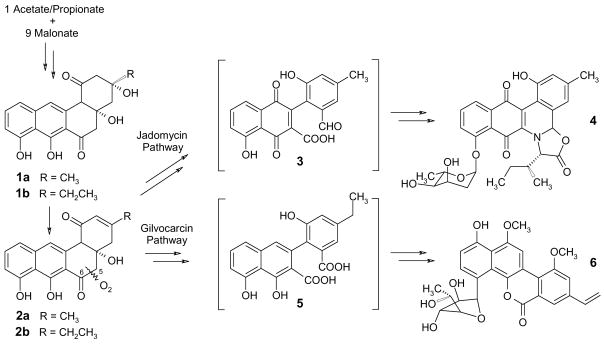
The biosyntheses of the gilvocarcins (e.g., gilvocarcin V, 6, scheme 1), and jadomycins (e.g., jadomycin B, 4) are both dominated by oxidative rearrangement cascades that lead from simple polyketide-derived angucyclinones to the unique tetracyclic benzo[d]naphtho[1,2-b]pyran-6-one and pentacyclic benz[b]oxazolophenanthridine backbones, respectively, which are typical and essential for these anticancer antibiotics. Previous work showed that UWM6 (1a) and prejadomycin (= 2,3-dehydro-UWM6, 2a) or their homologues 1b and 2b are the last isolable intermediates of these pathways and also serve as intermediates of various other angucycline group antibiotics.1–4 The immediately following multi-step oxygenation cascades, that include the key rearrangements of the jadomycin and gilvocarcin biosyntheses, remained widely obscure and were referred to as biosynthetic black box.2 For the jadomycin and the gilvocarcin pathways, two similar, but clearly different products of the oxidative 5,6-bond cleavage of 2 were suggested, namely a phenyl-naphthoquinone decorated with an acid and an aldehyde function (3) in the former and a phenyl-naphthol-diacid (5) for the latter.2, 4 The reactive aldehyde function in 3 appeared essential for the non-enzymatic incorporation of L-isoleucine into the 4-pathway,5–7 while an acid function in 5 seemed to be necessary for the lactone formation in the 6-pathway.4 In this communication, we present results of our further examination of these key reaction cascades, which suggest that jadomycin and gilvocarcin biosynthesis diverge later in the respective pathways than previously assumed, and that the 5,6-bond cleavage necessitates the formation of multi-oxygenase complexes consisting of at least 3 and 4 enzymes, respectively.
Scheme 1.
Key oxidative cleavage steps of the jadomycin and gilvocarcin biosyntheses, with the previously suggested different hypothetical intermediates 3 and 5 resulting from the oxidative 5,6-bond cleavage.
From previous work and alignment studies,2–4, 8–10 we concluded that the oxygenases JadF, JadG, and JadH are participating in the oxidative rearrangement of the jadomycin pathway, while their counterparts GilOIV, GilOII, and GilOI catalyze similar reactions in the gilvocarcin pathway. JadF/H and GilOIV/OI were suggested to prepare and perform the oxidative 5,6-bond cleavage, for which a Baeyer-Villiger oxidation is likely to serve as an essential key step.11, 12 JadG and GilOII showed high similarities to co-factor free monooxygenases such as DnrG or TcmH, which catalyze anthrone oxidation reactions.13–16 For the gilvocarcin pathway, we also considered GilR in this study, a putative oxidoreductase that has strong similarities with an FAD/FMN-depending dehydrogenase from the steffimycin biosynthesis and an oxidoreductase from the spinosad biosynthesis.17, 18 However, the exact role of any of these enzymes remained unclear, both in the gilvocarcin pathway as well as in any of the other pathways that require a GilR homologue.
To compare the jadomycin with the gilvocarcin enzymes, we used our previously generated oxygenase-inactivation mutants4, 19 along with newly generated single and double mutants, and carried out various gene complementation experiments (for the complete set of experiments, see Table 1 in the supporting information). These studies revealed that JadH and JadF were interchangeable with GilOI and GilOIV, respectively. For instance, the gilOI-minus mutant S. lividans (cosG9B3-OI−) could be complemented with JadH (but not with JadF or JadG) to reconstitute 6-biosynthesis, and gilvocarcin biosynthesis could only be restored in the gilOIV-minus mutant S. lividans (cosG9B3-OIV−) through complementation with jadF. However, jadG was not able to replace gilOII, pointing out somewhat different roles of these closely related enzymes in both pathways.
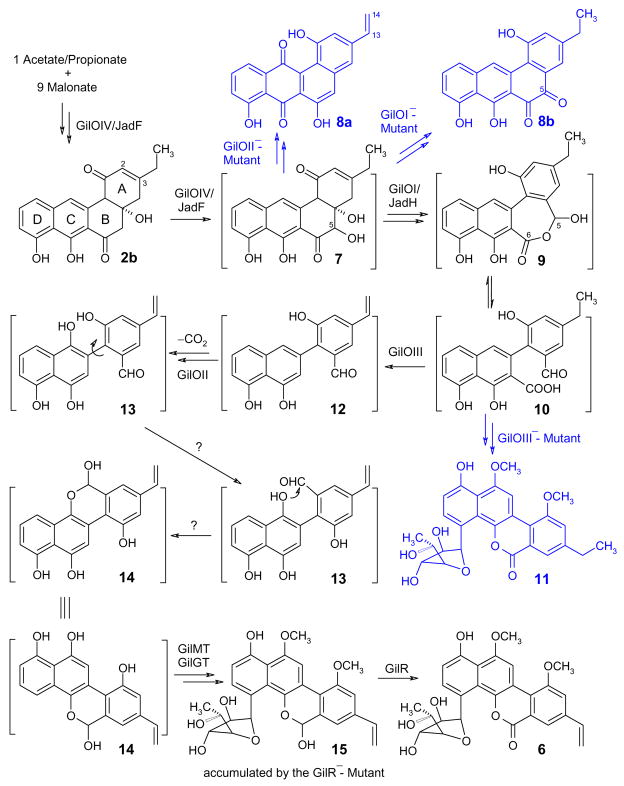
The newly generated gilOII- and gilR-minus mutant strains accumulated intriguing metabolites, which were isolated and the structures were investigated with NMR and mass spectrometry (for physico-chemical data of the new compounds and their derivatives, see supporting material). The gilOII-minus mutant S. lividans (cosG9B3-OII−) accumulated vinyl-dehydrorabelomycin (8a, scheme 2) as its major metabolite, an angucyclinone shunt product, which proves that GilOII partakes as an essential component of the C-C-bond cleavage process within the oxidative rearrangement cascade of the gilvocarcin pathway. This was not unexpected, since the jadG inactivation had also yielded angucycline and angucyclinone shunt products, suggesting a similar important role in the oxidative rearrangement of the jadomycin pathway.2 However, the vinyl group in molecule 8a reveals that the GilOIII-reaction, that establishes the vinyl group, occurs much earlier in the pathway than expected from the accumulation of gilvocarcin E (11) in the gilOIII-minus mutant. We now assume that the GilOIII-catalyzed reaction occurs as soon as the aromatization of the angucyclic ring A is complete or shortly afterwards (see scheme 2). The gilR-minus mutant S. lividans (cosG9B3-R−) accumulated pregilvocarcin V (15) as its major metabolite. Compound 15 appeared as a mixture of two diastereomeric compounds. Isolating each one of these from and re-injecting it into the HPLC showed the same mixture of two compounds in the same ratio, indicating that these two compounds were in equilibrium. Once a hemiacetal was suspected from the NMR data of the mixture, stirring in acidic methanol yielded the corresponding methylacetals, which were separated and characterized by NMR (supporting information) leading to the unambiguous elucidation of structure 15. The accumulation of hemiacetal 15 as a result of inactivating gilR shows that GilR acts as a dehydrogenase oxidizing hemicacetal 15 to the corresponding lactone, gilvocarcin V (6, scheme 2). In addition, GilR might also directly or indirectly regenerate the co-factor free GilOII, while the regeneration of the co-factor free JadG in the jadomycin pathway (and its analogues of the daunorubicin and tetracenomycin pathways) is achieved through establishing of their final quinone moiety (see 3 and 4) from a hydroquinone intermediate. Crossfeeding experiments using the gilOIV-minus mutant19 showed 15 to be clearly an intermediate of the gilvocarcin pathway, while 8a is not. This suggests that the GilR-reaction is the final step in gilvocarcin biosynthesis, following even the tailoring O-methylation and C-glycosylation reactions. The result is in good agreement with previous crossfeeding studies that showed that the ‘aglycon’ defucogilvocarcin V is not an intermediate of the gilvocarcin pathway.19
Scheme 2.
Revised gilvocarcin biosynthetic pathway (shunt products in blue). Oxygenases GilOIV and GilOI can be substituted by the jadomycin pathway enzymes JadF and JadH, respectively. GilOIV/GilOI/GilOII and a 4th so far unidentified enzyme (catalyzing steps marked with a “?”) form a multi-enzyme complex. The aldehyde function of the suggested C-C splitting product 10 is indirectly ‘visible’ in the hemicacetal function found in intermediate 15.
Finally, we complemented a minimal gene set establishing the formation of intermediate UWM6 (1a), accumulated on plasmid pWHM1238.1 In these experiments, S. lividans TK24 was transformed with pWHM1238 as well as with various other plasmids containing oxygenase genes from either the jadomycin or the gilvocarcin pathways (see Table 1, supporting information). The experiments revealed that the multi-oxygenase complexes Jad-F-G-H (on plasmid pJadFGH) or Gil-OI-OII-OIV (on pGilOIOIIOIV) could complement the UWM6-genes to reconstitute jadomycin A (= aglycon of 4) production, when isoleucine was added to the culture medium. However, these multi-oxygenase complexes could not reconstitute the typical backbone of the gilvocarcins, which shows that there is an enzyme missing that is most likely an essential component of the multi-oxygenase complex of the gilvocarcin pathway. This enzyme is probably responsible for both the rotation of the phenyl residue in 13 and the immediately following formation of hemiacetal 14 (scheme 2). Because the C-C-bond cleavage occurs in absence of either GilOIII (→ gilvocarcin E 11) or GilR (→ pregilvocarcin V 15), we have to exclude these two enzymes from partaking in the multi-oxygenase complex responsible for the 5,6 bond cleavage. However, the gilvocarcin cluster offers four candidate genes encoding enzymes with so far unknown function (gilN, gilL, gilV, and gilM), which we will explore in near future.
In summary, our studies shed some light into the enigmatic oxidative processes in jadomycin and gilvocarcin biosyntheses. The studies revealed that the oxidative rearrangement cascades in both pathways are catalyzed by multi-oxygenase complexes, and follow essentially identical paths up to the C-C-bond cleavage product 10, thus diverge much later than previously appreciated. Furthermore, GilR was identified as hemiacetal dehydrogenase, an enzyme catalyzing a lactone formation in a novel way.
Supplementary Material
Listing and experimental details of all gene complementation experiments, isolation and physicochemical properties of compounds 8a and 15, derivatization of 15, and crossfeeding experiments.
Acknowledgments
This work was supported by the NIH (grant CA 102102) and the Kentucky Lung Cancer Research Program. We thank the NMR and Mass Spectrometry Core Facilities of the University of Kentucky, and Professor C. R. Hutchinson (U. Wisconsin-Madison) for providing pWHM1238.
References and Notes
- 1.Kulowski K, Wendt-Pienkowski E, Han L, Yang KQ, Vining LC, Hutchinson CR. J Am Chem Soc. 1999;121:1786–1794. [Google Scholar]
- 2.Rix U, Wang C, Chen Y, Lipata FM, Remsing Rix LL, Greenwell LM, Vining LC, Yang K, Rohr J. Chembiochem. 2005;6:838–845. doi: 10.1002/cbic.200400395. [DOI] [PubMed] [Google Scholar]
- 3.Chen YH, Wang CC, Greenwell L, Rix U, Hoffmeister D, Vining LC, Rohr J, Yang KQ. J Biol Chem. 2005;280:22508–22514. doi: 10.1074/jbc.M414229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu T, Fischer C, Beninga C, Rohr J. J Am Chem Soc. 2004;126:12262–12263. doi: 10.1021/ja0467521. [DOI] [PubMed] [Google Scholar]
- 5.Rix U, Zheng J, Remsing Rix LL, Greenwell L, Yang K, Rohr J. J Am Chem Soc. 2004;126:4496–4497. doi: 10.1021/ja031724o. [DOI] [PubMed] [Google Scholar]
- 6.Jakeman DL, Farrell S, Young W, Doucet RJ, Timmons SC. Bioorg Med Chem Lett. 2005;15:1447–1449. doi: 10.1016/j.bmcl.2004.12.082. [DOI] [PubMed] [Google Scholar]
- 7.Jakeman DL, Graham CL, Reid TR. Bioorg Med Chem Lett. 2005;15:5280–5283. doi: 10.1016/j.bmcl.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 8.Fischer C, Lipata F, Rohr J. J Am Chem Soc. 2003;125:7818–7819. doi: 10.1021/ja034781q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang K, Han L, Ayer SW, Vining LC. Microbiology. 1996;142:123–132. doi: 10.1099/13500872-142-1-123. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, White RL, Vining LC. Microbiology. 2002;148:1091–1103. doi: 10.1099/00221287-148-4-1091. [DOI] [PubMed] [Google Scholar]
- 11.Rix U, Fischer C, Remsing LL, Rohr J. Nat Prod Rep. 2002;19:542–580. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- 12.Gibson M, Nur-e-alam M, Lipata F, Oliveira MA, Rohr J. J Am Chem Soc. 2005;127:17594–17595. doi: 10.1021/ja055750t. [DOI] [PubMed] [Google Scholar]
- 13.Ye J, Dickens ML, Plater R, Li Y, Lawrence J, Strohl WR. J Bacteriol. 1994;176:6270–80. doi: 10.1128/jb.176.20.6270-6280.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Summers RG, Wendt-Pienkowski E, Motamedi H, Hutchinson CR. J Bacteriol. 1993;175:7571–80. doi: 10.1128/jb.175.23.7571-7580.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung JY, Fujii I, Harada S, Sankawa U, Ebizuka Y. J Bacteriol. 2002;184:6115–6122. doi: 10.1128/JB.184.22.6115-6122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kendrew SG, Hopwood DA, Marsh EN. J Bacteriol. 1997;179:4305–4310. doi: 10.1128/jb.179.13.4305-4310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gullón S, Olano C, Abdelfattah MS, Braña AF, Rohr J, Méndez C, Salas JA. Appl Environ Microbiol. 2006;72:4172–4183. doi: 10.1128/AEM.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waldron C, Matsushima P, Rosteck PR, Jr, Broughton MC, Turner J, Madduri K, Crawford KP, Merlo DJ, Baltz RH. Chem Biol. 2001;8:487–499. doi: 10.1016/s1074-5521(01)00029-1. [DOI] [PubMed] [Google Scholar]
- 19.Liu T, Kharel MK, Fischer C, McCormick A, Rohr J. Chembiochem. 2006;7:1070–1077. doi: 10.1002/cbic.200600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Listing and experimental details of all gene complementation experiments, isolation and physicochemical properties of compounds 8a and 15, derivatization of 15, and crossfeeding experiments.