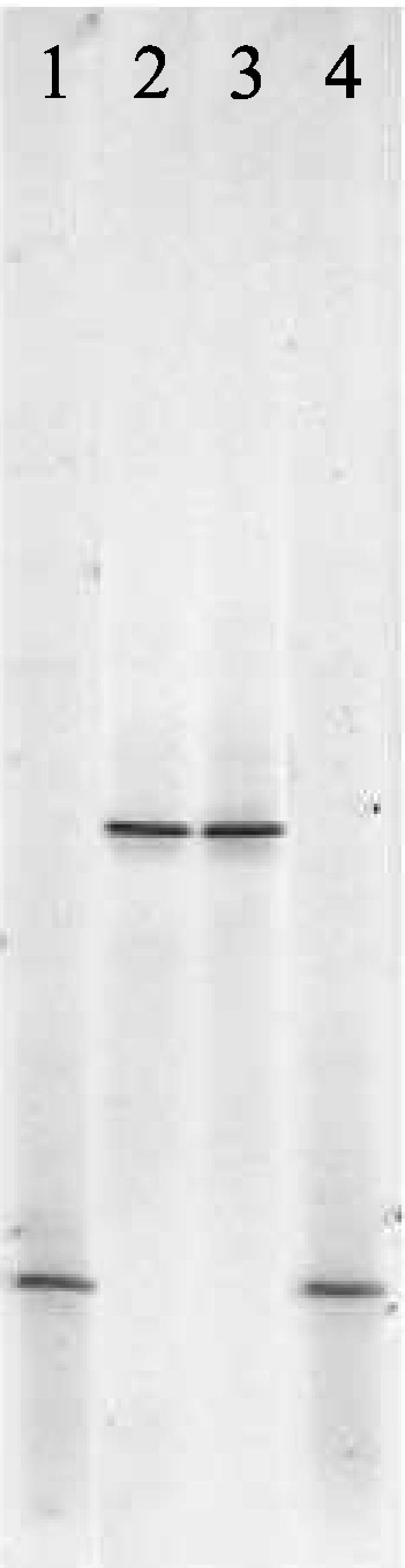
DGGE of M. bovigenitalium and Mycoplasma ovine/caprine serogroup 11. Lanes: 1 and 4, E. coli marker; 2, M. bovigenitalium; 3, Mycoplasma ovine/caprine serogroup 11.
To determine the robustness of DGGE for mycoplasma species identification, 20 strains of M. mycoides subsp. mycoides SC from Africa, Australia, and Europe were compared by using DGGE. The same DGGE profile was obtained with 22 of the 23 M. mycoides subsp. mycoides SC strains tested. The only strain that gave a different DGGE profile was a vaccine strain, T144 (results not shown). Previous studies have also indicated that the vaccine strains of M. mycoides subsp. mycoides SC are genotypically different from the field strains (8, 20). Clearly, further work on more strains of each species needs to be carried out in order to confirm that the same profiles are obtained.
This is the first study to detect and differentiate all major mycoplasmas of veterinary importance with a single test. DGGE of the 16S rRNA gene could differentiate almost all mycoplasmas within a host animal group. The application of this technique could be particularly useful for mycoplasmas that are difficult to culture, such as porcine mycoplasmas. In addition, mycoplasmas that are difficult to distinguish by using normal culture and serological techniques, including M. bovis and Mycoplasma agalactiae, can be differentiated (13).
As universal bacterial primers were used, the approach described here will not be limited to species that have already been characterized; novel and unknown species will also be detected. The disadvantage of this is that bacteria other than members of the Mollicutes will also generate a band on the DGGE gel, which may give confusing results unless the Mycoplasma organisms have been enriched by cultivation. This may be overcome by designing Mycoplasma-specific primers.
Although DGGE is rapid compared with traditional culture and serological techniques (typically taking less than 24 h compared with up to 2 weeks for serological or culture-based diagnosis), it is feasible that the process could be accelerated further by the use of DNA chip technology. The use of 16S oligonucleotide probes has already been applied to the detection of soil microorganisms (18), and there is increasing interest in its use for the detection of pathogenic bacteria (22).
Acknowledgments
We thank Defra for their continuing financial support.
REFERENCES
- 1.Ayling, R. D., R. A. J. Nicholas, and K. E. Johansson. 1997. Application of the polymerase chain reaction for the routine identification of Mycoplasma bovis. Vet. Rec. 141:307-308. [DOI] [PubMed] [Google Scholar]
- 2.Ball, H. J., and D. Finlay. 1998. Diagnostic application of monoclonal antibody (MAb)-based sandwich ELISAs. Methods Mol. Biol. 104:127-132. [DOI] [PubMed] [Google Scholar]
- 3.Bashiruddin, J. B., T. K. Taylor, and A. R. Gould. 1994. A PCR-based test for the specific identification of Mycoplasma mycoides subspecies mycoides SC. J. Vet. Diagn. Investig. 6:428-434. [DOI] [PubMed] [Google Scholar]
- 4.Fisher, S. G., and L. S. Lerman. 1983. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with theory. Proc. Natl. Acad. Sci. USA 80:1579-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia, M., M. W. Jackwood, M. Head, S. Levisohn, and S. H. Kleven. 1996. Use of species-specific oligonucleotide probes to detect Mycoplasma gallisepticum, M. synoviae, and M. iowae PCR amplification products. J. Vet. Diagn. Investig. 8:56-63. [DOI] [PubMed] [Google Scholar]
- 6.Gürtler, V., H. D. Barrie, and B. C. Mayall. 2001. Use of denaturing gradient gel electrophoresis to detect mutations in VS2 of the 16S-23S rDNA spacer amplified from Staphylococcus aureus isolates. Electrophoresis 22:1920-1924. [DOI] [PubMed] [Google Scholar]
- 7.Lerman, L. S., and C. Beldjord. 1999. Comprehensive mutation detection with denaturing gradient gel electrophoresis, p. 35-61. In R. G. H. Cotton, E. Edkins, and S. Forrest (ed.), Mutation detection. Oxford University Press, Inc., New York, N.Y.
- 8.March, J. B., J. Clark, and M. Brodlie. 2000. Characterization of strains of Mycoplasma mycoides subsp. mycoides small colony type isolated from recent outbreaks of contagious bovine pleuropneumonia in Botswana and Tanzania: evidence for a new biotype. J. Clin. Microbiol. 38:1419-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muthomi, E. K., and F. R. Rurangirwa. 1983. Passive haemagglutination and complement fixation as diagnostic tests for contagious caprine pleuropneumonia caused by the F-38 strain of mycoplasma. Res. Vet. Sci. 35:1-4. [PubMed] [Google Scholar]
- 10.Muyzer, G. 1999. DGGE/TGGE: a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 11.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholas, R. A. J. 1998. The veterinary significance of mycoplasmas. Methods Mol. Biol. 104:17-24. [DOI] [PubMed] [Google Scholar]
- 13.Nicholas, R. A. J., and S. E. Baker. 1998. Recovery of mycoplasmas from animals. Methods Mol. Biol. 104:37-43. [DOI] [PubMed] [Google Scholar]
- 14.Nicholas, R. A. J., L. A. Khan, B. Houshaymi, R. J. Miles. R. D. Ayling, H. Hotzel, and K. Sachse. 2002. Close phylogenetic and phenotypic relatedness between Mycoplasma ovine/caprine serogroup 11 and Mycoplasma bovigenitalium. Syst. Appl. Microbiol. 25:396-402. [DOI] [PubMed] [Google Scholar]
- 15.Nicholas, R. A. J., F. G. Santini, K. M. Clark, N. M. A. Palmer, P. D. Santis, and J. B. Bashiruddin. 1996. A comparison of serological tests and gross lung pathology for the detection of contagious bovine pleuropneumonia in two groups of Italian cattle. Vet. Rec. 139:89-93. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen, E. M., J. Engberg, V. Fussing, L. Petersen, C. H. Brogren, and S. L. On. 2000. Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J. Clin. Microbiol. 38:3800-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stark, K. D., J. Nicolet, and J. Frey. 1998. Detection of Mycoplasma hyopneumoniae by air sampling with a nested PCR assay. Appl. Environ. Microbiol. 64:543-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiaucourt, F., S. Lorenzon, A. David, J. J. Tulasne, and J. Domench. 1998. Vaccination against contagious bovine pleuropneumonia and the use of molecular tools in epidemiology. Ann. N. Y. Acad. Sci. 849:146-151. [DOI] [PubMed] [Google Scholar]
- 21.Weisburg, W. G., J. G. Tully, D. L. Rose, J. P. Petzel, H. Oyaizu, D. Yang, L. Mandelco, J. Sechrest, T. G. Lawrence, J. Van Etten, et al. 1989. A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 171:6455-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson, W. J., C. L. Strout, T. Z. DeSantis, J. L. Stilwell, A. V. Carrano, and G. L. Andersen. 2002. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes 16:119-127. [DOI] [PubMed] [Google Scholar]