Abstract
The aim of this study was to determine which human immunodeficiency virus type 1 (HIV-1) subtypes were circulating in Australia and to correlate the subtypes with risk factors associated with the acquisition of HIV-1 infection. DNA was extracted from peripheral blood mononuclear cells, and HIV-1 env genes were amplified and subtyped using heteroduplex mobility analysis, with selected samples sequenced and phylogenetic analysis performed. The HIV-1 env subtypes were determined for 141 samples, of which 40 were from female patients and 101 were from male patients; 13 samples were from children. Forty-seven patients were infected by homosexual or bisexual contact, 46 were infected through heterosexual contact, 21 were infected from injecting drug use (IDU), 13 were infected by vertical transmission, 8 were infected from nosocomial exposure, and 6 were infected by other modes of transmission, including exposure to blood products, ritualistic practices, and two cases of intrafamilial transmission. Five subtypes were detected; B (n = 104), A (n = 5), C (n = 17), E (CRF01_AE; n = 13), and G (n = 2). Subtype B predominated in HIV-1 acquired homosexually (94% of cases) and by IDU (100%), whereas non-subtype B infections were mostly seen in heterosexually (57%) or vertically (22%) acquired HIV-1 infections and were usually imported from Africa and Asia. Subtype B strains of group M viruses predominate in Australia in HIV-1 transmitted by homosexual or bisexual contact and IDU. However, non-B subtypes have been introduced, mostly acquired via heterosexual contact.
Human immunodeficiency virus (HIV) is characterized by a high degree of genetic variation and recombination. Based on phylogenetic analysis of env sequences, HIV type 1 (HIV-1) strains have been divided into three major groups: the M, or Main, group; the O, or Outlier, group; and the recently described N group (non-M, non-O) (10, 13, 19, 22, 31, 37, 40). The M group is further divided into at least 11 genetic subtypes, or clades (A to K), and 10 recombinant viruses, or circulating recombinant forms (CRF) (26). Numerous studies have shown that multiple subtypes of HIV-1 circulate both globally and locally (4, 12, 16, 23, 29, 36, 45, 48). Loose geographic clustering of HIV-1 env subtypes was apparent early in the HIV-1 pandemic, where HIV-1 subtypes were often limited to specific geographic regions or risk groups (4, 5, 23, 28, 35, 42; C. Williamson, S. Engelbrecht, M. Lambrick, E. J. van Rensburg, R. Wood, W. Bredell, and A. L. Williamson, Letter, Lancet 346:782, 1995). However, international travel, war, urbanization, and population migration, among other factors, have made these boundaries less obvious. As a consequence, diverse env subtypes and recombinant viruses are being found in previously homogenous backgrounds, such as the United States (2, 6, 8, 9, 20, 34, 44), obscuring the previous regional compartmentalization of subtypes.
HIV-AIDS was first described in Australia in the homosexual-bisexual population in the early 1980s. As of the end of 2001, 21,725 HIV infections and 8,810 cases of AIDS had been reported, although the overall prevalence remains low, with a national infection rate in the nonindigenous population of 5.51/100,000. The majority of infections are associated with homosexual-bisexual contact (78%), with smaller proportions associated with heterosexual contact (11%) and injecting drug use (IDU) (5%, plus another 4% also with homosexual-bisexual contact). As a consequence, more men (94%) than women (6%) are infected with HIV-1 in Australia (32).
Since the majority of HIV-1 infections in Australia occur in the homosexual-bisexual population, the predominant HIV-1 subtype is thought to be subtype B, especially as this subtype is predominant in North America, Europe, and neighboring Papua New Guinea and New Zealand (1, 14, 15, 17). However, HIV infection rates in other countries in the Asia-Pacific region (Thailand, Cambodia, Myanmar, India, and southern China, among others) continue to increase, and the incidence of non-B subtypes in these countries is high (21, 42, 45, 48). There are increasing numbers of people traveling abroad for pleasure, military deployment, and employment and increasing numbers of individuals entering Australia from other countries via immigration or repatriation or as refugees. Therefore, the possibility of non-B subtypes entering Australia is substantial. To address the paucity of data on HIV-1 genetic diversity in Australia, an assessment of HIV-1 group M subtypes circulating in Australia was made, correlating the circulating subtypes with risk factors for the acquisition of HIV-1 infection.
(This study was presented in part at the Seventh Conference on Retroviruses and Opportunistic Infections, San Francisco, Calif., 2000.)
MATERIALS AND METHODS
Patient samples.
Samples were randomly selected from 141 patients presenting between 1993 and 2002 to Westmead Hospital, the Parramatta Sexual Health Clinic, the New Children's Hospital at Westmead, and other local and interstate clinics. Epidemiological data collected included the country of origin of the infection, clinical stage of disease, sex of the patient, and risk factor for acquisition of HIV-1. One hundred and one (72%) samples were from males, and 40 (28%) were from females; 13 were from children. Risk factors associated with the acquisition of HIV are listed in Table 1. The study included 55 patients who acquired HIV-1 overseas and nine pairs of epidemiologically linked individuals. The latter group included two unrelated cohorts in which the index case acquired HIV-1 heterosexually but the mode of transmission to the linked family member remained unestablished (18).
TABLE 1.
Distribution of HIV-1 subtypes in Australia according to risk group, subtype, and geographic origin of infectiona
| Subtype (no.) | No. from risk group
|
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Homosexual-bisexual (n = 47)
|
Heterosexual (n = 46)
|
Vertical (n = 13)
|
IDU (n = 21)
|
Nosocomial (n = 8)
|
Other (n = 6)b
|
|||||||||||||||||||||||||||||||
| Asia | Africa | Europe | Americas | Australasia | Unknown | Asia | Africa | Europe | Americas | Australasia | Unknown | Asia | Africa | Europe | Americas | Australasia | Unknown | Asia | Africa | Europe | Americas | Australasia | Unknown | Asia | Africa | Europe | Americas | Australasia | Unknown | Asia | Africa | Europe | Americas | Australasia | Unknown | |
| A (5) | 3 | 1 | 1 | |||||||||||||||||||||||||||||||||
| B (104) | 1 | 1 | 2 | 40 | 9 | 2 | 8 | 1 | 2 | 7 | 1 | 20 | 7 | 3 | ||||||||||||||||||||||
| C (17) | 3 | 9 | 3 | 1 | 1 | |||||||||||||||||||||||||||||||
| E (13) | 2 | 1 | 8 | 1 | 1 | |||||||||||||||||||||||||||||||
| G (2) | 2 | |||||||||||||||||||||||||||||||||||
| Total (141) | 3 | 1 | 2 | 40 | 1 | 20 | 14 | 1 | 2 | 8 | 1 | 3 | 3 | 7 | 1 | 20 | 1 | 7 | 1 | 1 | 1 | 3 | ||||||||||||||
All samples were collected from individuals resident in Australia. The geographic regions listed identify where individuals reported HIV-1 acquisition; Americas includes North and South America, and Australasia includes Australia, Papua New Guinea, and New Zealand.
Other includes one individual who acquired HIV-1 through ritualistic practices in Africa, three who acquired HIV-1 through contaminated blood products, and two who acquired HIV nonsexually from other family members.
DNA preparation and amplification, HMA, and sequencing.
DNA was extracted from peripheral blood mononuclear cells using either cell lysis (38) or the QIAamp DNA Blood Mini Kit (Qiagen, Clifton Hill, Victoria, Australia). Envelope gene sequences (V1 to V5 or V3 to V5; primer sets ED5-ED12 and ES7-ES8, respectively) were amplified from proviral DNA using nested PCR (11, 12). Amplified fragments (V3 to V5; ES7-ES8) were then used for subtyping analysis by heteroduplex mobility analysis (HMA) as previously described (11). Amplified fragments (ES7-ES8; length, 703 bp) from samples identified by HMA as non-B subtypes (n = 26) were sequenced to confirm the subtype assignment by HMA. In 37 cases, a 284-bp fragment spanning the V3 loop of the envelope gene was generated using nested PCR (external primers V3ext-1 6959-6976 [5′-CAATGTACACATCGAATT-3′] and V3ext-2 7381-7362 [5′-ATTACAGTAGAAAAATTCCC-3′]; internal primers V3int-1 7011-7029 [5′-CGAGTCTAGCAGAAGAAGA-3′] and V3int-2 7331-7313 [5′-TGGGTCCCCTCCGAGGA-3′]) and sequenced (46, 47). As these 37 sequences were all subtype B on phylogenetic analysis, HMA was not performed.
Phylogenetic analysis.
Nucleotide sequences of 415 bp were aligned with consensus and reference sequences (obtained from the HIV Sequence Database at http://hiv-web.lanl.gov) from each HIV-1 subtype, including a consensus subtype O sequence. The reference sequences used in alignments (and their origins) included IBNG (Nigeria), SF170 (Rwanda), RW20 (Rwanda), AF082486 (Ukraine), U93665 (Ukraine), and U93611 (Russia) for subtype A; JRFL (United States), TH14 (Thailand), and SF162 (United States) for subtype B; BR025 (Brazil), MW959 (Malawi), and ZAM18A (Zambia) for subtype C; TH06 (Thailand), TH22 (Thailand), HIVU48278 (Uruguay), and CM240 (Thailand) for CRF01_AE; pBLV (Gabon) and RU131 (Russia) for subtype G; and BFP90 (Burkina Faso) for CRF06_cpx. Methods described by Learn et al. were followed to exclude possible sample contamination (27). Multiple sequence alignments were generated using CLUSTAL W (43). A neighbor-joining phylogenetic tree was constructed using a maximum-likelihood distance model. One hundred bootstrap replications were performed for each data set.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences discussed are AF400076 to AF400088 and AY081962 to AY081972.
RESULTS
A total of 141 samples were collected from 109 HIV-1-seropositive individuals and 32 people with AIDS, PCR amplified, and subtyped by HMA (Table 1). Forty-seven patients (33%) acquired HIV-1 through homosexual or bisexual contact, 46 (33%) acquired it through heterosexual contact, 21 (15%) acquired it from IDU, 13 (9%) acquired it by vertical transmission, 8 (6%) acquired it from nosocomial exposure—including four cases of transmission in a doctor's surgery in Australia (N. K. Saksena, J. Z. Song, D. E. Dwyer, and A. L. Cunningham, Abstr. Sixth Conference on Retroviruses and Opportunistic Infections, abstr. 280, 1999)—and 6 (4%) acquired it by other means, including blood products, ritualistic practices, and other family members. Six cases of transmission between heterosexual partners, one between homosexual partners, and two between IDU partners were identified. All pediatric cases were in babies born to HIV-1-infected mothers.
Five subtypes were detected, subtypes B, A, C, E, and G (Table 1), with the majority being subtype B (104 of 141; 74%). Subtype B was predominantly seen in homosexual-bisexual men (44 of 47; 93.6%), 91% of whom (40 of 44) acquired it in Australia. Injecting drug users were exclusively infected with subtype B, with all but one case acquired locally. In contrast, subtype B was detected in only 43% of heterosexually transmitted cases (20 of 46), and only 8 of the 20 (40%) were acquired locally. Seven of the nine (78%) vertically acquired cases of subtype B infection originated in Australia, and two were from Asia. Nineteen cases (18%) of subtype B infection were acquired overseas, in Asia (Thailand, Hong Kong, and Cambodia), the United States, South America, and Europe (mainly the United Kingdom) by homosexual-bisexual contact (4 cases), heterosexual contact (12 cases), vertical transmission (2 cases), and IDU (1 case).
Thirty-seven non-subtype B viruses were detected by HMA; subtype A (5 viruses), subtype C (17 viruses), subtype E (13 viruses), and subtype G (2 viruses). All non-B infections were acquired overseas or by direct contact with a foreign national within Australia, or vertically. Most non-B infections (34 of 37; 92%) were acquired heterosexually (26 cases), vertically (4 cases), or by other means (4 cases); only 3 cases of a non-B subtype virus (all subtype E from Thailand) were acquired by homosexual contact. Infection with a non-B subtype was strongly associated with heterosexual acquisition (P < 0.001; chi-square test) and overseas contact (P < 0.001; chi-square test). Subtype A infections originated from Africa (one each from Uganda, South Africa, and Africa unspecified) and Russia (two); subtype C originated from either India (three) or Africa (Zimbabwe, five; Ethiopia, two; one each from Swaziland, Somalia, Ghana, and Zambia; and Africa unspecified, three); subtype E originated from Southeast Asia (four each from Cambodia and Thailand; Philippines, three; and Asia unspecified, two); and subtype G originated from Africa (Burkina Faso, two).
Nucleotide analysis.
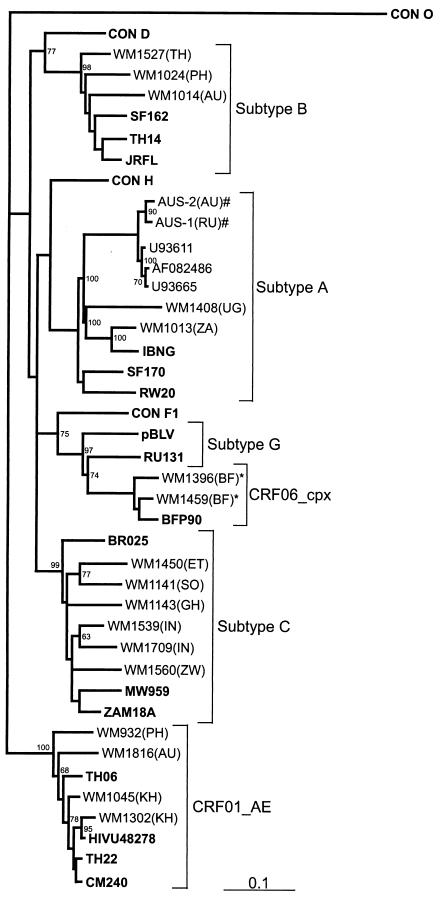
The V3 to V5 envelope regions of 19 non-subtype B and 8 subtype B viruses were sequenced to confirm the HMA assignments. Phylogenetic analysis of 19 sequences (8 were omitted due to inadequate length for analysis) indicated that there was 100% correlation between the HMA and sequence subtypes, as shown by others (4, 12) (Fig. 1). Intrasubtype genetic distances between the consensus and test sequences varied depending on the origin of infection. For instance, the two Indian subtype C sequences were phylogenetically distinct from African subtype C sequences, and the Cambodian E sequence was most similar to a CRF01_AE sequence from a Uruguayan peacekeeper who had been deployed in Cambodia. The subtype G sequences from Burkina Faso were from a heterosexual couple and have been further identified as an A/G/J recombinant or CRF, CRF06_cpx (33). V3 loop peptide sequences were deduced from all nucleotide sequences, and analysis of subtype B V3 loop nucleotide and peptide sequences did not confirm any epidemiological linkage or presence of “founder” viruses in the various patient groups (data not shown). The commonest V3 loop tetrapeptide observed was GPGR. The subtype B′ tetrapeptide motif GPGQ, commonly observed in injecting drug users in Bangkok (3, 42), was not seen in the four subtype B sequences generated from individuals who reported acquiring HIV-1 infection in Thailand or the other eight subtype B infections acquired elsewhere in Asia.
FIG. 1.
Phylogenetic tree constructed using a maximum-likelihood method from 415-bp nucleotide sequences corresponding to the C2-V3 regions of 19 patient samples and 20 HIV-1 reference and consensus sequences from the Los Alamos HIV-1 database representing 11 genetic subtypes (shown in boldface). Annotation of samples is as follows: WM indicates samples subtyped at Westmead Hospital, Sydney, Australia, and the following number indicates the sample code, with the country of origin indicated in parentheses (TH, Thailand; PH, Philippines; ZA, South Africa; UG, Uganda; RU, Russia; BF, Burkina Faso; IN, India; ET, Ethiopia; SO, Somalia; ZW, Zimbabwe; GH, Ghana; AU, Australia, and KH, Cambodia). Bootstrap values of >50% are indicated. Linked individuals are also indicated (*, CRF06_cpx, and #, subtype A).
DISCUSSION
The data confirm that subtype B is the predominant HIV-1 subtype circulating in Australia, as it was detected in almost all samples from homosexual-bisexual men, injecting drug users, cases acquired nosocomially, and recipients of blood products. Most subtype B infections acquired through heterosexual contact were imported from countries where subtype B is also prevalent, including the United States, the United Kingdom, and Thailand. Evidence of founder viruses and unique sequence differences between risk groups, particularly homosexual men and injecting drug users, have been demonstrated in other populations (24, 25, 41) but were not seen in this study.
In contrast, infections in the group with heterosexually acquired infections were predominantly non-B subtypes. They were usually found in individuals who had acquired HIV-1 overseas from various regions in Africa, Asia, and Eastern Europe or whose partner or mother had HIV-1 contact overseas. Australia's near neighbors, New Zealand and Papua New Guinea, also have predominantly subtype B viruses circulating, although subtype C has been detected in New Zealanders infected heterosexually from Africans (14, 15). Non-B subtype viruses will continue to be seen in Australia, as the incidence of heterosexually acquired HIV-1 is increasing in those who report sexual contact overseas and in indigenous Australians (39): 53.6% of new notifications from 1996 to 2001 of heterosexually acquired HIV-1 were for individuals (or their partners) from high-prevalence countries (30). Although the heterosexual population in Australia may not consider themselves as at risk of acquiring HIV-1, appropriate public health initiatives that target these groups may need review. These initiatives would need not only to educate individuals about the risks of acquiring HIV-1, particularly following sexual contact overseas, but to ensure that ongoing transmission to other partners or groups (who may also regard themselves as low risk) does not occur. The presence of multiple HIV-1 subtypes in Australia also has implications for local vaccine development, use of commercial viral-load assays, and antiretroviral drug resistance genotyping (7, 49).
Molecular surveillance of HIV subtypes in newly acquired cases of HIV-1 in Australia should be continued to monitor the emergence of new strains. The availability of antiretroviral drug resistance genotyping allows such monitoring (49), although non-subtype B viruses may circulate in risk groups or regions where such services are not accessible. As the various drug resistance databases contain predominantly subtype B sequences, non-subtype B viruses identified by such systems should be confirmed by HMA or sequencing of other regions of the HIV-1 genome. Surveillance in neighboring regions (for example, in Papua New Guinea and Indonesia, where explosive epidemics are under way, and in East Timor, where there has been significant societal disruption) is also needed to identify emerging epidemics and to ensure that appropriate public health interventions are implemented.
Acknowledgments
We thank Christine Brown, Jim Chew, Wayne Bolton, Hassan Naif, and Martyn French for assistance with samples and their analyses. The contributions of the many physicians around Australia who provided patient samples are appreciated.
This project was supported by Commonwealth AIDS Research Grant 956071.
REFERENCES
- 1.Alaeus, A. 2000. Significance of HIV-1 genetic subtypes. Scand. J. Infect. Dis. 32:455-463. [DOI] [PubMed] [Google Scholar]
- 2.Alaeus, A., T. Leitner, K. Lidman, and J. Albert. 1997. Most HIV-1 genetic subtypes have entered Sweden. AIDS 11:199-202. [DOI] [PubMed] [Google Scholar]
- 3.Amornkul, P. N., S. Tansuphasawadikul, K. Limpakarnjanarat, S. Likanonsakul, N. Young, B. Eampokalap, J. Kaewkungwal, T. Naiwatanakul, J. Von Bargen, D. J. Hu, and T. D. Mastro. 1999. Clinical disease associated with HIV-1 subtype B′ and E infection among 2104 patients in Thailand. AIDS 13:1963-1969. [DOI] [PubMed] [Google Scholar]
- 4.Bachmann, M. H., E. L. Delwart, E. G. Shpaer, P. Lingenfelter, R. Singal, J. I. Mullins, et al. 1994. Rapid genetic characterization of HIV type 1 strains from four World Health Organization-sponsored vaccine evaluation sites using a heteroduplex mobility assay. AIDS Res. Hum. Retrovir. 10:1345-1353. [DOI] [PubMed] [Google Scholar]
- 5.Bongertz, V., D. C. Bou-Habib, L. F. Brigido, M. Caseiro, P. J. Chequer, J. C. Couto-Fernandez, P. C. Ferreira, B. Galvao-Castro, D. Greco, M. L. Guimaraes, M. I. L. de Carvalho, M. G. Morgado, C. A. Oliveira, S. Osmanov, C. A. Ramos, M. Rossini, E. Sabino, A. Tanuri, M. Ueda, et al. 2000. HIV-1 diversity in Brazil: genetic, biologic, and immunologic characterization of HIV-1 strains in three potential HIV vaccine evaluation sites. J. Acquir. Immune Defic. Syndr. 23:184-193. [DOI] [PubMed] [Google Scholar]
- 6.Brodine, S. K., J. R. Mascola, P. J. Weiss, S. I. Ito, K. R. Porter, A. W. Artenstein, F. C. Garland, F. E. McCutchan, and D. S. Burke. 1995. Detection of diverse HIV-1 genetic subtypes in the USA. Lancet 346:1198-1199. [DOI] [PubMed] [Google Scholar]
- 7.Chew, C. B., B. L. Herring, F. Zheng, C. Browne, N. K. Saksena, A. L. Cunningham, and D. E. Dwyer. 1999. Comparison of three commercial assays for the quantification of HIV-1 RNA in plasma from individuals infected with different HIV-1 subtypes. J. Clin. Virol. 14:87-94. [DOI] [PubMed] [Google Scholar]
- 8.Contag, C. H., A. Ehrnst, J. Duda, A. B. Bohlin, S. Lindgren, G. H. Learn, and J. I. Mullins. 1997. Mother-to-infant transmission of human immunodeficiency virus type 1 involving five envelope sequence subtypes. J. Virol. 71:1292-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couturier, E., F. Damond, P. Roques, H. Fleury, F. Barin, J. B. Brunet, F. Brun-Vezinet, and F. Simon. 2000. HIV-1 diversity in France, 1996-1998. AIDS 14:289-296. [DOI] [PubMed] [Google Scholar]
- 10.De Leys, R., B. Vanderborght, M. Vanden Haesevelde, L. Heyndrickx, A. van Geel, C. Wauters, R. Bernaerts, E. Saman, P. Nijs, B. Willems, H. Taelman, G. van der Groen, P. Piot, T. Tersmette. J. G. Huisman, and H. van Heuverswyn. 1990. Isolation and partial characterization of an unusual human immunodeficiency retrovirus from two persons of west-central African origin. J. Virol. 64:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart, E. L., B. Herring, A. G. Rodrigo, and J. I Mullins. 1995. Genetic subtyping of human immunodeficiency virus using a heteroduplex mobility assay. PCR Methods Appl. 4:S202-S216. [DOI] [PubMed]
- 12.Delwart, E. L., E. G. Shpaer, F. E. McCutchan, J. Louwagie, H. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 13.Dumitrescu, O., M. L. Kalish, S. C. Kliks, C. I. Bandea, and J. A. Levy. 1994. Characterization of human immunodeficiency virus type 1 isolates from children in Romania: identification of a new envelope subtype. J. Infect. Dis. 169:281-288. [DOI] [PubMed] [Google Scholar]
- 14.Dwyer, D. E., Y. C. Ge, B. Wang, W. V. Bolton, J. G. McCormack, A. L. Cunningham, and N. K. Saksena. 1997. First human immunodeficiency virus type 1 sequences in the V3 region, nef and vpr genes from Papua New Guinea. AIDS Res. Hum. Retrovir. 13:625-627. [DOI] [PubMed] [Google Scholar]
- 15.Dwyer, D. E., B. L. Herring, Y. C. Ge, W. V. Bolton, R. B. Ellis-Pegler, M. Thomas, B. A. Schroeder, M. C. Croxon, A. L. Cunningham, J. I. Mullins, and N. K. Saksena. 1998. Human immunodeficiency virus type 1 subtypes B and C detected in New Zealand. AIDS Res. Hum. Retrovir. 14:1105-1108. [DOI] [PubMed] [Google Scholar]
- 16.Essex, M. 1999. Human immunodeficiency viruses in the developing world. Adv. Virus Res. 53:71-88. [DOI] [PubMed] [Google Scholar]
- 17.Foley, B., H. Pan, S. Buchbinder, and E. L. Delwart. 2000. Apparent founder effect during the early years of the San Francisco HIV type 1 epidemic (1978-1979). AIDS Res. Hum. Retrovir. 16:1463-1469. [DOI] [PubMed] [Google Scholar]
- 18.French, M. A., B. L. Herring, J. M. Kaldor, D. C. Sayer, V. Furner, C. C. de Chaneet, and D. E. Dwyer. 2003. Intrafamilial transmission of HIV-1 infection from individuals with unrecognised HIV-1 infection. AIDS 17:1977-1981. [DOI] [PubMed]
- 19.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. N. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 20.Gao, F., L. Yue, S. C. Hill, D. L. Robertson, A. H. Graves, M. S. Saag, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. HIV-1 sequence subtype D in the United States. AIDS Res. Hum. Retrovir. 10:625-627. [DOI] [PubMed] [Google Scholar]
- 21.Gayle, H. D., and G. L. Hill. 2001. Global impact of human immunodeficiency virus and AIDS. Clin. Microbiol. Rev. 14:327-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurtler, L. G., P. H. Hauser, J. Eberle, A. vonBrunn, S. Knapp, L. Zekeng, J. M. Tsague, and L. Kaptue. 1994. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J. Virol. 68:1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssens, W., A. Buve, and J. N. Nkengasong. 1997. The puzzle of HIV-1 subtypes in Africa. AIDS 11:705-712. [DOI] [PubMed] [Google Scholar]
- 24.Kuiken, C., R. Thakallapalli, A. Esklid, and A. de Ronde. 2000. Genetic analysis reveals epidemiologic patterns in the spread of human immunodeficiency virus. Am. J. Epidemiol. 152:814-822. [DOI] [PubMed] [Google Scholar]
- 25.Kuiken, C. L., M. T. Cornelissen, F. Zorgdrager, S. Hartman, A. J. Gibbs, and J. Goudsmit. 1996. Consistent risk group-associated differences in human immunodeficiency virus type 1 vpr, vpu and V3 sequences despite independent evolution. J. Gen. Virol. 77:783-792. [DOI] [PubMed] [Google Scholar]
- 26.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. Mellors, J. Mullins, J. Sodroski, and S. E. Wolinksy. 2000. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N. Mex.
- 27.Learn, G. H., B. T. Korber, B. Foley, B. H. Hahn, S. M. Wolinsky, and J. I. Mullins. 1996. Maintaining the integrity of human immunodeficiency virus sequence databases. J. Virol. 70:5720-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Limpakarnjanarat, K., K. Ungchusak, T. D. Mastro, N. L. Young, C. Likhityingvara, O. Sangwonloy, B. G. Weniger, C. P. Pau, and T. J. Dondero. 1998. The epidemiological evolution of HIV-1 subtypes B and E among heterosexuals and injecting drug users in Thailand, 1992-1997. AIDS 12:1108-1109. [PubMed] [Google Scholar]
- 29.Louwagie, J., W. Janssens, J. Mascola, L. Heyndrickx, P. Hegerich, G. van der Goen, F. E. McCutchan, and D. S. Burke. 1995. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J. Virol. 69:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald, A. 2002. Newly diagnosed HIV infection in Australia attributed to heterosexual contact. Aust. HIV Surv. Rep. 18:1-6. [Google Scholar]
- 31.Myers, G., B. Korber, B. Hahn, K. T. Jeang, J. Mellors, F. E. McCutchan, L. E. Henderson, and G. N. Pavlakis. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N. Mex.
- 32.National Centre in HIV Epidemiology and Clinical Research. 2002. The National HIV Database. Aust. HIV Surv. Rep. 18:1-23. [Google Scholar]
- 33.Oelrichs, R. B., C. Workman, T. Laukkanen, F. E. McCutchan, and N. J. Deacon. 1998. A novel subtype A/G/J recombinant full-length HIV type 1 genome from Burkina Faso. AIDS Res. Hum. Retrovir. 14:1495-1500. [DOI] [PubMed] [Google Scholar]
- 34.Op de Coul, E. L. M., V. V. Lukashov, G. J. J. J. van Doornum, J. Goudsmit, and R. A. Coutinho. 1998. Multiple HIV-1 subtypes present amongst heterosexuals in Amsterdam 1988-1996: no evidence for spread of non-B subtypes. AIDS 12:1253-1255. [DOI] [PubMed] [Google Scholar]
- 35.Ou, C.-Y., Y. Takebe, C. C. Luo, M. Kalish, W. Auwanit, C. Bandea, N. de la Torre, J. L. Moore, G. Schochetman, S. Yamazaki, H. D. Gayle, N. L. Young, and B. G. Weniger. 1992. Wide distribution of two subtypes of HIV-1 in Thailand. AIDS Res. Hum. Retrovir. 8:1471-1472. [DOI] [PubMed] [Google Scholar]
- 36.Paladin, F. J. E., O. T. Monzon, H. Tsuchie, M. R. A. Aplasca, G. H. Learn, and T. Kurimura. 1998. Genetic subtypes of HIV-1 in the Philippines. AIDS 12:291-300. [DOI] [PubMed] [Google Scholar]
- 37.Peeters, M., A. Gueye, S. Mboup, F. Bibollet-Ruche, E. Ekaza, C. Mulanga, R. Ouedrago, R. Gandji, P. Mpele, G. Dibanga, B. Koumare, M. Saidou, E. Esu-Williams, J. P. Lombart, W. Badombena, N. Luo, M. Vanden Haesevelde, and E. Delaporte. 1997. Geographical distribution of HIV-1 group O viruses in Africa. AIDS 11:493-498. [DOI] [PubMed] [Google Scholar]
- 38.Rogers, M. F., C.-Y. Ou, M. Rayfield, P. A. Thomas, E. E. Schoenbaum, E. Abrams, K. Krasinski, P. A. Selwyn, J. Moore, A. Kaul, et al. 1989. Use of the polymerase chain reaction for early detection of the proviral sequences of human immunodeficiency virus in infants born to seropositive mothers. N. Engl. J. Med. 320:1649-1654. [DOI] [PubMed] [Google Scholar]
- 39.Savage, J. 2001. HIV infection and indigenous Australians. Microbiol. Aust. 22:26-27. [Google Scholar]
- 40.Simon, F., P. Mauclere, P. Roques, I. Loussert-Ajaka, M. C. Muller-Trutwin, S. Saragosti, M. C. Georges-Courbot, F. Barre-Sinoussi, and F. Brun-Vezinet. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032-1037. [DOI] [PubMed] [Google Scholar]
- 41.Stoeckli, T. C., I. Steffen-Klopfstein, P. Erb, T. M. Brown, M. L. Kalish, et al. 2000. Molecular epidemiology of HIV-1 in Switzerland: evidence for a silent mutation in the C2V3 region distinguishing intravenous drug users from homosexual men. J. Acquir. Immune Defic. Syndr. 23:58-67. [DOI] [PubMed] [Google Scholar]
- 42.Subbarao, S., K. Limpakarnjanarat, T. D. Mastro, J. Bhumisawasdi, P. Warachit, C. Jayavasu, N. L. Young, C. C. Luo, N. Shaffer, M. L. Kalish, and G. Schochetman. 1998. HIV type 1 in Thailand, 1994-1995: Persistence of two subtypes with low genetic diversity. AIDS Res. Hum. Retrovir. 14:319-327. [DOI] [PubMed] [Google Scholar]
- 43.Thompson, J. D., D. J. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson, M. M., and R. Najera. 2001. Travel and the introduction of human immunodeficiency virus type 1 non-B subtype genetic forms into Western countries. Clin. Infect. Dis. 32:1732-1737. [DOI] [PubMed] [Google Scholar]
- 45.Tsuchie, H., T. S. Saraswathy, M. Sinniah, B. Vijayamalar, J. Maniar, O. T. Monzon, R. T. Santana, F. J. E. Paladin, C. Wasi, P. Thongcharoen, M. M. Hossain, O. Yamada, S. Kageyama, T. Kitamura, and T. Kurimura. 1995. HIV-1 variants in South and South-East Asia. Int. J. STD AIDS 6:117-120. [DOI] [PubMed] [Google Scholar]
- 46.Wang, B., Y. C. Ge, P. Palasanthiran, S. H. Xiang, J. Ziegler, D. E. Dwyer, C. Randle, D. Dowton, A. Cunningham, and N. K. Saksena. 1996. Gene defects clustered at the C-terminus of the vpr gene of HIV-1 in long-term nonprogressing mother-child pair: in vivo evolution of vpr quasispecies in blood and plasma. Virology 223:224-232. [DOI] [PubMed] [Google Scholar]
- 47.Wang, B., Y. C. Ge, P. Palasanthiran, J. Ziegler, W. Bolton, S. H. Xiang, D. E. Dwyer, A. L. Cunningham, and Nitin K. Saksena. 1997. HIV type 1 V3 loop sequences derived from peripheral blood of transmitting mothers, their infants, and nontransmitting mothers differ in their crown octapeptide motifs. AIDS Res. Hum. Retrovir. 13:275-279. [DOI] [PubMed] [Google Scholar]
- 48.Wasi, C., B. Herring, S. Raktham, S. Vanichseni, T. D. Mastro, N. L. Young, H. Rubsamen-Waigmann, H. von Briesen, M. Kalish, C. C. Luo, A. Baldwin, J. I. Mullins, E. L. Delwart, J. Esparza, W. L. Heyward, and S. Osmanov. 1995. Determination of HIV-1 subtypes in injecting drug users in Bangkok, Thailand, using peptide-binding enzyme immunoassay and heteroduplex mobility assay: evidence of increasing infection with HIV-1 subtype E. AIDS 9:843-849. [DOI] [PubMed] [Google Scholar]
- 49.Yahi, N., J. Fantini, C. Tourres, N. Tivoli, N. Koch, and C. Tamalet. 2001. Use of drug resistance sequence data for the systematic detection of non-B human immunodeficiency virus type 1 (HIV-1) subtypes: how to create a sentinel site for monitoring the genetic diversity of HIV-1 at a country scale. J. Infect. Dis. 183:1311-1317. [DOI] [PubMed] [Google Scholar]